Abstract
MITF (microphthalmia-associated transcription factor) represents a melanocytic lineage-specific transcription factor whose role is profoundly extended in malignant melanoma. Over the last few years, the function of MITF has been tightly connected to plasticity of melanoma cells. MITF participates in executing diverse melanoma phenotypes defined by distinct gene expression profiles. Mutation-dependent alterations in MITF expression and activity have been found in a relatively small subset of melanomas. MITF activity is rather modulated by its upstream activators and suppressors operating on transcriptional, post-transcriptional and post-translational levels. These regulatory mechanisms also include epigenetic and microenvironmental signals. Several transcription factors and signaling pathways involved in the regulation of MITF expression and/or activity such as the Wnt/β-catenin pathway are broadly utilized by various types of tumors, whereas others, e.g., BRAFV600E/ERK1/2 are more specific for melanoma. Furthermore, the MITF activity can be affected by the availability of transcriptional co-partners that are often redirected by MITF from their own canonical signaling pathways. In this review, we discuss the complexity of a multilevel regulation of MITF expression and activity that underlies distinct context-related phenotypes of melanoma and might explain diverse responses of melanoma patients to currently used therapeutics.
Keywords: BRAFV600E, Melanoma cancer plasticity, Gene regulation, MicroRNA, MITF, Oncogene, Transcription factor
Introduction
Melanocytes are pigment-producing cells whose differentiation, proliferation and survival largely depend on MITF (microphthalmia-associated transcription factor), a melanocyte-specific transcription factor [for review 1, 2]. Melanoma is a melanocyte-derived tumor in which MITF dependence is retained [for review 3]; thus, MITF represents a lineage-restricted regulator that operates in normal cells, and its activity is also used by malignant cells. Enforced expression of MITF in immortalized melanocytes [4] or neural crest progenitor cells [5] when introduced together with melanoma-specific BRAFV600E suggests MITF’s role as a melanoma addiction oncogene. MITF is recognized as a driver of melanoma progression [for review 6], but its role in suppression of invasion and metastasis has been also shown [7–10]. By activating the expression of almost one hundred genes, MITF can regulate multiple biological processes in melanoma cells such as differentiation, proliferation, migration, and senescence [11–13; for review 14, 15]. MITF also exerts pro-survival role by activating the expression of anti-apoptotic genes including BCL2A1, BCL2 and BIRC7 (ML-IAP/livin) [for review 16, 17]. Recent studies implicate MITF in energy metabolism and organelle biogenesis [18; for review 19]. This variety of often mutually exclusive cellular programs driven by MITF stands for distinct phenotypes of melanoma cells [12, 20, 21; for review 22, 23]. MITF is also recognized as a major regulator in a “phenotypic switching” concept explaining a high plasticity of melanoma cells [20, 21, 24–27; for review 22, 28]. Therefore, better understanding of the intracellular mechanisms underlying a contextual regulation of MITF is of utmost importance. In this review, we focus on melanoma-related mechanisms underlying the regulation of MITF expression and activity.
Gene structure and transcriptional regulation of MITF
In human, MITF locus is mapped to chromosome 3 and spans 229 kbp. MITF encodes a b-HLH-Zip (basic helix-loop-helix leucine zipper) transcription factor that belongs to the MYC superfamily. Together with TFEB, TFEC and TFE3, MITF constitutes the MiT (microphthalmia) family of transcription factors [29]. All of them share a common b-HLH-Zip dimerization motif containing a positively charged fragment involved in DNA binding, and a transactivation domain (TAD) [29]. As a result of differential usage of alternative promoters, a single MITF gene produces several isoforms including MITF-A [30], MITF-B [31], MITF-C [32], MITF-D [33], MITF-E [34], MITF-H [35], MITF-J [36], MITF-Mc [37] and MITF-M [38, 39]. These isoforms differ in their N-termini encoded by exon 1, and show tissue-specific pattern of expression. The expression of the shortest isoform MITF-M (a 419-residue protein) is limited to melanocytes and melanoma cells [39; for review 40]. MITF-Mdel, a variant of MITF-M harboring two in-frame deletions within the exons 2 and 6, has been identified as restrictedly expressed in these cells [41]. MITF contains two TADs responsible for its transcriptional activity; however, a functional domination of the TAD at N-terminus over that one at C-terminus has been reported [42]. MITF binds to DNA as a homodimer or heterodimer with one of the MiT proteins [29], but does not form heterodimers with other b-HLH-Zip transcription factors such as MYC, MAX and USF, despite a common ability to bind to the palindromic CACGTG E-box motif [43]. It was shown that the heptad repeat register of the leucine zipper in MITF is broken by a three-residue insertion that generates a kink in one of the two zipper helices, which limits the ability of MITF to form dimers only with those bHLHZip transcription factors that contain the same type of insertion [43]. Functionally, the MITF-binding sites in the promoters of target genes involve E-box: CA[C/T]GTG and M-box, extended E-box with an additional 5′-end flanking thymidine nucleotide: TCATGTGCT [for review 44].
Genetic alterations in MITF and alternative splicing
Some genetic alterations have been associated with MITF. Initially, high-density single nucleotide polymorphism arrays revealed the MITF amplification in up to 20 % of melanomas, with higher incidence among metastatic melanoma samples [4]. This aberration correlated with decreased overall patient survival [4]. However, in a recent study involving targeted-capture deep sequencing, no copy gains at the MITF locus have been found in a panel of melanoma metastases [45]. Genetic abnormalities related to MITF also include single base substitutions in the regions encoding its functional domains [46]. These somatic mutations, however, do not affect the DNA-binding ability of MITF in melanoma cells [47]. Recently, two independent studies have identified a rare oncogenic MITFE318K variant representing a gain-of-function allele for MITF that is present in patients with familial melanoma and a small fraction of sporadic melanomas [48, 49]. MITF E318K has been described as a medium-penetrance gene in melanoma associated with multiple primary melanomas developed in its carriers [50, 51], and as predisposing to renal carcinoma as well [48].
Alternative splicing is another mechanism of MITF regulation in melanoma. Two spliced variants of MITF, MITF(+) containing an internal six-amino acid fragment encoded by exon 6a and MITF(−) that lacks this fragment, have been described. These two variants possess different activity, with anti-proliferative property of MITF(+). This effect is tightly related to the interaction between the N-terminal fragment of MITF(+) with its specific hexapeptide [52]. Activation of the MEK1-ERK2 (extracellular signal-regulated kinase 2) pathway, independently of the mutational status of BRAF and NRAS, has been indicated as a mechanism underlying the expression of MITF splice variants [53]. Additionally, the quantification of these variants in a panel of 86 melanoma samples revealed the apparently increased expression of MITF(−) in metastatic melanomas [53].
Transcriptional activators of MITF
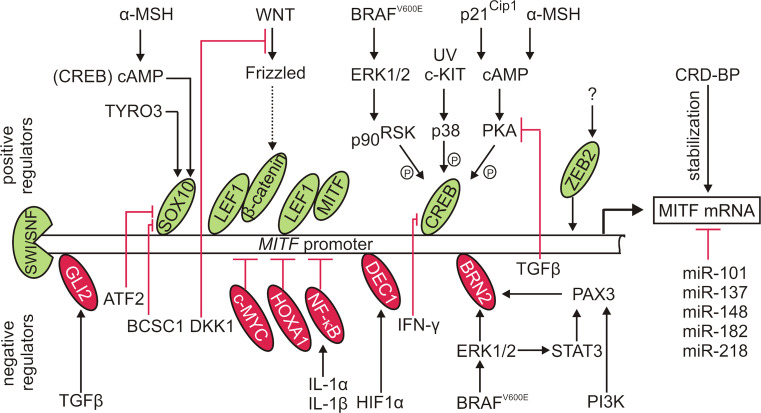
The transcriptional control of MITF is governed by a number of transcription factors and their regulators associated with signaling pathways involved in diverse cellular processes (Fig. 1). SOX10 (sex-determining region Y-box 10)-responsive element was found between −264 and −266 in the MITF promoter [54]. In addition, an activating frameshift or non-sense mutations in SOX10 have been identified in melanoma cells, and MITF and SOX10 have been found mutated in a mutually exclusive manner [46]. The nuclear localization of SOX10 is maintained by a protein tyrosine kinase TYRO3 [55]. SOX10 also cooperates with CREB (cAMP response element-binding protein) in the responsiveness of MITF to α-MSH (α-melanocyte-stimulating hormone)-cAMP signaling. This constitutes a tightly restricted mechanism of regulation due to a ubiquitous expression of CREB and a cell type-limited expression of SOX10 [56]. CREB is targeted by a number of regulators that promote its phosphorylation at Ser133, thus activating CREB-dependent transcription (Fig. 1) [57; for review 58]. It has been demonstrated that p38, activated by either UV (ultraviolet radiation) or receptors, e.g., KIT, phosphorylates CREB and promotes its binding to the MITF promoter [57]. p21Cip1, a cell cycle inhibitor, has been identified as a CREB co-factor involved in cAMP-dependent MITF expression in melanoma [59]. MITF expression can be also mediated by the complex of two key effectors of the Wnt (wingless-type) signaling pathway, LEF1 (lymphoid enhancer-binding factor 1) and β-catenin [20, 60]. In contrast to β-catenin, a phenotype-specific expression of LEF1 has been shown in melanoma cells limiting LEF1/β-catenin-dependent MITF transcription to a defined cellular context [20]. Importantly, MITF can cooperate with LEF1 as a non-DNA-binding coactivator to enhance its own expression [60]. It has been also demonstrated that mediators of α-MSH/cAMP/PKA (protein kinase A) signaling can redirect β-catenin to the CREB-specific promoters to activate transcription of CREB target genes including MITF [61]. Most recently, a transcription factor involved in epithelial–mesenchymal transition [62; for review 63, 64], ZEB2 (zing finger E-box binding protein 2) has been shown to activate MITF expression, and a ZEB2 loss that resulted in a decreased MITF level and several MITF-dependent target genes was associated with melanoma progression [65]. In contrast to activating potential of ZEB2 on the MITF promoter, ZEB1 has been found to directly repress MITF expression in retinal pigment epithelium [66]. Thus, the role of ZEB1 in the context of MITF expression in melanoma needs to be elucidated. On the level of chromatin remodeling, it has been demonstrated that a SWI/SNF complex containing BRM (Brahma) or BRG1 (Brahma-related gene 1) promotes MITF expression [67].
Fig. 1.
Transcriptional and post-transcriptional regulation of MITF expression. The variety of MITF regulators, activated by diverse signaling pathways often modified in melanoma, comprises a number of transcription factors either promoting MITF expression (positive regulators; shown in green) or inhibiting its transcription (negative regulators; shown in red). Upstream regulators of these transcription factors may indirectly affect MITF level. A correlation is also observed between MITF level and activity of transcription factors, e.g., NF-κB, not operating on MITF promoter. Moreover, a chromatin-remodeling complex SWI/SNF promotes MITF expression. In addition, MITF transcript can be either negatively regulated by miRs, or stabilized by the association with CRD-BP
Transcriptional repressors of MITF
Several transcription factors have been identified as direct repressors of MITF (Fig. 1). Inverse expression of GLI2 (glioma-associated oncogene family member 2) and MITF-M, relating to the mutually exclusive transcriptional programs, has been observed in melanoma cells [26]. GLI2 is a Kruppel-like transcription factor activated by TGFβ (transforming growth factor β) [26]. Furthermore, a GLI2 binding site was identified in the −334/−296 region of the MITF promoter confirming direct inhibitory activity of GLI2 towards MITF [68]. The contribution of PAX3 (paired box 3) to MITF expression represents another melanoma-specific mechanism. Although positive regulation of MITF by PAX3 in melanocytes is well described [69], PAX3 is thought to function independently of MITF [70] or even play a repressive role on MITF expression in melanoma [21]. PAX3 is activated by PI3 K (phosphatidylinositol 3-kinase) [71] and STAT3 (signal transducer and activator of transcription 3) [72] in melanoma cells, and a negative PAX3-dependent regulation of MITF expression is mediated by BRN2 encoded by POU3F2 [21, 71]. Notably, BRN2-mediated repression of MITF transcription represents a mechanism distinguishing between melanoma cells and melanocytes due to the lack of BRN2 expression in the latter, which might be explained by the involvement of melanoma-specific BRAFV600E in BRN2 upregulation [73]. Moreover, inverse expression of MITF and BRN2 was shown in vitro [8] and in vivo [7]. The antagonistic MITF and PAX3 expression has been proposed as a switch model in which MITF and miR-211, residing in the sixth intron of TRPM1, can activate one cellular program while suppressing another one driven by PAX3 and BRN2 [9, 21, 74]. MITF repression is also mediated by DEC1 (differentially expressed in chondrocytes protein 1) whose recruitment to the MITF promoter is regulated by HIF1 (hypoxia-inducible factor 1) [75]. As HIF1α is a MITF target [76] and can be expressed in melanoma cells not only under hypoxic conditions [77], this mechanism constitutes an interesting negative feedback loop regulating MITF expression.
Several proteins have been found to indirectly suppress MITF expression by acting as upstream inhibitors of positive regulators of MITF expression (Fig. 1). Independently of its effect on GLI2, TGFβ inhibits PKA that otherwise promotes CREB-dependent MITF transcription [68]. DKK1 (Dickkopf-1), a secreted inhibitor of the Wnt/β-catenin pathway, has been shown to suppress both MITF expression and the MITF-dependent differentiation program [78]. Accordingly, both DKK1 expression and secretion have been substantially reduced in the multicellular anchorage-independent melanospheres showing high expression of MITF and numerous MITF target genes [79]. ATF2 (activating transcription factor 2) displays inhibitory activity towards SOX10 both in melanocytes and melanoma cells, resulting in a decreased MITF transcript level [80]. A co-immunoprecipitation approach confirmed selective affinity of BCSC1 (breast cancer suppressor candidate-1) to SOX10, but not other MITF regulators such as CREB, and down-regulated MITF mRNA level was observed upon BCSC-1 overexpression [81]. MITF expression is also affected by IFN-γ (interferon γ) that inhibits CREB binding to the MITF promoter by inducing the association of CBP (CREB binding protein) with STAT1 [82].
Regulation of MITF transcript stability
Melanoma cells show high expression of CRD-BP (coding region determinant-binding protein), an mRNA-binding protein that has been found to stabilize MITF transcript [83]. MITF transcript is also under control of several small non-coding RNAs, microRNAs (miRs), which promote mRNA degradation or suppress protein synthesis via binding to 3′-UTR of a target transcript [23]. miR-137, located in the locus 1p22, negatively regulates MITF [84, 85]. No mutations have been found in the putative miR-137-binding sites in the MITF mRNA 3′-UTR, however, miR-137 possesses a 15-bp tandem repeat in the pre-miR-137 sequence that alters the processing and function of miR-137 in melanoma cell lines [84]. In metastatic melanoma samples, MITF transcript has been determined as a target of miR-182-mediated degradation [86]. miR-182 is a member of the miR cluster residing in a chromosomal locus (7q31–34) frequently amplified in melanomas. Interestingly, overexpression of MITF has been related to the suppression of the miR-182-dependent pro-invasive effect [86]. p53-dependent miR-182 has been also found to down-regulate MITF in uveal melanoma [87]. The 3′-UTR of MITF transcript is also targeted by miR-148 [88], miR-101 [89] and miR-218, and inverse correlation between MITF and miR-218 has been observed in melanocytes and melanoma cell lines [90]. Notably, exosome-dependent miR exchange between melanoma cells may influence MITF transcript level as well [for review 91].
Regulation of MITF protein level and activity
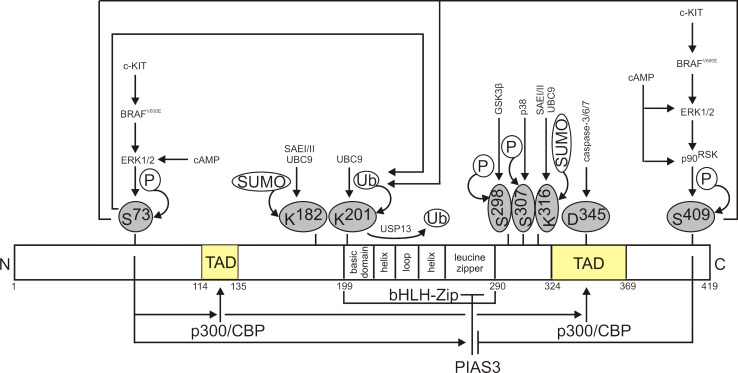
The transcriptional activity of MITF depends on its post-translational modifications and availability of co-operating partners (Fig. 2). MITF can be regulated by phosphorylation maintained by ERK1/2 (at Ser73), p90RSK (at Ser409) [92], GSK3β (at Ser298) [93, 94] and p38 (at Ser307) [95]. In general, phosphorylation enhances transcriptional activity of MITF [94, 95; for review 96]. The phosphorylation at Ser73 promotes the interaction with a MITF co-factor, histone acetyl transferase p300/CBP within the transactivation domain of MITF [92]. On the other hand, this modification promotes the binding of PIAS3 (protein inhibitor of activated STAT3) that involves the N-terminal fragment of PIAS3 and the leucine zipper of MITF [97, 98]. Interaction with PIAS3 leads to the attenuation of MITF transcriptional activity. This effect is, however, inhibited when MITF is phosphorylated at Ser409 in a p90RSK-dependent manner [97]. The phosphorylation of MITF at Ser73, a residue located within a degradation-promoting PEST sequence, is also a prerequisite to the MITF proteasome-dependent turnover [99] e.g., in response to ultraviolet C radiation [100]. Lys201 has been identified as a site of UBC9-mediated ubiquitylation of MITF [99]. Proteasome-mediated MITF protein degradation has also been observed after double phosphorylation at Ser73 and Ser409 [92]. An unphosphorylatable mutant at Ser73/Ser409 has been very stable but transcriptionally incompetent [92], indicating that signals promoting transcriptional activity and degradation of MITF protein are coupled in melanoma cells. Both phosphorylations promoting MITF degradation depend on melanoma-specific BRAFV600E causing the enhanced activation of MAPK (MAP kinase)/ERK1/2 pathway [101]. Deubiquitinase USP13 (ubiquitin-specific protease 13) has been linked to the protection of MITF from proteasomal degradation in melanoma cells [102]. MITF can be also processed by the effector caspases. It has been demonstrated in melanoma cells that MITF-derived C-terminal peptide cleaved by these proteases has a pro-apoptotic function [103].
Fig. 2.
A schematic domain structure of MITF-M protein, a melanocyte/melanoma-specific isoform, and its key regulatory mechanisms. MITF-M comprises 419 amino acids. The functional domains of MITF-M common for all isoforms are encoded by the exons from 2 to 9. Phosphorylation enhances the transcriptional activity of MITF. However, this modification may also promote proteasome-dependent degradation of MITF, or enhance interaction between MITF and p300/CBP. MITF can be subjected to USP13-mediated deubiquitylation, thus preventing MITF from proteasomal degradation. In addition, MITF can be a target for other modifications, including SUMOylation and caspase-mediated cleavage
MITF activity is also modulated by SUMOylation at two lysine residues, Lys182 and Lys316 [104]. SUMOylation of MITF depends on an E1 SUMO-activating heterodimeric enzyme SAEI/SAEII and E2 SUMO-conjugating enzyme UBC9 [104] (Fig. 2). It has been concluded that this modification plays an essential role in the regulation of MITF activity, and non-SUMOylatable MITF mutants displayed increased transcriptional activity on distinct sets of target genes [48, 49, 104, 105]. It has been also indicated that PIAS3 can promote MITF SUMOylation at both SUMOylation sites [105]; however, this observation has not been clearly supported by another report [104]. A recent study on melanoma patients bearing the MITFE318K variant [51], in which a point mutation occurs at the consensus binding site for SUMOylation [105], supports the conclusion that this substitution does not affect MITF protein stability and nuclear localization.
Besides aforementioned regulations, MITF activity also depends on the availability of co-operating partners such as p300/CBP [92]. p300/CBP is a versatile regulator that links transcription factors bound to DNA with a basal transcriptional machinery, thus promoting the assembly of pre-initiation complex [for review 106]. Immunofluorescence and immunoprecipitation studies have demonstrated that MITF can physically interact with BRM and BRG1, and depending on the type of SWI/SNF complex composed of either the BRG1 or BRM subunit, different sets of MITF-dependent genes are activated [107]. This, however, is not an exclusive role of the BRG1-containing SWI/SNF complex in melanoma since its MITF-independent activity has been shown as well [108]. SOX10 can synergistically activate the MITF-dependent genes as demonstrated for MET [109]. It has been also shown that MITF can redirect β-catenin from the Wnt signaling pathway, and engage it to the activation of MITF-dependent genes [110]. Thus, SWI/SNF complex, SOX10 and β-catenin can function not only as activators of MITF expression (Fig. 1), but also as its co-factors. HINT1 (histidine triad nucleotide-binding protein 1) has been identified as an inhibitor of transcriptional activity of MITF acting through binding the chromatin at the MITF sites [111]. Moreover, since HINT1 expression is lost in primary melanomas [111], it may support a role of MITF in melanomagenesis.
The mechanisms behind the regulation of MITF level and activity are still being explored. Most recently, very interesting correlations have been observed between activity of MITF and other transcription factors, probably not operating on the MITF promoter (Fig. 1). HOXA1 (homeobox transcription factor 1) has been identified as a potent inhibitor of MITF expression whose activity might be concomitant with the activation of the TGFβ pathway [112]. An interesting repression mechanism has been reported in melanoma cells expressing IL-1R (interleukin-1 receptor). The stimulation of melanoma cells with interleukin-1α or 1β resulted in the reduction of MITF expression, and it has been suggested that this process is NF-κB-dependent [113]. Accordingly, a suppressive role of the NF-κB signaling on MITF level has been reported [114]. A reverse correlation has been observed between MYC-related chromosomal copy number gains in 8q24 and MITF expression [115]. In contrast, suppression of ETV1 (E twenty-six variant 1) was associated with a decreased level of MITF protein [116]. Whether c-MYC and ETV1 can act as direct regulators of MITF remains to be elucidated. Other poorly characterized regulators have been associated with MITF level, and the detailed mechanisms of their actions need to be clarified. PRMT5 (protein arginine methyltransferase 5) is an enzyme involved in post-translational protein modifications. PRMT5 expression has been found to be increased in melanoma, and siRNA-mediated depletion of PRMT5 resulted in a substantial decrease in the level of MITF protein indicating a positive regulatory effect [117]. A similar influence on the MITF level has been demonstrated for PDE4D (phosphodiesterase subtype 4D), and PDE4D-depleted cells have shown a decreased MITF transcript level [118].
Final conclusions
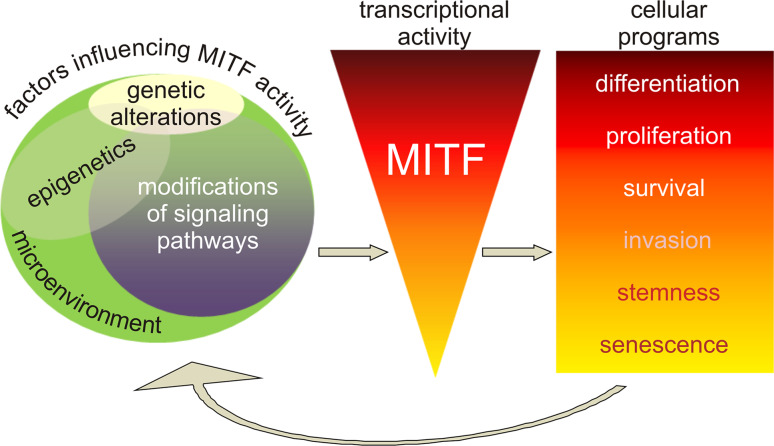
MITF operates within a wide range of activity levels determining melanoma cell fate (Fig. 3) [20, 24–27; for review 22, 23, 28]. Melanoma cells expressing MITF at high level can either differentiate or proliferate. Low activity of MITF is related to stem cell-like or invasive potential. Finally, long-term MITF suppression drives cell senescence. Although genetic alterations, including mutations and amplification of MITF, are found in melanoma samples [4, 46, 48, 49], fluctuating MITF activity in melanoma cells is rather due to microenvironmental cues, critical epigenetic states and modifications of upstream signaling pathways [7, 8, 10, 79, 104–138]. Different combinations of those factors determine transcriptional activity of MITF which in turn contributes to diverse cellular capabilities. This may explain a variable MITF expression across melanoma specimens but also between different areas of individual tumor samples reflecting both inter-tumoral heterogeneity and diversity of melanoma cell subpopulations comprising a tumor mass [123, 124; for review 125].
Fig. 3.
MITF expression and activity in melanoma cells are determined by genetic alterations, epigenetics, changes in upstream signaling pathways and microenvironment. Different combinations of those factors result in varied transcriptional activity of MITF which contributes to diverse cellular programs from differentiation and proliferation at high level of MITF activity to stemness and senescence at the lowest one. However, the outcome is not stable and can be modified by fluctuations in microenvironment-dependent critical epigenetic states and signaling pathways. Several MITF-dependent feedback mechanisms are also determined
MITF does not exhibit a druggable target, and MITF-aimed approaches are thought to be rather based on the modulation of its upstream regulatory pathways [126, 127]. A number of natural and synthetic compounds have been used to modify MITF activity. A few natural compounds that either reduced or increased MITF transcript level in heterogeneous patient-derived melanoma populations were identified in our laboratory [128]. A dietary flavonoid fisetin that targets Wnt/β-catenin pathway has substantially reduced MITF expression and influenced MITF-dependent cellular processes [129]. Downregulation of MITF at the transcriptional level was observed for ciglitazone that also showed anti-melanoma effects in vivo [130]. Hirsein A reduced the expression of MITF by modulating the expression of diverse components of MAPK signaling pathway [131]. Several studies have linked a high MITF level with the resistance to MAPK-pathway inhibitors [132, 133; for review 134–136]. MITF targets are up-regulated by MAPK-pathway inhibitors [137], and enforced expression of MITF in BRAFV600E melanoma cells promotes resistance towards inhibitors of RAF, MEK and ERK [for review 134]. As MITF expression can be reduced by histone deacetylase inhibitors (HDACi) [138], combined HDAC and MAPK inhibition was shown to prevent MITF-driven resistance in melanoma cells [132]. Another study suggests, however, that intrinsically resistant melanomas can be characterized by low expression/activity of MITF accompanied by enhanced activity of NF-κB signaling, and BRAF inhibition in MITF-high, drug-sensitive cells induces a transition to the MITF-low/NF-κB–high state [114]. Most recent findings that modulation of MITF activity can drive phenotype switching in vivo, and abrogating MITF activity in melanoma leads to tumor regression, but a low level of wild-type MITF is oncogenic [27] indicate that further studies are needed.
Acknowledgments
Research in the authors’ laboratory is supported by Grant 2011/01/B/NZ4/04921 from the National Science Centre.
References
- 1.Park HY, Kosmadaki M, Yaar M, Gilchrest BA. Cellular mechanisms regulating human melanogenesis. Cell Mol Life Sci. 2009;66(9):1493–1506. doi: 10.1007/s00018-009-8703-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011;24(3):411–421. doi: 10.1111/j.1755-148X.2011.00834.x. [DOI] [PubMed] [Google Scholar]
- 3.Haq R, Fisher DE. Biology and clinical relevance of the microphthalmia family of transcription factors in human cancer. J Clin Oncol. 2011;29(25):3474–3482. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- 4.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 5.Kumar SM, Dai J, Li S, Yang R, Yu H, Nathanson KL, Liu S, Zhou H, Guo J, Xu X. Human skin neural crest progenitor cells are susceptible to BRAF(V600E)-induced transformation. Oncogene. 2013;33(7):832–841. doi: 10.1038/onc.2012.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regad T. Molecular and cellular pathogenesis of melanoma initiation and progression. Cell Mol Life Sci. 2013;70(21):4055–4065. doi: 10.1007/s00018-013-1324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, Bonvin E, Goding C, Sahai E. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res. 2009;69(20):7696–7977. doi: 10.1158/0008-5472.CAN-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurber AE, Douglas G, Sturm EC, Zabierowski SE, Smit DJ, Ramakrishnan SN, Hacker E, Leonard JH, Herlyn M, Sturm RA. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene. 2011;30(27):3036–3048. doi: 10.1038/onc.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R, Levy C. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol. 2014;134(2):441–451. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 10.Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, Bahadoran P, Lacour JP, Tartare-Deckert S, Bertolotto C, Ballotti R. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31(19):2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- 11.Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsdteinsdottir K, Schepsky A, Dummer R, Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21(6):665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 12.Giuliano S, Cheli Y, Ohanna M, Bonet C, Beuret L, Bille K, Loubat A, Hofman V, Hofman P, Ponzio G, Bahadoran P, Ballotti R, Bertolotto C. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res. 2010;70(9):3813–3822. doi: 10.1158/0008-5472.CAN-09-2913. [DOI] [PubMed] [Google Scholar]
- 13.Widmer DS, Cheng PF, Eichhoff OM, Belloni BC, Zipser MC, Schlegel NC, Javelaud D, Mauviel A, Dummer R, Hoek KS. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 2012;25(3):343–353. doi: 10.1111/j.1755-148X.2012.00986.x. [DOI] [PubMed] [Google Scholar]
- 14.Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, Ohgami N, Tamura H, Yamanoshita O, Kawamoto Y, Furukawa K, Kato M. Molecular network associated with MITF in skin melanoma development and progression. J Skin Cancer. 2011;2011:730170. doi: 10.1155/2011/730170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys. 2014;S0003–9861(14):00270–00277. doi: 10.1016/j.abb.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman ML, Czyz M. Pro-survival role of MITF in melanoma. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.319. [DOI] [PubMed] [Google Scholar]
- 17.Hartman ML, Czyz M. Anti-apoptotic proteins on guard of melanoma cell survival. Cancer Lett. 2013;331(1):24–34. doi: 10.1016/j.canlet.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, Wargo JA, Song JS, Fisher DE, Arany Z, Widlund HR. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23(3):1–14. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci. 2014;71(13):2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichhoff OM, Weeraratna A, Zipser MC, Denat L, Widmer DS, Xu M, Kriegl L, Kirchner T, Larue L, Dummer R, Hoek KS. Differential LEF1 and TCF4 expression is involved in melanoma cell phenotype switching. Pigment Cell Melanoma Res. 2011;24(4):631–642. doi: 10.1111/j.1755-148X.2011.00871.x. [DOI] [PubMed] [Google Scholar]
- 21.Eccles MR, He S, Ahn A, Slobbe LJ, Jeffs AR, Yoon HS, Baguley BC. MITF and PAX3 play distinct roles in melanoma cell migration; outline of a “genetic switch” theory involving MITF and PAX3 in proliferative and invasive phenotypes of melanoma. Front Oncol. 2013;3:229. doi: 10.3389/fonc.2013.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goding CR. A picture of Mitf in melanoma immortality. Oncogene. 2011;30(20):2304–2306. doi: 10.1038/onc.2010.641. [DOI] [PubMed] [Google Scholar]
- 23.Bell RE, Levy C. The three m’s: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 2011;24(6):1088–1106. doi: 10.1111/j.1755-148X.2011.00931.x. [DOI] [PubMed] [Google Scholar]
- 24.Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006;20(24):3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheli Y, Giuliano S, Botton T, Rocchi S, Hofman V, Hofman P, Bahadoran P, Bertolotto C, Ballotti R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and differentiated progeny. Oncogene. 2011;30(20):2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- 26.Javelaud D, Alexaki VI, Pierrat MJ, Hoek KS, Dennler S, Van Kempen L, Bertolotto C, Ballotti R, Saule S, Delmas V, Mauviel A. GLI2 and M-MITF transcription factors control exclusive gene expression programs and inversely regulate invasion in human melanoma cells. Pigment Cell Melanoma Res. 2011;24(5):932–943. doi: 10.1111/j.1755-148X.2011.00893.x. [DOI] [PubMed] [Google Scholar]
- 27.Lister JA, Capper A, Zeng Z, Mathers ME, Richardson J, Paranthaman K, Jackson IJ, Patton EE. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J Invest Dermatol. 2014;134:133–140. doi: 10.1038/jid.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23(6):746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 29.Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 30.Amae S, Fuse N, Yasumoto K, Sato S, Yajima I, Yamamoto H, Udono T, Durlu YK, Tamai M, Takahashi K, Shibahara S. Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem Biophys Res Commun. 1998;247(3):710–715. doi: 10.1006/bbrc.1998.8838. [DOI] [PubMed] [Google Scholar]
- 31.Udono T, Yasumoto K, Takeda K, Amae S, Watanabe K, Saito H, Fuse N, Tachibana M, Takahashi K, Tamai M, Shibahara S. Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim Biophys Acta. 2000;1491(1–3):205–219. doi: 10.1016/s0167-4781(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 32.Fuse N, Yasumoto KI, Takeda K, Amae S, Yoshizawa M, Udono T, Takahashi K, Tamai M, Tomita Y, Tachibana M, Shibahara S. Molecular cloning of cDNA encoding a novel microphthalmia-associated transcription factor isoform with a distinct amino-terminus. J Biochem. 1999;126(6):1043–1051. doi: 10.1093/oxfordjournals.jbchem.a022548. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Yasumoto K, Kawaguchi N, Udono T, Watanabe K, Saito H, Takahashi K, Noda M, Shibahara S. Mitf-D, a newly identified isoform, expressed in the retinal pigment epithelium and monocyte-lineage cells affected by Mitf mutations. Biochim Biophys Acta. 2002;1574(1–3):15–23. doi: 10.1016/s0167-4781(01)00339-6. [DOI] [PubMed] [Google Scholar]
- 34.Oboki K, Morii E, Kataoka TR, Jippo T, Kitamura Y. Isoforms of mi transcription factor preferentially expressed in cultured mast cells of mice. Biochim Biophys Res Commun. 2002;290(4):1250–1254. doi: 10.1006/bbrc.2002.6332. [DOI] [PubMed] [Google Scholar]
- 35.Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8(3):256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 36.Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347(1):73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Takemoto CM, Yoon YJ, Fisher DE. The identification and functional characterization of a novel mast cell isoform of the microphthalmia-associated transcription factor. J Biol Chem. 2002;277(33):30244–30252. doi: 10.1074/jbc.M201441200. [DOI] [PubMed] [Google Scholar]
- 38.Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 39.Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8(3):251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 40.Widlund HR, Fisher DE. Microphthalmia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22(20):3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Radfar S, Liu S, Riker AI, Khong HT. Mitf-Mdel, a novel melanocyte/melanoma-specific isoform of microphthalmia-associated transcription factor-M as a candidate biomarker for melanoma. BMC Med. 2010;8:14. doi: 10.1186/1741-7015-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vachtenheim J, Drdova B. A dominant negative mutant of microphthalmia transcription factor (MITF) lacking two transactivation domains suppresses transcription mediated by wild type MITF and a hyperactive MITF derivative. Pigment Cell Res. 2004;17(1):43–50. doi: 10.1046/j.1600-0749.2003.00108.x. [DOI] [PubMed] [Google Scholar]
- 43.Pogenberg V, Ogmundsdottir MH, Bergsteinsdottir K, Schepsky A, Phung B, Deineko V, Milewski M, Steingrimsson E, Wilmanns M. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012;26(23):2647–2658. doi: 10.1101/gad.198192.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocytic lineage. Genes Dev. 2000;14(14):1712–1728. [PubMed] [Google Scholar]
- 45.Harbst K, Lauss M, Cirenajwis H, Winter C, Howlin J, Torngren T, Kvist A, Nodin B, Olsson E, Hakkinen J, Jirstrom K, Staaf J, Lundgren L, Olsson H, Ingvar C, Gruvberger-Saal SK, Saal LH, Jonsson G. Molecular and genetic diversity in the metastatic process of melanoma. J Pathol. 2014;233(1):39–50. doi: 10.1002/path.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, Vemula S, Burrell AS, Agrawal NS, Lin JC, Banister CE, Buckhaults P, Rosenberg SA, Bastian BC, Pavan WJ, Samuels Y. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;229(4):435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grill C, Bergsteinsdottir K, Ogmundsdottir MH, Pogenberg V, Schepsky A, Wilmanns M, Pingault V, Steingrimsson E. MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum Mol Genet. 2013;22(21):4357–4367. doi: 10.1093/hmg/ddt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d’Hayer B, Mohamdi H, Remenieras A, Maubec E, de la Fouchardiere A, Molinie V, Vabres P, Dalle S, Poulalhon N, Martin-Denavit T, Thomas L, Andry-Benzaquen P, Dupin N, Boitier F, Rossi A, Perrot JL, Labeille B, Robert C, Escudier B, Caron O, Brugieres L, Saule S, Gardie B, Gad S, Richard S, Couturier J, Teh BT, Ghiorzo P, Pastorino L, Puig S, Badenas C, Olsoon H, Ingvar C, Rouleau E, Lidereau R, Bahadoran P, Vielh P, Corda E, Blanche H, Zelenika D, Galan P, Aubin F, Bachollet B, Becuwe C, Berthet P, Bignon YJ, Bonadona V, Bonafe JL, Bonnett-Dupeyron MN, Cambazard F, Chevrant-Breton J, Coupier I, Dalac S, Demange L, d’Incan M, Dugast C, Faivre L, Vincent-Fetita L, Gauthier-Villars M, Gilbert B, Grange F, Grob JJ, Humbert P, Janin N, Joly P, Kerob D, Lasset C, Leroux D, Levang J, Limacher JM, Livideanu C, Longy M, Lortholary A, Stoppa-Lyonnet D, Mansard S, Mansuy L, Marrou K, Mateus C, Maugard C, Meyer N, Nogues C, Souteyrand P, Venat-Bouvet L, Zattara H, Chaudru V, Lenoir GM, Lathrop M, Davidson I, Avril MF, Demenais F, Ballotti R, Bressac-de-Paillerets B, French Familial Melanoma Study Group A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, Taylor JC, Duffy DL, Holohan K, Dutton-Regester K, Palmer JM, Bonazzi V, Stark MS, Symmons J, Law MH, Schmidt C, Lanagan C, O’Connor L, Holland EA, Schmidt H, Maskiell JA, Jetann J, Ferguson M, Jenkins MA, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Whiteman DC, Pharoah PD, Easton DF, Dunning AM, Newton-Bishop JA, Montgomery GW, Martin NG, Mann GJ, Bishop DT, Tsao H, Trent JM, Fisher DE, Hayward NK, Brown KM. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480(7375):99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghiorzo P, Pastorino L, Queirolo P, Bruno W, Tibiletti MG, Nasti S, Andreotti V, Paillerets BB, Bianchi Scarra G, Genoa Pancreatic Cancer Study Group Prevalence of the MITF E318 K germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res. 2013;26(2):259–262. doi: 10.1111/pcmr.12047. [DOI] [PubMed] [Google Scholar]
- 51.Sturm RA, Fox C, McClenahan P, Jagirdar K, Ibarrola-Villava M, Banan P, Abbott NC, Ribas G, Gabrielli B, Duffy DL, Soyer HP. Phenotypic characterization of nevus and tumor patterns in MITF E318 K mutation carrier melanoma patients. J Invest Dermatol. 2014;134(1):141–149. doi: 10.1038/jid.2013.272. [DOI] [PubMed] [Google Scholar]
- 52.Bismuth K, Maric D, Arnheiter H. MITF and cell proliferation: the role of alternative splice forms. Pigment Cell Res. 2005;18(5):349–359. doi: 10.1111/j.1600-0749.2005.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Primot A, Mogha A, Corre S, Roberts K, Debbache J, Adamski H, Dreno B, Khammari A, Lesimple T, Mereau A, Goding CR, Galibert MD. ERK-regulated differential expression of the Mitf 6a/b splicing isoforms in melanoma. Pigment Cell Melanoma Res. 2010;23(1):93–102. doi: 10.1111/j.1755-148X.2009.00652.x. [DOI] [PubMed] [Google Scholar]
- 54.Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem. 2000;275(40):30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- 55.Zhu S, Wurdak H, Wang Y, Galkin A, Tao H, Li J, Lyssiotis CA, Yan F, Tu BP, Miraglia L, Walker J, Sun F, Orth A, Schulz PG, Wu X. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proc Natl Acad Sci USA. 2009;106(40):17025–17030. doi: 10.1073/pnas.0909292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huber WE, Price ER, Widlund HR, Du J, Davies IJ, Wegner M, Fisher DE. A tissue-restricted cAMP transcriptional response: sOX10 modulates alpha-melanocyte-stimulating hormone-triggered expression of microphthalmia-associated transcription factor in melanocytes. J Biol Chem. 2003;278(46):45224–45230. doi: 10.1074/jbc.M309036200. [DOI] [PubMed] [Google Scholar]
- 57.Saha B, Singh SK, Sarkar C, Bera R, Ratha J, Tobin DJ, Bhadra R. Activation of the Mitf promoter by lipid-stimulated activation of p38-stress signaling to CREB. Pigment Cell Res. 2006;19(6):595–605. doi: 10.1111/j.1600-0749.2006.00348.x. [DOI] [PubMed] [Google Scholar]
- 58.Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15(8):2583–2587. doi: 10.1158/1078-0432.CCR-08-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sestakova B, Ondrusova L, Vachtenheim J. Cell cycle inhibitor p21/WAF1/CIP1 as a cofactor of MITF expression in melanoma cells. Pigment Cell Melanoma Res. 2010;23(2):238–251. doi: 10.1111/j.1755-148X.2010.00670.x. [DOI] [PubMed] [Google Scholar]
- 60.Saito H, Yasumoto KI, Takeda K, Takahashi K, Fukuzaki A, Orikasa S, Shibahara S. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J Biol Chem. 2002;277(32):28787–28794. doi: 10.1074/jbc.M203719200. [DOI] [PubMed] [Google Scholar]
- 61.Bellei B, Pitisci A, Catricala C, Larue L, Picardo M. Wnt/beta-catenin signaling is stimulated by alpha-melanocyte-stimulating hormone in melanoma and melanocyte cells: implication in cell differentiation. Pigment Cell Melanoma Res. 2010;24(2):309–325. doi: 10.1111/j.1755-148X.2010.00800.x. [DOI] [PubMed] [Google Scholar]
- 62.Tatari MN, De Craene B, Soen B, Taminau J, Vermassen P, Goossens S, Haigh K, Cazzola S, Lambert J, Huylebroeck D, Haigh JJ, Berx G. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell Mol Life Sci. 2014;71(18):3599–3609. doi: 10.1007/s00018-014-1589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci. 2012;69(15):2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A, Postigo A. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69(20):3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denecker G, Vandamme N, Akay O, Koludrovic D, Taminau J, Lemeire K, Gheldof A, De Craene B, Van Gele M, Brochez L, Udupi GM, Rafferty M, Balint B, Gallagher WM, Ghanem G, Huylebroeck D, Haigh J, van den Oord J, Larue L, Davidson I, Marine JC, Berx G. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014;21(8):1250–1261. doi: 10.1038/cdd.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Ye F, Li Q, Tamiya S, Darling DS, Kaplan HJ, Dean DC. Zeb1 represses Mitf and regulates pigment synthesis, cell proliferation, and epithelial morphology. Invest Ophthalmol Vis Sci. 2009;50(11):5080–5088. doi: 10.1167/iovs.08-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vachtenheim J, Ondrusova L, Borovansky J. SWI/SNF chromatin remodeling complex is critical for the expression of microphthalmia-associated transcription factor in melanoma cells. Biochem Biophys Res Commun. 2010;392(3):454–459. doi: 10.1016/j.bbrc.2010.01.048. [DOI] [PubMed] [Google Scholar]
- 68.Pierrat MJ, Marsaud V, Mauviel A, Javelaud D. Expression of microphthalmia-associated transcription factor (MITF), which is critical for melanoma progression, is inhibited by both transcription factor GLI2 and transforming growth factor β. J Biol Chem. 2012;287(22):17996–18004. doi: 10.1074/jbc.M112.358341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang G, Li Y, Nishimura EK, Xin H, Zhou A, Guo Y, Dong L, Denning MF, Nickoloff BJ, Cui R. Inhibition of PAX3 by TGF-beta modulates melanocyte viability. Mol Cell. 2008;32(4):554–563. doi: 10.1016/j.molcel.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 70.He S, Li CG, Slobbe L, Glover A, Marshall E, Baguley BC, Eccles MR. PAX3 knockdown in metastatic melanoma cell lines does not reduce MITF expression. Melanoma Res. 2011;21(1):24–34. doi: 10.1097/CMR.0b013e328341c7e0. [DOI] [PubMed] [Google Scholar]
- 71.Bonvin E, Falletta P, Shaw H, Delmas V, Goding CR. A phosphatidylinositol 3-kinase-PAX3 axis regulates Brn-2 expression in melanoma. Mol Cell Biol. 2012;32(22):4674–4683. doi: 10.1128/MCB.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu F, Cao J, Wu J, Sullivan K, Shen J, Ryu B, Xu Z, Wei W, Cui R. Stat3-targeted therapies overcome the acquired resistance to vemurafenib in melanomas. J Invest Dermatol. 2013;133(8):2041–2049. doi: 10.1038/jid.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodall J, Carreira S, Denat L, Kobi D, Davidson I, Nuciforo P, Sturm RA, Larue L, Goding CR. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res. 2008;68(19):7788–7794. doi: 10.1158/0008-5472.CAN-08-1053. [DOI] [PubMed] [Google Scholar]
- 74.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24(3):525–537. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 75.Feige E, Yokoyama S, Levy C, Khaled M, Igras V, Lin RJ, Lee S, Widlund HR, Granter SR, Kung AL, Fisher DE. Hypoxia-mediated transcriptional repression of the melanoma-associated oncogene MITF. Proc Natl Acad Sci USA. 2011;108(43):E924–E933. doi: 10.1073/pnas.1106351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Busca R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribere L, Bertolotto C, Virolle T, Barbry P, Pouyssegur J, Ponzio G, Ballotti R. Hypoxia-inducible factor 1α is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol. 2005;170(1):49–59. doi: 10.1083/jcb.200501067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mills CN, Joshi SS, Niles RM. Expression and function of hypoxia inducible factor-1 alpha in human melanoma under non-hypoxic conditions. Mol Cancer. 2009;8:104. doi: 10.1186/1476-4598-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang I, Park JH, Park HS, Choi KA, Seol KC, Oh SI, Kang S, Hong S. Neural stem cells inhibit melanin production by activation of Wnt inhibitors. J Dermatol Sci. 2013;72(3):274–283. doi: 10.1016/j.jdermsci.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 79.Hartman ML, Talar B, Noman Z, Gajos-Michniewicz A, Chouaib S, Czyz M. Gene expression profiling identifies microphthalmia-associated transcription factor (MITF) and Dickkopf-1 (DKK1) as regulators of microenvironment-driven alterations in melanoma phenotype. PLoS One. 2014;9(4):e95157. doi: 10.1371/journal.pone.0095157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shah M, Bhoumik A, Goel V, Dewing A, Breitwieser W, Kluger H, Krajewski S, Krajewska M, Dehart J, Lau E, Kallenberg DM, Jeong H, Eroshkin A, Bennett DC, Chin L, Bosenberg M, Jones N, Ronai ZA. A role for ATF2 in regulating MITF and melanoma development. PLoS Genet. 2010;6(12):e1001258. doi: 10.1371/journal.pgen.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anghel SI, Correa-Rochal R, Budinska E, Boligan KF, Abraham S, Colombetti S, Fontao L, Mariotti A, Rimoldi D, Ghanem GE, Fisher DE, Levy F, Delorenzi M, Piguet V. Breast cancer suppressor candidate-1 (BCSC-1) is a melanoma tumor suppressor that down regulates MITF. Pigment Cell Melanoma Res. 2012;25(4):482–487. doi: 10.1111/j.1755-148X.2012.01018.x. [DOI] [PubMed] [Google Scholar]
- 82.Son J, Kim M, Jou I, Park KC, Kang HY. IFN-γ inhibits basal and α-MSH-induced melanogenesis. Pigment Cell Melanoma Res. 2014;27(2):201–208. doi: 10.1111/pcmr.12190. [DOI] [PubMed] [Google Scholar]
- 83.Craig EA, Spiegelman VS. Inhibition of coding region determinant binding protein sensitizes melanoma cells to chemotherapeutic agents. Pigment Cell Melanoma Res. 2011;25(1):83–87. doi: 10.1111/j.1755-148X.2011.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68(5):1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 85.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmuller SB. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133(3):768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 86.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci USA. 2009;106(6):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan D, Dong XD, Chen X, Yao S, Wang L, Wang J, Wang C, Hu DN, Qu J, Tu L. Role of microRNA-182 in posterior uveal melanoma: regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One. 2012;7(7):e40967. doi: 10.1371/journal.pone.0040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E. miR-148 regulates Mitf in melanoma cells. PLoS One. 2010;5(7):e11574. doi: 10.1371/journal.pone.0011574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luo C, Merz PR, Chen Y, Dickes E, Pscherer A, Schadendorf D, Eichmuller SB. MiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2. Cancer Lett. 2013;341(2):240–247. doi: 10.1016/j.canlet.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 90.Guo J, Zhang JF, Wang WM, Cheung FW, Lu YF, Ng CF, Kung HF, Liu WK. MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression. RNA Biol. 2014;11(6):732–741. doi: 10.4161/rna.28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gajos-Michniewicz A, Duechler M, Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347(1):29–37. doi: 10.1016/j.canlet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Wu M, Hemesath TJ, Takemoto CM, Horstmann WA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000;14(3):301–312. [PMC free article] [PubMed] [Google Scholar]
- 93.Takeda K, Takemoto C, Kobayashi I, Watanabe A, Nobukuni Y, Fisher DE, Tachibana M. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum Mol Genet. 2000;9(1):125–132. doi: 10.1093/hmg/9.1.125. [DOI] [PubMed] [Google Scholar]
- 94.Terragni J, Nayak G, Banerjee S, Medrano JL, Graham JR, Brennan JF, Sepulveda S, Cooper GM. The E-box binding factors Max/Mnt, MITF, and USF1 act coordinately with FoxO to regulate expression of proapoptotic and cell cycle control genes by phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3 signaling. J Biol Chem. 2011;286(42):36215–36227. doi: 10.1074/jbc.M111.246116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J Biol Chem. 2002;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- 96.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12(9):406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 97.Levy C, Sonnenblick A, Razin E. Role played by microphthalmia transcription factor phosphorylation and its zip domain in its transcriptional inhibition by PIAS3. Mol Cell Biol. 2003;23(24):9073–9080. doi: 10.1128/MCB.23.24.9073-9080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levy C, Lee YN, Nechushtan H, Schueler-Furman O, Sonnenblick A, Hacohen S, Razin E. Identifying a common molecular mechanism for inhibition of MITF and STAT3 by PIAS3. Blood. 2006;107(7):2839–2845. doi: 10.1182/blood-2005-08-3325. [DOI] [PubMed] [Google Scholar]
- 99.Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res. 2000;255(2):135–143. doi: 10.1006/excr.2000.4803. [DOI] [PubMed] [Google Scholar]
- 100.Liu F, Singh A, Yang Z, Garcia A, Kong Y, Meyskens FL., Jr MiTF links Erk1/2 kinase and p21CIP1/WAF1 activation after UVC radiation in normal human melanocytes and melanoma cells. Mol Cancer. 2010;9:214. doi: 10.1186/1476-4598-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One. 2008;3(7):e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun. 2011;2:414. doi: 10.1038/ncomms1421. [DOI] [PubMed] [Google Scholar]
- 103.Larribere L, Hilmi C, Khaled M, Gaggioli C, Bille K, Auberger P, Ortonne JP, Ballotti R, Bertolotto C. The cleavage of microphthalmia-associated transcription factor, MITF, by caspases plays an essential role in melanocyte and melanoma cell apoptosis. Genes Dev. 2005;19(17):1980–1985. doi: 10.1101/gad.335905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18(4):265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem. 2005;280(1):146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 106.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013;70(21):3989–4008. doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2010;29(1):81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ondrusova L, Vachtenheim J, Reda J, Zakova P, Benkova K. MITF-independent pro-survival role of BRG1-containing SWI/SNF complex in melanoma cells. PLoS ONE. 2013;8(1):e54110. doi: 10.1371/journal.pone.0054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mascarenhas JB, Littlejohn EL, Wolsky RJ, Young KP, Nelson M, Salgia R, Lang D. PAX3 and SOX10 activate MET receptor expression in melanoma. Pigment Cell Melanoma Res. 2010;23(2):225–237. doi: 10.1111/j.1755-148X.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schepsky A, Bruser K, Gunnarsson GJ, Goodall J, Hallsson JH, Goding CR, Steingrimsson E, Hecht A. The microphthalmia-associated transcription factor Mitf interacts with beta-catenin to determine target gene expression. Mol Cell Biol. 2006;26(23):8914–8927. doi: 10.1128/MCB.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Genovese G, Ghosh P, Li H, Rettino A, Sioletic S, Cittadini A, Sgambato A. The tumor suppressor HINT1 regulates MITF and β-catenin transcriptional activity in melanoma cells. Cell Cycle. 2012;11(11):2206–2215. doi: 10.4161/cc.20765. [DOI] [PubMed] [Google Scholar]
- 112.Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, Creighton CJ, Chin L, Scott KL. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33(8):1017–1026. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kholmanskikh O, van Baren N, Brasseur F, Ottaviani S, Vanacker J, Arts N, van der Bruggen P, Coulie P, De Plaen E. Interleukins 1α and 1β secreted by some melanoma cell lines strongly reduce expression of MITF-M and melanocyte differentiation antigens. Int J Cancer. 2010;127:1625–1636. doi: 10.1002/ijc.25182. [DOI] [PubMed] [Google Scholar]
- 114.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, Frederick DT, Barzily-Rokni M, Straussman R, Haq R, Fisher DE, Mesirov JP, Hahn WC, Flaherty KT, Wargo JA, Tamayo P, Garraway LA. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4(7):816–827. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pouryazdanparast P, Brenner A, Haghighat Z, Guitart J, Rademaker A, Gerami P. The role of 8q24 copy number gains and C-MYC expression in amelanotic cutaneous melanoma. Mod Pathol. 2012;25(9):1221–1226. doi: 10.1038/modpathol.2012.75. [DOI] [PubMed] [Google Scholar]
- 116.Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, Baker AC, Nazarian RM, Vijayendran KG, Sellers WR, Hahn WC, Duncan LM, Rubin MA, Fisher DE, Garraway LA. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70(5):2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nicholas C, Yang J, Peters SB, Bill MA, Baiocchi RA, Yan F, Sif S, Tae S, Gaudio E, Wu X, Grever MR, Young GS, Lesinski GB. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27Kip1 . PLoS One. 2013;8(9):e74710. doi: 10.1371/journal.pone.0074710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin DC, Xu L, Ding LW, Sharma A, Liu LZ, Yang H, Tan P, Vadgama J, Karlan BY, Lester J, Urban N, Schummer M, Doan N, Said JW, Sun H, Walsh M, Thomas CJ, Patel P, Yin D, Chan D, Koeffler HP. Genomic and functional characterizations of phosphodiesterase subtype 4D in human cancers. Proc Natl Acad Sci USA. 2013;110(15):6109–6114. doi: 10.1073/pnas.1218206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perego M, Tortoreto M, Tragni G, Mariani L, Deho P, Carbone A, Santinami M, Patuzzo R, Mina PD, Villa A, Pratesi G, Cossa G, Perego P, Daidone MG, Alison MR, Parmiani G, Rivoltini L, Castelli C. Heterogenous phenotype of human melanoma cells with in vitro and in vivo features of tumor-initiating cells. J Invest Dermatol. 2010;130(7):1877–1886. doi: 10.1038/jid.2010.69. [DOI] [PubMed] [Google Scholar]
- 120.Ramgolam K, Lauriol J, Lalou C, Lauden L, Michel L, de la Grange P, Khatib AM, Aoudjit F, Charron D, Alcaide-Loridan C, Al-Daccak R. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS One. 2011;6(4):e18784. doi: 10.1371/journal.pone.0018784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sztiller-Sikorska M, Koprowska K, Jakubowska J, Zalesna I, Stasiak M, Duechler M, Czyz M. Sphere formation and capacity of melanoma cells is affected by the microenvironment. Melanoma Res. 2012;22(3):215–224. doi: 10.1097/CMR.0b013e3283531317. [DOI] [PubMed] [Google Scholar]
- 122.Ghislin S, Deshayes F, Lauriol J, Middendorp S, Martins I, Al-Daccak R, Alcaide-Loridan C. Plasticity of melanoma cells induced by neural cell crest conditions and three-dimensional growth. Melanoma Res. 2012;22(3):184–194. doi: 10.1097/CMR.0b013e328351e7c4. [DOI] [PubMed] [Google Scholar]
- 123.Ennen M, Keime C, Kobi D, Mengus G, Lipsker D, Thibault-Carpentier C, Davidson I. Single-cell expression signatures reveal melanoma cell heterogeneity. Oncogene. 2014 doi: 10.1038/onc.2014.262. [DOI] [PubMed] [Google Scholar]
- 124.Somasundaram R, Villanueva J, Herlyn M. Intratumoral heterogeneity as a therapy resistance mechanism: role of melanoma subpopulations. Adv Pharmacol. 2012;65:335–359. doi: 10.1016/B978-0-12-397927-8.00011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12(5):349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 126.Haq R, Fisher DE. Targeting melanoma by small molecules: challenges ahead. Pigment Cell Melanoma Res. 2013;26(4):464–469. doi: 10.1111/pcmr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shtivelman E, Davies MQ, Hwu P, Yang J, Lotem M, Oren M, Flaherty KT, Fisher DE. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5(7):1701–1752. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sztiller-Sikorska M, Koprowska K, Majchrzak K, Hartman M, Czyz M. Natural compounds’ activity against cancer stem-like or fast-cycling melanoma cells. PLoS One. 2014;9(3):e90783. doi: 10.1371/journal.pone.0090783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Syed DN, Afaq F, Maddodi N, Johnson JJ, Sarfaraz S, Ahmad A, Setaluri V, Mukhtar H. Inhibition of human melanoma cell growth by the dietary flavonoid fisetin is associated with disruption of Wnt/β-catenin signaling and decreased Mitf levels. J Invest Dermatol. 2011;131(6):1291–1299. doi: 10.1038/jid.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Botton T, Puissant A, Cheli Y, Tomic T, Giuliano S, Fajas L, Deckert M, Ortonne JP, Bertolotto C, Tartare-Deckert S, Ballotti R, Rocchi S. Ciglitazone negatively regulates CXCL1 signaling through MITF to suppress melanoma growth. Cell Death Differ. 2011;18(1):109–121. doi: 10.1038/cdd.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Villareal MO, Han J, Ikuta K, Isoda H. Mechanisms of Mitf inhibition and morphological differentiation effects of hirsein A on B16 melanoma cells revealed by DNA microarray. J Dermatol Sci. 2012;67(1):26–36. doi: 10.1016/j.jdermsci.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 132.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, Garraway LA. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504(7478):138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen G, Davies MA. Targeted therapy resistance mechanisms and therapeutic implications in melanoma. Hematol Oncol Clin N Am. 2014;28:523–536. doi: 10.1016/j.hoc.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 134.Roesch A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene. 2014 doi: 10.1038/onc.2014.249. [DOI] [PubMed] [Google Scholar]
- 135.Smith MP, Ferguson J, Arozarena I, Hayward R, Marais R, Chapman A, Hurlstone A, Wellbrock C. Effect of SMURF2 targeting on susceptibility to MEK inhibitors in melanoma. J Natl Cancer Inst. 2013;105(1):33–46. doi: 10.1093/jnci/djs471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, Hodis E, Rosenberg M, McKenna A, Cibulskis K, Farlow D, Zimmer L, Hillen U, Gutzmer R, Goldinger SM, Ugurel S, Gogas HJ, Egberts F, Berking C, Trefzer U, Loquai C, Weide B, Hassel JC, Gabriel SB, Carter SL, Getz G, Garraway LA, Schadendorf D, Dermatologic Cooperative Oncology Group of Germany The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Frederick DT, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, Mitra D, Boni A, Newton LP, Liu C, Peng W, Sullivan RJ, Lawrence DP, Hodi FS, Overwijk WW, Lizee G, Murphy GF, Hwu P, Flaherty KT, Fisher DE, Wargo JA. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res. 2013;19(5):1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yokoyama S, Feige E, Poling LL, Levy C, Widlund HR, Khaled M, Kung AL, Fisher DE. Pharmacological suppression of MITF expression via HDAC inhibitors in the melanocyte lineage. Pigment Cell Melanoma Res. 2008;21(4):457–463. doi: 10.1111/j.1755-148X.2008.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]