Abstract
In a previous study, we found an unknown element that caused growth inhibition after its copy number increased in the 3′ region of DIE2 in Saccharomyces cerevisiae. In this study, we further identified this element and observed that overexpression of a small protein (sORF2) of 57 amino acids encoded in this region caused growth inhibition. The transcriptional response and multicopy suppression of the growth inhibition caused by sORF2 overexpression suggest that sORF2 overexpression inhibits the ergosterol biosynthetic pathway. sORF2 was not required in the normal growth of S. cerevisiae, and not conserved in related yeast species including S. paradoxus. Thus, sORF2 (designated as OTO1) is an orphan ORF that determines the specificity of this species.
Introduction
We previously analyzed the copy number limits of most of the protein-coding genes in the budding yeast Saccharomyces cerevisiae using the genetic tug-of-war (gTOW) method [1]. In the gTOW method, the copy number of a plasmid containing a target gene (with its native promoter and terminator region) is increased on basis of the selection bias of the leu2d gene [2,3]. The copy number of the empty plasmid exceeds 100 in the leucine-negative condition. If the target gene has a copy number limit of <100, the plasmid copy number reflects the copy number limit.
When a target gene has the low copy number limit, we consider that overexpression of the protein encoded by the target gene (i.e., the annotated open reading frame (ORF)) results in growth inhibition. However, elements other than the target gene in the DNA fragment could determine the low copy number. For example, increasing the copy number of a DNA element, overexpression of an RNA element, or overexpression of an unannotated protein could result in growth inhibition.
To test this possibility, we previously analyzed the low limit genes by introducing a frameshift mutation to disrupt each annotated ORF and we isolated 10 DNA fragments where frameshift mutations in the annotated ORFs still obtained low copy number limits [1]. We also dissected the fragments and isolated four DNA fragments with unknown elements that determined the low copy number limits. Thus, we isolated a 600-base pair (bp) DNA fragment that contained the 3′ region of DIE2, which resulted in a low copy number limit (Frag5 in Fig. 1A) [1].
Fig 1. Isolation of the element responsible for the low copy number limit in the DIE2 region.
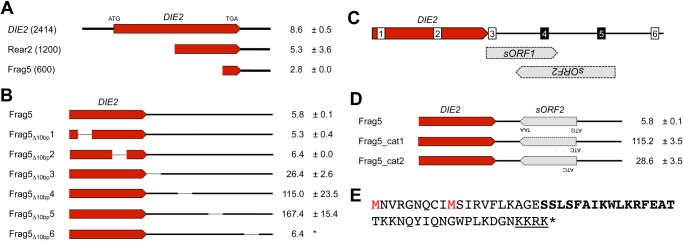
- Copy number limits of DNA fragments from the DIE2 region. The data were obtained from our previous study [1].
- Copy number limits of DNA fragments (Frag5 in A) with serial 10-bp deletions every 100 bp. The asterisk indicates that only single experiment was performed.
- Locations of the small ORFs (sORF1 and sORF2) in the 3′ region of DIE2. The numbers indicate the 10-bp deletions analyzed in B. The deletions shown in white did not affect the toxicity of the DNA fragment, whereas the deletion shown in black disrupted the toxicity.
- Copy number limits of DNA fragments with ATG to ATC substitutions in sORF2.
- Amino acid sequence of sORF2. The substituted methionines (ATG codons) in C are shown in red. A potential NLS sequence is underlined, and an amino acid sequence predicted to construct a helical structure is shown in bold letters.
In this study, we further analyzed this region and showed that expression of a small ORF encoding 58 codons caused growth inhibition.
Results and Discussion
Isolation of the element responsible for low copy number limits in the DIE2 region
To isolate the specific element responsible for the low copy number limit in the 3′ region of DIE2, we introduced a series of 10-bp deletions in every 100 bp of Frag5 and measured their copy number limits. As shown in Fig. 1B, deletions of two sites in the downstream region of DIE2 increased the copy number limit to >100. As shown in Fig. 1C, two small ORFs of >100 bp are encoded in Frag5 (denoted as sORF1 and sORF2). Both of these two 10-bp deletions disrupted sORF2, which indicates that sORF2 might be responsible for the low copy number limit of Frag5.
To disrupt sORF2 alone, we introduced mutations to change the potential start codons (ATG) of sORF2 into ATC. The results obtained are shown in Fig. 1D. Frag5 with a mutation that changed the first ATG codon of sORF2 into ATC possessed a copy number limit of >100. Frag5 with a mutation in the second ATG had a higher limit than the original Frag5, but the limit was still low (28.6 ± 3.5). This result strongly suggests that overexpression of the protein encoded by sORF2 causes growth inhibition when its copy number is increased in the 3′ region of DIE2. Fig. 1E shows the amino acid sequence of sORF2.
High level expression of sORF2 driven by the GAL1 promoter inhibits cellular growth
To confirm whether sORF2 overexpression alone caused growth inhibition, we tried to express sORF2 from the GAL1 promoter (P GAL1). As shown in Fig. 2A, yeast cells that harbored the P GAL1 –sORF2 plasmid did not grow on galactose plates. Next, we observed the growth inhibition process using time-lapse microscopic imaging. As shown in Fig. 2B, at the time point when the induction of P GAL1 -GFP was observed, each cell that expressed sORF2 ceased its proliferation and a large void structure was present. These results indicate that the high level expression of sORF2 inhibited cellular growth.
Fig 2. Genetic analyses of sORF2.
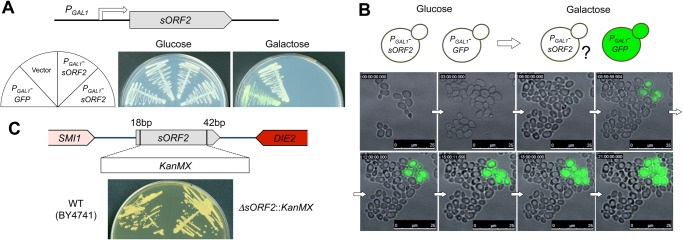
- Overexpression of sORF2 from the GAL1 promoter (P GAL1). The construct used in this experiment is shown. Cells with pTOW-PGAL1-sORF2 (P GAL1 -sORF2) were streaked onto SC-glucose and SC-galactose plates. Two independent plasmid clones were analyzed. pTOW40836 (Vector) was used as an empty vector control and pTOW-PGAL1-GFP (P GAL1 -GFP) was used to monitor the P GAL1 induction.
- Time-lapse imaging of cells after the induction of sORF2. The cells with pTOW-PGAL1-sORF2 (P GAL1 -ORF2) and pTOW-PGAL1-GFP (P GAL1 -GFP) were cultured in SC-glucose mixed at a ratio of 10:1 and then cultivated in SC-galactose medium. P GAL1 -GFP was used to monitor the induction of P GAL1. The cellular images shown were obtained every 5min. A movie is available as S1 Movie.
- Deletion of sORF2. The construct used to delete sORF2 from the chromosome is shown. The strain with sORF2 deleted was streaked onto a YPD agar plate. The strain BY4741 was used as a wild-type control.
sORF2 is not required for the normal growth of S. cerevisiae
To test whether sORF2 is required for the growth of S. cerevisiae, we disrupted sORF2 by replacing it with a kanamycin resistance gene cassette (KanMX), as shown in Fig. 2C. The ΔsORF2::KanMX cells exhibited the same growth as the wild-type cells in normal growth conditions (YPD, 30°C; Fig. 2C).
Increasing the copy number of the sORF2-containing DNA fragment induces the expression of ergosterol synthesis genes
We performed transcriptome analysis (RNAseq) to analyze the cellular response after the overexpression of sORF2. We compared the mRNA expression profiles of cells that harbored the vector plasmids and the plasmid containing the DIE2 3′ fragment (Rear2, Fig. 1A). Tables 1 and 2 show the genes with significantly different expression levels.
Table 1. Genes with higher expression levels in cells that harbored pTOW-Rear2 compared with the control cells.
| Name gene | Brief description* |
|---|---|
| DAN1 | Cell wall mannoprotein |
| DAN4 | Cell wall mannoprotein |
| ERG1 | Squalene epoxidase |
| ERG3 | C-5 sterol desaturase |
| ERG11 | Lanosterol 14-alpha-demethylase |
| ERG25 | C-4 methyl sterol oxidase |
| EXG1 | Major exo-1,3-beta-glucanase of the cell wall |
| FUN30 | Snf2p family member with ATP-dependent chromatin remodeling activity |
| MAK16 | Essential nuclear protein |
| PBI1 | Putative protein of unknown function |
| RNR1 | Major isoform of large subunit of ribonucleotide-diphosphate reductase |
| RPA12 | RNA polymerase I subunit A12.2 |
| SFG1 | Nuclear protein putative transcription factor |
| TIR3 | Cell wall mannoprotein |
| TIR4 | Cell wall mannoprotein |
| TPO2 | Polyamine transporter of the major facilitator superfamily |
| YJR005C-A | Putative protein of unknown function |
*Saccharomyces genome database: http://www.yeastgenome.org
Table 2. Genes with lower expression levels in cells that harbored pTOW-Rear2 compared with the control cells.
| Name gene | Brief description* |
|---|---|
| ADH4 | Alcohol dehydrogenase isoenzyme type IV |
| ADY2 | Acetate transporter required for normal sporulation |
| BTN2 | v-SNARE binding protein |
| DSF1 | Putative mannitol dehydrogenase |
| ECM23 | Non-essential protein of unconfirmed function |
| ENA1 | P-type ATPase sodium pump |
| FMP43 | Highly conserved subunit of mitochondrial pyruvate carrier |
| FMP45 | Integral membrane protein localized to mitochondria |
| HXT6 | High-affinity glucose transporter |
| HXT7 | High-affinity glucose transporter |
| ISF1 | Serine-rich, hydrophilic protein |
| JEN1 | Monocarboxylate/proton symporter of the plasma membrane |
| NCE103 | Carbonic anhydrase |
| PHO89 | Plasma membrane Na+/Pi cotransporter |
| PUT1 | Proline oxidase |
| RGI2 | Protein of unknown function |
| SMA1 | Protein of unknown function involved in prospore membrane assembly |
| SPG1 | Protein required for high temperature survival during stationary phase |
| SPG4 | Protein required for high temperature survival during stationary phase |
| SPL2 | Protein with similarity to cyclin-dependent kinase inhibitors |
| TMA10 | Protein of unknown function that associates with ribosomes |
| YBR285W | Putative protein of unknown function |
| YGR067C | Putative protein of unknown function |
| YLR307C-A | Putative protein of unknown function |
| YNL194C | Integral membrane protein |
| YNL195C | Protein of unknown function |
*Saccharomyces genome database: http://www.yeastgenome.org
We analyzed the enriched genes based on gene ontology (GO) terms. The genes with higher expression levels in the cells that harbored the pTOW-Rear2 plasmid were significantly enriched in terms of genes involved in the ergosterol biosynthesis pathway (p = 2.2e−4). Eight genes (DAN1, DAN4, ERG1, ERG3, ERG11, ERG25, TIR3, and TIR4) with higher expression levels were identified as genes that could be induced by treatment with ketoconazole [4]. Ketoconazole is known to inhibit the ergosterol biosynthetic pathway [5]; thus, sORF2 overexpression appeared to affect this pathway. The genes with lower expression levels were not significantly enriched with respect to GO terms. They however contained many genes encoding transporters and membrane proteins, such as ADY2, ENA1, FMP43, FMP45, HXT6, HXT7, JEN1, PHO89, SMA1, and YNL194C, suggesting that sORF2 overexpression modulates the expression of membrane proteins.
Expression analysis of sORF2
We analyzed the RNAseq data to determine whether sORF2 is transcribed. As shown in Fig. 3A, transcript reads containing sORF2 were not detected in the mRNAs from BY4741 that harbored an empty vector pTOWug2–836, whereas a large number of transcript reads were detected in the mRNAs that harbored pTOW-Rear2.
Fig 3. Expression analysis of sORF2.
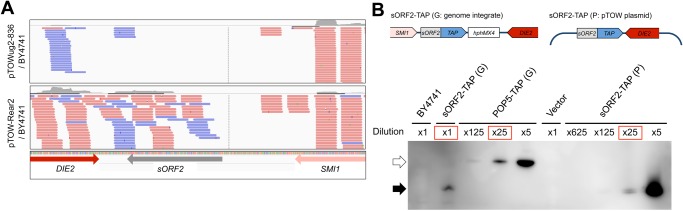
- RNAseq analysis of the sORF2 region of the strain BY4741 with the control vector (pTOWug2–836) and pTOW-Rear2. Parts of the detected reads are shown. The locations of DIE2, sORF2, and SMI1 are indicated.
- Western blot analysis of sORF2 using TAPtag. Expression of sORF2-TAP from the genomic region or plasmids was detected using peroxidase anti-peroxidase soluble complex. BY4714 is a negative control strain without any TAP-tagged protein expressed. Vector is another negative control, in which BY4741 harbors an empty vector (pTOWug2–836). Cells of BY4741, sORF2-TAP (genome), and POP5-TAP (genome) were cultivated in YPD medium; cells of Vector and sORF2-TAP (plasmid) were cultivated in SC—Ura medium. Dilution indicates the fold-dilution of the cellular lysate applied to the gel. Red-squared dilutions were used to calculate the expression levels of TAP-tagged proteins. The white arrowhead indicates the expected molecular weight of Pop5-TAP protein (39.6kDa), and the black arrowhead indicates the one of sORF2-TAP (27.1 kDa). Structures of sORF2-TAP constructs are shown.
To test whether sORF2 was translated, we attached the tandem affinity purification (TAP) tag to sORF2 and attempted to detect the TAP-tagged sORF2 by Western blotting. As shown in Fig. 3B, sORF2-TAP expressed from its genomic region was detected, and the expression of sORF2-TAP from the plasmid was highly increased. The expression of sORF2 from its genomic region was detected in the log phage cell lysate, but not in the post-log phase lysate (S1 Fig.). The expression was not increased under mating conditions (S1 Fig.).
We further estimated the expression level of sORF2-TAP in comparison to the expression level of a reference protein Pop5-TAP, whose protein copy number was previously determined (2230 copies/cell) [6]. As the result, the expression level of sORF2-TAP from its genomic region was estimated to be 45 copies/cell, which corresponds to the level of lowly expressed proteins [6]. The estimated expression level of sORF2-TAP from the plasmid was 1938 copies/cell. It should be noted that the copy number limit of the plasmid that contained the sORF2-TAP DNA fragment was >100 (data not shown). This suggests that the small size of sORF2 itself is required to inhibit growth.
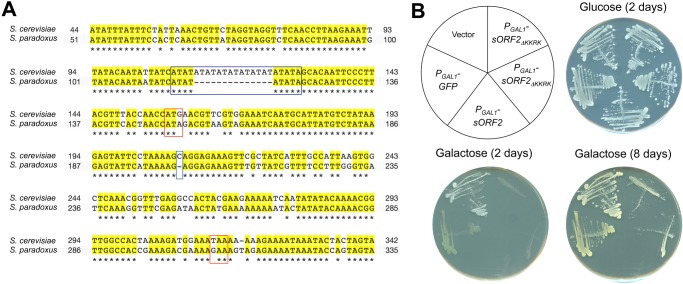
Currently, we do not know the reason why we could not detect the mRNA of sORF2 expressed from its genomic region by our RNAseq analysis above. Although it is possible that integrating TAP-tag sequence and a marker gene stimulated the expression of sORF2, the result still suggests that there is an expression potential from the sORF2 locus. Supporting this idea, there is a TA repeat in the upstream region of sORF2, which provides potential binding sites for transcriptional factors such as the TATA-binding protein Spt15 (S2 Fig.). Notably, the TA repeat is far shorter in the corresponding genomic region of S. paradoxus, which lacks sORF2 (Fig. 4A). These binding sites might function as promoters for sORF2.
Fig 4. Structural analysis of sORF2.
- Alignment of the sORF2 regions of S. cerevisiae and S. paradoxus. Identical nucleotides are shown in yellow. ATG and STOP codons of sORF2 are shown in red. A TATA repeat and deletion in the S. paradoxus sequence are indicated in blue. The image is a snapshot from the fungal sequence alignment of SGD (http://www.yeastgenome.org/cache/fungi/YGR229C.html). The nucleotide numbers indicate the positions relative to the stop codon of SMI1.
- Overexpression of sORF2 without the potential NLS (sORF2 ΔKKRK). The construct used in this experiment is shown. Cells with pTOW-PGAL1-sORF2 (P GAL1 -sORF2) or pTOW-PGAL1-sORF2ΔKKRK (P GAL1 -sORF2 ΔKKRK) were streaked onto SC-glucose and SC-galactose plates and incubated for indicated days. pTOW40836 (Vector) was used as an empty vector control and pTOW-PGAL1-GFP (P GAL1 -GFP) was used to monitor the P GAL1 induction.
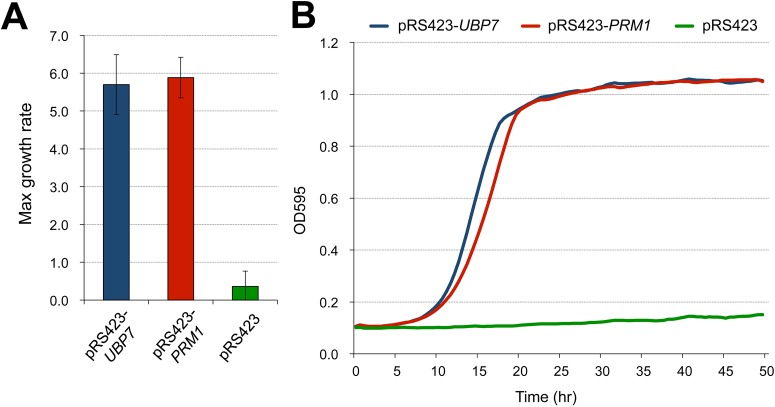
Multicopy UBP7 and PRM1 suppress the growth inhibition caused by the high copy number sORF2-containing DNA fragment
To further elucidate the molecular mechanism responsible for growth inhibition by sORF2, we attempted to isolate multicopy suppressors of the growth inhibition caused by high copy number pTOW-Rear2. As shown in Fig. 5, we isolated two multicopy suppressors, UBP7 and PRM1. UBP7 encodes a ubiquitin protease (UBPs) that controls protein degradation [7]. PRM1 encodes a pheromone-regulated membrane protein, which is involved in membrane fusion during mating [8]. PRM1 is known to have a genetic interaction with ERG genes [9,10]. This result also suggests the involvement of sORF2 in the ergosterol biosynthetic pathway.
Fig 5. Multicopy suppressors of growth inhibition after increasing the copy number in the DIE2 3′ fragment.
- Maximum growth rate of BY4741 cells that harbored both pTOW-Rear2 and the suppressor plasmids (pRS423-UBP7 and pRS423-PRM1, and the empty vector, pRS423) in SC—Ura—His medium. The averages and standard deviations from six independent experiments are shown.
- Growth curves of the BY4741 cells that harbored both pTOW-Rear2 and the suppressor plasmids in SC—Ura—His medium. One representative data is shown from each experiment.
Structural analysis of sORF2
In order to speculate the molecular function of sORF2, we performed some bioinformatics analyses. We first performed the BLAST search toward the protein sequences stored at NCBI database (http://blast.ncbi.nlm.nih.gov/), but we could not obtain any significantly similar protein.
The corresponding ORF was not conserved in any closely-related yeast species (S. paradoxus, S. bayanus, S. mikatae, S. castellii, and S. kudriavzevii). Fig. 4A shows the comparison of the corresponding genomic locus from S. cerevisiae and S. paradoxus (most closely-related species to S. cerevisae), as an example.
During the structural analysis, we noticed that sORF2 contained a consensus sequence of nuclear localization signals (K-K/R-X-K/R) [11] at its C-terminal (underlined in Fig. 1E). To test if this potential nuclear localization signal (NLS) is important for the toxicity of sORF2, we overexpressed sORF2 without the sequence (sORF2ΔKKRK). As shown in Fig. 4B, yeast cells that harbored the P GAL1 –sORF2 ΔKKRK plasmid grew on galactose plates, but much slower than the cells with the empty vector or P GAL1 –GFP plasmids. This result indicates that the potential NLS is partly required (but not essential) for the toxicity of sORF2.
We next tried to predict the secondary and tertiary structure of sORF2 using a protein homology/analogy recognition engine, Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/). The analysis predicted that there was a helical structure in the middle of the protein (shown in bold letters in Fig. 1E) based on its similarity with two template proteins (d1k78a1 and d6paxa1) with the confidence scores > 70 (the prediction results are summarized in S3 Fig.). Because the template proteins were both structurally classified into DNA/RNA-binding 3-helical bundle (Fold), homeodomain-like (superfamily), and paired domain (family), sORF2 might have DNA/RNA binding activity.
sORF2 (OTO1/YGR228C-A) as an orphan ORF
In this study, we obtained evidence that overexpression of a small ORF of 58 codons (sORF2) encoded within the 3′ region of DIE2 causes growth inhibition. Our results also suggest that sORF2 overexpression affects the ergosterol synthetic pathway. Based on the fact that sORF2 has a potential NLS and a helical structure involved in DNA/RNA binding, sORF2 might function through its nuclear function such as transcriptional regulation.
sORF2 was not identified in previous studies that aimed to detect small ORFs based on their expression and evolutionary conservation [12–15]. In fact, sORF2 is not conserved in the corresponding genomic region of the most closely-related yeast species S. paradoxus (Fig. 4A). We thus think that sORF2 is an orphan ORF (ORFan) [16, 17], which distinguishes species by functioning in species-specific cellular situations, and propose its name as OTO1 (ORFan toxic when overexpressed) with its locus name YGR228C-A.
Our gTOW approach might be useful for isolating other ORFans. In fact, we had isolated three more genomic loci potentially contain unannotated toxic elements when the copy numbers were increased [1].
Materials and Methods
Strains and growth conditions
BY4741 (MATa his3Δ0 leu2 Δ0 met15 Δ0 ura3 Δ0) [18] was used as the host yeast strain to test the toxicity of DIE2 fragments and sORF2. The sORF2 deletion strain was created, as follows: The genomic region of sORF2 (from ATG to stop) in BY4743 (MATa/α his3 Δ1/his3 Δ1 leu2 Δ0/leu2 Δ0 LYS2/lys2 Δ0 met15 Δ0/MET15 ura3 Δ0/ura3 Δ0) [18] was replaced by the KanMX6 cassette using a DNA fragment, which was amplified by PCR with the primers OHM0969 and OHM0970 using pKT127 [19] as a template. The strains were sporulated, and the tetrads were dissected. After genotypic analysis of the tetrads, haploid deletion strains were isolated. The sORF2-TAP strain was created, as follows: A sORF2-TAP fragment was amplified by PCR with the primers OHM1030 and OHM1032 using pTOW-sORF2-TAP. A hphMX4 fragment was amplified by PCR with primers the OHM1031 and OH1033 using pAG34 [20]. Both fragments were introduced into BY4741 to integrate sORF2-TAP-hphMX4 into the genomic region of sORF2. BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) [18] was used for a mating partner of BY4741 with sORF2-TAP-hphMX4.
Yeast cells were grown in standard growth conditions [21]. The PCR primers used to amplify the DNA fragments employed in strain construction are listed in S1 Table.
Plasmids used in this study
The plasmids used in this study are listed in Table 3. The plasmids were constructed on the basis of the homologous recombination activity of yeast cells [22]. The PCR primers used to amplify the DNA fragments, which were employed in plasmid construction are listed in S1 Table.
Table 3. Plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| pTOWug2–836 | Amp R, ColE1ori, 2μori, URA3-yEGFP, leu2d | [1] |
| pTOW40836 | Amp R, ColE1ori, 2μori, URA3, leu2d | [3] |
| pTOW-Rear2 | DIE2 3′ region (Rear2) cloned into pTOWug2–836 | [1] |
| pTOW-Frag5 | DIE2 3′ region (Frag5) cloned into pTOWug2–836 | [1] |
| pTOW-Frag5Δ10bp1 | pTOW-Frag5 containing 10-bp deletion (16–25) | This study |
| pTOW-Frag5Δ10bp2 | pTOW-Frag5 containing 10-bp deletion (116–125) | This study |
| pTOW-Frag5Δ10bp3 | pTOW-Frag5 containing 10-bp deletion (216–225) | This study |
| pTOW-Frag5Δ10bp4 | pTOW-Frag5 containing 10-bp deletion (312–321) | This study |
| pTOW-Frag5Δ10bp5 | pTOW-Frag5 containing 10-bp deletion (416–425) | This study |
| pTOW-Frag5Δ10bp6 | pTOW-Frag5 containing 10-bp deletion (516–525) | This study |
| pTOW-Frag5_cat1 | First ATG of sORF2 changed into ATC in pTOW-Frag5 | This study |
| pTOW-Frag5_cat2 | Second ATG of sORF2 changed into ATC in pTOW-Frag5 | This study |
| pTOW-PGAL1-GFP | P GAL1 -yEGFP-T GAL1 cloned into pTOW40836 | This study |
| pTOW-PGAL1-sORF2 | P GAL1 -sORF2-T GAL1 cloned into pTOW40836 | This study |
| pTOW-PGAL1-sORF2 ΔKKRK | P GAL1 -sORF2 ΔKKRK -T GAL1 cloned into pTOW40836 | This study |
| pTOW-sORF2-TAP | TAPtag inserted into the C-terminal of sORF2 of pTOW-Frag5 | This study |
| pRS423ks | Amp R, ColE1ori, 2μori, HIS3 | [1] |
| pRS423-UBP7 | UBP7 cloned into pRS423ks | This study |
| pRS423-PRM1 | PRM1 cloned into pRS423ks | This study |
Measurement of the plasmid copy number limit
The copy number limits of plasmids were measured as described in our previous study [1]. Briefly, DNA from yeast cells grown in SC—Ura, SC—Ura—Leu, or SC—Ura—His medium were extracted, and the relative plasmid copy number compared with the genomic DNA in the DNA solution was measured using real-time PCR. HIS3, LEU2, and LEU3 genes were detected as indicators of the plasmid copy number for pRS423ks, pTOWug2–836/40836, and genomic DNA, respectively. More than two independent experiments were performed for each experiment otherwise stated.
RNAseq analysis
Yeast cells that harbored pTOWug2–836, pTOW40836, and pTOW-Rear2 were cultivated in SC—Ura medium until the mid-log phase, and RNA from each culture was then isolated using the hot phenol method [23]. A cDNA library was prepared using a SureSelect strand-specific RNA library preparation kit (G9691A, Agilent), and sequencing was performed using an Illumina Hiseq2500 with TruSeq SBS kit v3-HS. The software connected to GenomeSpace (http://www.genomespace.org) was used for the sequence data analysis. The sequence data were analyzed using TopHat (ver. 6) and Cufflink/cuffdiff (ver.4) on the GenePattern platform (http://genepattern.broadinstitute.org), with sacCer3 for gene annotation (http://genome.ucsc.edu/cgi-bin/hgTables). First, we isolated genes that differed significantly (FDR < 0.5) between pTOWug2–836 and pTOW-Rear2 (Comp1), pTOW40836 and pTOW-Rear2 (Comp2), pTOWug2–836 and pTOW40836 (Comp3), and the pTOW-Rear2 duplicates (Comp4). Next, we prepared a gene list from genes isolated in Comp1 or Comp2, but not in Comp3 or Comp4 (summarized in S4 Fig. and S2 Table). The Integrative Genomics Viewer (IGV2.3, http://www.broadinstitute.org/igv/) was used to visualize the sequence reads shown in Fig. 3A. The GO, publication, and pathway enrichments were analyzed using YeastMine (http://yeastmine.yeastgenome.org).
Microscopic observation
Cells were cultivated in SC—Ura medium until the mid-log phase and the cells were then transferred to SC-galactose—Ura medium, before being applied to a PDMS microfluidic chamber (YC-1, Warner instruments). Cellular images were acquired every 5 min using a Leica DM6000 B microscope. GFP fluorescence was determined using a GFP filter cube (excitation filter 470/40 and emission filter 525/50).
Western blot analysis
Western blotting was performed as described previously [24]. Briefly, proteins extracted from the 0.25 OD600 cells (with indicated fold dilutions) cultivated in the indicated medium were separated by SDS-PAGE and transferred onto a PVDF membrane. The TAP-tagged protein was then detected using peroxidase anti-peroxidase soluble complex (P1901l, Sigma-Aldrich). The chemiluminescent image was taken and the intensity of each band was measured using the LAS-4000 image analyzer (GE Healthcare).
Multicopy suppressor screening
A multicopy plasmid library where most of the genes in S. cerevisiae were cloned into pRS423ks (our laboratory stock) was introduced into yeast strains that harbored pTOW-Rear2. Next, the colonies were grown on SC—Ura—His plates and then replica-plated onto SC—Ura—Leu—His plates. The plasmids were recovered from the colonies grown on SC—Ura—Leu—His and the DNA sequences of inserts in the plasmids were determined. The suppressor activities of the isolated candidates were re-evaluated by measuring the growth of the cells that harbored both pTOW-Rear2 and the suppressor plasmids in SC—Ura—His medium. Cellular growth was measured by monitoring OD595 every 30 min using a microplate reader (Infinite F200, TECAN). The maximum growth rate was calculated as described previously [2, 3].
Supporting Information
(XLSX)
(XLSX)
Expression of sORF2-TAP from the genomic region under indicated conditions were detected using peroxidase anti-peroxidase soluble complex. Cellular lysates from the 0.0625 OD600 cells were loaded. To create mating conditions, BY4741 with sORF2-TAP-hphMX4 cells were mixed with BY4742 cells on a YPD agar plate and incubated for 2 hours in prior to prepare of the cellular lysate. BY4741 with sORF2-TAP-hphMX4 cells were cultivated in YPD medium to prepare log phase cells and post-log phase cells. The cellular density of the cultures were shown as OD600.
(TIF)
The image is a snapshot from the YeTFaSCo analysis (http://yetfasco.ccbr.utoronto.ca). The arrowhead indicates sORF2.
(TIF)
The 3D structures, summaries, and alignments are shown. The images were snapshots of displayed on the Phyre2 website (http://www.sbg.bio.ic.ac.uk/phyre2/).
(TIF)
We first isolated genes showing significant difference (FDR < 0.5) between; pTOWug2–836 and pTOWug2-Rear2 (Comp1), pTOW40836 and pTOWug2-Rear2 (Comp2), pTOWug2–836 and pTOW40836 (Comp3), and between pTOWug2-Rear2 duplicates (Comp4). We then made a gene list, which contained true positives, from isolated genes in Comp1 or Comp2, but neither in Comp3 nor Comp4 (see the Venn diagram).
(TIF)
(MOV)
Data Availability
All RNAseq data files are available from the DDBJ Sequence Read Archive (accession number DRA002585).
Funding Statement
This research was supported partly by MEXT KAKENHI (B, Challenging Exploratory Research, and 221S0002), the Asahi Glass Foundation, and grants-in-aid for strategic research promotion from Okayama University. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Research. 2013;23(2):300–11. 10.1101/gr.146662.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moriya H, Shimizu-Yoshida Y, Kitano H. In vivo robustness analysis of cell division cycle in Saccharomyces cerevisiae (vol 2, pg 7, 2006). Plos Genetics. 2006;2(12):2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moriya H, Makanae K, Watanabe K, Chino A, Shimizu-Yoshida Y. Robustness analysis of cellular systems using the genetic tug-of-war method. Molecular Biosystems. 2012;8(10):2513–22. 10.1039/c2mb25100k [DOI] [PubMed] [Google Scholar]
- 4. Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, et al. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem. 2003;278(37):34998–5015. [DOI] [PubMed] [Google Scholar]
- 5. Kelly SL, Arnoldi A, Kelly DE. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans. 1993;21(4):1034–8. [DOI] [PubMed] [Google Scholar]
- 6. Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. [DOI] [PubMed] [Google Scholar]
- 7. Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381(9–10):981–92. [DOI] [PubMed] [Google Scholar]
- 8. Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151(3):719–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J Cell Biol. 2008;180(4):813–26. 10.1083/jcb.200705076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aguilar PS, Heiman MG, Walther TC, Engel A, Schwudke D, Gushwa N, et al. Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating. Proc Natl Acad Sci U S A. 2010;107(9):4170–5. 10.1073/pnas.0914094107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dingwall C, Robbins J, Dilworth SM, Roberts B, Richardson WD. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol. 1988;107(3):841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oshiro G, Wodicka LM, Washburn MP, Yates JR, Lockhart DJ, Winzeler EA. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12(8):1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kessler MM, Zeng Q, Hogan S, Cook R, Morales AJ, Cottarel G. Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res. 2003;13(2):264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar A, Harrison PM, Cheung KH, Lan N, Echols N, Bertone P, et al. An integrated approach for finding overlooked genes in yeast. Nat Biotechnol. 2002;20(1):58–63. [DOI] [PubMed] [Google Scholar]
- 15. Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16(3):365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li QR, Carvunis AR, Yu H, Han JD, Zhong Q, Simonis N, et al. Revisiting the Saccharomyces cerevisiae predicted ORFeome. Genome Res. 2008;18(8):1294–303. 10.1101/gr.076661.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15(9):759–62. [DOI] [PubMed] [Google Scholar]
- 18. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–32. [DOI] [PubMed] [Google Scholar]
- 19. Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21(8):661–70. [DOI] [PubMed] [Google Scholar]
- 20. Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14):1541–53. [DOI] [PubMed] [Google Scholar]
- 21. Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 22. Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25(2):451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. [DOI] [PubMed] [Google Scholar]
- 24. Sasabe M, Shintani S, Kintaka R, Kaizu K, Makanae K, Moriya H. Evaluation of the lower protein limit in the budding yeast Saccharomyces cerevisiae using TIPI-gTOW. Bmc Systems Biology. 2014;8 10.1186/1752-0509-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Expression of sORF2-TAP from the genomic region under indicated conditions were detected using peroxidase anti-peroxidase soluble complex. Cellular lysates from the 0.0625 OD600 cells were loaded. To create mating conditions, BY4741 with sORF2-TAP-hphMX4 cells were mixed with BY4742 cells on a YPD agar plate and incubated for 2 hours in prior to prepare of the cellular lysate. BY4741 with sORF2-TAP-hphMX4 cells were cultivated in YPD medium to prepare log phase cells and post-log phase cells. The cellular density of the cultures were shown as OD600.
(TIF)
The image is a snapshot from the YeTFaSCo analysis (http://yetfasco.ccbr.utoronto.ca). The arrowhead indicates sORF2.
(TIF)
The 3D structures, summaries, and alignments are shown. The images were snapshots of displayed on the Phyre2 website (http://www.sbg.bio.ic.ac.uk/phyre2/).
(TIF)
We first isolated genes showing significant difference (FDR < 0.5) between; pTOWug2–836 and pTOWug2-Rear2 (Comp1), pTOW40836 and pTOWug2-Rear2 (Comp2), pTOWug2–836 and pTOW40836 (Comp3), and between pTOWug2-Rear2 duplicates (Comp4). We then made a gene list, which contained true positives, from isolated genes in Comp1 or Comp2, but neither in Comp3 nor Comp4 (see the Venn diagram).
(TIF)
(MOV)
Data Availability Statement
All RNAseq data files are available from the DDBJ Sequence Read Archive (accession number DRA002585).