Abstract
The role of infection in erythropoietic dysfunction is poorly understood. In children with P. falciparum malaria, the by-product of hemoglobin digestion in infected red cells (hemozoin) is associated with the severity of anemia which is independent of circulating levels of the inflammatory cytokine tumor necrosis alpha (TNF-α). To gain insight into the common and specific effects of TNF-α and hemozoin on erythropoiesis, we studied the gene expression profile of purified primary erythroid cultures exposed to either TNF-α (10ng/ml) or to hemozoin (12.5μg/ml heme units) for 24 hours. Perturbed gene function was assessed using co-annotation of associated gene ontologies and expression of selected genes representative of the profile observed was confirmed by real time PCR (rtPCR). The changes in gene expression induced by each agent were largely distinct; many of the genes significantly modulated by TNF-α were not affected by hemozoin. The genes modulated by TNF-α were significantly enriched for those encoding proteins involved in the control of type 1 interferon signalling and the immune response to viral infection. In contrast, genes induced by hemozoin were significantly enriched for functional roles in regulation of transcription and apoptosis. Further analyses by rtPCR revealed that hemozoin increases expression of transcription factors that form part of the integrated stress response which is accompanied by reduced expression of genes involved in DNA repair. This study confirms that hemozoin induces cellular stress on erythroblasts that is additional to and distinct from responses to inflammatory cytokines and identifies new genes that may be involved in the pathogenesis of severe malarial anemia. More generally the respective transcription profiles highlight the varied mechanisms through which erythropoiesis may be disrupted during infectious disease.
Introduction
Although recent efforts to reduce the number of deaths due to malaria have had some success, there is still an estimated 1.2 billion people at high risk of infection worldwide with almost half a million deaths occurring in children. The majority of these infections are due to P. falciparum and P. vivax [1]. Most mortality is caused by P. falciparum or mixed infections of Plasmodia including falciparum where the majority of hospital admissions in endemic regions are of children under the age of four [2, 3].
In young infants in holo-endemic regions of Africa, the predominant syndrome of severe malaria is severe malarial anemia (reviewed in [4, 5]). The recently observed elevated levels of hepcidin in patients with acute P. falciparum malaria suggest that the reduced bioavailability of iron contributes to developing severe anemia [6, 7]. Severe malarial anemia is due not only to increased hemolysis of infected and non-infected red blood cells but also to a striking degree of abnormal development of erythroid precursors in acute and chronic infection [8, 9] and an inadequate erythropoietic response in spite of elevated levels of erythropoietin (Epo) [9–11]. The distribution of erythroid precursors in the cell-cycle is also abnormal with an increased number of cells in the G2 phase compared with normal controls [12, 13].
Severe malaria is characterized by elevated levels of the inflammatory cytokine TNF-α [14, 15] which is thought to be produced following phagocytosis of malarial pigment (hemozoin) by macrophages [16]. Hemozoin is formed in the food vacuole of developing intra-erythrocytic parasites, as toxic heme remaining after digestion of hemoglobin forms a crystalline dimer of α hematin, complexed with lipid and protein. Hemozoin crystals closely resemble β hematin, consisting of a ferric ion within a protoporphyrin IX ring structure [17]. Hemozoin released after the lysis of infected red blood cells is heterogeneous and associated with proteins, nucleic acids, and host- and parasite- derived lipids including products from lipid peroxidation such as 4-hydroxy-2-nonenal (HNE) [18, 19].
Although the link between TNF-α and bone marrow suppression in anemia of chronic disease such as rheumatoid arthritis is well documented [20], the inadequate response of the bone marrow during severe malarial anemia can be attributed to factors other than TNF-α. In clinical studies of children with malarial anemia, the proportion of circulating monocytes containing hemozoin and levels of plasma hemozoin were associated with anemia and reticulocyte suppression, independently of the level of circulating cytokines, including TNF-α. Furthermore, histologic examination of the bone marrow of children who have died from malaria shows that pigmented erythroid and myeloid precursors are associated with the degree of abnormal erythroid development [9]. Taken together, these observations provide compelling evidence for independent inhibition of erythropoiesis by hemozoin and TNF-α.
Extensive examination of clinical bone marrow samples is difficult for ethical reasons but in vitro studies have shown that suppression of erythroid precursor expansion by TNF-α is mediated by induction of nuclear factor kappa B (NF-κB) that inhibits erythroid-specific gene expression [21] and reviewed in [22]. Addition of extracted hemozoin to TNF-α enhances inhibition by TNF-α alone [9]. However hemozoin can induce apoptosis in the absence of inflammatory cytokines [23] and HNE can cause cell cycle arrest in erythroid progenitors [24].
These observations suggest that hemozoin and TNF-α exert their inhibitory effects on erythroblasts via different molecular pathways, but may act additively on developing erythroid cells. To better understand the pathology of severe malarial anemia we wanted to determine if there are molecular signatures for dyserythropoiesis and/or inadequate erythropoiesis that can distinguish between the effects of hemozoin and TNF-α, and to understand if their inhibitory effects in terms of modulating gene expression are overlapping or largely distinct. To do this, we used gene expression profiling by microarray to compare the transcriptome of purified erythroblasts incubated with either TNF-α or hemozoin extracted from P. falciparum cultures.
Results
Development of erythroid cultures exposed to hemozoin or TNF-α
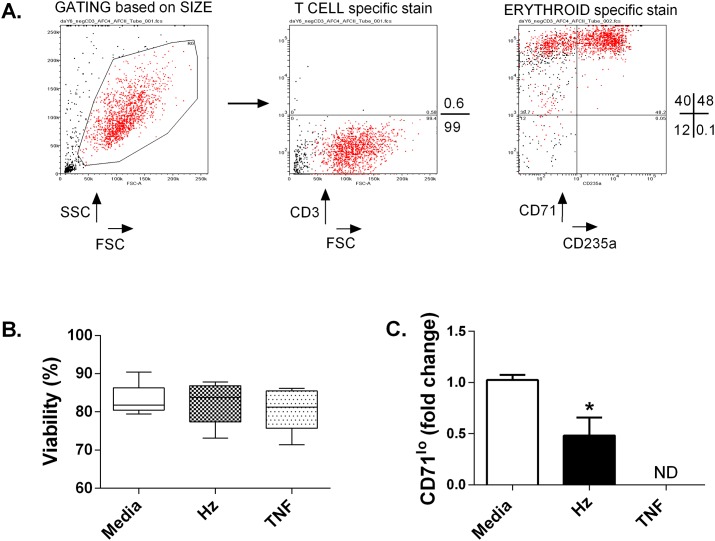
The viable erythroblasts used for microarray comprised 97% early pro-erythroblasts (CD71highCD235a-) and the more mature intermediate stage of erythroid development (CD71highCD235a+) with negligible contamination from other lineages (Fig. 1a and S1 Dataset). These erythroblasts were isolated from cultures derived from different donors and incubated with either 12.5 μg/ml heme equivalents of hemozoin or 10ng/ml TNF-α, doses previously shown to inhibit erythroblasts derived from adult peripheral blood [9, 23]. The viability of erythroid cells derived from different donor cultures used for microarray and qPCR were unchanged at 24 hours (Fig. 1b), but after 6 days the proportion of erythroid progenitors (CD235a+) that had matured and were expressing low levels of CD71 (CD71lo) (Fig. 1c) was reduced by 40% in cultures treated with hemozoin. This supports previous observations that this dose and preparation of hemozoin can inhibit erythroid expansion and development and so warranted further investigation to determine if the mechanism(s) that underlie its effect differ from those induced by TNF-α.
Fig 1. Representative example of samples used for microarray and rtPCR.
The purity of erythroblast populations treated with either TNF-α or hemozoin was determined by flow cytometry (A). The forward scatter (FSC) and side scatter (SSC) of live cells is shown in red. The proportion of cells (% of total events acquired) expressing lymphoid (CD3) and erythroid markers (CD71-CD235a+ and CD71+CD235a+) is represented by values to the right of each plot. The average response to treatment of all samples after 24 hours in which viability was assessed (B) or after 6 days when maturation was assessed by comparing the proportion of committed erythroid progenitors (CD235a+) that express low levels of CD71 (CD71lo) (C). Error bars show standard error of the mean of responses from 4 different donors; ND, not determined due to insufficient cell recovery. *p≤0.05 when compared with media alone (two tailed student’s test).
Gene expression profiles in erythroid cultures exposed to hemozoin or TNF-α
Erythroblasts incubated with either 12.5 μg/ml heme equivalents of hemozoin or 10ng/ml TNF-α were harvested after 24 hours and total RNA extracted. Following confirmation of inhibition by hemozoin or TNF-α after 6 days. Biotinylated fragmented cRNA was synthesized and hybridized to Affymetrix GeneChip Human Genome arrays (HG-U133_plus _2.0).
Data was normalized using RMA [25] and filtered to select probes with a signal of at least 100 in any one sample. To examine changes in response to hemozoin and TNF-α exposure, the dataset was filtered for genes changing greater than 1.5-fold in response to hemozoin and was hierarchically clustered by sample and by gene together with data previously acquired from erythroblasts sorted and grouped by stage of erythroid development using cell surface markers (CD36, CD71 and CD235a) and morphology [26]. The list of differentially expressed genes is given in Supporting Information 1 (S1 Dataset).
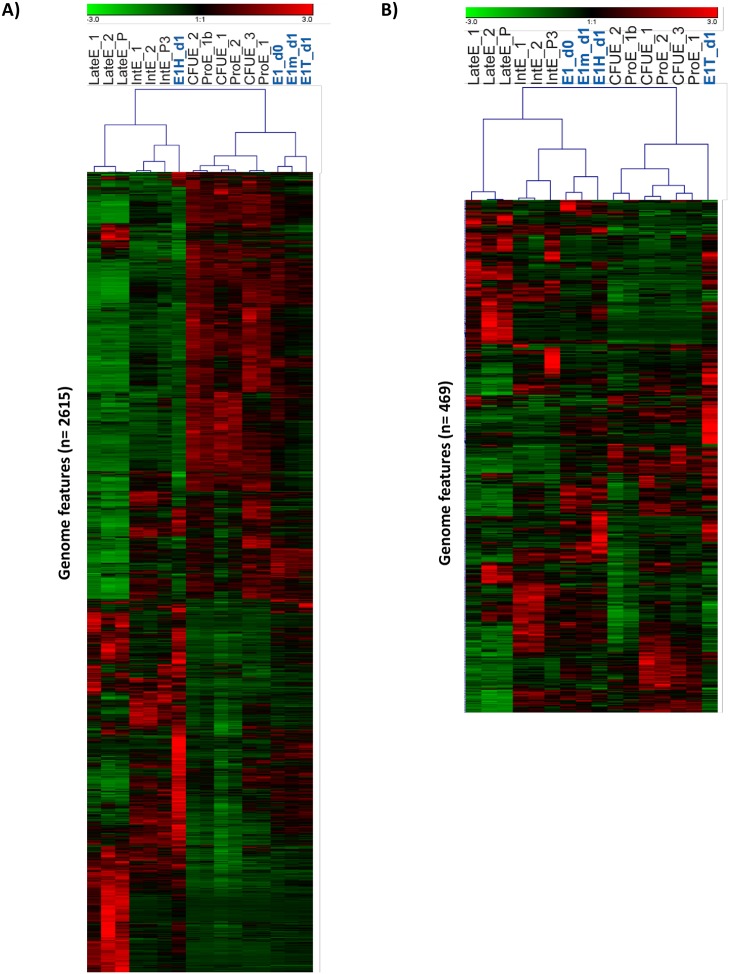
When considering all genes irrespective of differential regulation, treated and untreated samples clustered together, closest to the intermediate stage of primary samples (S1 Fig.). However when considering only those genes regulated by hemozoin, the separation of the gene expression profiles of hemozoin- and TNF-treated erythroblasts in the first bifurcation of the sample clustering dendrogram suggested that the gene expression program unfolding in response to hemozoin is quite different from that of untreated and TNF-treated counterparts (Fig. 2a). Similarly, repeating this analysis centered on the genes modulated by TNF-α treatment clustered the TNF-treated cells away from their untreated counterparts (Fig. 2b). Notably, in both cases genes differentially regulated by the agent under study are not similarly affected by the other agent; i.e. cells treated with hemozoin cluster together with untreated cells when examining the TNF-α differential genes, and vice versa.
Fig 2. Hierarchical clustering.
Expression of genes modulated >1.5x by hemozoin in erythroid cells before treatment (E1_d0), after incubation for 24 hours with hemozoin (E1H_d1) or TNF-α (E1T_d1) and without treatment in culture medium (E1m_d1) are shown, together with previously described sorted populations of erythroblasts [26]. Hierarchical clustering by gene and experiment (Pearson correlation) is shown centred around these hemozoin- modulated genes (A) or TNF-α- modulated genes (B). CFUE, colony forming unit erythroid FACsorted; ProE, pro- erythroblast FACsorted; Int, Intermediate erythroblast FACsorted; LateE, Late erythroblast sorted; E1, magnetic bead sorted erythroblasts; m, medium; T, TNF-α; H, hemozoin.
Functional associations between differentially regulated genes and biological processes
We first examined differential gene expression within each treatment group by comparing genes that were up- or down-regulated by at least 1.5 fold in response to each treatment. The statistical significance of functional associations between genes and biological processes within treatment groups were then determined using Genecodis-3 Hypergeometric analyses, corrected using a false determination rate calculation [27].
Response of erythroid cultures to TNF-α
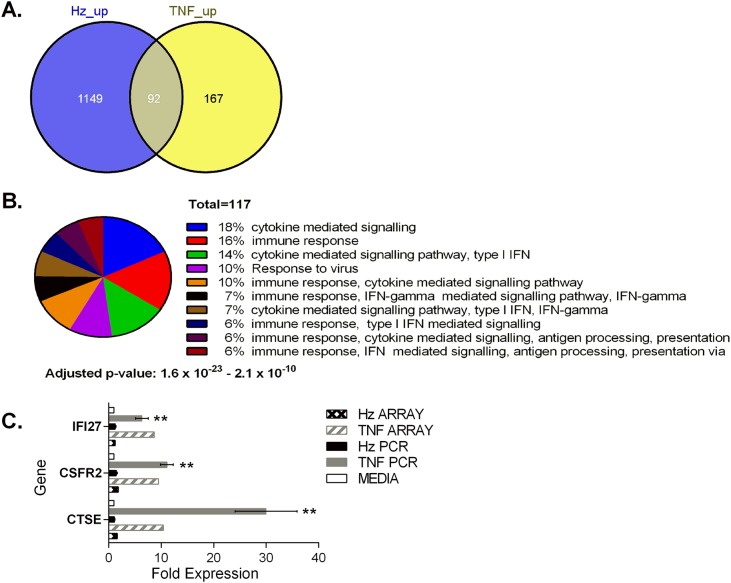
The most striking finding was that the majority of genes up-regulated by TNF-α were not differentially expressed in erythroblasts incubated with hemozoin (Fig. 3a and Table A in S1 Dataset), consistent with the clustering analysis. Of the 259 genes up-regulated by TNF-α approximately 64% of these were not significantly induced by hemozoin. As expected, gene ontology analysis revealed that more than 80% of the genes induced in erythroblasts treated with TNF-α are involved in innate and adaptive immune responses to infection (p<10-15) (Fig. 3b and Table B in S1 Dataset). Only 8% of the 1241 genes up-regulated by hemozoin were induced in erythroblasts treated with TNF-α. Amongst the up-regulated genes associated with immune function are Cathepsin E (CTSE), colony stimulating factor receptor 2 (CSF2RB) and interferon α-inducible protein 27 (IFI27). CTSE and IFI27 are involved in proteolysis of antigens and cytokine mediated signaling respectively. CSF2RB is the signaling sub-unit (βc) for the IL-3, IL-5 and GMCSF family of receptors reviewed in [28] and is required for cell growth, adaptive immune responses and myeloid differentiation. However further analysis by qPCR using erythroblasts from additional donors showed that up-regulation of these TNF-α up genes by hemozoin was insignificant when compared with the up-regulation induced by the pro-inflammatory cytokine TNF-α (Fig. 3c).
Fig 3. TNF-α induces up-regulation of genes distinct to those induced by hemozoin.
Venn analysis illustrates the overlap in genes up-regulated following treatment of primary erythroid cultures. Primary cultures of erythroblasts were established from peripheral blood and basophilic (CD71+CD235a- and CD71+CD235a+) erythroblasts isolated as described previously [23]. Erythroblasts were incubated with 10ng/ml TNF-α or 12.5μg/ml hematin units of hemozoin for 24 hours (A). Pie chart showing the number of up-regulated genes associated with biological processes described per concurrent annotation following treatment with TNF-α. The number of genes with annotations that fall within the hypothetical p values shown are listed, together with the proportion that they represent (B). Confirmation of distinct patterns of up-regulation by TNF-α observed in microarray by rtPCR using TaqMan gene expression assays (C). Fold expression compared with cells cultured in media alone is shown. TNF, TNF-α; Hz, hemozoin. Statistical analysis of rtPCR data was performed using the two tailed student’s t test. ** p<0.05 when compared with media or hemozoin. Fold expression determined from the original array data is shown for comparison.
Response of erythroid cultures to hemozoin
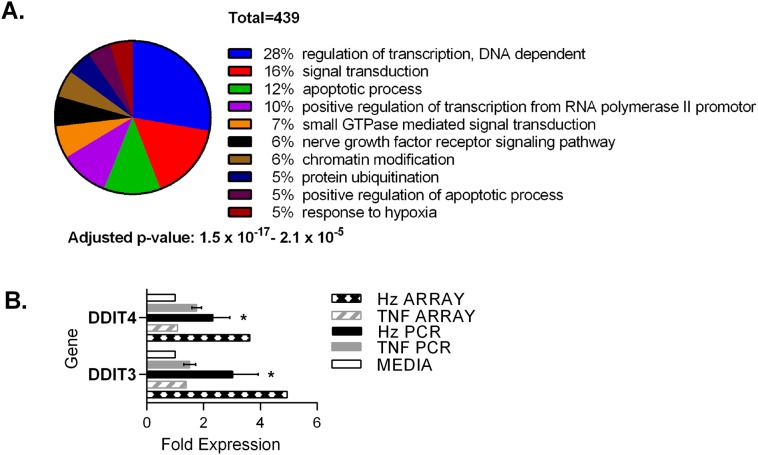
Statistical analyses showed that the majority of genes up-regulated by hemozoin are required for control of gene expression (p = 10-17) e.g. transcription factors controlling red cell differentiation and apoptosis whilst others were enriched in processes required for survival during cellular stress, neurotrophin tyrosine receptor kinase signaling and protein ubiquitination (p<10-7) (Fig. 4a and Table D in S1 Dataset). Genes up-regulated by TNF-α were not significantly associated with these gene ontology categories (Table B in S1 Dataset).
Fig 4. Up-regulated genes induced by hemozoin.
Pie chart showing the number of up-regulated genes associated with biological processes following treatment with hemozoin (A). Confirmation of observations in microarray data by rtPCR using TaqMan gene expression assays (B). Fold expression compared with cells cultured in media alone is shown. TNF, TNF-α; Hz, hemozoin. Statistical analysis of rtPCR data was performed using the two tailed student’s t test. * p≤0.05 when compared with media alone. Fold expression determined from the original array data is shown for comparison.
Inhibition of erythropoiesis by hemozoin
We wanted to identify differential expression that could explain inhibition of erythropoiesis by hemozoin. An example of up-regulation that could alter erythroid gene expression and differentiation is DDIT3, the C/EBP homologous protein (CHOP), a transcription factor that also mediates growth arrest and apoptosis as part of the integrated stress response to misfolded proteins in the ER (reviewed in [29]). Of note, also induced by hemozoin was the related gene DDIT4 (DNA damage-induced transcript 4 (REDD1) which can be induced by p53 in response to DNA damage and also forms part of the integrated stress response [30]. Further rtPCR examination of induced expression in erythroblasts from four donors confirmed significant induction of DDIT3 and DDIT4 transcripts by hemozoin which was absent in cultures incubated with TNF-α (Fig. 4b).
Down-regulation of genes in reponse to TNF-α or hemozoin
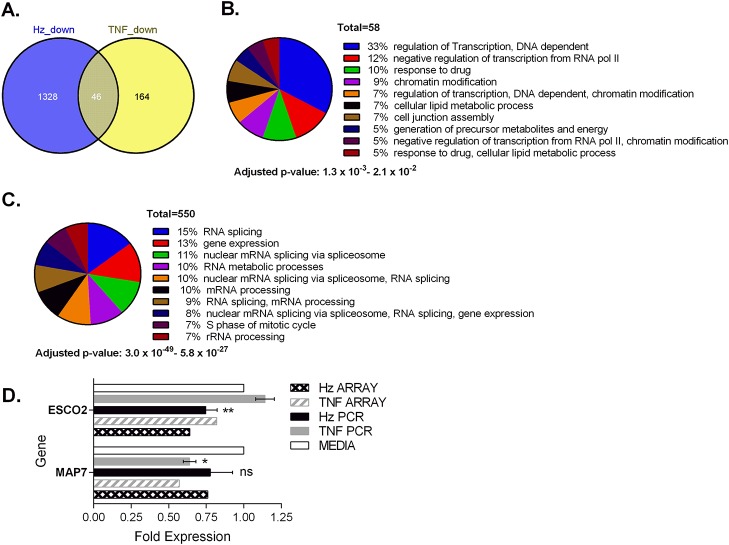
Of the 210 genes down-regulated by TNF-α, 78% were not significantly modulated by hemozoin and returned weak functional associations with relatively insignificant corrected p values above 10-3 (Figs. 5a, 5b and Table F in S1 Dataset). Conversely, statistical analysis of the 1328 genes down-regulated by hemozoin returned annotations that revealed a significant enrichment of genes known to encode for proteins involved in RNA splicing, ribosome biogenesis and the mitotic cell cycle. All of these predictions had corrected hypothetical probabilities of 10-10 or less (Fig. 5c and Table G in S1 Dataset). Genes down-modulated by both hemozoin and TNF-α were weakly associated with ontologies including cell cycle and nuclear mRNA splicing via the spliceosome with p values > 10-3 (Table J in S1 Dataset) possibly suggesting some degree of coalescence of these down-regulated programs at the functional level.
Fig 5. Hemozoin induces down-regulation of genes distinct to those affected by TNF-α.
Venn analysis illustrates the overlap in genes down-regulated following treatment of erythroid cultures (A). Pie charts that show the number of down-regulated genes associated with biological processes described per concurrent annotation following treatment with TNF-α (B) or hemozoin (C). The number of genes with annotations that fall within the hypothetical p values shown are listed together with the proportion that they represent. Microarray observations were confirmed by rtPCR for microtubule associated protein 7 (MAP7) and establishment of cohesion 2 (ESCO2) (D). Statistical analysis of rtPCR data was performed using the two tailed student’s t test. ** p<0.05 when compared with media or TNF-α, * p≤ 0.05 when compared with media alone; ns, non-significant compared with media.
Regulation of the spliceosome apparatus is closely linked to the cell cycle [31] and since hemozoin and its by-products are known to induce cell cycle arrest, we were interested to know if treatment of cells with hemozoin or TNF-α affected genes known to be important during mitosis. The microarray data showed that a gene required for stabilisation of the cytoskeleton during mitosis, microtubule associated protein 7 (MAP7/ensconsin) [32, 33], was reduced almost 2 fold by TNF-α and reduced 1.5 fold by hemozoin. The expression of another gene active during mitosis ESCO2 (establishment of sister chromatid cohesion 2) was reduced 1.5 fold by hemozoin, but remained unaffected by TNF-α. Further confirmatory rtPCR on treated cultures derived from 4 donors confirmed that MAP7 was more markedly and significantly reduced by TNF-α (Fig. 5d). Conversely ESCO2 was significantly reduced by hemozoin (p = 0.029) but remained unchanged or increased with TNF-α.
Discussion
The analysis of gene expression in erythroid cells described here provides, for the first time, a mechanistic insight into the different pathways that can lead to dyserythropoiesis and erythroid death during a malaria infection. This has been achieved by comparison of carefully temporally- and phenotypically-defined populations of erythroid cells exposed to TNF-α. Notably, hemozoin treatment that reduced viability and maturation of erythroblasts did not induce expression of TNF-α or ontologically-linked target genes in the transcriptional data, supporting the idea that although both agents can induce dyserythropoiesis, this is achieved via largely independent transcriptional programs.
There were at least seven times as many genes uniquely regulated by hemozoin as there were by TNF-α. However, in contrast to TNF-α few of the observed gene expression changes modulated by hemozoin were directly associated with immune function. Interestingly, changes in the transcriptome that indicate increased DNA damage and misfolding of proteins in the ER were prominent. This is consistent with previous observations that describe increased oxidative stress, apoptosis and/or cell cycle arrest in erythroid progenitors incubated with hemozoin or lipids associated with hemozoin, such as 4-hydroxy-nonenal (HNE) [23, 24].
The transcriptional profile following treatment of erythroblasts with either TNF-α or hemozoin shares some overlap with previous observations in peripheral blood of children with acute malaria [34] and in murine models of malaria [35]. In these studies the gene expression signature would be influenced by multiple cell types exposed to different components of the parasite’s life- cycle.
By using highly-enriched and purified erythroid cultures in which all viable responsive cells comprised hematopoietic erythroid precursors (Fig. 1a; S1 Tables), we were able to focus on the effects of hemozoin on erythroid gene expression without the confounding contribution from gene expression signatures of other contaminating cell lineages. Previously, a similar study has observed less potent inhibition of cell expansion by hemozoin but a greater effect on erythroid commitment when compared with TNF-α [36]. Although the erythroblast stage of development, culture conditions and hemozoin preparation used in this study differ from those described in the work presented here these observations are also consistent with the suggestion that distinct molecular pathways are affected by hemozoin and TNF-α.
In our study we have used physiologically relevant quantities of native hemozoin and TNF-α that were capable of inducing modest cell death. This aimed to mimic the scenario in the bone marrow where macrophages and neutrophils are unable to scavenge pigment due to inhibition by ingested trophozoites [19]. Indeed a recent study has shown that free hemozoin is present in the bone marrow which would support this hypothesis [37]. Since the majority of responsive cells in our cultures were erythroblasts, the role of macrophages would have been minimal (S1 Tables) such that the entry of hemozoin into erythroblasts would likely be via endocytic pits [38]. It is possible that with non-physiological higher doses, the effects of hemozoin on the erythroid transcriptome might differ from that described here, and might yield higher fold changes in gene expression but with the risk of inducing non-specific effects. It could be argued that pooling both early CD235a- and more mature CD235a+ proliferating CD71+ progenitors provided a less distinct pattern of gene expression than would be seen if these populations were separated. A future approach to achieve this beyond the scope of this study would be to use a fluorescence activated sorter to separate these populations.
Notwithstanding these points, we were able to identify and confirm distinguishable gene expression program changes induced by each of the two toxins on erythroblasts that may discriminate between the pathophysiological features of anemia due to inflammation, and those due to P. falciparum infection.
Previous work shows that four days treatment with TNF-α inhibits erythroid development by changing the balance of expression of the cross-antagonistic lineage-affiliated transcription factors GATA-1 (erythroid) and PU.1 (myeloid), thereby reducing the expression of GATA-responsive genes such as those encoding the Epo receptor and the hemoglobin α and β chains [39]. Here, we did not observe these changes, which may be partly due to differences in the timing of the experiment, or possibly that the probes used on the array may not optimally detect these genes [40]. However we did observe reversal of the expected pattern of erythroid gene expression in TNF-treated erythroid cells, in that expression of CSF2RB was increased, where normally it is decreased during erythropoiesis. Moreover, the pro-apoptotic gene IFI27 was induced by TNF-α when normally it should diminish during erythropoiesis [41]. These observations are consistent with activation of NF-κB downstream of TNF-α binding to TNFR1 [42]. The induction of CSF2RB by TNF-α could potentially result in formation of the tissue protective form of the Epo receptor or the receptor for GMCSF that supports myeloid differentiation [28, 43, 44]. Any one of these changes could arrest development of the erythroid precursors used in this study and prevent their progression to mature stages.
During erythropoiesis in vitro expression of both DDIT4 and DDIT3 increases from CFU-e through to the intermediate and late stages of erythroid differentiation, and ESCO2 drops significantly at the latest stage of development [41]. After 24 hours in culture with hemozoin, we found that mRNA transcripts for DDIT4 and DDIT3 were significantly increased and the transcripts for ESCO2 were prematurely decreased, compared with media controls or cells treated with TNF-α. These changes would therefore be expected to contribute to the clustering of pro-erythroblasts towards more mature intermediate erythroblasts following incubation with hemozoin for 24 hours (see Fig. 2).
We have previously shown that hemozoin mediates mitochondrial depolarization and apoptosis in erythroblasts [23] which can be mediated by increases in ROS and DNA damage respectively. Our observations by microarray, confirmed by rtPCR, are consistent with this. DDIT4 can increase the cell’s sensitivity to ROS resulting in cell death [30]. During cellular stress, especially after misfolding of proteins in the ER, DDIT3 accumulates in the nucleus where it transactivates the proapoptotic activating protein 1 (AP-1) complex [45]. DDIT3 can also inhibit the induction of Bcl2 [46] which allows accumulation of Bax and Bak in mitochondrial membranes leading to mitochondrial permeability and apoptosis. ESCO2 is required for cohesion during replication and repair of double strand breaks caused by DNA damage [47]. Loss of function mutations in ESCO2 result in hypersensitivity to DNA damaging reagents and reduced cell expansion [48] which may be mediated by activation of caspase 8 following G2 arrest [49]. The observed down-regulation of ESCO2 transcripts shown here therefore agrees with the previously described cell cycle arrest and induction of cleaved caspase 8 by hemozoin [23, 24].
The expression of DDIT4 is consistent with up regulation of p53 activity. Indeed induced expression of TP53INP1 (p53 induced nuclear protein 1) which mediates apoptosis [50] and reduced expression of TP53BP1 (tumor protein p53 binding protein) required for the repair of double stranded breaks in DNA [51], was seen in erythroblasts treated with hemozoin (Tables C and G in S1 Dataset). Furthermore, we observed down-modulation of the minichromosome maintenance complex (MCM) family of genes [52] by hemozoin and not TNF-α supporting an increase in p53 activity due to DNA damage [53] (Table K in S1 Dataset).
Other investigators have shown that the highly reactive component of hemozoin, HNE, can also alter expression of genes required for cell cycle checkpoint signalling and repair of DNA damage in a macrophage cell line [54]. In contrast to the work described here, they also observed up-regulation of pro-inflammatory cytokines such as TNF-α in response to HNE and IL-1β in response to β-hematin. This difference could be attributed to one or all of the following: differential gene expression programs extant in macrophages and erythroid cells driving differing responses; intrinsic properties of the macrophage cell line used; the maturation step using lipopolysaccharide that also activates the innate immune response; or the dose of HNE used which was equivalent to 100μM native hemozoin. The work described here uses 10 fold less native hemozoin on primary cultures of erythroblasts and thus, we believe, would more closely reflect the responses of developing erythroid cells. We (S2 Fig.) and others [55] have observed induction of DDIT3 protein by HNE. These observations suggest that HNE may in part contribute to the induction of integrated stress response by hemozoin in erythroblasts and therefore warrants further investigation in the context of malarial anemia.
The gene expression profile of primary erythroblasts infected with the early ring stage of P. falciparum has also revealed transcriptional changes in the host associated with regulation of reactive oxygen species and ER stress [56]. Of particular note is that genes required for cleavage and polyadenylation of pre-mRNA were specifically up-regulated in response to P. falciparum. The authors suggest this could provide nutrient and metabolic factors to the parasite. As such the transcriptional changes described would therefore include host responses to the parasite developing within the erythroblast. In this study we used an isolated component from later stages of the parasite’s life cycle to exclude direct effects of the parasite on erythropoiesis.
Taken together, the gene expression described in these studies, suggest multiple factors that can contribute to ineffective erythropoiesis in severe malarial anemia: hemozoin-induced repression of macrophage function; secretion of pro-inflammatory cytokines from macrophages; and perturbation of erythroblast metabolism to reduce red cell output from the bone marrow.
By comparing the effects of the parasite product hemozoin with the inflammatory mediator TNF-α on primary erythroblasts, we were able to distinguish between some of the effects of the host’s immune response to infection and that of hemozoin on the erythroid transcriptome. We found that largely distinct molecular programs were induced by hemozoin from those induced by TNF-α. The response to hemozoin was associated more with defects in the DNA repair response and a greater activity within the ER stress response pathway similar to that seen in macrophages incubated with HNE [54].
Conclusion
The work described here therefore provides further insight into the mechanisms that can mediate severe malarial anemia by identifying components of the integrated stress response pathway that can be investigated further to better understand the etiology of bone marrow suppression due to P. falciparum infection. Future studies will therefore target specific steps within the stress response and DNA repair pathway of erythroblasts and macrophages to reverse inhibition of erythropoiesis.
Methods
All tissue culture grade reagents were obtained from (Sigma, Poole, UK) unless otherwise stated. Peripheral blood mononuclear cells (PBMCs) were obtained from blood donated to the National Health Service Blood and Transplant (NHSBT; www.nhsbt.nhs.uk) following written consent and approval of their use by the National Health Service Oxfordshire Regional Ethical Committee.
Tissue Culture
The 3D7 strain of P. falciparum was up to 10% parasitemia at 1–2% hematocrit as described previously [57, 58]. Erythroid cultures were grown from PBMCs in a two-stage system [59]. Briefly, PBMCs were isolated from a buffy coat (National Health Service Blood and Transplant, Bristol, UK) and early erythroblasts were expanded for one week in 10% conditioned medium (CM) from the bladder cancer cell line 5637, 10% non-heat inactivated FBS (SLI, Salisbury, UK) and 1μg/ml Cyclosporin A (Sandoz Pharmaceuticals, Surrey, UK) in α-modified MEM. Erythroid cells were washed twice before transfer into phase II medium comprising α-MEM with 1U/ml recombinant Epo (Ortho Biotech Janssen Cilag Ltd, Bucks, UK), 10ng/ml SCF (R&D Systems, Abingdon, UK), 0.3 mg/ml holo-transferrin (MP Biomedical, London, UK), 1μM dexamethasone (Faulding Pharmaceuticals plc, Warwickshire, UK), 3% non-heat inactivated FBS (SLI) and 1% deionized fraction V BSA. Erythroid precursors differentiate into mature erythroblasts and hemoglobinized normoblasts over 14 days. On day 6 of this second phase the cultures are enriched for erythroid progenitors using magnetic bead isolations (Miltenyi Biotec). Briefly CD3+ cells were depleted by a LD column and CD71+ cells selected from the excluded fraction using an LS column. Eluted cells (>90% viable) from the final step were incubated in freshly prepared Phase II medium with hemozoin or TNF-α for 24 hours before harvesting for RNA extraction. Lineage content of cells used for each experiment is shown in supporting information (S1 Tables).
Isolation and preparation of malarial pigment
Mycoplasma-free P. falciparum (3D7) cultures were enriched for trophozoites and schizonts over 60% Percoll [60] and lysed in 8μg/ml digitonin for 10’ on ice. Centrifugation at 16400xg for 10’ at 4°C was followed by sterile sonication (Soni prep 150, MSE) in 2% SDS Tris pH8 for 2s at amplitude of 10 microns on ice. Hemozoin was obtained after 4–5 washes in 2% SDS Tris pH8, one wash in 1% Triton X-100 and finally three washes in 100-fold volumes of PBS, before storage under nitrogen at -80°C. Before use all pigment preparations were sonicated for 10–20s as described above.
Quantification of pigment
The concentrations of hemozoin were determined following depolymerization in 20mM NaOH for 2 hours at room temperature [61]. The absorbance at 405nm was compared with known concentrations of α-hematin and the concentration determined as hematin equivalents/ml.
RNA extraction, biotinylation of cRNA preparation and hybridization
Total RNA using 7.5 x105 cells per treatment from one donor was extracted using Trizol (Invitrogen, Paisley, UK) or the RNEasy Mini kit (Qiagen) in accordance with the manufacturer’s instructions. An additional phenol-chloroform step was included for preparations isolated using Trizol. RNA integrity was evaluated using an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Only RNA with RIN>9 was processed further for microarray. Any contribution of RNA from dead cells would have resulted in poor RNA integrity with a RIN considerably lower than 9. Biotinylated fragmented cRNA was synthesized from 100ng of pooled RNA using the 2-Cycle cDNA Synthesis and the 2-Cycle Target Labelling and Control Reagent kits (Affymetrix, Santa Clara, CA), and hybridized to GeneChip Human Genome U133_Plus_2.0 arrays (Affymetrix) following the manufacturer’s recommendations.
Analysis of microarray data
Cell intensity calculation and scaling was performed using GeneChip Operating Software (GCOS) (Affymetrix) and data analysed by the Robust Multiarray Average (RMA) method of Irrizarry [25] using the Bioconductor suite packages in R (www.bioconductor.org). Probes with low signal intensity in all samples were filtered out and the resulting probes analysed for differential expression in Excel (Microsoft). Expression data for genes of interest were hierarchically clustered by gene and by experiment using Average linkage and Pearson correlation with Genesis (Graz University of Technology) [62]. GeneCodis was used for gene ontology mapping and obtaining functional output from differentially expressed genes. This stringent analysis provides gene ontologies which are significantly over-represented in the list of modulated genes, to detect programs of gene expression changes relevant to key biological processes. Corrected hypothetical p values are generated after the removal of redundant terms and inclusion of modules that clearly define different biological functions. Gene names were determined from Affymetrix HGU133_plus_2.0 probe identifiers (PIDs) using BioDB.net. Hierarchical clustering was performed by gene and by experiment using Pearson correlation in Genesis. The array data has been submitted to GEO with Accession number GSE65577.
RT PCR
cDNA was synthesized from 100ng RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Paisely, UK) following DNAse treatment where applicable. Real time PCR was performed using TaqMan Universal PCR Master Mix and validated gene expression assays (Life Technologies) (Table 1). TaqMan cycling conditions used were according to the manufacturer’s instructions. Expression data for each transcript was normalized to that for the reference gene Ribosomal Protein L13A (RPL13A) [63], which did not change expression during erythroid development [26] nor upon exposure to TNF-α or hemozoin (See gene lists in S1 Dataset). Fold changes compared with media controls were determined using the comparative Ct method.
Table 1. Taqman gene expression assays used for RT- PCR.
| Gene Symbol | Context sequence for inventoried assay | Ref Seq | Assay ID | Amplicon length |
|---|---|---|---|---|
| CTSE | CTACATGAGCAGTAACCCAGAAGGT | NM_148964.1 | Hs00157213_m1 | 57 |
| CSFR2 | TGACCCAGCATGTCCAGCCTCCTGA | NM_000395.2 | Hs00166144_m1 | 73 |
| DDIT3 | AACCTGAGGAGAGAGTGTTCAAGAA | NM_004083.4 | Hs01090850_m1 | 78 |
| DDIT4 | CGGAGGAAGACACGGCTTACCTGGA | NM_019058.2 | Hs01111686_g1 | 68 |
| ESCO2 | TGAAGTGTGACAGCCTTTTACACTT | NM_001017420.2 | Hs00411577_m1 | 136 |
| IFI27 | TGCCCCTGGCCAGGATTGCTACAGT | NM_001130080.1 | Hs00271467_m1 | 63 |
| MAP7 | GAGAAAGAAGCGACTTGAGGAGATT | NM_003980.3 | Hs01009609_m1 | 91 |
| RPL13A | AGACTGGGAAGATGCACAACCAAGG | NM_012423.2 | Hs01926559_g1 | 105 |
The inventoried target sequence and NCBI reference sequence (Ref Seq) for each amplicon is shown.
Flow cytometry
Erythroid cell maturation was determined by measuring expression of the transferrin receptor (CD71) and glycophorin A (CD235a) with the appropriate mAbs (Beckman Coulter plc, High Wycombe, UK and DAKO Cytomation Ltd, Ely, UK). Monocytes and macrophages were identified with murine anti-CD14 (Serotec, Oxford, UK), using appropriate isotype controls. Cells were stained in 0.5% BSA, 0.05% sodium azide in PBS (staining buffer) for 30’ at 4°C, washed twice and fixed in 2% paraformaldehyde. Live cells were identified from FSC and SSC profiles of 7-AAD (Becton Dickinson, Oxford, UK) negative unfixed cells. The absolute number of erythroid cells was determined by multiplying the viable cell count by the proportion of CD71+ and/or CD235a+ cells acquired. Countbright fluorescent beads (Invitrogen, Paisley, UK) were also used to determine erythroid numbers in culture. Cells were washed and analyzed on the same day by flow cytometry on a FACSCalibur or LSR II machine (Becton Dickinson) at a flow rate of <1500 events/s. Events were analyzed with WinMDI (http://facs.scripps.edu/software.html) and Weasel software (http://www.wehi.edu.au/cytometry/WEASEL.html).
Supporting Information
Lists of genes up-regulated and down-regulated by each treatment; tables showing the fold changes in expression determined by microarray and derivation of co-annotations with associated gene ontologies.
(XLSX)
Experimental data described in text was clustered with data previously obtained from primary erythroblasts sorted according to stage of development (26).
(TIF)
Induction of CHOP protein in pro- and intermediate erythroblasts by (A) hemozoin and (B) HNE. Response to thapsigargin (Sigma T9033; induces Ca2+ efflux from mitochondria at 3μM) is shown as positive control. (C) The proportion of annexin positive 7AAD+ erythroblasts induced by hemozoin extracted as described in materials and methods (Hz_per) is comparable to hemozoin extracted following schizont release in culture (Hz_scr). *p<0.05 compared with medium controls using an un-paired t-test.
(TIF)
(PDF)
Acknowledgments
We would like to thank Mr Hoi Pat Tsang and Mr Michel Theron for their technical assistance; Dr Kathryn Robson and Dr Climent Casals-Pascual for their constructive comments during the preparation of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files. Array data have been deposited to GEO with Accession number GSE65577.
Funding Statement
This article summarizes independent research funded by the National Institute for Health Research (NIHR) under its Programme Grants for Applied Research Programme (Grant Reference Number RP-PG-0310-1004). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. World Malaria Report 2013. WHO Press, 2013.
- 2. Okiro EA, Kazembe LN, Kabaria CW, Ligomeka J, Noor AM, Ali D, et al. Childhood malaria admission rates to four hospitals in Malawi between 2000 and 2010. PloS one. 2013;8(4):e62214 Epub 2013/05/03. 10.1371/journal.pone.0062214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Douglas NM, Lampah DA, Kenangalem E, Simpson JA, Poespoprodjo JR, Sugiarto P, et al. Major burden of severe anemia from non-falciparum malaria species in Southern Papua: a hospital-based surveillance study. PLoS medicine. 2013;10(12):e1001575; discussion e. Epub 2013/12/21. 10.1371/journal.pmed.1001575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weatherall DJ, Miller LH, Baruch DI, Marsh K, Doumbo OK, Casals-Pascual C, et al. Malaria and the red cell. Hematology (Am Soc Hematol Educ Program). 2002:35–57. [DOI] [PubMed] [Google Scholar]
- 5. Lamikanra AA, Brown D, Potocnik A, Casals-Pascual C, Langhorne J, Roberts DJ. Malarial anemia: of mice and men. Blood. 2007;110(1):18–28. [DOI] [PubMed] [Google Scholar]
- 6. Portugal S, Carret C, Recker M, Armitage AE, Goncalves LA, Epiphanio S, et al. Host-mediated regulation of superinfection in malaria. Nat Med. 2011;17(6):732–7. Epub 2011/05/17. 10.1038/nm.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casals-Pascual C, Huang H, Lakhal-Littleton S, Thezenas ML, Kai O, Newton CR, et al. Hepcidin demonstrates a biphasic association with anemia in acute Plasmodium falciparum malaria. Haematologica. 2012;97(11):1695–8. Epub 2012/06/13. 10.3324/haematol.2012.065854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abdalla S, Weatherall DJ, Wickramasinghe SN, Hughes M. The anaemia of P. falciparum malaria. Br J Haematol. 1980;46(2):171–83. [DOI] [PubMed] [Google Scholar]
- 9. Casals-Pascual C, Kai O, Cheung JO, Williams S, Lowe B, Nyanoti M, et al. Suppression of erythropoiesis in malarial anemia is associated with hemozoin in vitro and in vivo. Blood. 2006. [DOI] [PubMed] [Google Scholar]
- 10. Phillips RE, Pasvol G. Anaemia of Plasmodium falciparum malaria. Baillieres Clin Haematol. 1992;5(2):315–30. [DOI] [PubMed] [Google Scholar]
- 11. Menendez C, Fleming AF, Alonso PL. Malaria-related anaemia. Parasitol Today. 2000;16(11):469–76. [DOI] [PubMed] [Google Scholar]
- 12. Abdalla SH, Wickramasinghe SN, Weatherall DJ. The deoxyuridine suppression test in severe anaemia following Plasmodium falciparum malaria. Trans R Soc Trop Med Hyg. 1984;78(1):60–3. [DOI] [PubMed] [Google Scholar]
- 13. Dormer P, Dietrich M, Kern P, Horstmann RD. Ineffective erythropoiesis in acute human P. falciparum malaria. Blut. 1983;46(5):279–88. [DOI] [PubMed] [Google Scholar]
- 14. Kurtzhals JA, Adabayeri V, Goka BQ, Akanmori BD, Oliver-Commey JO, Nkrumah FK, et al. Low plasma concentrations of interleukin 10 in severe malarial anaemia compared with cerebral and uncomplicated malaria. Lancet. 1998;351(9118):1768–72. [DOI] [PubMed] [Google Scholar]
- 15. Othoro C, Lal AA, Nahlen B, Koech D, Orago AS, Udhayakumar V. A low interleukin-10 tumor necrosis factor-alpha ratio is associated with malaria anemia in children residing in a holoendemic malaria region in western Kenya. J Infect Dis. 1999;179(1):279–82. [DOI] [PubMed] [Google Scholar]
- 16. Giribaldi G, Prato M, Ulliers D, Gallo V, Schwarzer E, Akide-Ndunge OB, et al. Involvement of inflammatory chemokines in survival of human monocytes fed with malarial pigment. Infect Immun. 2010;78(11):4912–21. Epub 2010/08/25. 10.1128/IAI.00455-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pagola S, Stephens PW, Bohle DS, Kosar AD, Madsen SK. The structure of malaria pigment beta-haematin. Nature. 2000;404(6775):307–10. [DOI] [PubMed] [Google Scholar]
- 18. Goldie P, Roth EF Jr., Oppenheim J, Vanderberg JP. Biochemical characterization of Plasmodium falciparum hemozoin. Am J Trop Med Hyg. 1990;43(6):584–96. Epub 1990/12/01. [DOI] [PubMed] [Google Scholar]
- 19. Schwarzer E, Kuhn H, Valente E, Arese P. Malaria-parasitized erythrocytes and hemozoin nonenzymatically generate large amounts of hydroxy fatty acids that inhibit monocyte functions. Blood. 2003;101(2):722–8. [DOI] [PubMed] [Google Scholar]
- 20. Papadaki HA, Kritikos HD, Valatas V, Boumpas DT, Eliopoulos GD. Anemia of chronic disease in rheumatoid arthritis is associated with increased apoptosis of bone marrow erythroid cells: improvement following anti-tumor necrosis factor-alpha antibody therapy. Blood. 2002;100(2):474–82. [DOI] [PubMed] [Google Scholar]
- 21. Rusten LS, Jacobsen SE. Tumor necrosis factor (TNF)-alpha directly inhibits human erythropoiesis in vitro: role of p55 and p75 TNF receptors. Blood. 1995;85(4):989–96. Epub 1995/02/15. [PubMed] [Google Scholar]
- 22. Morceau F, Dicato M, Diederich M. Pro-inflammatory cytokine-mediated anemia: regarding molecular mechanisms of erythropoiesis. Mediators Inflamm. 2009;2009:405016 Epub 2009/01/01. 10.1155/2009/405016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lamikanra AA, Theron M, Kooij TW, Roberts DJ. Hemozoin (malarial pigment) directly promotes apoptosis of erythroid precursors. PloS one. 2009;4(12):e8446 10.1371/journal.pone.0008446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skorokhod OA, Caione L, Marrocco T, Migliardi G, Barrera V, Arese P, et al. Inhibition of erythropoiesis in malaria anemia: role of hemozoin and hemozoin-generated 4-hydroxynonenal. Blood. 2010;116(20):4328–37. 10.1182/blood-2010-03-272781 [DOI] [PubMed] [Google Scholar]
- 25. Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003:249–64. [DOI] [PubMed] [Google Scholar]
- 26. Merryweather-Clarke AT, Atzberger A, Soneji S, Gray N, Clark K, Waugh C, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011;117(13):e96–108. 10.1182/blood-2010-07-290825 [DOI] [PubMed] [Google Scholar]
- 27. Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic acids research. 2012;40(Web Server issue):W478–83. Epub 2012/05/11. 10.1093/nar/gks402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hercus TR, Thomas D, Guthridge MA, Ekert PG, King-Scott J, Parker MW, et al. The granulocyte-macrophage colony-stimulating factor receptor: linking its structure to cell signaling and its role in disease. Blood. 2009;114(7):1289–98. Epub 2009/05/14. 10.1182/blood-2008-12-164004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim R, Emi M, Tanabe K, Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11(1):5–13. Epub 2005/12/24. [DOI] [PubMed] [Google Scholar]
- 30. Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Molecular cell. 2002;10(5):995–1005. [DOI] [PubMed] [Google Scholar]
- 31. Blencowe BJ. Splicing regulation: the cell cycle connection. Curr Biol. 2003;13(4):R149–51. Epub 2003/02/21. [DOI] [PubMed] [Google Scholar]
- 32. Bulinski JC, Odde DJ, Howell BJ, Salmon TD, Waterman-Storer CM. Rapid dynamics of the microtubule binding of ensconsin in vivo. J Cell Sci. 2001;114(Pt 21):3885–97. Epub 2001/11/24. [DOI] [PubMed] [Google Scholar]
- 33. Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(14):5391–6. Epub 2006/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Griffiths MJ, Shafi MJ, Popper SJ, Hemingway CA, Kortok MM, Wathen A, et al. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005;191(10):1599–611. [DOI] [PubMed] [Google Scholar]
- 35. Sexton AC, Good RT, Hansen DS, D'Ombrain MC, Buckingham L, Simpson K, et al. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J Infect Dis. 2004;189(7):1245–56. [DOI] [PubMed] [Google Scholar]
- 36. Awandare GA, Kempaiah P, Ochiel DO, Piazza P, Keller CC, Perkins DJ. Mechanisms of erythropoiesis inhibition by malarial pigment and malaria-induced proinflammatory mediators in an in vitro model. Am J Hematol. 2011;86(2):155–62. Epub 2011/01/26. 10.1002/ajh.21933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aguilar R, Magallon-Tejada A, Achtman AH, Moraleda C, Joice R, Cistero P, et al. Molecular evidence for the localization of Plasmodium falciparum immature gametocytes in bone marrow. Blood. 2014;123(7):959–66. Epub 2013/12/18. 10.1182/blood-2013-08-520767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Betin VM, Singleton BK, Parsons SF, Anstee DJ, Lane JD. Autophagy facilitates organelle clearance during differentiation of human erythroblasts: evidence for a role for ATG4 paralogs during autophagosome maturation. Autophagy. 2013;9(6):881–93. Epub 2013/03/20. 10.4161/auto.24172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grigorakaki C, Morceau F, Chateauvieux S, Dicato M, Diederich M. Tumor necrosis factor alpha-mediated inhibition of erythropoiesis involves GATA-1/GATA-2 balance impairment and PU.1 over-expression. Biochem Pharmacol. 2011;82(2):156–66. Epub 2011/04/20. 10.1016/j.bcp.2011.03.030 [DOI] [PubMed] [Google Scholar]
- 40. Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21(7):818–21. Epub 2003/06/10. [DOI] [PubMed] [Google Scholar]
- 41.Human Erythroblast Maturation (HEM) Database [Internet]. 2011 [cited September 27th 2011]. Available from: https://cellline.molbiol.ox.ac.uk/eryth/index.html.
- 42. Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. Epub 2009/03/24. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- 43. Bohmer RM. IL-3-dependent early erythropoiesis is stimulated by autocrine transforming growth factor beta. Stem cells (Dayton, Ohio). 2004;22(2):216–24. [DOI] [PubMed] [Google Scholar]
- 44. Goodman JW, Hall EA, Miller KL, Shinpock SG. Interleukin 3 promotes erythroid burst formation in "serum-free" cultures without detectable erythropoietin. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(10):3291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ubeda M, Vallejo M, Habener JF. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Molecular and cellular biology. 1999;19(11):7589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Molecular and cellular biology. 2001;21(4):1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science (New York, NY. 2007;317(5835):245–8. [DOI] [PubMed] [Google Scholar]
- 48. Gordillo M, Vega H, Trainer AH, Hou F, Sakai N, Luque R, et al. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Human molecular genetics. 2008;17(14):2172–80. 10.1093/hmg/ddn116 [DOI] [PubMed] [Google Scholar]
- 49. Monnich M, Kuriger Z, Print CG, Horsfield JA. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PloS one. 2011;6(5):e20051 Epub 2011/06/04. 10.1371/journal.pone.0020051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Y, Zhang E, Berggreen C, Jing X, Osmark P, Lang S, et al. Survival of pancreatic beta cells is partly controlled by a TCF7L2-p53-p53INP1-dependent pathway. Human molecular genetics. 2012;21(1):196–207. Epub 2011/10/04. 10.1093/hmg/ddr454 [DOI] [PubMed] [Google Scholar]
- 51. Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–73. Epub 2006/12/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bailis JM, Forsburg SL. MCM proteins: DNA damage, mutagenesis and repair. Curr Opin Genet Dev. 2004;14(1):17–21. Epub 2004/04/28. [DOI] [PubMed] [Google Scholar]
- 53. Scian MJ, Carchman EH, Mohanraj L, Stagliano KE, Anderson MA, Deb D, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27(18):2583–93. Epub 2007/11/06. [DOI] [PubMed] [Google Scholar]
- 54. Schrimpe AC, Wright DW. Comparative analysis of gene expression changes mediated by individual constituents of hemozoin. Chem Res Toxicol. 2009;22(3):433–45. Epub 2009/02/05. 10.1021/tx8002752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin MH, Yen JH, Weng CY, Wang L, Ha CL, Wu MJ. Lipid peroxidation end product 4-hydroxy-trans-2-nonenal triggers unfolded protein response and heme oxygenase-1 expression in PC12 cells: Roles of ROS and MAPK pathways. Toxicology. 2014;315:24–37. Epub 2013/12/03. 10.1016/j.tox.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 56. Tamez PA, Liu H, Wickrema A, Haldar K. P. falciparum modulates erythroblast cell gene expression in signaling and erythrocyte production pathways. PloS one. 2011;6(5):e19307 Epub 2011/05/17. 10.1371/journal.pone.0019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trager W, Jensen JB. Human malaria parasites in continuous culture. Science (New York, NY. 1976;193(4254):673–5. [DOI] [PubMed] [Google Scholar]
- 58. Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, et al. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357(6380):689–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fibach E, Manor D, Oppenheim A, Rachmilewitz EA. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73(1):100–3. [PubMed] [Google Scholar]
- 60. Inger L, Hedvig P, Martha S, Artur S, Mats W, editors. Methods In Malaria Research. Fourth ed: MR4.ATCC; 2004. [Google Scholar]
- 61. Sullivan DJ Jr., Gluzman IY, Russell DG, Goldberg DE. On the molecular mechanism of chloroquine's antimalarial action. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–8. Epub 2002/02/12. [DOI] [PubMed] [Google Scholar]
- 63. Jesnowski R, Backhaus C, Ringel J, Lohr M. Ribosomal highly basic 23-kDa protein as a reliable standard for gene expression analysis. Pancreatology. 2002;2(4):421–4. Epub 2002/07/26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists of genes up-regulated and down-regulated by each treatment; tables showing the fold changes in expression determined by microarray and derivation of co-annotations with associated gene ontologies.
(XLSX)
Experimental data described in text was clustered with data previously obtained from primary erythroblasts sorted according to stage of development (26).
(TIF)
Induction of CHOP protein in pro- and intermediate erythroblasts by (A) hemozoin and (B) HNE. Response to thapsigargin (Sigma T9033; induces Ca2+ efflux from mitochondria at 3μM) is shown as positive control. (C) The proportion of annexin positive 7AAD+ erythroblasts induced by hemozoin extracted as described in materials and methods (Hz_per) is comparable to hemozoin extracted following schizont release in culture (Hz_scr). *p<0.05 compared with medium controls using an un-paired t-test.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Array data have been deposited to GEO with Accession number GSE65577.