Abstract
BRAF mutations can be found in various solid tumors. But accurate and reliable screening for BRAF mutation that is compatible for clinical application is not yet available. In this study, we used an automated immunohistochemistry (IHC) staining coupled with mouse monoclonal anti-BRAF V600E (VE1) primary antibody to screen the BRAF V600E mutation in 779 tumor cases, including 611 colorectal carcinomas (CRC), 127 papillary thyroid carcinomas (PTC) and 41 malignant melanomas. Among the 779 cases, 150 cases were positive for BRAF (V600E) staining, including 38 (of 611, 6%) CRCs, 102 (of 127, 80%) PTCs and 10 (of 41, 24%) malignant melanomas. Sanger sequencing and real-time PCR confirmed the sensitivity and specificity of IHC staining for the V600E mutation are 100% and 99%, respectively. Therefore, our study demonstrates that the fully automated IHC is a reliable tool to determine BRAF mutation status in CRC, PTC and melanoma and can be used for routine clinical screen.
The v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) gene encodes a serine/threonine protein kinase that is belonged to the Mitogen Activated Protein Kinase cascade (MAPK) signaling pathway. Since the discovery in 2002, BRAF mutations have been found in various solid tumors, including thyroid carcinoma, malignant melanoma and colorectal carcinoma1. The most common BRAF mutation is the T1799A transversion, which results the substitution of glutamic acid for valine at amino acid 600 (V600E) and leads to constitutive activation of BRAF2,3,4.
Due to poor response to convention chemotherapy, melanoma has a poor prognosis. Recent development of Vemurafenib that specifically targets BRAF(V600E) mutation have yield promising results5. The diagnostic test that is able to recognize melanoma patients harboring mutant BRAF allow the identification of patients who can benefit from Vemurafenib treatment6,7,8,9,10. BRAF is also important in the development of colorectal carcinoma (CRC). The progression of CRC relies on oncogenic activation of signaling pathways downstream of the EGFR, including BRAF mutation11,12,13. Papillary thyroid carcinoma (PTC) is the most common type of thyroid carcinoma, accounting for more than 80% of the thyroid carcinoma. Many works have shown that a high prevalence of BRAF mutations was found in PTC14,15. The rate of BRAF mutation increased significantly over a 15-year period16.
Currently, Sanger sequencing and real-time PCR are the clinical methods that are used to detect BRAF mutations in diagnostic laboratories, including selecting melanoma patients eligible to Vemurafenib treatment. However, Sanger sequencing and real-time PCR have significant disadvantages. Both methods are expensive and time-consuming, which limited their clinical application. Immunohistochemistry (IHC) is a technique that is readily available in pathology laboratories, and it is relatively cheap, efficient and suitable as a screening tool. Recently, several studies have demonstrated that a BRAF V600E mutation–specific monoclonal antibody (clone VE1) could detect the V600E mutated BRAF protein in different carcinomas. Yet some researchers believe that IHC is not a valid surrogate for sequencing to detect V600E mutated BRAF in CRC17,18,19,20. Hence, the optimal method to detect BRAF mutations in cancers remains to be determined. Here we report a novel and fully automated IHC assay to screen the BRAF V600E mutation in Chinese patients with CRC, PTC and melanoma. The sensitivity and specifity of this novel IHC assay are 100% and 99% respectively when compared with Sanger sequencing and real-time PCR for the detection of BRAF V600E mutation.
Results
Immunohistochemistry
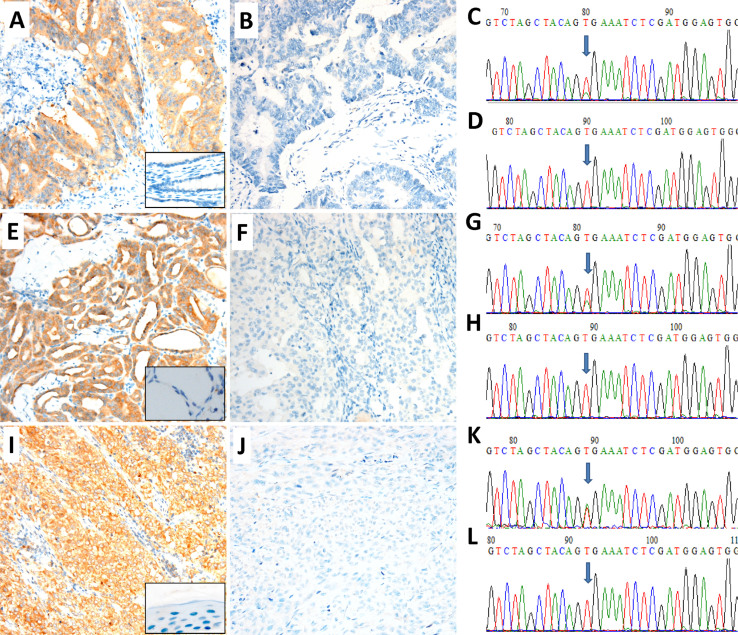
Ventana IHC assay using BRAF V600E (VE1) mouse monoclonal primary antibody was performed to screen for the BRAF V600E mutation in 779 patients, including 611 cases of CRC, 127 cases of PTC and 41 cases of malignant melanoma. Among the 779 cases, 150 cases were positive for BRAF (V600E) staining, including 38 cases (of 611, 6%) of CRC, 102 (of 127, 80%) cases of PTC and 10 (of 41, 24%) cases of malignant melanoma (Figure 1).
Figure 1. Detection of BRAF mutation in colorectal carcinoma (CRC), papillary thyroid carcinoma (PTC) and melanoma by immunochemistry (IHC) and Sanger sequencing.
Representative images of positive (A, E, I) and negative (B, F, J) for BRAF expression by VE1 IHC. Boxes in A, E, I show the negative controls from their corresponding non-tumor tissues. C, G and K images show a c.1799T > A (p.V600E) point mutation (arrow) of the BRAF gene. D, H and L images show the BRAF mutation (V600E) negative. BRAF Ventana VE1 IHC assay revealed strong expression in BRAF mutation positive patients and no expression in BRAF mutation negative patients in colorectal carcinoma (A–D), papillary thyroid carcinoma (E–H) and melanoma (I–L), respectively. Original magnification ×200.
Molecular analyses
A total of 349 patients were analyzed for BRAF mutation by both Sanger sequencing and real-time PCR (Cobas 4800 BRAF V600 Mutation Test), including 181 cases of CRC, 127 cases of PTC and 41 cases of malignant melanoma. Of the 349 tumors, 148 harbored T1799A mutation (p.V600E) of the BRAF gene by both Sanger sequencing and Cobas 4800 BRAF V600 Mutation Test, including 38 cases of CRC, 100 cases of PTC and 10 cases of malignant melanoma (Figure 1). No other mutation beyond V600E were detected in the exon 15 of BRAF gene. The results of Sanger sequencing and Cobas 4800 BRAF V600 Mutation Test matched to each other in all tested tumors (Table 1).
Table 1. Correlation of BRAF mutation detection between Sanger sequencing and real-time PCR.
| real-time PCR | ||||
|---|---|---|---|---|
| positive | negative | Total | ||
| Sanger sequencing | positive | 148 | 0 | 148 (42%) |
| negative | 0 | 201 | 201 (58%) | |
| Total | 148 (42%) | 201 (58%) | 349 (100%) | |
BRAF real-time PCR using Cobas 4800 BRAF V600 Mutation Test.
Comparison of immunohistochemistry and molecular analyses
As shown in Table 2, 150 patients with Ventana IHC BRAF V600E mutation were positive and 148 patients were positive by molecular assays. All patients with Ventana IHC BRAF V600E mutation negative were negative by molecular genetic techniques. Two PTCs with BRAF mutation positive by Ventana IHC were found to be negative by molecular assays. The details of these two discrepant cases for BRAF mutation detection were shown in Table 3. The sensitivity of BRAF Ventana IHC was measured as the proportion of the IHC positive cases in the molecular assays positive cases (148/148). The specificity of BRAF Ventana IHC was determined as the proportion of the IHC negative cases in the molecular assays negative cases (199/201). Therefore, the sensitivity and specificity of BRAF Ventana IHC for BRAF V600E mutation detection were 100% and 99%, respectively.
Table 2. Correlation of BRAF mutation detection between IHC and molecular assays.
| BRAF molecular assays | ||||
|---|---|---|---|---|
| positive | negative | Total | ||
| IHC | positive | 148 | 2 | 150 (43%) |
| negative | 0 | 199 | 199 (57%) | |
| Total | 148 (42%) | 201 (58%) | 349 (100%) | |
IHC, immunohistochemistry; BRAF molecular assays using Sanger sequencing and Cobas 4800 BRAF V600 Mutation Test.
Table 3. List of discrepant cases for BRAF mutation detection between IHC and molecular assays.
| Sample ID | Sex | Age | Histologic diagnosis | IHC | molecular assays |
|---|---|---|---|---|---|
| 256 | F | 50 | Papillary thyroid carcinoma | Positive | Negative |
| 307 | F | 36 | Papillary thyroid carcinoma | Positive | Negative |
Discussion
In this study, we have performed a fully automated IHC analysis to detect the V600E mutated BRAF protein using the Ventana BRAF V600E (VE1) mouse monoclonal primary antibody combined with the Optiview DAB IHC detection kit and compared it on matched samples with conventional molecular methods. Using Sanger sequencing and real-time PCR results as the reference, the sensitivity and specificity of VE1 immunohistochemistry for the BRAF V600E mutation are 100% and 99%, respectively. The BRAF Ventana IHC assay is a fully automated IHC test that can provide laboratory professionals and pathologists with a sensitive and specific standardized test for BRAF V600E mutation in formalin fixed paraffin embedded (FFPE) tissues. The interpretation of the results is clear. The negative and positive samples can be easily distinguished without the need of a subjective IHC scoring system based on staining intensity or percentage of positively stained cells.
The BRAF mutation is a promising diagnostic and prognostic marker and is also an important indicator for targeted therapy by BRAF V600E specific inhibitors. Accurate and reliable screening for BRAF mutation would be highly desired. In this study, we screened BRAF mutation status by Sanger sequencing, real-time PCR and immunohistochemistry. The Cobas 4800 BRAF V600 Mutation Test is the FDA-approved companion diagnostic for Vemurafenib, which consists of a real-time quantitative PCR step with two primers that amplify a 116 base pair fragment of the exon 15 of BRAF (containing codon 600). However, the real-time PCR cannot identify all BRAF mutation types. It can detect the main mutation types. Though it is designed to detect BRAF V600E (c.1799T > A) mutation, this kit also has some degree of cross-reactivity with V600K (c.1798_1799GT > AA) and other less common mutations such as V600E2 (c.1799_1800TG > AA), V600R (c.1798_1799GT > AG) and V600D (c.1799_1800TG > AC)21. Usually the sensitivity of the real-time PCR is higher than Sanger sequencing, although Sanger sequencing can detect all types of BRAF mutation. Discrepancies between these two assays have been previously reported. However, in our study, the results of BRAF mutation from the Sanger sequencing and Cobas 4800 BRAF V600 Mutation Test 100% correlates to each other, and no discrepancies were observed. The main reason for this high concordance might be the quality control of tumor samples. The H&E slides of all tumor tissues were evaluated for tumor content before proceeding to molecular analysis. When the proportion of tumor cells was less than 50%, the slides were marked for subsequent tumor macrodissection to enrich tumor cell populations before testing. Immunohistochemistry is a simple, rapid and relatively inexpensive method for detection of BRAF mutation. We performed immunohistochemistry using the Ventana BRAF V600E (VE1) mouse monoclonal primary antibody on Ventana Benchmark IHC automated strainers in combination with the OptiView DAB IHC detection kit in CRC, PTC and melanoma samples, and the results show highly sensitivity (100%) and specificity (99%). Our results demonstrated BRAF IHC is a simple, accurate and reliable screening method, which can be used routinely for BRAF mutation analyses in clinics.
Most importantly, the present study has used the fully automated IHC method to detect BRAF V600E mutation in various carcinomas including CRC, PTC and melanoma. Recently published studies exploring the VE1 antibody have also shown a high specificity and sensitivity, but only focus on single tumor type or small sample size22,23,24. In this study, we screen the BRAF V600E mutation in 779 patients, including 611 CRCs, 127 PTCs and 41 malignant melanoma. Of the 779 cases, 349 cases were also independently screened by Sanger sequencing and Cobas 4800 BRAF V600 Mutation Test, including 181 CRCs, 127 PTCs and 41 malignant melanomas. Only 2 PTCs with BRAF mutation positive by VE1 IHC were found to be negative by molecular genetic techniques. Although molecular genetic techniques are the standard method to detect BRAF mutations, it requires high tumor contents, special equipment and skilled operator. Our study demonstrated that IHC would be a useful tool for BRAF mutation screening in CRC, PTC and melanoma and potentially other tumor types.
Methods
Patients
The study was approved by the medical ethical committee of Cancer Institute and Hospital, Chinese Academy of Medical Sciences. The methods were carried out in accordance with the approved guidelines. The informed consents were obtained from all patients. A total of 779 patients were enrolled in this study, including 611 cases of CRC, 127 cases of PTC and 41 cases of malignant melanoma. All patients were treated at the Cancer Hospital, Chinese Academy of Medical Sciences, from July 2010 to June 2014. A total of 181 CRC samples and all cases of PTC and melanoma screened for BRAF mutation by IHC were also analyzed by both molecular methods. All cases were formalin-fixed and paraffin-embedded specimens. A pathologist evaluated the H&E slides for tumor content to make sure the proportion of tumor cells was more than 50%, otherwise, the slides were marked for subsequent tumor dissection to enrich tumor cell populations before proceeding to molecular analysis.
Immunohistochemistry
IHC for BRAF protein expression was performed on 4 μm-thick sections of formalin-fixed, paraffin-embedded tissues, using the Ventana BRAF V600E (VE1) Mouse Monoclonal Primary Antibody on Ventana Benchmark IHC automated slide strainer in combination with the OptiView DAB IHC detection kit. The specimens were fixed in 10% neutral buffered formalin for 24–48 hours. Negative and positive controls were included in each round of analysis. Briefly, after deparaffinization, the slides were pretreated with cell conditioning 1 for 64 minutes for antigen unmasking and followed by pre-primary antibody peroxidase inhibition. The slides were then incubated with the VE1 antibody at 37°C for 16 minutes, and counterstained with hematoxylin II for 4 minutes and bluing reagent for 4 minutes. The staining pattern for anti-BRAF V600E (VE1) antibody is cytoplasmic staining of tumor cells. The cases of cytoplasmic staining in tumor cells are positive when the anti-BRAF V600E (VE1) antibody is used and no staining when the negative control is selected.
Sanger Sequencing
Genomic DNA was extracted from paraffin-embedded tumor tissues using QIAamp® DNA Mini Kit (Qiagen, Germany), according to the manufacturer's instructions. Quality and concentration of the DNA samples were examined by NanoDrop (Thermo). The primers used to amplify BRAF exon 15 were as follows: forward 5′-TCATAATGCTTGCTCTGATAGGA-3′ and reverse 5′-GGCCAAAAATTTAATCAGTGGA-3′. Polymerase chain reaction (PCR) was carried as following: a final volume of 25 μl containing purified genomic DNA (100 ng/μl) 1 μl, 10 × ABI buffer 2.5 μl, MgCl2 (25 mM) 1.5 μl, dNTP (2.5 mM) 2 μl, ABI AmpliTaq Gold DNA Taq polymerase 0.125 μl (5 U/μl), forward primer and reverse primer (10 μM) 1 μl, after denaturation at 95°C for 10 minutes, 38 amplification cycles at 95°C for 30 s, 56°C for 30 s, 72°C for 45 s, and elongation at 72°C for 10 minutes. The PCR products sequencing were performed with ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer's instructions. The sequencing primers were the same as the PCR primers. Sequencing reactions were electrophoresed on an ABI 3500XL genetic analyzer (Applied Biosystems). Sequence data were analyzed using an ABI 3500XL DNA Analyzer (Applied Biosystems).
Real-time PCR
The real-time PCR was performed using the Cobas 4800 BRAF V600 Mutation Test Kit. DNA was adjusted to a fixed concentration and added to the detection mixture. The target DNA was then amplified and detected on the Cobas z 480 analyzer using the amplification and detection reagents provided in the Cobas 4800 BRAF V600 Mutation Test Kit. Tests follow the Cobas 4800 system Operator's Manual Software Version 2.0 for Cobas 4800 BRAF V600 Mutation Test for detailed instructions for the BRAF workflow steps. All runs and specimen validation were performed by the Cobas 4800 software.
Author Contributions
T.Q., J.Y. and N.L. designed experiments, T.Q. and J.Y. conducted experiments and data analysis, and wrote the paper. H.L. and W.H. performed the pathological diagnosis. T.Q., L.G., W.H., Y.L., L.S. and W.L. performed the experiments. All authors reviewed the manuscript.
Acknowledgments
We thank Dr Shan Zha from Institute for Cancer Genetics, Columbia University for critical editing for this manuscript. This work was supported by a grant from Youth Backbone Program (to Jianming Ying) of Cancer Hospital, CAMS, and the Technology Foundation for Selected Overseas Chinese Scholar, Ministry of Personnel of China.
References
- Davies H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). [DOI] [PubMed] [Google Scholar]
- Cohen Y. et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst 95, 625–627 (2003). [DOI] [PubMed] [Google Scholar]
- Dhomen N. & Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am 23, 529–545 (2009). [DOI] [PubMed] [Google Scholar]
- Ilie M. et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 24, 742–748 (2013). [DOI] [PubMed] [Google Scholar]
- Ascierto P. A. et al. The role of BRAF V600 mutation in melanoma. J Transl Med 10, 85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves W. O. et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn 15, 220–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchadkar R., Paraiso K. H. & Smalley K. S. Targeting mutant BRAF in melanoma: current status and future development of combination therapy strategies. Cancer J 18, 124–131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkenau H. T., Kefford R. & Long G. V. Targeting BRAF for patients with melanoma. Br J Cancer 104, 392–398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV1 W. J., Capper D., Preusser M., Zhang Y. E., Thompson J. F., Kefford R. F., von Deimling A. & Scolyer R. A. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol 37, 61–65 (2013). [DOI] [PubMed] [Google Scholar]
- Richter A. et al. A multisite blinded study for the detection of BRAF mutations in formalin-fixed, paraffin-embedded malignant melanoma. Sci Rep 3, 1659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siena S., Sartore-Bianchi A., Di Nicolantonio F., Balfour J. & Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst 101, 1308–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D. J. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38, 787–793 (2006). [DOI] [PubMed] [Google Scholar]
- Markowitz S. D. & Bertagnolli M. M. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 361, 2449–2460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer 12, 245–262 (2005). [DOI] [PubMed] [Google Scholar]
- de Biase D. et al. High-sensitivity BRAF mutation analysis: BRAF V600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. J Clin Endocrinol Metab 99, E1530–1538 (2014). [DOI] [PubMed] [Google Scholar]
- Mathur A. et al. Higher rate of BRAF mutation in papillary thyroid cancer over time: a single-institution study. Cancer 117, 4390–4395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adackapara C. A., Sholl L. M., Barletta J. A. & Hornick J. L. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 63, 187–193 (2013). [DOI] [PubMed] [Google Scholar]
- Day F. et al. A mutant BRAF V600E-specific immunohistochemical assay: correlation with molecular mutation status and clinical outcome in colorectal cancer. Target Oncol (2014) 10.1007/s11523-014-0319-8. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursault L. et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One 8, e70826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie M. I. et al. Diagnostic value of immunohistochemistry for the detection of the BRAF(V600E) mutation in papillary thyroid carcinoma: comparative analysis with three DNA-based assays. Thyroid 24, 858–866 (2014). [DOI] [PubMed] [Google Scholar]
- Angulo B., Lopez-Rios F. & Gonzalez D. A new generation of companion diagnostics: cobas BRAF, KRAS and EGFR mutation detection tests. Expert Rev Mol Diagn 14, 517–524 (2014). [DOI] [PubMed] [Google Scholar]
- Colomba E. et al. Detection of BRAF p.V600E mutations in melanomas: comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagn 15, 94–100 (2013). [DOI] [PubMed] [Google Scholar]
- Chen Q. J. et al. Immunohistochemistry as a quick screening method for clinical detection of BRAF(V600E) mutation in melanoma patients. Tumour Biol 35, 5727–5733 (2014). [DOI] [PubMed] [Google Scholar]
- Lasota J. et al. Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the imunohistochemical approach. Am J Surg Pathol 38, 1235–1241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]