Abstract
Campylobacter infections are a major cause of diarrhoea world-wide and two of the antimicrobials used for their control (erythromycin and ciprofloxacin) have been losing efficacy in recent years. In a sample of 174 genotyped isolates from the stools of patients with severe diarrhoea in Qatar, collected between 2005 and 2012, 63.2% showed resistance to ciprofloxacin, 8.6% to erythromycin, 0.57% to chloramphenicol and all were sensitive to gentamycin. While 33.9% of isolates were sensitive to all four antimicrobials, 59.8% were resistant to at least one, 6.3% were resistant to two and none showed resistance to three antimicrobials. There was no host sex- or age-dependence among isolates resistant to ciprofloxacin and erythromycin and no significant variation was found with the region of origin of the patients. All isolates were screened for the presence of 3 virulence factors (ciaB, cadF and cdtB) and two stress-response factors (htrB and clpP), all of which were present in more than 50% of the isolates. Host sex-, age- and region of origin-dependent variations in prevalence were found for some of these factors. Data analysis for the combination of virulence factors and their effect on antimicrobial resistance indicated that the prevalence of resistance to both erythromycin and ciprofloxacin was higher in isolates harbouring ciaB but not clpP. Prevalence of resistance to ciprofloxacin was similar in clpP positive and negative isolates also possessing htrB, while for htrB-negative isolates prevalence was higher in the absence of clpP. These results are discussed and their implications are highlighted.
Introduction
Campylobacter spp. are the leading cause of bacterial gastroenteritis worldwide [1]. These zoonotic bacteria can colonize humans with a relatively low infective dose and have potentially life-threating sequelae. Campylobacter spp. are suspected of causing disease via several mechanisms such as cytotoxin production, intestinal cell invasion, and adherence and translocation. The production of cytotoxin after intestinal colonization by ingested organisms arrests the cellular cycle, ultimately leading to cell death and thereby inducing diarrhoea [2]. The ability to invade intestinal cells results in damage to the mucosal surface cells of the jejunum, ileum and colon [3]. Finally, Campylobacter has been reported to translocate extra-intestinally, crossing the epithelium and gaining access to other parts of the body [4].
The drug of choice for the treatment of human Campylobacter infections is erythromycin (member of the macrolide family), followed by ciprofloxacin (fluoroquinolone family) and tetracycline [5], however, a dramatic increase in Campylobacter jejuni resistance to fluoroquinolones has been observed in the USA since the mid-1990s [5]. Initially, during the period of 1982–1992, no isolates of C. jejuni showed resistance to fluoroquinolone, and only 2% were found to be resistant to erythromycin among 142 patients [5]. By 2001, resistance to fluoroquinolone had developed, and was widespread (40.5% among 47 C. jejuni isolates) [5]. This rise of resistance to fluoroquinolone in human C. jejuni isolates has been linked to the licensing of fluoroquinolone use in poultry production [6]. Enrofloxacin, a related fluoroquinolone, was licensed in the early 1990s in Europe and Asia, and soon afterwards a high incidence of resistant C. jejuni isolates was reported from animal sources, for example from Spain (99% C. jejuni isolated from broiler farms), while 72% of human isolates were found to show resistance in the same period [7]. A similar trend was observed in Taiwan, where 92% of C. jejuni isolated from chickens and 52% of human isolates were found to be resistant to fluoroquinolones by 1998–2008 [8,9]. Fluoroquinolones are just one example, as increased antimicrobial resistance has been documented also for other commonly used antimicrobials, including clindamycin, erythromycin, streptomycin, and tetracycline [10].
Several mechanisms for antimicrobial resistance have been suggested for Campylobacter differing between the drugs involved. For fluoroquinolones the main resistance mechanism is probably a mutation in the gyrA gene, which encodes part of the DNA gyrase [11], while tetracycline resistance is thought to be typically mediated through the tetO gene [12,13]. For macrolides, a mutation in the target ribosome binding site mediates resistance [14]. Compared with infection by antimicrobial-susceptible strains, antimicrobial-resistant strains of Campylobacter have been reported to be associated with a longer duration of clinical symptoms [15,16]. Ciprofloxacin-resistant C. jejuni strains colonize poultry intestinal tracts better than isogenic susceptible C. jejuni strains even in the absence of fluoroquinolone selection pressure [17], indicating an association between resistance and the ability to colonize.
In addition, several genes have been linked to enhanced virulence of Campylobacter but the most important ones are Campylobacter invasion protein B (ciaB), cytolethal distending toxin B (cdtB) which disrupts mucosal barriers by causing host cell death, Campylobacter adhesin to fibronectin F (cadF), and the heat survival and stress-response proteins htrB and clpP. It is well-documented that there is an association between certain Campylobacter virulence genes and the pattern of clinical infection [18,19]. However, virulence may also be involved in modulating the expression of resistance to antimicrobial agents and such an association of antimicrobial resistance with virulence genes has already been noted in bacterial pathogens including Escherichia coli and Enterococcus faecalis [20,21] but not yet for Campylobacter.
To our knowledge, no data are available from Qatar regarding the status of antimicrobial resistance in Campylobacter. Herein, we first analyzed a nine-year dataset from the Hamad Medical Corporation (HMC) from which we extracted prevalence data on the most common enteric bacterial pathogens encountered in recent years in the stools of subjects with severe bloody diarrhoea in Qatar, in order to assess the relative importance of Campylobacter in the local population. We then investigated the prevalence and association of antimicrobial resistance and several virulence factors among C. jejuni isolates from symptomatic patients presenting as outpatients at the HMC hospital in order to enable knowledge based informed decisions relevant to the continuously changing dynamics of this society to be made by local health professionals and policy makers [22].
Materials and Methods
Clinical specimens
Ethical approval was obtained by Hamad Medical Corporation, Medical Research Center (Research protocol #13334/13). Consent was not required as the data were analyzed anonymously. All outpatients with severe and bloody diarrhoea are routinely screened at the HMC hospital for the presence of enteric bacteria using U.K. standards for microbiology investigations [23].
As part of the current work, data on the number of subjects screened and positives for each infectious agent were made available for the period 2005 to 2013, and these data are presented as prevalence values, in order to show the relative frequencies of enteric bacteria among subjects with severe diarrhoea in Qatar.
Further work, however, focused on Campylobacter. For this we exploited 216 cryopreserved (−80°C) specimens from earlier years as well as across the period for which we have presented prevalence values. Although we attempted to grow the bacterium from all 216 specimens known to be positive for Campylobacter, only 174 (n = 101 for 2005 to 2008 and n = 73 for 2009 to 2012) produced colonies of bacteria that could be genotyped and assessed for sensitivity to antimicrobials.
Specimens were tested for the presence of Campylobacter spp. using standard culturing techniques [24]. Specimens were streaked directly on Campyfix media supplemented with Skirrow Campylobacter selective antibiotic mixture (Oxiod) and incubated at 42°C under microaerobic conditions for 48 hours. The isolates were identified as C. jejuni based on the colony morphology, catalase, oxidase, hippurate hydrolysis, and H2S (TSI) biochemical tests, in addition to resistance to nalidixic acid and sensitivity towards cephalothin. A total of a 174 specimens identified positive for C. jejuni were analyzed. DNA was extracted directly from bacterial culture using a Mericon DNA extraction (Qiagen) kit. Specimens were confirmed to be C. jejuni by real-time PCR using a previously published primers/probe pair detecting the 16S rRNA gene [25].
Presence of potential virulence factors genes in Campylobacter isolates
Real-time PCR was used to detect the presence of three of the most important Campylobacter virulence factors. The genes are ciaB, cadF, and cdtB which are involved mainly in adhesion and invasion [26]. Screening for another two genes that enhance the survival of the bacteria in the environment and the host (htrB and clpP) was performed. These five genes will be referred to as virulence factors from this point onwards. The primers (Table 1) were designed using primer design software (Primer3) based on available sequences in GenBank. The primers were tested on C. jejuni ATCC 33560 strain to confirm their efficiency. For each reaction, the following conditions were used: A total of 7μl of Sybrselect (Life Technologies) master mixture was used in each reaction along with 0.3μl primer mixture, 2.5μl specimen DNA, and 10.2μl molecular grade water. RT-PCR cycles consisted of holding for 2 minutes at 95°C, followed by 40 cycles at 95°C for 15 sec, 55°C for 35 sec, and 72°C for 1 minute. Initially, specimens having positive Ct values were confirmed by agarose gel electrophoresis for each of the genes targeted in this study. Specimens were run in duplicates.
Table 1. Primers for virulence genes and stress response genes used in this study.
| Genes | Accession number | Primers | Sequence |
|---|---|---|---|
| cadF | U87559.1 | Forward | 5′- TAT GGT GTA GAA AAA AGT CGC ATC A-3′ |
| Reverse | 5′- ATC CGC TCT ACC TTC TTT AGT GTC A-3′ | ||
| cdtB | CP008787.1 | Forward | 5′- AAT GCA AGC TGA AGA AGT GAT TGT-3′ |
| Reverse | 5′- AGC ATC ATT TCC ATT GCG AAT-3′ | ||
| ciaB | AF114831.1 | Forward | 5′- CAA CTT TAT ATT TGC ACT CCG ATG-3′ |
| Reverse | 5′- GGA ACG ACT TGA GCT GAG AAT AAA C-3′ | ||
| clpP | CP008787.1 | Forward | 5′- TGG GAG CAT TTT TGC TTA GTT G-3′ |
| Reverse | 5′- CTC CAC CTA AAG GTT GAT GAA TCA T-3′ | ||
| htrB | AL111168.1 | Forward | 5′- CGC ACC CAA TTT GAC ATA GAA C-3′ |
| Reverse | 5′- TTT TTA GAG CGC TTA GCA TTT GTC T-3′ |
Testing for antimicrobial resistance
The susceptibility of each C. jejuni isolate to erythromycin, ciprofloxacin, gentamicin and chloramphenicol was determined using the E-test (bioMerieux, Durham USA) and read after 24 hours incubation in a microaerophilic environment at 42°C. The Minimum Inhibitory Concentrations (MIC) breakpoints of erythromycin (susceptible ≤8μg/ml), and ciprofloxacin (susceptible ≤1μg/ml) were used as recommended by Clinical Laboratory Standards Institute approved guidelines [27] for infrequently isolated or fastidious bacteria, and for gentamicin (susceptible ≤4μg/ml) and chloramphenicol (susceptible ≤8μg/ml), as recommended for non-Enterobacteriaceae [28].
Statistical analysis
Throughout, data are presented as prevalence values (%) with 95% confidence limits (in columns in tables, in parenthesis in the text and as error bars on figures), calculated as described by Rohlf and Sokal [29], employing bespoke software. For sample sizes in excess of 1000 we used the Poisson distribution if the number of positives was 100 or fewer, and for those over 100 we used the Gaussian distribution. All calculations were carried out on numbers based to at least three decimal places, but are rounded to the nearest single decimal place or two decimal places as relevant in the text and tables.
The first data-set used for analysis comprised 26,140 subjects screened for enteric bacteria. These data were analysed using maximum likelihood on log-linear analysis of contingency tables, implemented in IBM-SPSS version 22, with YEAR (9 levels, corresponding to 2005 to 2013) and the presence/absence of each infectious agent in turn (INFECTION- present or absent) as a binary factor.
The second data-set comprised 174 isolates that were positive for Campylobacter and were genotyped as well as assessed for sensitivity to each of the four antimicrobials used in the study. These were derived from 104 male and 70 female subjects, for whom age was recorded in months if under 1 year-old, and then in years for all older subjects. Most came from children under three years of age (62.6%), and hence to allow analysis with age of host taken into consideration, they were allocated to 5 age classes as follows: age class 1 = children less than 1 year-old (for males n = 22, mean age 0.47; for females n = 12, mean age = 0.53); age class 2 = 1 year-old children (for males n = 24, for females n = 20); age class 3 = 2 year-old children (for males n = 21, for females n = 10); age class 4 = 3–12 year-old (for males n = 26, mean age = 5.2; for females n = 17, mean age = 6.7); age class 5 = 19–75 year-old (for males n = 11, mean age = 48.7; for females n = 11, mean age = 40.6).
The subjects were allocated to five geographical regions of origin because they came from 24 different countries as follows: Qatar (n = 72), Arabian Peninsula (total n = 33; Iran n = 7; Iraq n = 1; Jordan n = 4; Oman n = 3; Palestine n = 7; Saudi Arabia n = 1; Syria n = 1; Yemen n = 9), Asia (total n = 42; Afghanistan n = 1; Bangladesh n = 2; India n = 16; Indonesia n = 1; Pakistan n = 20; Philippines n = 1; Sri Lanka n = 1), Africa (total n = 21; Egypt n = 11; Eritrea n = 1; Morocco n = 1; Sudan n = 6; Tunisia n = 2) and elsewhere (total n = 6; Canada n = 2; United Kingdom n = 1; United States n = 3).
For analysis of presence/absence of sensitivity to specific antimicrobials, and presence/absence of virulence factors we used maximum likelihood methods on log-linear analysis of contingency tables with SEX (2 levels, male and female), AGE (5 age classes as detailed above), REGION (region of origin at 5 levels as detailed above) and PRESENCE/ABSENCE of either resistance to a specified antibiotic or of a specific virulence factor (binary factor). Full factorial models were reduced to minimum sufficient models by step-wise deletion of non-significant terms until only significant effects remained (backward selection procedure). All Chi-squared values cited refer to these significant terms in minimum sufficient models, with all other relevant factors taken into consideration.
In specific cases we also employed 2x2 and 2xn Chi-squared test to test specific hypotheses as indicated in the text, using the methods described previously [30].
Results
Prevalence of enteric bacteria in stool specimens of outpatients with diarrhoea
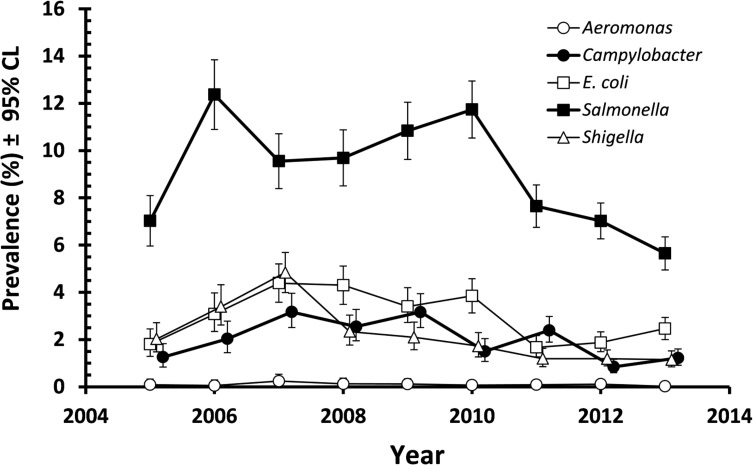
Fig. 1 shows the annual prevalence of pathogenic enteric bacteria in the period 2005 to 2013, in 26,140 subjects whose stools were examined because they had severe and bloody diarrhoea. The prevalence of Salmonella was the highest across the whole period (8.6% [8.28–8.96]) but there was significant variation between years (YEAR*INFECTION, χ 2 8 = 161.2, P< 0.001). Peak prevalence was recorded in 2006 and this was followed by a consistent decline over the following years. E. coli (2.8% [2.64–3.04]), Shigella (2.8% [1.84–2.18]) and Campylobacter (1.9% [1.73–2.06]) all showed lower but similar prevalence over the whole of this period, again with some variation between years, which in all three cases was significant (χ 2 8 = 94.9, 139.1 and 97.5, respectively and P<0.001 in all cases). As with Salmonella, there was a gradual decline in cases in the later years of the period (Fig. 1). Across the whole period, 26 cases of Aeromonas, 7 of Vibrio, and 3 of Yersinia were also detected. A detailed breakdown of the number of cases of Aeromonas, Campylobacter, E. coli, Salmonella, Shigella, Vibrio and Yersinia positive cases is provided as supplementary information (See S1 Table).
Fig 1. Prevalence of enteric bacteria in the stools of outpatients reporting to the MHC with severe diarrhoea in the period 2005–2013.
The sample sizes for 2005–2013 were 2220, 1916, 2460, 2394, 2528, 2726, 3347, 4373 and 4176, respectively.
Factors affecting resistance of Campylobacter to antimicrobials
Gentamycin and chloramphenicol
All specimens were sensitive to gentamycin and only one specimen showed resistance to chloramphenicol (prevalence of resistance = 0.57% [0.052–4.198]), so these were not analysed further, other than when the specimen that was resistant to chloramphenicol was included in the analysis of multiple-drug resistance.
Ciprofloxacin
The most resistance was observed to ciprofloxacin (63.2% [54.02–71.63]), and this was similar for isolates from both host sexes (Table 2), across all age classes (Table 2; range 59.1% to 68.2%), and among isolates from subjects from the five regions (Table 2; range 57.6% to 72.1%). No significant effects were detected for the effect of SEX, AGE or REGION of origin, or any interactions between these factors.
Table 2. The effect of host sex, age and country of origin on resistance of Campylobacter isolates to antimicrobials.
| Class | Ciprofloxacin | Erythromycin | |||
|---|---|---|---|---|---|
| n | % | 95% CL | % | 95% CL | |
| Males | 104 | 61.5 | 54.43–68.16 | 7.7 | 4.65–12.40 |
| Females | 70 | 65.7 | 54.14–75.91 | 10.0 | 4.82–19.14 |
| Age class 1 | 34 | 64.7 | 49.84–77.54 | 11.8 | 4.92–24.50 |
| Age class 2 | 44 | 59.1 | 41.97–74.63 | 6.8 | 1.53–20.36 |
| Age class 3 | 31 | 67.7 | 53.55–79.82 | 9.7 | 3.77–21.51 |
| Age class 4 | 43 | 60.5 | 43.58–75.58 | 7.0 | 1.65–20.37 |
| Age class 5 | 22 | 68.2 | 45.35–84.82 | 9.1 | 1.64–29.07 |
| Qatari | 72 | 59.7 | 47.95–70.83 | 11.1 | 5.45–20.71 |
| Arabian Peninsula | 33 | 57.6 | 42.87–71.27 | 3.0 | 0.39–12.95 |
| Asia | 42 | 73.8 | 57.28–86.05 | 7.1 | 1.78–20.39 |
| Africa | 21 | 61.9 | 40.33–80.26 | 14.3 | 4.01–35.43 |
| Elsewhere | 6 | 66.7 | 27.14–93.71 | 0 | 0–41.13 |
Erythromycin
Resistance to erythromycin was rarer (8.6% [4.57–15.32]), also similar for isolates from both sexes (Table 2) and across all five age classes (Table 2; range 7.0% to 11.8%). However, there were some discrepancies between regions of origin of subjects (Table 2; 0% to 14.3%), but these were not significant.
Multiple drug resistance
While 33.9% [25.81–43.10] of specimens were sensitive to all four antimicrobials, 59.8% [50.55–68.54] were resistant to at least one and 6.3% [3.07–12.27] were resistant to two. None showed resistance to 3 or all four antimicrobials.
Factors affecting prevalence of virulence factors
All five virulence factors were present in more than 50% of specimens (Table 3). The most frequent was cdtB and the least frequent cadF. Table 3 shows prevalence by geographic origin of the hosts.
Table 3. Prevalence of virulence factors among isolates from subjects from different geographical regions and in the combined dataset.
| Region of origin | ciaB | cadF | cdtB | htrB | clpP | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| % | 95% CL | % | 95% CL | % | 95% CL | % | 95% CL | % | 95% CL | |
| Qatar | 76.4 | 64.99–85.14 | 63.9 | 52.14–74.33 | 90.3 | 81.02–95.43 | 56.9 | 45.16–68.07 | 81.9 | 71.28–89.43 |
| Arabian Peninsula | 66.7 | 52.03–79.31 | 51.5 | 36.83–66.11 | 87.9 | 75.31–94.77 | 87.9 | 75.31–94.77 | 84.8 | 71.35–93.03 |
| Asia | 71.4 | 54.87–84.12 | 52.4 | 35.77–68.22 | 83.3 | 67.34–92.65 | 71.4 | 54.87–84.12 | 92.9 | 79.61–98.22 |
| Africa | 61.9 | 40.33–80.26 | 61.9 | 40.33–80.26 | 81.0 | 59.68–93.21 | 57.1 | 35.44–76.73 | 66.7 | 44.90–84.10 |
| Elsewhere | 83.3 | 41.14–99.14 | 100 | 58.87–100 | 100 | 58.87–100 | 100 | 58.87–100 | 83.3 | 41.14–99.14 |
| Combined | 71.8 | 62.98–79.36 | 59.8 | 50.55–68.54 | 87.4 | 80.12–92.48 | 67.8 | 58.64–75.80 | 83.3 | 75.34–89.21 |
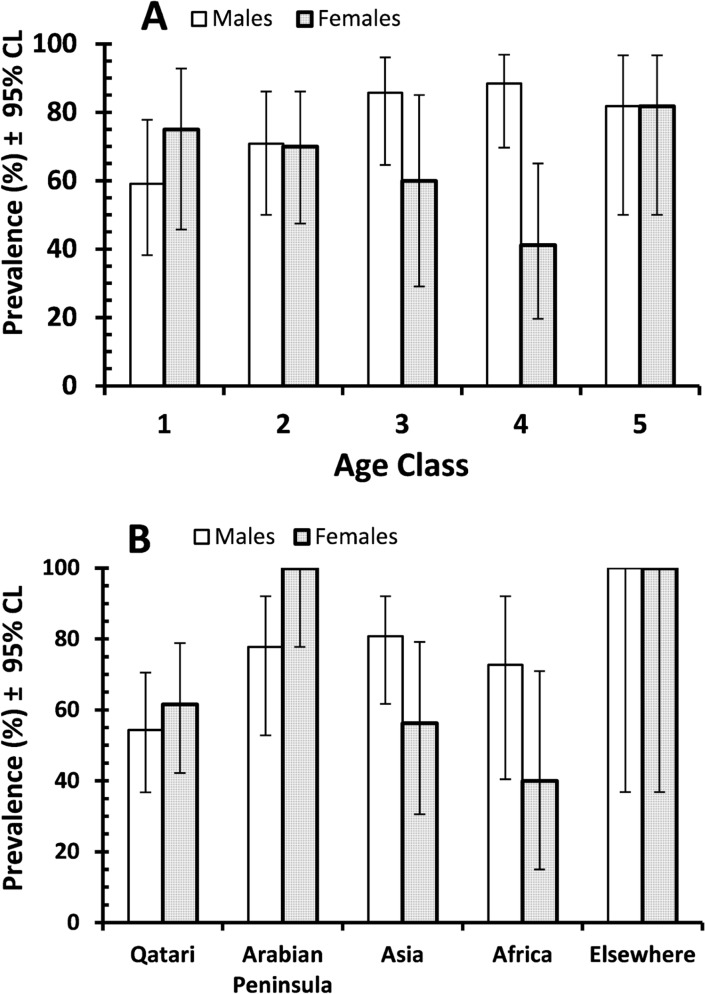
For ciaB there was a significant interaction between SEX and AGE on prevalence (χ 2 4 = 10.8, P = 0.029), as illustrated in Fig. 2-A. The prevalence of ciaB increased with the age of males, but among females prevalence was lower in age classes 3 and 4. There was no difference between isolates from subjects from different regions.
Fig 2. Prevalence of the virulence factor ciaB (A) and htrB (B) among male and female subjects in each of the age classes (A) and from each of the five regions of origin used for analysis (B).
The sample sizes in A were as follows: for the 104 male subjects, 22, 24, 21, 26 and 11, and for the 70 female subjects 12, 20, 10, 17 and 11 for age classes 1–5 respectively in both cases. For the age span and average age of each age class see Materials and Methods. The sample sizes in B were as follows: for the 104 male subjects, 46, 18, 26, 11 and 3, and for the 70 female subjects 26, 15, 16, 10 and 3 for the five regions of origin (Qatar, Arabian Peninsula, Asia, Africa and elsewhere) respectively in both cases. For further details of countries of origin in each region see Materials and Methods.
The prevalence of cadF only differed between the sexes (males = 67.3% [60.24–73.68]; females = 48.6% [36.94–60.23];χ 2 1 = 6.09, P = 0.014), and there was no effect of AGE or REGION of origin (Table 3) and no interactions between these factors.
None of the factors affected the prevalence of cdtB among the specimens, but there was a significant interaction between SEX and REGION of origin on the prevalence of htrB (Fig. 2-B; χ 2 4 = 10.7, P = 0.03). While in Qatar and the Arabian peninsula prevalence of htrB showed female bias, in Asia and Africa the bias was in favour of the male subjects.
The prevalence of clpP also varied markedly between sexes with a male bias (males = 89.4% [84.24–93.20]; females = 74.3% [63.05–83.16];χ 2 1 = 6.8, P = 0.009) and there was a significant interaction between AGE and REGION (χ 2 16 = 29.3, P = 0.022), which we did not explore further.
Relationship between virulence factors and resistance to ciprofloxacin and erythromycin
Although the data in Table 4 show some bias in prevalence of virulence factors in some cases, none of these were significant and therefore none of the virulence factors were clearly linked to resistance to either drug. The nearest to significance was the discrepancy in the prevalence of htrB among ciproxin sensitive and resistant isolates, for which there was a difference of 13.3% between resistant isolates compared with those that were sensitive (Table 4). However, analysis revealed that this was just the wrong side of the cut-off for significance, so this is indicative of a link but is not conclusive.
Table 4. Prevalence of virulence factors among erythromycin sensitive and resistance isolates, and among ciprofloxacin sensitive and resistant isolates.
| Virulence factor | Erythromycin | Ciprofloxacin | ||||||
|---|---|---|---|---|---|---|---|---|
| Sensitive (n = 159) | Resistant (n = 15) | Sensitive (n = 64) | Resistant (n = 110) | |||||
| % | 95% CL | % | 95% CL | % | 95% CL | % | 95% CL | |
| ciaB | 71.7 | 63.24–78.90 | 73.3 | 46.58–90.33 | 67.2 | 56.10–76.73 | 74.5 | 67.78–80.41 |
| cadF | 61.0 | 52.25–69.02 | 46.7 | 22.23–70.60 | 54.7 | 43.54–65.27 | 62.7 | 55.44–69.51 |
| cdtB | 88.1 | 81.26–92.69 | 80.0 | 53.43–94.31 | 89.1 | 80.12–94.49 | 86.4 | 80.64–90.73 |
| htrB | 69.2 | 60.55–76.73 | 53.3 | 29.40–77.77 | 59.4* | 48.25–69.92 | 72.7 | 65.73–78.77 |
| clpP | 84.9 | 77.62–90.32 | 66.7 | 39.68–85.83 | 85.9 | 76.31–92.31 | 81.8 | 75.53–86.85 |
*2x2 Chi-squared test, χ 2 1 = 3.3, P = 0.059
Relationship between virulence factors and isolates of Campylobacter showing sensitivity or resistance to 1 or 2 antimicrobials
Table 5 summarises the prevalence of each virulence factor among the isolates that were either sensitive to any one or two of the antimicrobials. No isolate was resistant to three antimicrobial and no significant effects were detected.
Table 5. Prevalence of virulence markers among isolates which are sensitive or resistant to 1 or 2 antimicrobials.
| Virulence factor | Sensitive to all antimicrobials (n = 59) | Resistant to 1 antimicrobials (n = 104) | Resistant to 2 antimicrobials (n = 11) | Statistical analysis | |||
|---|---|---|---|---|---|---|---|
| % | 95% CL | % | 95% CL | % | 95% CL | ||
| ciaB +ve | 67.8 | 57.11–76.98 | 73.1 | 66.27–78.96 | 81.8 | 50.00–96.66 | χ22 = 0.82; P = 0.66 = NS |
| ciaB—ve | 32.2 | 23.02–42.89 | 26.9 | 21.04–33.73 | 18.2 | 3.34–50.00 | |
| cadF +ve | 55.9 | 45.19–66.13 | 62.5 | 55.39–69.13 | 54.5 | 26.46–80.04 | χ 2 2 = 0.742; P = 0.69 = NS |
| cadF-ve | 44.1 | 33.87–54.81 | 37.5 | 30.87–44.61 | 45.5 | 19.96–73.54 | |
| cdtB +ve | 89.8 | 81.58–94.80 | 86.5 | 80.99–90.78 | 81.8 | 50.00–96.66 | χ 2 2 = 0.44; P = 0.80 = NS |
| cdtB—ve | 10.2 | 5.20–18.42 | 13.5 | 9.22–19.01 | 18.2 | 3.34–50.00 | |
| htrB +ve | 61.0 | 50.30–70.76 | 72.1 | 65.28–78.06 | 63.6 | 33.29–86.49 | χ 2 2 = 2.16; P = 0.34 = NS |
| htrB—ve | 39.0 | 29.24–49.70 | 27.9 | 21.94–34.72 | 36.4 | 13.51–66.71 | |
| clpP +ve | 86.4 | 77.47–92.37 | 83.7 | 77.76–88.37 | 63.6 | 33.29–86.49 | χ 2 2 = 2.45; P = 0.25 = NS |
| clpP-ve | 13.6 | 7.63–22.53 | 16.3 | 11.63–22.24 | 36.4 | 13.51–66.71 | |
All statistical tests here were 2x3 Chi-squared tests
Combinations of virulence factors and their effects on resistance to ciprofloxacin and erythromycin
We first examined pairwise combinations of the five virulence factors and their effects on resistance to both antimicrobials. Of the ten possible combinations, in the case of erythromycin prevalence of resistance was lower in those showing both factors compared with those with neither factor in eight out of ten cases, but the degree of reduction in prevalence of resistance ranged only from −1.9% (ciaB and htrB) to −10.3% (cadF and htrB). In two cases prevalence of resistance was marginally higher in those isolates expressing both virulence factors (ciaB and cdtB by 7.1%; ciaB and clpP by 5.4%), but in no case was the difference in prevalence of resistance among isolates that were negative for both factors and those expressing both, significant. Six combinations showed increased resistance to ciprofloxacin, ranging from 7.5% (cadF and clpP) to 19.2% (cadF and htrB), the latter being just outside the cut-off for significance (2x2 Chi-squared test, χ 2 1 = 3.47, P = 0.062). Four combinations showed lower resistance ranging from −4.5% (ciaB and cdtB) to −15.7% (cdtB and clpP) and none were significant.
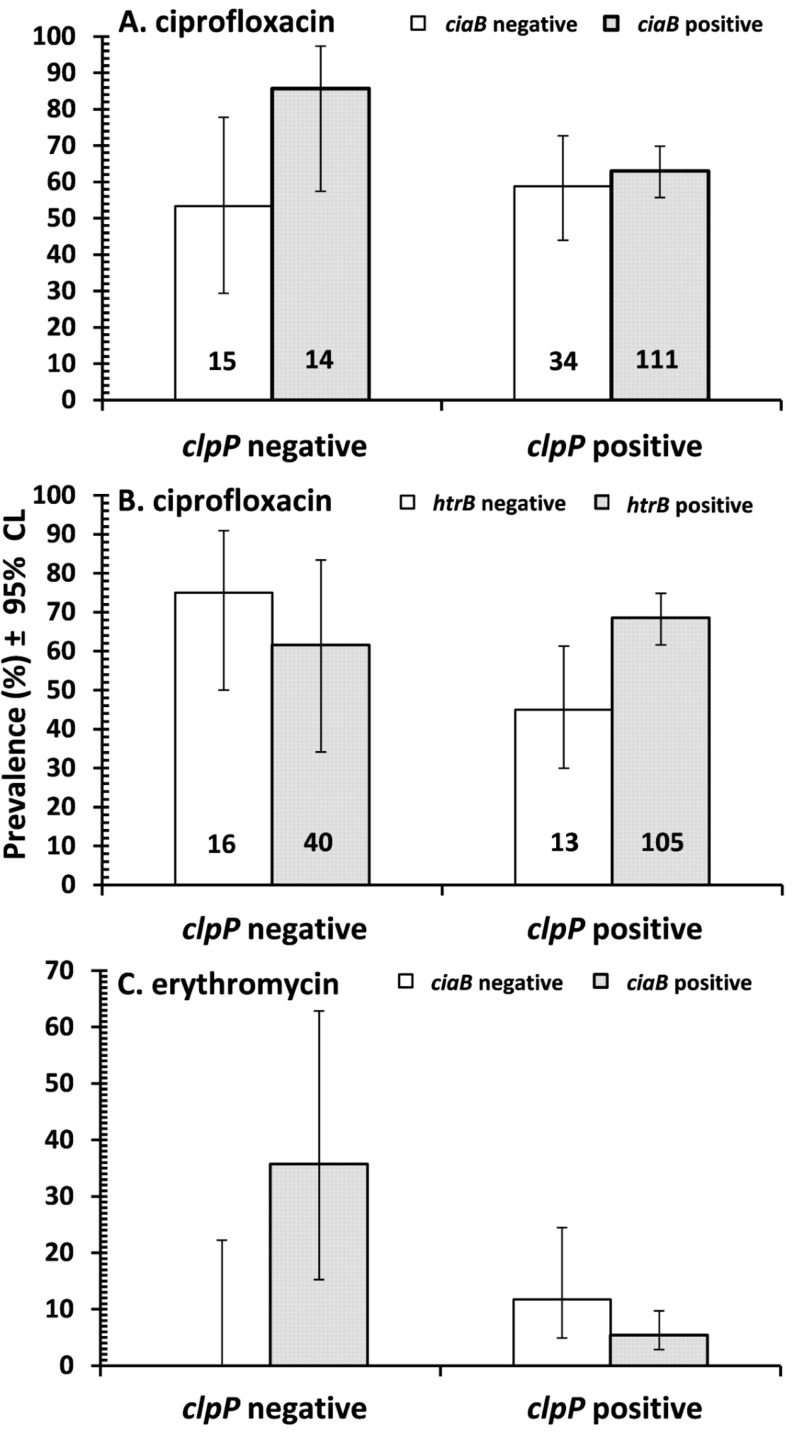
Analysis by fitting log-linear models indicated three significant interactions. Fig. 3-A shows that prevalence of resistance to ciprofloxacin was very similar in ciaB negative isolates whether they expressed clpP or not, while for ciaB positive isolates prevalence of resistance was higher in the absence of clpP (χ 2 1 = 5.2, P = 0.023). Fig. 3-B shows that while in clpP negative isolates prevalence of resistance to ciprofloxacin was similar whether they expressed htrB or not, among clpP positive isolates prevalence of resistance was higher when htrB was also present (χ 2 1 = 6.9, P = 0.002). Finally in Fig. 3-C it can be seen that among clpP negative isolates prevalence of resistance to erythromycin was higher in ciaB positive isolates, while in clpP positive isolates prevalence of resistance was generally lower and only differed marginally between isolates that were positive or negative for ciaB (χ 2 1 = 9.5, P = 0.002).
Fig 3. Prevalence of resistance to ciprofloxacin (A and B) and erythromycin (C) in Campylobacter isolates which were positive or negative for the clpP virulence factor, in the presence/absence of CiaB (A and C) and htrB (B).
Sample sizes are given on the columns in A and B, and for C the sample sizes were the same as in A.
Discussion
Diarrhoeal diseases remain a leading cause for morbidity and mortality worldwide despite advances in our understanding of pathogen transmission patterns. The nine-year dataset we analyzed from HMC for 26,140 subjects who presented with clinical gastrointestinal signs showed that a group of bacterial pathogens are responsible for a significant proportion of these cases. Indeed, Salmonella, Campylobacter, E. coli, or Shigella were involved collectively in at least 15% of the cases. Prevalence of these pathogens varied over the years, with a clear decline in most cases towards the end of the study period. This year-to-year variation in prevalence emphasizes the importance of the surveillance program HMC is implementing to monitor this group of pathogens. The analysis of the nine-year data set also confirmed the importance of C. jejuni as a common enteric pathogen in Qatar.
As the next step in this study, we investigated the prevalence of antimicrobial resistance and its association with the presence of virulence and environment stress response genes in C. jejuni isolates from symptomatic individuals. As erythromycin is the drug of choice for the treatment of C. jejuni, it was reassuring to find that antimicrobial resistance to this drug was relatively low, as only 8.6% of the isolates were resistant. On the other hand, the relatively high level of resistance to ciprofloxacin (63.2%), regardless of the country of origin of the patient, is alarming as this fluoroquinolone is indicated for the treatment of a variety of critical infections in adults [31]. Fluoroquinolones are considered critical drugs for the treatment of humans by the World Health Organization and the veterinary fluoroquinolones, enrofloxacin and danofloxacin, are commonly used antimicrobial drugs in food-producing animals and are thought to play a role in the spread of resistance to human isolates [32,33]. A high rate of resistance has also been reported in the United Arab Emirates where 85.4% of isolates were resistant to this antimicrobial [34], while isolates from other countries have shown lower rates of prevalence, e.g. 40% in Poland and 2% in Australia were reported to be resistant [35,36]. As the use of fluoroquinolones in food-producing animals is permitted in the UAE and Qatar, while it is banned in Australia, this difference in the levels of resistance could be attributable to the continued use of these antimicrobials. Further assessment of the rate of antimicrobial resistance among C. jejuni isolates from animals, in addition to concurrent genetic relationships, should determine if this is truly the case in Qatar.
As for the presence of virulence factors, all five factors investigated here were present in more than 50% of the specimens, with cdtB being the most frequently encountered and cadF the least. cadF is an adhesin and fibronectin-binding protein involved in the process of invasion by inducing structural re-arrangement through microfilament manipulation [37]. Since all the Campylobacter isolates investigated here were obtained from symptomatic patients and only 59.8% carried this gene, it is likely that the presence or absence of this one gene is not enough to set up infection or abolish it. The low prevalence of cadF contrasts with several other studies that have found this virulence factor in nearly all of the Campylobacter isolates that were examined [38,39,40,41]. Campylobacter strains that have a cadF deletion mutation are unable to colonize in chickens and their internalization ability into INT 407 intestinal cells is compromised [37]. However, clearly isolates in which we did not detect the gene survive in human subjects, as evidenced in our study, but because of the methods employed here we cannot eliminate the possibility that a variant of the gene, that was not detected by our PCR based methodology, was present and capable of mediating invasion of the host (see below for further discussion). Nevertheless, there is no current information on any variants of this gene, with a different nucleotide base sequence, that would not have been detected by our PCR primers.
The cytotoxin produced by C. jejuni can damage nuclear DNA, causing the cell cycle to arrest in the G2 or M phase [42]. Cytolethal distending toxin is also involved in the production of pro-inflammatory cytokines such as IL-8 from intestinal epithelial cells in vitro [43] which might explain the role it plays in pathogenesis. Several studies have indicated that the prevalence of cdtB in isolates from poultry or humans exceeds 90% [41], which is in agreement with our findings and was expected, given that the isolates studied here were all from clinical cases.
The ciaB gene is critical for invasion of epithelial cells as mutants lacking this gene are not able to invade cell lines in vitro [44]. As C. jejuni lacks a type III secretion system, this protein is secreted through a flagellar export system [45], but again in this study we did not detect ciaB in 28.2% of the isolates studied. Although ciaB was shown to be important for invasion using in vitro systems, in this study human isolates that lack this gene still caused clinical signs, indicating that even in its absence C. jejuni can cause disease.
C. jejuni is likely to encounter a wide range of temperatures during a contamination cycle and must therefore be able to adapt and respond to these temperatures. In contrast to other foodborne pathogens, C. jejuni does not possess genes encoding cold-shock proteins such as cspA which might explain why it cannot grow at low temperatures [1,46]. On the other hand, C. jejuni utilizes heat-shock proteins, including the ATP-dependent proteases clpP, which repair and prevent damage caused by accumulation of unfolded proteins [47,48]. Our study detected an interesting interaction between the presence/absence of this stress response gene and ciaB. In the case of resistance to both ciprofloxacin and erythromycin, isolates that did not express clpP, but were positive for ciaB, showed a higher prevalence of resistance. How the gene products of these two genes interact to bring this interaction about is not known, but it is clearly of interest if we are to achieve a comprehensive understanding of the mechanisms of the evolution and maintenance of antimicrobial resistance in Campylobacter infections.
The C. jejuni htrB gene encodes an acyltransferase that contributes to lipid A synthesis [49,50] and is conserved in C. jejuni. It is similar to the htrB gene of Salmonella typhimurium, E. coli and Haemophilus influenzae. The high prevalence of this gene in this group of clinical specimens might indicate its importance for the survival of this bacterium inside the host, but, nevertheless a proportion (32.2%) of our isolates did not express this gene, and here also we detected an interaction with clpP on prevalence of resistance to ciprofloxacin. While htrB positive isolates showed similar prevalence of resistance irrespective of whether they expressed clpP or not, among htrB negative isolates resistance was higher in the absence of clpP. Again, this suggests complex underlying background genetic and epistatic interactions.
Whilst we did not find any evidence of significant variation in the extent of resistance to ciprofloxacin or erythromycin that was host age-, sex- or region of origin-dependent, we did find that the presence of some of the virulence and stress factors on isolates was dependent on these host factors. For example, the prevalence of isolates expressing ciaB increased with the age in male subjects, but among females the prevalence was lower in subjects aged 2–12 year-old. A sex effect was also noticed for cadF which was more prevalent in males than females. As cadF is involved in adhesion to the intestinal wall of the host while ciaB is thought to be involved in cellular invasion of Campylobacter, it is likely that the different sex and age effects are linked to the different habitats for the bacteria in hosts of varying age and sex. Moreover, these sex- and age-linked effects on the expression of virulence/stress factors may in part explain the known predominance of Campylobacter among male subjects, which has been attributed to physiological differences between the sexes [51].
Although the findings in this study show a high prevalence of some of the virulence factors investigated, such as cdtB and ciaB, it is important to emphasize that the presence or absence of these genes is indicative, but may not predict precisely how virulent a particular Campylobacter isolate may be in vivo and further studies have to be carried out to reach a definitive conclusion. As already intimated above, it is also important to be aware that a negative result by RT-PCR could be attributed not just to an absence of the gene in question but possibly also to sequence variation at the primer binding site, perhaps indicating a variant of the same molecule with either identical or perhaps different biological activity. This is an important limitation of the molecular methods employed here, relying entirely on the amplification of the targeted region, and therefore the data should be interpreted carefully.
Positive and negative associations between virulence genes and antimicrobial resistance have been found in several bacterial pathogens including E. coli and E. faecalis [21,52]. A mutation in the gyrA gene in C. jejuni on the other hand enhances its ability to colonize chickens and therefore survival [17]. Another clear example is the effect of streptomycin resistance on the growth of Salmonella, as resistant strains have poor growth on rich media because they cannot express the rpoS gene which is important for regulation of environmental stress responses [53]. As the presence of antimicrobial resistance and potential virulence factors are both important in their own right, further investigation of the basis of synergistic and antagonistic interactions between their presence/absence in clinical isolates might provide new insight into the nature of Campylobacter pathogenesis. Indeed, methicillin resistance of Staphylococci, for example, has been used over the years as a marker to classify the different isolates as it is a critical factor in predicting not only the outcome of infection but also the fitness level of the isolate [54]. As for E. coli, a positive relationship has been established between multiple drug resistance and the severity and duration of clinical symptoms [55].
Finally, in this study we have provided for the first time data on the prevalence of resistance to two of the most popular and widely used antimicrobials for the treatment of Campylobacter, among isolates of this bacterium from subjects in Qatar. We have also quantified the frequency of five key virulence factors and detected some interactions between these and the expression of antimicrobial resistance. The exact nature of the underling mechanisms and the significance of these effects and their various interactions for the adaptability of C. jejuni is not yet clear. In view of the importance of Campylobacter as a disease causing organism in humans, and its frequent occurrence among the Qatari population, further work is warranted urgently and we hope to have provided here a springboard for such research in the years ahead.
Supporting Information
(XLSX)
Acknowledgments
We would like to thank the Biomedical Research Centre at Qatar University for providing facilities for this work. We thank also Dr. Gheyath Nasrallah, Mr. Jamal Mohamed and Jaafer Pakari for their advice and support with components of this work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This publication was made possible by a National Priority Research Program grant number NPRP4-1283-3-327 from the Qatar National Research Fund (http://www.qnrf.org/), a member of Qatar Foundation, and Hamad Medical Corporation (http://www.hmc.org.qa) grant #13334/13. The statements made herein are solely the responsibility of the authors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moore JE, Corcoran D, Dooley JS, Fanning S, Lucey B, Matsuda DA, et al. Campylobacter. Vet Res. 2005; 36: 351–382. [DOI] [PubMed] [Google Scholar]
- 2. Cover TL, Perez-Perez GI, Blaser MJ. Evaluation of cytotoxic activity in fecal filtrates from patients with Campylobacter jejuni or Campylobacter coli enteritis. FEMS Microbiol Lett. 1990; 58: 301–304. [DOI] [PubMed] [Google Scholar]
- 3. Szymanski CM, Gaynor EC. How a sugary bug gets through the day: recent developments in understanding fundamental processes impacting Campylobacter jejuni pathogenesis. Gut Microbes 2012; 3: 135–144. 10.4161/gmic.19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007; 5: 665–679. [DOI] [PubMed] [Google Scholar]
- 5. Nachamkin I, Ung H, Li M. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA,1982–2001. Emerg Infect Dis. 2002; 8: 1501–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jimenez A, Velazquez JB, Rodriguez J, Tinajas A, Villa TG. Prevalence of fluoroquinolone resistance in clinical strains of Campylobacter jejuni isolated in Spain. J Antimicro Chemoth. 1994; 33: 188–190. [DOI] [PubMed] [Google Scholar]
- 7. Saenz Y, Zarazaga M, Lantero M, Gastanares MJ, Baquero F, et al. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997–1998. Antimicrob Agents Ch. 2000; 44: 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liao CH, Chuang CY, Huang YT, Lee PI, Hsueh PR. Bacteremia caused by antimicrobial resistant Campylobacter species at a medical center in Taiwan, 1998–2008. J Infect. 2012; 65: 392–399. 10.1016/j.jinf.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 9. Li CC, Chiu CH, Wu JL, Huang YC, Lin TY. Antimicrobial susceptibilities of Campylobacter jejuni and coli by using E-test in Taiwan. Scand J Infect Dis. 1998; 30: 39–42. [DOI] [PubMed] [Google Scholar]
- 10. Hou FQ, Sun XT, Wang GQ. Clinical manifestations of Campylobacter jejuni infection in adolescents and adults, and change in antibiotic resistance of the pathogen over the past 16 years. Scand J Infect Dis. 2012; 44: 439–443. 10.3109/00365548.2011.652163 [DOI] [PubMed] [Google Scholar]
- 11. Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006; 8: 1967–1971. [DOI] [PubMed] [Google Scholar]
- 12. Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter . Biomed Res Int. 2013; 2013: 340605 10.1155/2013/340605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iovine NM. Resistance mechanisms in Campylobacter jejuni . Virulence 2013; 4: 230–240. 10.4161/viru.23753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perez-Boto D, Herrera-León S, Garcia-Pena FJ, Abad-Moreno JC, Echeita MA. Molecular mechanisms of quinolone, macrolide, and tetracycline resistance among Campylobacter isolates from initial stages of broiler production. Avian Pathol. 2014; 43: 176–182. 10.1080/03079457.2014.898245 [DOI] [PubMed] [Google Scholar]
- 15. Helms M, Simonsen J, Olsen KE, Mølbak K. Adverse health events associated with antimicrobial drug resistance in Campylobacter species: a registry-based cohort study. J Infect Dis. 2005; 191: 1050–1055. [DOI] [PubMed] [Google Scholar]
- 16. Nelson JM, Smith KE, Vugia DJ, Rabatsky-Ehr T, Segler SD, Kassenborg HD, et al. Prolonged diarrhea due to ciprofloxacin-resistant Campylobacter infection. J Infect Dis. 2004; 190: 1150–1157. [DOI] [PubMed] [Google Scholar]
- 17. Luo N, Pereira S, Sahin O, Lin J, Huang S, Michewl L, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A. 2005; 102: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Al-Mahmeed A, Senok AC, Ismaeel AY, Bindayna KM, Tabbara KS, Botta GA. Clinical relevance of virulence genes in Campylobacter jejuni isolates in Bahrain. J Med Microbiol. 2006; 55: 839–843. [DOI] [PubMed] [Google Scholar]
- 19. Rozynek E, Dzierzanowska-Fangrat K, Jozwiak P, Popowski J, Korsak D, Dzierzanowska D. Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J Med Microbiol. 2005; 54: 615–619. [DOI] [PubMed] [Google Scholar]
- 20. Bagger-Skjøt L, Sandvang D, Frimodt-Møller N, Lester CH, Olsen KE, Porsbo LJ, et al. Association between antimicrobial resistance and virulence genes in Escherichia coli obtained from blood and faeces. Scand J Infect Dis. 2007; 39: 724–727. [DOI] [PubMed] [Google Scholar]
- 21. McGowan-Spicer LL, Fedorka-Cray PJ, Frye JG, Meinersmann RJ, Barrett JB, Jackosn CR. Antimicrobial resistance and virulence of Enterococcus faecalis isolated from retail food. J Food Prot. 2008; 71: 760–769. [DOI] [PubMed] [Google Scholar]
- 22. Abu-Madi MA, Behnke JM, Doiphode SH. Intestinal parasitic infections among long-term-residents and settled immigrants in Qatar in the period 2005 to 2011. Am J Trop Med Hyg. 2013; 88: 1185–1195. 10.4269/ajtmh.13-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. England PH. UK Standards for Microbiology Investigations Investigations of Faecal Specimens for Enteric Pathogens. London: Public Health England; 2014. [Google Scholar]
- 24. Garcia LS, Isenberg HD. Clinical microbiology procedures handbook; Isenberg HD, editor. Washington, DC: ASM Press; 2010. [Google Scholar]
- 25. Wiemer D, Loderstaedt U, von Wulffen H, Priesnitz S, Fischer M, Tannich E, et al. (2011) Real-time multiplex PCR for simultaneous detection of Campylobacter jejuni, Salmonella, Shigella and Yersinia species in fecal samples. Int J Med Microbiol. 2011; 301: 577–584. 10.1016/j.ijmm.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 26. Chansiripornchai N, Sasipreeyajan J. PCR detection of four virulence-associated genes of Campylobacter jejuni isolates from Thai broilers and their abilities of adhesion to and invasion of INT-407 cells. J Vet Med Sci. 2009; 71: 839–844. [DOI] [PubMed] [Google Scholar]
- 27. CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Inferequently Isolated or Fastidious Bacteria; Approved guidelines-second edition. Pennsylvania, USA: Clinical and Laboratory standards Institute, 2010. 10.1016/j.vetmic.2010.01.010 [DOI] [Google Scholar]
- 28.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Pennsylvania, USA: Clinical and Laboratory Standards Institute, 2014.
- 29. Rohlf FJ, Sokal RR. Statistical Tables (3rd Edition). San Francisco: W.H.Freeman and Company; 1995. [Google Scholar]
- 30. Barnard C, Gilbert F, McGregor P. Asking question in Biology. London: Pearson Eduaction Ltd. 2007. [Google Scholar]
- 31. Dryden MS, Gabb RJ, Wright SK. Empirical treatment of severe acute community-acquired gastroenteritis with ciprofloxacin. Clin Infect Dis. 1996; 22: 1019–1025. [DOI] [PubMed] [Google Scholar]
- 32. Deckert A, Valdivieso-Garcia A, Reid-Smith R, Tamblyn S, Seliske P, Irwin R, et al. Prevalence and antimicrobial resistance in Campylobacter spp. isolated from retail chicken in two health units in Ontario. J Food Prot. 2010; 73: 1317–1324. [DOI] [PubMed] [Google Scholar]
- 33. McDermott PF, Bodeis SM, English LL, White DG, Walker RD, Zhao S, et al. Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J Infect Dis. 2002; 185: 837–840. [DOI] [PubMed] [Google Scholar]
- 34. Sonnevend A, Rotimi VO, Kolodziejek J, Usmani A, Nowotny N, Pál T. High level of ciprofloxacin resistance and its molecular background among Campylobacter jejuni strains isolated in the United Arab Emirates. J Med Microbiol. 2006; 55: 1533–1538. [DOI] [PubMed] [Google Scholar]
- 35. Rozynek E, Dzierzanowska-Fangrat K, Korsak D, Konieczny P, Wardak S, Szych J, et al. Comparison of antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from humans and chicken carcasses in Poland. J Food Prot. 2008; 71: 602–607. [DOI] [PubMed] [Google Scholar]
- 36. Unicomb LE, Ferguson J, Stafford RJ, Ashbolt R, Kirk MD, Becker NG, et al. Low-level fluoroquinolone resistance among Campylobacter jejuni isolates in Australia. Clin Infect Dis. 2006; 42: 1368–1374. [DOI] [PubMed] [Google Scholar]
- 37. Monteville MR, Yoon JE, Konkel ME. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 2003; 149: 153–165. [DOI] [PubMed] [Google Scholar]
- 38. Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999; 37: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pearson BM, Pin C, Wright J, I'Anson K, Humphrey T, Wells JM. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 2003; 554: 224–230. [DOI] [PubMed] [Google Scholar]
- 40. Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001; 11: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003; 52: 345–348. [DOI] [PubMed] [Google Scholar]
- 42. Whitehouse CA, Balbo PB, Pesci EC, Cottle DL, Mirabito PM, Pickett CL. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect Immun. 1998; 66: 1934–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hickey TE, McVeigh AL, Scott DA, Michielutti RE, Bixby A, Carroll SA, et al. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect Immun. 2000; 68: 6535–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001; 183: 1607–1616. [DOI] [PubMed] [Google Scholar]
- 45. Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, et al. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004; 186: 3296–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chan KF, Le Tran H, Kanenaka RY, Kathariou S. Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4 degrees C). Appl Environ Microbiol. 2001; 67: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Konkel ME, Kim BJ, Klena JD, Young CR, Ziprin R. Characterization of the thermal stress response of Campylobacter jejuni . Infect Immun. 1998; 66: 3666–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stintzi A. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J Bacteriol. 2003; 185: 2009–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, et al. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J Biol Chem. 2002; 277: 327–337. [DOI] [PubMed] [Google Scholar]
- 50. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000; 403: 665–668. [DOI] [PubMed] [Google Scholar]
- 51. Strachan NJ, Watson RO, Novik V, Hofreuter D, Ogden ID, Galán JE. Sexual dimophism in campylobacteriosis. Epidemiol Infect. 2008; 136: 1492–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Adib N, Ghanbarpour R, Solatzadeh H, Alizade H. Antibiotic resistance profile and virulence genes of uropathogenic Escherichia coli isolates in relation to phylogeny. Trop Biomed. 2014; 31: 17–25. [PubMed] [Google Scholar]
- 53. Paulander W, Maisnier-Patin S, Andersson DI. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (sigmaS). Genetics 2009; 183: 539–546. 10.1534/genetics.109.106104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ender M, McCallum N, Adhikari R, Berger-Bächi B. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus . Antimicrob Agents Chemother. 2004; 48: 2295–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Momtaz H, Dehkordi FS, Hosseini MJ, Sarshar M, Heidari M. Serogroups, virulence genes and antibiotic resistance in Shiga toxin-producing Escherichia coli isolated from diarrheic and non-diarrheic pediatric patients in Iran. Gut Pathog. 2013; 5: 39 10.1186/1757-4749-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.