Abstract
Mitogen-activated protein kinases (MAPKs) are important transducers of external signals for cell growth, survival and other cellular responses including cell differentiation. Several MAPK cascades are known with the MEK1/2-ERK1/2, JNK, and p38MAPKs receiving most attention, but the role of MEK5-ERK5 in intracellular signaling deserves more scrutiny, as this pathway transmits signals that can complement ERK/2 signaling. We hypothesized that the ERK5 pathway plays a role in the control of monocytic differentiation, which is disturbed in myeloid leukemia. We therefore examined the cellular phenotype and key molecular events which occur when human myeloid leukemia cells, acute (AML) or chronic (CML), are forced to differentiate by vitamin D derivatives (VDDs). This study was performed using established cell lines HL60 and U937, and primary cultures of blasts from 10 patients with ML. We found that ERK5 and its direct downstream target transcription factor MEF2C are upregulated by 1,25D in parallel with monocytic differentiation. Further, inhibition of ERK5 activity by specific pharmacological agents BIX02189 and XMD8-92 alters the phenotype of these cells by reducing the abundance of the VDD-induced surface monocytic marker CD14, and concomitantly increasing surface expression of the general myeloid marker CD11b. Similar results were obtained when the expression of ERK5 was reduced by siRNA or short hairpin (sh) RNA. ERK5 inhibition resulted in an expected decrease in MEF2C activation. We also found that in AML the transcription factor C/EBPβ is positively regulated, while C/EBPα is negatively regulated by ERK5. These findings provide new understanding of dysregulated differentiation in human myeloid leukemia.
Keywords: MAPKs, ERK5, vitamin D, MEF2C, C/EBP, Myeloid leukemia
Introduction
The ability of transcription factors to regulate cell differentiation and lineage selection has been dramatically demonstrated by the induction of pluripotent stem cells (iPSCs) as the result of transfection of just a few transcription factors into adult cells, such as fibroblasts, to create cells similar to the embryonic stem cells (Takahashi et al., 2007; Yu et al., 2007). Thus de-differentiated cells can then differentiate into multiple cell lineages to produce adult cells with various phenotypes and functions [e.g., (Lohle et al., 2012)]. During normal cell differentiation, additional transcription factors become sequentially activated in a cell type-specific fashion when the imprinted genetic and epigenetic programs interact with signals provided by environmental clues. In neoplastic cells differentiation is impeded and aberrant, and it is hoped that these differences can be exploited for cancer therapy.
Accordingly, numerous studies have been directed to the potential improvements in cancer treatment by including agents which induce differentiation of malignant cells in the treatment regimens, based on observations of cell differentiation in neoplastic cells (Breitman et al., 1980; Friend et al., 1987; Pierce and Verney, 1961; Sachs, 1978; Schubert et al., 1971). Indeed, all-trans retinoic acid (ATRA) has been an effective differentiation agent in the treatment of Acute Promyelocytic Leukemia (APL), the subtype FAB M3 of AML and induces complete remissions in up to 95% of patients with APL, many of which are durable (Wang and Chen, 2008). The efficacy of ATRA is due to the presence of a hybrid gene PML-RARα and its variants in APL cells, which encode a transcription factor-like fusion protein that binds with enhanced affinity to sites on the cell’s DNA, blocking transcription and differentiation of granulocytes (Grignani et al., 1993). The mechanisms include dissociation of nuclear co-repressor (NCoR) molecules and recruitment of histone acetylases (HATs) which open the chromatin structure and relieve transcriptional repression, as well as PML-RARα degradation which can significantly contribute to the therapeutic effects of ATRA (Wang and Chen, 2008). However, so far no clinically effective differentiation agent has been identified for the treatment of the other sub-types of AML. Thus, attention has continued to be directed to the elucidation of signal transduction mechanisms and transcription factors that can be targeted for therapeutic interventions by normalizing the malignant cell phenotype.
Multiple pathways are known which transmit environmental signals which interact with genetic and epigenetic programs to turn on transcription factors for various lineages of cell differentiation, and many of these are aberrantly expressed and are abnormally functioning in neoplastic cells. Among these, mitogen-activated protein kinases (MAPKs) including ERK1/2, JNK and p38MAPK, and the PI3K pathway have been of interest as targets for selective killing of cancer cells (Chang et al., 2003; Friday and Adjei, 2008; Santarpia et al., 2012). Increased c-jun expression during monocytic differentiation of human leukemia cells induced by 1,25-dihydroxyvitamin D3 (1,25D) or the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) has been reported in early studies (Sherman et al., 1990), and further work showed that c-jun expression could be mediated by the RAF/ERK pathway (Gaynor et al., 1991; Pulverer et al., 1991; Sherman et al., 1990). Additional research revealed that c-jun can be phosphorylated and thus activated by either JNK2 or ERK1/2 (Pulverer et al., 1991), and that c-jun is required for monocytic differentiation of human leukemia HL60 cells, functioning as a part of the AP-1 transcription factor dimer, particularly in the initial stages of differentiation induced by 1,25D (Wang and Studzinski, 2001; Wang and Studzinski, 2006).
In the early studies, Ras-transmitted growth factor signaling was considered to take place largely by the RAF-MEK1/2-ERK1/2 pathway [reviewed by (Morris et al., 2013; Santarpia et al., 2012)]. More recently, ERK5 was recognized as a component of a parallel MAPK pathway, with MEK5-ERK5 as the principal module (Zhou et al., 1995), and it has been suggested that some of the regulatory signaling previously attributed to ERK1/2 was actually due to ERK5 (Wang and Tournier, 2006). Deletion of erk5 and its upstream regulator mek5 genes in mice showed that the ERK5 cascade is not redundant with ERK1/2, and is essential for normal cardiovascular development (Regan et al., 2002; Sohn et al., 2002; Yan et al., 2003), and in some species plays a role in neuronal survival and differentiation (Cavanaugh, 2004; Nishimoto et al., 2005; Wang and Tournier, 2006). Further, ERK5 appears to mediate the actions of oncogenes in some cancers including breast (Esparis-Ogando et al., 2002; Song et al., 2004) and prostate (Mehta et al., 2003). Although the role of ERK5 in myeloid leukemias has not been previously well studied, it is important to note that a well-documented downstream effector of ERK5, the transcription factor MEF2C, is a key regulator of myeloid cell fate in mice, by influencing the cell fate decisions between monocyte and granulocyte differentiation (Schuler et al., 2008).
The importance of understanding the signaling pathways and activation of transcription factors in AML cells lies in the lack of satisfactory treatment that can be offered to most patients with this disease. Currently, in adults the majority of AML cases are incurable with five-year survival approximately 20% (http://seer.cancer.gov/statfacts/html/amyl.html). In children, the prognosis is somewhat better, but despite 90% initial remission rate approximately 40% of the pediatric patients with AML relapse (Kaspers and Creutzig, 2005). Thus, there is the continued challenge to improve the therapy for AML, and one approach is to recognize new targets for the pharmacological eradication of the disease, which may supplement the conventional cytotoxic therapy.
It seems reasonable to suggest that ERK5 is such a target in AML, since its functions include the aforementioned oncogenic effects, along with the stimulation of cell proliferation and cell survival in other cell types [reviewed in (Alvarez-Fernandez et al., 2013; Charni et al., 2009; Drew et al., 2012; Roberts et al., 2010)]. In this study, we have identified for the first time an additional function for ERK5, namely the positive regulation of monocytic differentiation in human AML cells, shown both in established cultures and in AML blasts ex vivo. Most importantly, we demonstrate that in human AML cells ERK5 regulates C/EBPβ, which controls CD14 expression, and thus directly promotes monocytic differentiation.
Materials and Methods
Reagents and Immunochemicals
1,25D was a kind gift from Dr. Milan Uskokovic (Bioxell). Doxercalciferol (1α-hydroxyvitamin D2; 1-D2) was purchased from Sigma-Aldrich (St. Louis, MO). The following antibodies: P-MEK5 (Ser311/Thr315, sc-135702), MEK5 (sc-10795), P-MEF2C (Thr300, SC-130201), and Crk-L (sc-319) were obtained from Santa Cruz Biotechnology (Dallas, TX). P-C/EBPβ (Thr235, #3084), P-C/EBPα (Thr222/226, #2844), MEF2C (#5030), ERK5 (#3372), P-ERK5 (Thr187/Tyr220, #3371), P-ERK1/2 (Thr202/Tyr204, #4370), anti-rabbit (#7074) and anti-mouse (#7076) secondary antibodies conjugated to HRP were purchased from Cell Signaling Technologies (Danvers, MA). The pharmacological inhibitors of ERK5 kinase MEK5 (BIX02189), and ERK5 autophosphorylation (XMD8-92) were purchased from Selleck Chemicals (Houston, TX) and Santa Cruz Biotechnology Inc., respectively. The MEK1/2 inhibitor PD98059 was obtained from Cell Signaling Technologies. Nitrocellulose membranes were purchased from GE Healthcare (Pittsburgh, PA). The vitamin D derivatives (VDDs) were dissolved in ethanol and kinase inhibitors in DMSO.
Cell lines, cell culture, and cell proliferation assays
HL60-G cells (FAB M2), subcloned from HL60 cells derived from a patient with promyeloblastic leukemia, and U937 monoblastic cells (FAB M4), derived from a patient with histiocytic lymphoma, were cultured in suspension under conditions standard in this laboratory (Wang et al., 2010). Routine microbiology testing for Mycoplasma was performed by selective culture techniques. For experiments involving kinase inhibitors, cells (0.1 × 106/ml) were pre-treated with these agents or 0.1% DMSO (vehicle) for 1 h before the addition of VDDs or 0.1% ethanol, followed by incubation for another 1–96 h. Cell number and viability were estimated on the basis of the trypan blue exclusion assay, by enumerating live and dead cells in a Vi-Cell XR cell viability analyzer (Beckman Coulter).
Ex vivo experiments with freshly isolated patient blasts
Leukemic cells were obtained from patients in Robert Wood Johnson University Hospital with newly diagnosed AML following informed consent, in accordance with an IRB approved clinical protocol. All patients met the criteria for the diagnosis, based on WHO, and the diagnosis was confirmed in each case by a formal histopathologic review. The study population comprised 6 male subjects and 4 female subjects with a median age of 63.7 years old. According to FAB classification of AML, there was one patient with subtype M1, four patients with M2, one patient with M3, one patient with M4, one patient with M5, and two with CML. Mononuclear cells were separated from the specimens of peripheral blood by Ficoll (Histopaque ®-1077, Sigma-Aldrich) gradient centrifugation (Zhang et al., 2010), and cultured with inhibitors and VDDs for 7 days in RPMI plus 10% bovine calf serum.
Knock down of ERK5 expression
siRNA targeting ERK5 and generic scrambled control siRNA were purchased from IDT (Coralville, Iowa). siRNAs were transfected at a final concentration of 20 nM for 48 h using Endo-Porter delivery reagent from Gene Tools Inc (Philomath, OR).before exposure to other compounds. Short hairpin (sh) shRNA ERK5 lentiviral particles or control shRNA lentiviral particles were purchased from Santa Cruz Biotechnology Inc. HL60 or U937 cells were stably transfected with ERK5 shRNA or control shRNA lentiviral particles according to the manufacturer’s instructions. Briefly, after 24 h transfection, cells were cultured in selection medium containing 2 ug/ml puromycin for 3 weeks before single clones were isolated. Individual puromycin-resistant colonies were isolated during drug selection and established as individual clones for further analysis. Transient or stably transfected cells were seeded at a concentration of 3 × 105 cells/ml and exposed to the specified agents for the indicated times. The cells were examined for the reduction of ERK5 expression at both mRNA and protein levels.
Determination of differentiation markers
The expression of cell surface markers of myeloid differentiation was determined by dual labeling of the cells with 10 μL of PBS containing 0.5 μg anti-CD11b MO1-FITC and 0.5 μg anti-CD14 My4-RD-1 (Beckman Coulter) followed by two-parameter flow cytometric analysis, as described previously (Wang et al., 2010). Monocytic phenotype was further confirmed by cytochemical staining for monocytic serine esterase activity, usually referred to as nonspecific esterase (NSE), as described previously (Wang et al., 2003).
Immunoblot analysis
This was performed using whole cell extracts, as described previously (Wang et al., 2010). The protein bands were visualized using a chemiluminescence Pierce® ECL system (Thermo Scientific), and the optical density of each band was quantitated using an ImageQuant 5.0 (Molecular Dynamics).
Real-time PCR (qRT-PCR)
RNA was extracted from cells using TRIzol® Reagent (Invitrogen, Carlsbad, CA). The procedures were previously described (Wang et al., 2010). Briefly, 5–10 × 106 cells were homogenized in 1 ml Trizol and incubated for 5 min at RT. After addition of chloroform followed by centrifugation at 14,000 rpm, the aqueous phase was transferred to a fresh tube. The RNA was precipitated by mixing with 500 μL isopropyl alcohol, and incubated 10 min at RT. RNA pellet was obtained by centrifugation, the supernatant discarded, and the pellet washed with 75% ethanol in DEPC water, then briefly dried for 10 min. The RNA pellet was then dissolved in 50 μL DEPC water. The forward and reverse primers sequences used, respectively, were: CD11b, 5′-GCAAGTGTCTGTGTGCAAGTGTGT-3′ and 5′-TCAGTGGAGAGAAGCTGCTGTGTT-3′, CD14, 5′-AACTCCCTCAATCTGTCGTTCGCT-3′ and 5′-GGGCAAAGGGTTGAATTGGTCGAA-3′, C/EBPβ, 5′-GTTCTTGACGTTCTTCGGCCG-3′ and 5′-TGGACAAGCACAGCGACGAGT-3′; C/EBPα, 5′-GTTAGCCATGTGGTAGGAGAC-3′ and, 5′-CCCAGCCGTTAGTGAAGAGT-3′; ARP0 (acidic ribosomal protein p0), 5′-AGATGCAGCAGATCCGCAT-3′ and 5′-GTGGTGATACCTAAAGCCTG-3′; RPII (RNA Pol II), 5′-GCACCACGTCCAATGACAT-3′, and 5′-GTGCGGCTGCTTCCATAA-3′. For reverse transcription, samples were incubated in an Eppendorf PCR system at 42°C for 15 min, then 99°C for 5 min, and 5°C for 5 min. For real-time PCR, relative quantification of target cDNA was performed using a Roche LightCycler instrument with Faststart DNA Masterplus Syber Green I kit (Hoffmann-La Roche) and gene-specific primers. The parameters for PCR were as follows: 45 cycles of 1) 95°C for 15 s, 2) 58°C for 10 s, and 3) 72°C for 15 s followed by an incremental increase (0.1°C/s) of temperature from 65 °C to 95°C to analyze the melting curve of the products. A mixture of forward and reverse primers at a final concentration of 1.5 μM was used to detect different mRNAs. ARP0 and RPII were used as internal controls.
Statistics
Each cell line experiment was performed at least three times and the results were expressed as percentages (mean ± S.D.) of the vehicle controls. Significance of the differences between mean values was assessed by a two-tailed Student’s t-test. A p value less than 0.05 was considered to be statistically significant. All computations were performed using Microsoft EXCEL or GraphPad Prism ver. 6 (GraphPad, La Jolla, CA).
Results
1. Upregulation of ERK5 activation by 1,25D parallels the induction of full differentiation phenotype in AML cells
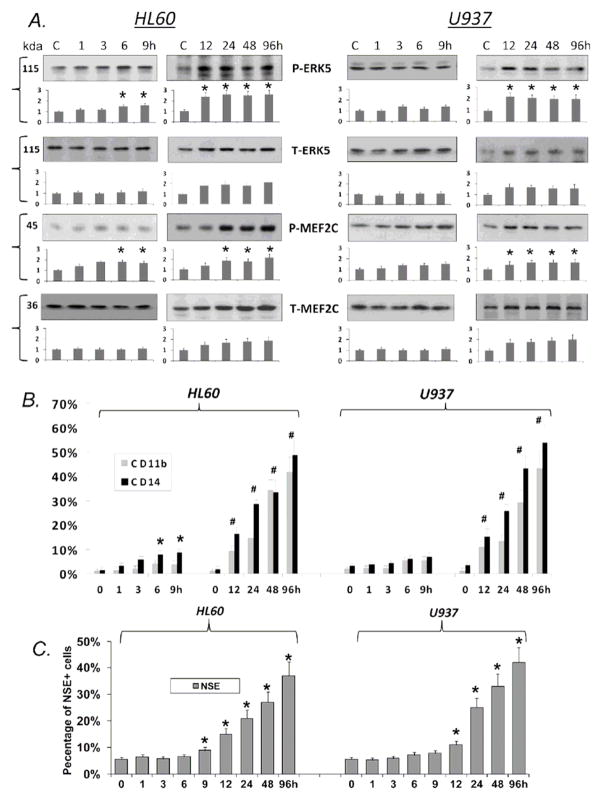
We have previously shown that the exposure of human AML cell to 1 nM 1,25D, a near physiological level of this vitamin/hormone, upregulates the expression of ERK5 (Wang et al., 2010) and influences differentiation of these cells, but the relative occurrence of these events was not established. To determine whether upregulation of ERK5 contributes to the initiation or is a consequence of differentiation, we compared the time at which ERK5 activation could be detected at the protein level with the time at which several differentiation markers could be detected in two AML sublines. As shown in Fig. 1, in the more rapidly differentiating HL60 cells, ERK5 activation was significantly (p<0.05) increased at 6 h after 1,25D administration, similar to the detection of the monocytic surface marker CD14, but before the general myeloid marker CD11b or the cytoplasmic monocytic marker NSE could be visualized. On the other hand, in U937 cells which differentiate more slowly in the presence of 1,25D, activation of ERK5 was not detected until 12 h after 1,25D exposure, approximately when CD14 was elevated. In both cell lines, the general myeloid marker CD11b was increased at 12 h, showing that individual parameters of differentiation can be dissociated in a cell type-specific manner. Taking together the parallel appearance of the differentiation phenotype with the activation of ERK5, and an earlier ERK5/MEF2C activation in HL60 cells in which 1,25D induces monocytic phenotype earlier than in U937 cells, our data suggest that the ERK5 pathway is an important driver of the differentiating effect of 1,25D in AML cells. Interestingly, ERK5 appears to have a permissive rather than a driving role in differentiation, as is activation did not continue to increase once it reached a threshold that allowed differentiation to be initiated.
Fig 1. Upregulation of ERK5 activation by 1,25D parallels the induction of differentiation phenotypes in AML cells.
A. Western blots illustrating the time course of the expression and activation by 1,25D (1 nM) of ERK5, and its direct target MEF2C, in two subtypes of human myeloid leukemia cells. Beneath each blot a bar graph shows the mean values +/− SE, n=3. The short term (1–9h) experiment was set up separately from the longer term (12–96h), thus signal intensity was compared to the corresponding Untreated control value, set as 1.0 on the vertical scale. Statistical significance of the difference from untreated cells (C) is shown by the asterisk (*), p< 0.05, n=3.
B. Time course of 1,25D -induced appearance of surface markers of differentiation measured by Flow Cytometry. In the short term experiments only the monocytic marker CD14 significantly increases, denoted by the asterisk (*), p< 0.05. In the long term experiment both the general myeloid marker CD11b and CD14 significantly increase, denoted by (#) p< 0.05. Other details as above.
C. Cytochemical demonstration of the appearance of the monocyte-specific esterase NSE induced by 1,25D. Note that significant positivity of this differentiation marker is reached in 9h in HL60 cells, but at 12h in U937 cells. Details and symbols as above.
2. Pharmacological inhibition of ERK5 activation modifies 1,25D-induced differentiation of AML cell lines and inhibits the ERK5 downstream target MEF2C
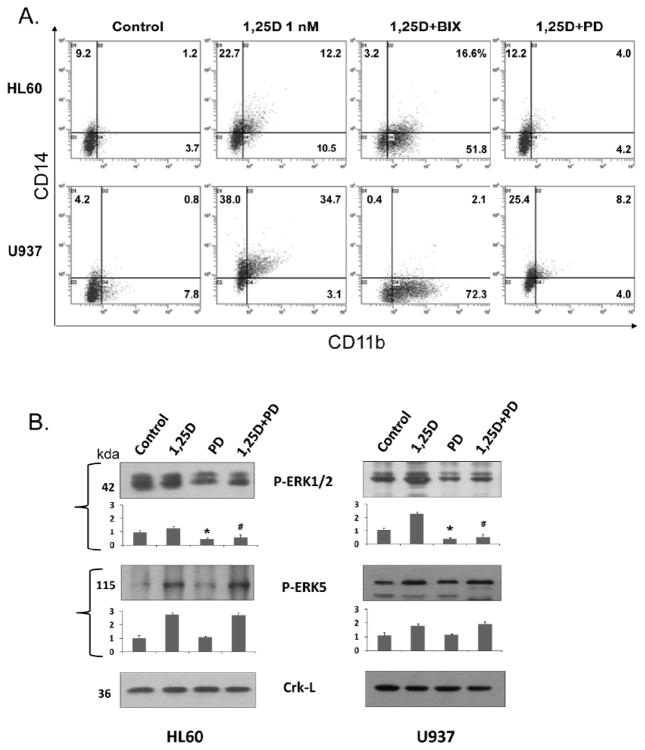
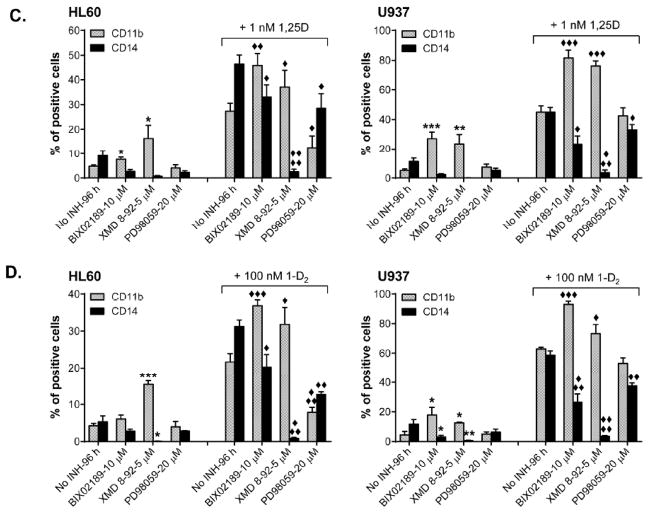
To confirm that ERK5 is functionally involved in 1,25D-induced differentiation of AML cells we employed specific pharmacological inhibitors of MEK5 kinase activity BIX02189 (Tatake et al., 2008), and of ERK5 autophosphorylation XMD8-92 (Yang et al., 2010). HL60 and U937 cells were pre-treated with 10 μM BIX02189 or 5 μM XMD8-92 for 1 h prior to the addition of 1 nM 1,25D followed by cell incubation for an additional 96 h. The extent of myeloid differentiation was then determined by the expression of a marker of monocytic differentiation CD14 and a marker of general myeloid differentiation CD11b. As exemplified in Fig. 2A and summarized in Fig. 2C, incubation of the cells with a low concentration of 1,25D resulted in a moderate expression of both surface markers. Pretreatment with MEK5 and ERK5 inhibitors led to a marked reduction in both the basal and 1,25D-induced CD14 expression. Interestingly, this was accompanied by a significant concomitant increase in the basal CD11b levels and a strong potentiation of 1,25D-stimulated expression of this marker. XMD8-92 was generally more potent than BIX02189 in changing the surface antigen expression pattern. For comparison, we used the widely employed specific inhibitor of MEK1/2 (20 μM PD98059) which caused a general decrease in the expression of both CD11b and CD14, while inhibiting only the ERK1/2 activation (Fig. 2A, B). This shows that ERK5 pathway has effects on differentiation distinct from the ERK1/2 pathway. The above experiments were then repeated using a synthetic VDD (1-D2) as an inducer of differentiation with essentially similar results (Fig. 2D). MEK5/ERK5 inhibitors produced decreases in the percentages of CD14-positive cells which were associated with elevated percentages of CD11b-positive cells, and there was also a general reduction in the levels of both surface markers in the presence of a MEK1/2 inhibitor (Fig. 2D).
Fig 2. Reciprocal modulation of VDD-induced expression of CD11b and CD14 differentiation markers by MEK5/ERK5 inhibitors in AML cell lines.
A. Primary data exemplifying distinct effects of MEK5/ERK5 and MEK1/2 inhibitors, BIX 02189 and PD 98059, respectively, on the basal and 1,25D-induced CD11b and CD14 levels. HL60 or U937 cells were pretreated with either BIX02189 (10 uM) or PD 98059 (20 uM) for 1 h, then 1,25D (1 nM) was added for an additional 96 h. CD11b and CD14 expression was determined by flow cytometry.
B. Western blots illustrating in AML cells the selective inhibition of ERK1/2 activation and expression by 20 uM PD 98059 under conditions used for experiments shown in panel A. Asterisks (*) show the statistically significant differences from the control group, and hash marks (#) the statistically significant differences from the 1.25D alone group, p< 0.05; n=3. Bars show mean values +/− SE.
C. Summarized differentiation data, obtained as shown in panel A. Data collected using the ERK5 auto-phosphorylation inhibitor XMD8-92 (5 μM) are also included. *, p < 0.05; and **, p < 0.01 versus control. ◆, p < 0.05; and ◆◆, p < 0.01 versus 1,25D alone. Data are the means +/− SE of 4–7 independent experiments.
D. Summarized data, obtained using the vitamin D analog doxercalciferol (1-D2; 100 nM) as the differentiation inducer. *, p< 0.05; and **, p < 0.01 versus control. ◆, p < 0.05; and ◆◆, p < 0.01 versus 1-D2 alone. Data are the means +/− SE of 3–5 independent experiments.
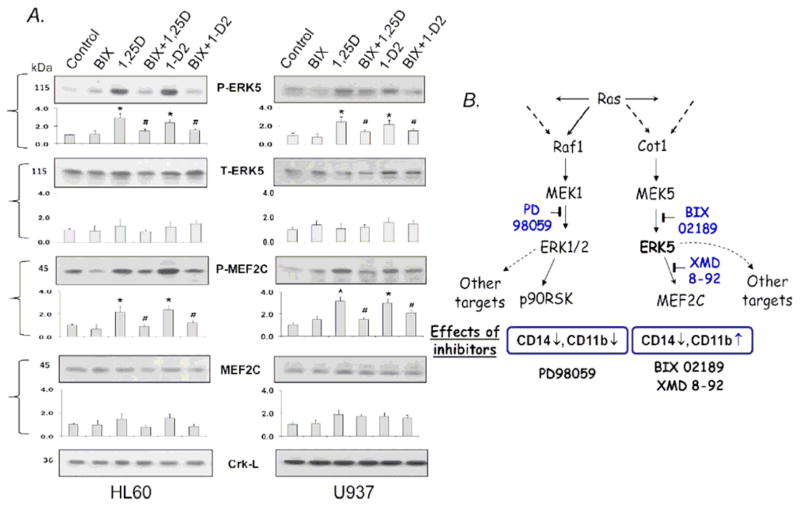
In an attempt to gain insight into the molecular events which underlie these phenotypic changes, we examined the effects of BIX2189 on the levels of total and activated by phosphorylation ERK5 and also MEF2C, previously reported to be the transcription factor immediately downstream from ERK5 in several cell types (Kato et al., 1997; Tatake et al., 2008; Yan et al., 2001), though this was not investigated in hematopoietic cells. As shown in Fig. 3A, 1,25D and its analog 1-D2 increased the protein and phosphorylation levels of both these signaling proteins in the two cell lines studied here, and BIX02189 reduced these levels. Importantly, these experiments confirmed the expected inhibition of ERK5 phosphorylation by the MEK5 inhibitor BIX02189, while the levels of total Crk-L, a loading control, were unchanged. Since phosphorylation can affect protein stability, changes in total ERK5 and MEF2C proteins following an exposure to BIX02189 were also observed, and the significance of this remains to be investigated.
Fig 3. BIX 02189 inhibits the VDD-activation of ERK5 and the phosphorylation of its direct target MEF2C.
A. HL60 and U937 cells were pretreated with ERK5 inhibitor BIX 02189 (10 uM) for 1 h, then 1,25D (1 nM) or doxercalciferol (100 nM; 1-D2) were added for 96 h. The total and phosphorylated protein levels in the ERK5 cascade were determined by Western blot analysis, Crk-L was used as a protein loading control. BIX= BIX 02189. Beneath each blot a bar graph shows the mean values +/− SE, n=3. Asterisks (*) show the statistically significant differences from the control group, and hash marks (#) the statistically significant differences from the 1.25D alone group, p< 0.05; n=3.
B. An outline of ERK5 and ERK1/2 signaling showing that in AML cells these two pathways signal differentiation in overlapping but distinct ways, and inhibitors of ERK1/2 or ERK5 pathways have different effects on the phenotype in AML cells.
The results of experiments with BIX02189, XMD-92 and PD098059 combined with the existing literature (Drew et al., 2012; Wang and Tournier, 2006) suggest that the ERK5 pathway and the related ERK1/2 MAPK pathway are not redundant, as they have different effects on the expression of the differentiation marker CD11b. This implies that both these MAPK pathways are important for VDD-induced differentiation, and an outline of this signaling is depicted in Fig. 3B.
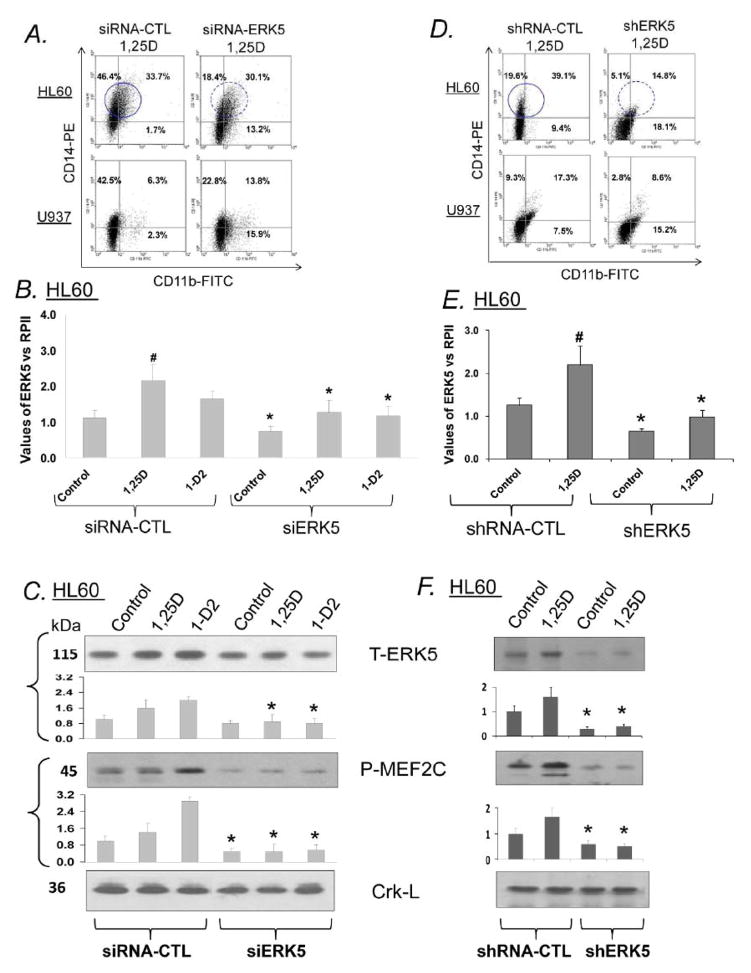
3. Knock-down of ERK5 expression modifies 1,25D-induced differentiation and reduces activation of the ERK5 pathway in HL60 cells
To complement the results with pharmacological inhibitors of ERK5 and confirm specificity, we first used transfection with silencing oligoribonucleotides (siERK5) which knock down ERK5 expression and observed the resulting changes in the phenotype and MEF2C activation in HL60 cells. Fig. 4A shows that the phenotypic changes elicited by siERK5 were similar to those seen following treatment with BIX02189: a decrease in the surface expression of CD14 accompanied by an increase in CD11b. Determination of ERK5 mRNA (Fig. 4B) and ERK5 protein (Fig. 4C) levels showed that the increases in ERK5 levels induced by 1,25D and 1-D2 were abrogated by siERK5, while the basal levels of cellular ERK5 were less markedly, but significantly, decreased as well (Fig. 3B). Activating phosphorylation of ERK5 by 1,25D was also decreased by siERK5, and the phosphorylation of the ERK5 immediate downstream target MEF2C was markedly reduced, below it basal levels (Fig. 4C). Thus, both the inhibition of ERK5 phosphorylation by BIX02189 and a reduction of ERK5 levels by siERK5 inhibit the activity of the ERK5 pathway and alter monocytic cell phenotype.
Fig 4. ERK5 knockdown (KD) modifies 1,25D-induced differentiation and ERK5 expression in AML cells.
A. Exposure of 1,25D-treated AML cells to ERK5 siRNA oligonucleotides (siERK5) inhibits the expression of CD14 monocytic differentiation marker, but promotes increases in the CD11b general myeloid cell marker. Circled areas illustrate that the intensity of the monocytic differentiation marker CD14 is decreased by the ERK5 KD.
B. siERK5 abrogates the VDD-induced increase in ERK5 in HL60 cells at both transcriptional (RT-qPCR, bottom bar chart) and protein (Western blots, upper panel) levels. The hash mark (#) shows that 1,25D significantly increased ERK5 mRNA level when compared to untreated control. Asterisks (*) show the statistically significant differences from the corresponding siRNA groups. p < 0.05; n=3. Bars represent mean values +/− SE.
C. Exposure of 1,25D-treated cells to siERK5 abrogates the increases in ERK5 and markedly reduces the levels P-MEF2C, a downstream target of ERK5. The levels of Crk-L, a loading control, were not significantly altered.
D. E. Exposure of AML cells stably transfected with non-targeting shRNA or shERK5 to cells treated with a vehicle control or with 1 nM 1,25D produces results similar to those obtained with siERK5. Details as described for panels A and B. Note that in the shERK5 experiments the vitamin D analog 1-D2 was not used.
F. Western blots of HL60 cells stably transfected with non-targeting shRNA and shERK5. The shERK5 reduced both the basal and 1,25D-induced ERK5 signaling. Asterisks (*) show the statistically significant differences from the corresponding non-targeting shRNA control groups. p < 0.05; n=3. Bars represent mean values +/− SE.
Because hematopoietic cells are difficult to transfect, we achieved only partial (approximately 40–60%) knock down of ERK5 using siRNA, we repeated the above experiment with 1,25D using cells stably expressing ERK5 shRNA and obtained essentially the same results (Fig. 4D–F). This further confirms findings with BIX 02189, and our conclusion that the ERK5 pathway participates in 1,25D-induced monocytic differentiation.
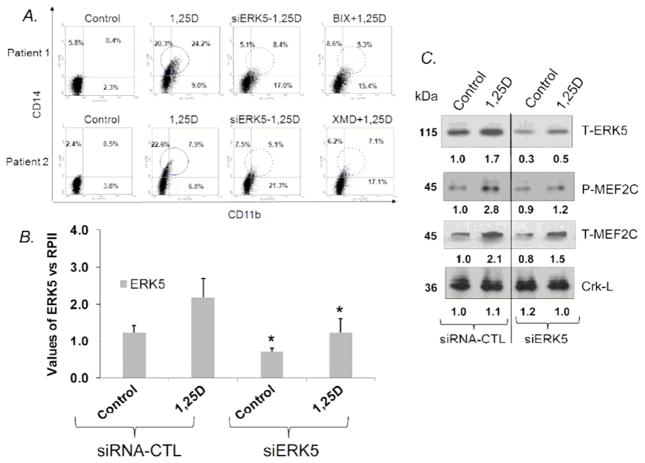
4. Inhibition of MEK5/ERK5 pathway or knock-down of ERK5 expression reduces the 1,25D-induced phosphorylation of downstream transcription factors in AML cells ex vivo
To determine the more direct disease relevance of the AML model systems provided by the established cell lines, we also examined ex vivo ten samples of freshly obtained blasts from 8 AML and 2 CML patients. Since in most cases the numbers of available cells were limited, we have not included the administration of the analog 1-D2, and the molecular studies were also limited. We were able to show, however, that in the samples examined siERK5, as well as the pharmacological inhibitors of ERK5, produced phenotypic changes in AML blasts which were similar to the changes seen in established cultures, i.e., the reduced 1,25D-induced CD14 expression, and increased CD11b expression (Fig. 5A). We have confirmed that siRNA was highly effective in knocking down ERK5 levels at both mRNA (Fig. 5B) and protein levels (Fig. 5C). Sufficient material was also available to show by Western blotting that 1,25D increases the levels of total and phosphorylated MEF2C transcription factor, suggesting its importance in 1,25D-induced monocytic differentiation of AML cells (Fig 5C). These results demonstrate that our data obtained in studies of MEF2C using established AML cell lines reflect the situation in at least some samples of primary AML blasts. Additional experiments performed using CML cells showed essentially the same results, with BIX02189, XMD8-92 and siERK5 inhibiting the expression of CD14, while enhancing the expression of CD11b induced by 1,25D. However, the increase in CD11b expression was less apparent than in AML blasts, consistent with partial granulocytic differentiation of the majority of circulating CML cells (data not shown).
Fig 5. Inhibition of the MEK5/ERK5 pathway modifies 1,25D-induced differentiation in AML cells obtained directly from patients (ex vivo).
A. Pharmacological inhibitor of MEK5 kinase activity, BIX 02189, and of ERK5 auto-phosphorylation, XMD8-92, have effects similar to siERK5 on changes in the phenotype of AML blasts in primary culture induced to differentiate by 1,25D.
B. In AML cells in primary culture, as in cell lines, ERK5 mRNA levels are significantly increased after exposure to 1,25D, and transfection of siERK5 significantly inhibits both the basal and 1,25D-induced ERK5 mRNA expression. Asterisks (*) show the statistically significant differences from the corresponding siRNA control groups. p < 0.05; bars represent mean values +/− SE.
C. ERK5 protein expression and the ERK5 downstream target activated by 1,25D in AML blasts from patient 1 are inhibited by siERK5. Note that the phosphorylation/activation of the known ERK5 downstream target, MEF2C, is markedly reduced by siERK5. There was insufficient protein quantity in other primary AML samples to replicate this experiment. Crk-L was used as a protein loading control.
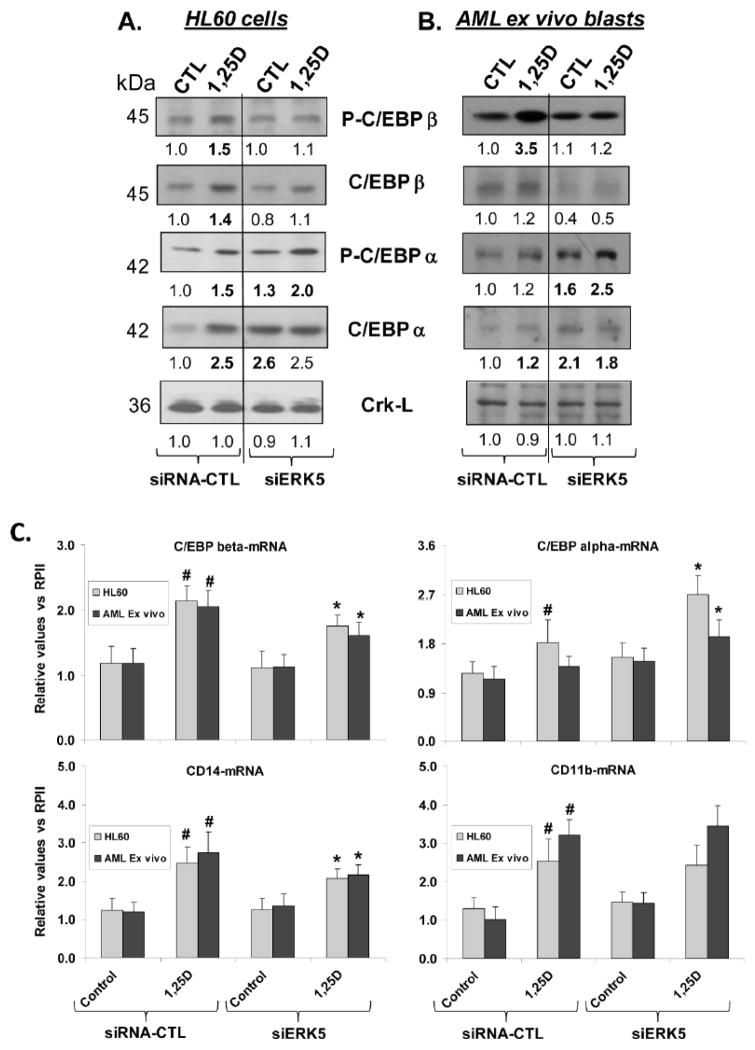
5. ERK5 is a positive regulator of 1,25D-induced C/EBPβ, but a negative regulator of C/EBPα in AML cells
Previous work in this and other laboratories has shown that members of the C/EBP transcription factor family, particularly C/EBPβ and C/EBPα play important roles in differentiation of AML cells (Ji and Studzinski, 2004; Studzinski et al., 2005), but their upstream regulation has not been clear. In this study, we found that the modulation by 1,25D of the levels of phosphorylated C/EBPβ and C/EBPα shows similarity when examined in ex vivo blasts or in HL60 cells, a representative AML cell line (Fig. 6, A and B). The similarity lies in a 1,25D-induced, ERK5-dependent, robust increase in the levels of phosphorylated C/EBPβ in both HL60 and in ex vivo AML blasts. However, siERK5 has a converse effect on 1,25D-induced C/EBPα phosphorylation, which is increased by siERK5 in both HL60 cells and in ex vivo blasts (Fig. 6A, and B), in contrast to the decrease seen in C/EBPβ. These data indicate that ERK5 has an enhancing effect on activation of C/EBPβ, but a negative/dampening effect on C/EBPα activation.
Fig 6. Distinct modulation of the transcription factors C/EBPa and C/EBPb by siERK5.
A. HL60 cells treated with 1,25D show increased levels of the major form of C/EBP isoform studied here (C/EBP beta LAP*, also known as C/EBP beta1), and of C/EBP alpha (p42), as well as of their phosphorylated forms. However, exposure to siERK5 reduces the expression of C/EBP beta (LAP*), but increases the phosphorylation of C/EBP alpha p42.
B. Similar effects of siERK5 are seen in AML blasts ex vivo as in HL60 cells. Because of the scarcity of cells obtained from patients, replication of Westerns was not possible in this experiment.
C. Tight correlation between the expression of C/EBP beta and CD14 mRNA expression and the responses to ERK5 KD in HL60 and AML ex vivo cells. 1,25D increases the expression of mRNAs for both C/EBP alpha and C/EBP beta, but siERK5 reduces the 1,25D-induced increase in C/EBP beta, while it further increases the expression of C/EBP alpha. There is no correlation between C/EBP alpha and CD11b, but the expression of C/EBP beta mRNA correlates well with CD14 mRNA. Hash marks (#) show that 1,25D significantly increased mRNA levels when compared to the untreated control. Asterisks (*) show the statistically significant differences from the corresponding siRNA groups following exposure to siERK5. p < 0.05; bars represent mean values +/− SE. or ex vivo cells, replicates were from the same patient.
6. There is a tight correlation between C/EBPβ and CD14 mRNA expression and their changes following ERK5 knockdown in AML cells
As is well known, CD14 is a monocyte/macrophage differentiation marker and is strongly up -regulated during hematopoiesis as well as during induced differentiation of AML cells [e.g., (Pan et al., 1999)]. It is also known that the promoter of CD14 has a C/EBP enhancer-binding element which is directly involved in the regulation of CD14 expression, as demonstrated in myelomonoblastic U937 cells (Pan et al., 1999). However, it is not well established which C/EBP isoform is responsible for CD14 expression, as both C/EBPα and C/EBPβ expression increase during monocytic differentiation, as reported in literature, and seen in Fig. 6C. We therefore compared the expression at mRNA level of these two isoforms of C/EBP with the expression of CD14 and CD11b mRNA. As shown in Fig. 6C, experimental modulation of C/EBP isoform levels of expression in HL60 and ex vivo AML cells showed a strict correlation between the levels of C/EBPβ mRNA and CD14 mRNA with regard to 1,25D-induced increases, and their partial abrogation by siERK5. In contrast, while both C/EBPα mRNA and CD11b mRNA levels were also increased by 1,25D, C/EBPα mRNA levels were further increased by siERK5, but CD11b mRNA was not altered by the ERK5 knockdown. This suggests that C/EBPα does not have any influence on CD11b expression, which has been reported to be controlled by the transcription factor PU.1 (Wang and Friedman, 2002). Thus our results are consistent with signaling of monocytic differentiation of AML cells by the 1,25D --- ERK5 ---- C/EBPβ → CD14 pathway.
Discussion
ERK5 is expressed ubiquitously and is evolutionarily conserved (Caffrey et al., 1999), but its expression patterns vary in different mammalian tissues (Avruch, 2007; Drew et al., 2012; Kamakura et al., 1999; Roberts et al., 2009; Song et al., 2004). In view of this well documented cell-type specificity of ERK5 expression and presumably function, it is important to determine to what extent the results of prior studies of ERK5 in cell types other than hematopoietic, or those obtained in murine models, apply to ERK5 in human AML cells.
We have focused on 1,25D-induced monocytic differentiation of human myeloid leukemia cells, both in established culture and in primary culture of freshly obtained AML blast cells, a system of importance in translating the knowledge of the underlying molecular events to the clinic, as a potential component of therapy. We found for the first time that the transcription factors MEF2C, C/EBPβ and C/EBPα, all known to participate in monocytic or myeloid differentiation (Marcinkowska et al., 2006; Natsuka et al., 1992; Radomska et al., 1998; Schuler et al., 2008; Wang and Studzinski, 2006), are regulated by ERK5 in human AML. This is of considerable interest, since the role of ERK5 in cell differentiation has been rarely studied, and apart from the demonstration of the role of ERK5 in neuronal differentiation (Liu et al., 2006; Nishimoto et al., 2005), the main interest in ERK5 was its role in the regulation of cell survival and proliferation [e.g., ref (Perez-Madrigal et al., 2012)]. Such studies in cells of hematological origin included the demonstration that ERK5 is essential for growth of leukemic cells in vivo (Garaude et al., 2006), that Hodgkin lymphoma cells show a constitutive activation of the ERK5 pathway (Nagel et al., 2007), and that in multiple myeloma inhibition of ERK5 affects cell proliferation and sensitizes the cells to apoptosis (Carvajal-Vergara et al., 2005). The most germane to the present study are the reports that ERK5 is needed for survival of CML cells with Bcr/Abl translocation (Buschbeck et al., 2005), and that Fms-like tyrosine kinase 3 (FLT3) activates MEK5/ERK5 in human AML cell lines with mutated FLT3 (FLT3-ITD) (Razumovskaya et al., 2011). However, in these studies the focus was also on the effect of ERK5 inhibition on cell survival, not differentiation.
We have also noted for the first time that ERK5 is required for optimal monocytic differentiation of AML cells when these cells are exposed to 1,25D or to 1α-hydroxyvitamin D2, an analog which has FDA approval for human use in kidney dialysis patients (Dennis and Albertson, 2006) (Fig. 1). When ERK5 activity is specifically inhibited, or its expression reduced, the expression of the VDD-induced CD14 surface marker, specific for the monocytic phenotype in hematopoietic cells (Goyert et al., 1988), is markedly reduced. Reciprocally, the expression of the general myeloid marker CD11b increases when ERK5 activity is inhibited by pharmacological inhibitors or by siERK5, evident both in cell lines and blasts ex vivo (Figs 2, 4 and 5). While the exact sequence of events requires further extensive studies, it is likely that the ERK5 regulation of CD14 expression is mediated directly at transcriptional level by C/EBPβ, as this transcription factor is known to activate the CD14 promoter in monocytic differentiation (Pan et al., 1999), and C/EBPβ is upregulated and activated by 1,25D under ERK5 control (Fig. 6). In contrast, phosphorylation of C/EBPα, another factor crucial for myelopoiesis (Radomska et al., 1998), also upregulated during 1,25D-induced differentiation of AML cells, is inhibited by the ERK5 pathway (Fig. 6), perhaps to inhibit granulocytic differentiation when the cells are exposed to 1,25D. Further, in the differentiation system studied here the control of CD11b expression appears to be mechanistically different from the control of CD14 expression, as at mRNA level CD11b expression was not altered by siERK5 (Fig. 6C), even though the surface expression of CD11b is regulated by ERK5 (Figs 2, 4 and 5). Thus, it seems likely that CD11b protein levels are kept in check by protein stability or subcellular localization influenced by the ERK5 pathway, while CD14 protein levels are positively regulated by the ERK5 pathway at the transcriptional level. This suggests that the ERK5 pathway serves to direct hematopoietic lineage selection towards the monocytic phenotype, and that the role of MEF2C, its immediate downstream target, in this pathway should be investigated.
Taken together, the studies reported here constitute a significant step forward in elucidating the complex relationships between signaling pathways and cell fate, and thus may provide a better basis for design of improvements in targeted therapy of AML.
Acknowledgments
Contract grant Sponsors: The National Cancer Institute NIH; the American Institute for Cancer Research, the Israel Science Foundation.
Contract grant numbers: 5R01CA44722-22; 10A049; 635/11
This research was supported by NIH grant 5R01CA44722-22 from the National Cancer Institute (to GPS), by the American Institute for Cancer Research grant #10A049 (to GPS and MD), and by the Israel Science Foundation grant 635/11 (to MD).
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.24513]
The authors have no conflict of interest to disclose.
References
- Alvarez-Fernandez S, Ortiz-Ruiz MJ, Parrott T, Zaknoen S, Ocio EM, San Miguel J, Burrows FJ, Esparis-Ogando A, Pandiella A. Potent antimyeloma activity of a novel ERK5/CDK inhibitor. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2118. [DOI] [PubMed] [Google Scholar]
- Avruch J. MAP kinase pathways: the first twenty years. Biochim Biophys Acta. 2007;1773(8):1150–1160. doi: 10.1016/j.bbamcr.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc Natl Acad Sci U S A. 1980;77(5):2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Hofbauer S, Di Croce L, Keri G, Ullrich A. Abl-kinase-sensitive levels of ERK5 and its intrinsic basal activity contribute to leukaemia cell survival. EMBO Rep. 2005;6(1):63–69. doi: 10.1038/sj.embor.7400316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey DR, O’Neill LA, Shields DC. The evolution of the MAP kinase pathways: coduplication of interacting proteins leads to new signaling cascades. J Mol Evol. 1999;49(5):567–582. doi: 10.1007/pl00006578. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Tabera S, Montero JC, Esparis-Ogando A, Lopez-Perez R, Mateo G, Gutierrez N, Parmo-Cabanas M, Teixido J, San Miguel JF, Pandiella A. Multifunctional role of Erk5 in multiple myeloma. Blood. 2005;105(11):4492–4499. doi: 10.1182/blood-2004-08-2985. [DOI] [PubMed] [Google Scholar]
- Cavanaugh JE. Role of extracellular signal regulated kinase 5 in neuronal survival. Eur J Biochem. 2004;271(11):2056–2059. doi: 10.1111/j.1432-1033.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, Franklin RA, McCubrey JA. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17(7):1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- Charni S, Aguilo JI, Garaude J, de Bettignies G, Jacquet C, Hipskind RA, Singer D, Anel A, Villalba M. ERK5 knockdown generates mouse leukemia cells with low MHC class I levels that activate NK cells and block tumorigenesis. J Immunol. 2009;182(6):3398–3405. doi: 10.4049/jimmunol.0803006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis VC, Albertson GL. Doxercalciferol treatment of secondary hyperparathyroidism. Ann Pharmacother. 2006;40(11):1955–1965. doi: 10.1345/aph.1G523. [DOI] [PubMed] [Google Scholar]
- Drew BA, Burow ME, Beckman BS. MEK5/ERK5 pathway: the first fifteen years. Biochim Biophys Acta. 2012;1825(1):37–48. doi: 10.1016/j.bbcan.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparis-Ogando A, Diaz-Rodriguez E, Montero JC, Yuste L, Crespo P, Pandiella A. Erk5 participates in neuregulin signal transduction and is constitutively active in breast cancer cells overexpressing ErbB2. Mol Cell Biol. 2002;22(1):270–285. doi: 10.1128/MCB.22.1.270-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday BB, Adjei AA. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14(2):342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- Friend C, Zajac-Kaye M, Holland JG, Pogo BG. Depletion of sodium butyrate from the culture medium of Friend erythroleukemia cells undergoing differentiation. Cancer Res. 1987;47(2):378–382. [PubMed] [Google Scholar]
- Garaude J, Cherni S, Kaminski S, Delepine E, Chable-Bessia C, Benkirane M, Borges J, Pandiella A, Iniguez MA, Fresno M, Hipskind RA, Villalba M. ERK5 activates NF-kappaB in leukemic T cells and is essential for their growth in vivo. J Immunol. 2006;177(11):7607–7617. doi: 10.4049/jimmunol.177.11.7607. [DOI] [PubMed] [Google Scholar]
- Gaynor R, Simon K, Koeffler P. Expression of c-jun during macrophage differentiation of HL-60 cells. Blood. 1991;77(12):2618–2623. [PubMed] [Google Scholar]
- Goyert SM, Ferrero E, Rettig WJ, Yenamandra AK, Obata F, Le Beau MM. The CD14 monocyte differentiation antigen maps to a region encoding growth factors and receptors. Science. 1988;239(4839):497–500. doi: 10.1126/science.2448876. [DOI] [PubMed] [Google Scholar]
- Grignani F, Fagioli M, Ferrucci PF, Alcalay M, Pelicci PG. The molecular genetics of acute promyelocytic leukemia. Blood Rev. 1993;7(2):87–93. doi: 10.1016/s0268-960x(05)80018-9. [DOI] [PubMed] [Google Scholar]
- Ji Y, Studzinski GP. Retinoblastoma protein and CCAAT/enhancer-binding protein beta are required for 1,25-dihydroxyvitamin D3-induced monocytic differentiation of HL60 cells. Cancer Res. 2004;64(1):370–377. doi: 10.1158/0008-5472.can-03-3029. [DOI] [PubMed] [Google Scholar]
- Kamakura S, Moriguchi T, Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J Biol Chem. 1999;274(37):26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- Kaspers GJ, Creutzig U. Pediatric acute myeloid leukemia: international progress and future directions. Leukemia. 2005;19(12):2025–2029. doi: 10.1038/sj.leu.2403958. [DOI] [PubMed] [Google Scholar]
- Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. Embo J. 1997;16(23):7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cundiff P, Abel G, Wang Y, Faigle R, Sakagami H, Xu M, Xia Z. Extracellular signal-regulated kinase (ERK) 5 is necessary and sufficient to specify cortical neuronal fate. Proc Natl Acad Sci U S A. 2006;103(25):9697–9702. doi: 10.1073/pnas.0603373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohle M, Hermann A, Glass H, Kempe A, Schwarz SC, Kim JB, Poulet C, Ravens U, Schwarz J, Scholer HR, Storch A. Differentiation efficiency of induced pluripotent stem cells depends on the number of reprogramming factors. Stem Cells. 2012;30(3):570–579. doi: 10.1002/stem.1016. [DOI] [PubMed] [Google Scholar]
- Marcinkowska E, Garay E, Gocek E, Chrobak A, Wang X, Studzinski GP. Regulation of C/EBPbeta isoforms by MAPK pathways in HL60 cells induced to differentiate by 1,25-dihydroxyvitamin D3. Exp Cell Res. 2006;312(11):2054–2065. doi: 10.1016/j.yexcr.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta PB, Jenkins BL, McCarthy L, Thilak L, Robson CN, Neal DE, Leung HY. MEK5 overexpression is associated with metastatic prostate cancer, and stimulates proliferation, MMP-9 expression and invasion. Oncogene. 2003;22(9):1381–1389. doi: 10.1038/sj.onc.1206154. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, Dinunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, Xiao L, Shipps G, Hruza A, Wang J, Kelly J, Paliwal S, Xiaolei G, Boga SB, Zhu L, Daublain P, Zhang L, Lutterbach BA, Pelletire MR, Philippar U, Siliphaivanh P, Witter D, Kirschmeier P, Bishop WR, Hicklin D, Gilliland G, Jayaraman L, Zawel L, Fawell SE, Samatar AA. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- Nagel S, Burek C, Venturini L, Scherr M, Quentmeier H, Meyer C, Rosenwald A, Drexler HG, MacLeod RA. Comprehensive analysis of homeobox genes in Hodgkin lymphoma cell lines identifies dysregulated expression of HOXB9 mediated via ERK5 signaling and BMI1. Blood. 2007;109(7):3015–3023. doi: 10.1182/blood-2006-08-044347. [DOI] [PubMed] [Google Scholar]
- Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79(2):460–466. [PubMed] [Google Scholar]
- Nishimoto S, Kusakabe M, Nishida E. Requirement of the MEK5-ERK5 pathway for neural differentiation in Xenopus embryonic development. EMBO Rep. 2005;6(11):1064–1069. doi: 10.1038/sj.embor.7400515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Hetherington CJ, Zhang DE. CCAAT/enhancer-binding protein activates the CD14 promoter and mediates transforming growth factor beta signaling in monocyte development. J Biol Chem. 1999;274(33):23242–23248. doi: 10.1074/jbc.274.33.23242. [DOI] [PubMed] [Google Scholar]
- Perez-Madrigal D, Finegan KG, Paramo B, Tournier C. The extracellular-regulated protein kinase 5 (ERK5) promotes cell proliferation through the down-regulation of inhibitors of cyclin dependent protein kinases (CDKs) Cell Signal. 2012;24(12):2360–2368. doi: 10.1016/j.cellsig.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Pierce GB, Jr, Verney EL. An in vitro and in vivo study of differentiation in teratocarcinomas. Cancer. 1961;14:1017–1029. doi: 10.1002/1097-0142(196109/10)14:5<1017::aid-cncr2820140516>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353(6345):670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18(7):4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razumovskaya E, Sun J, Ronnstrand L. Inhibition of MEK5 by BIX02188 induces apoptosis in cells expressing the oncogenic mutant FLT3-ITD. Biochem Biophys Res Commun. 2011;412(2):307–312. doi: 10.1016/j.bbrc.2011.07.089. [DOI] [PubMed] [Google Scholar]
- Regan CP, Li W, Boucher DM, Spatz S, Su MS, Kuida K. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc Natl Acad Sci U S A. 2002;99(14):9248–9253. doi: 10.1073/pnas.142293999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts OL, Holmes K, Muller J, Cross DA, Cross MJ. ERK5 and the regulation of endothelial cell function. Biochem Soc Trans. 2009;37(Pt 6):1254–1259. doi: 10.1042/BST0371254. [DOI] [PubMed] [Google Scholar]
- Roberts OL, Holmes K, Muller J, Cross DA, Cross MJ. ERK5 is required for VEGF-mediated survival and tubular morphogenesis of primary human microvascular endothelial cells. J Cell Sci. 2010;123(Pt 18):3189–3200. doi: 10.1242/jcs.072801. [DOI] [PubMed] [Google Scholar]
- Sachs L. Control of normal cell differentiation and the phenotypic reversion of malignancy in myeloid leukaemia. Nature. 1978;274(5671):535–539. doi: 10.1038/274535a0. [DOI] [PubMed] [Google Scholar]
- Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Humphreys S, Jacob F, de Vitry F. Induced differentiation of a neuroblastoma. Dev Biol. 1971;25(4):514–546. doi: 10.1016/0012-1606(71)90004-2. [DOI] [PubMed] [Google Scholar]
- Schuler A, Schwieger M, Engelmann A, Weber K, Horn S, Muller U, Arnold MA, Olson EN, Stocking C. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood. 2008;111(9):4532–4541. doi: 10.1182/blood-2007-10-116343. [DOI] [PubMed] [Google Scholar]
- Sherman ML, Stone RM, Datta R, Bernstein SH, Kufe DW. Transcriptional and post-transcriptional regulation of c-jun expression during monocytic differentiation of human myeloid leukemic cells. J Biol Chem. 1990;265(6):3320–3323. [PubMed] [Google Scholar]
- Sohn SJ, Sarvis BK, Cado D, Winoto A. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J Biol Chem. 2002;277(45):43344–43351. doi: 10.1074/jbc.M207573200. [DOI] [PubMed] [Google Scholar]
- Song H, Jin X, Lin J. Stat3 upregulates MEK5 expression in human breast cancer cells. Oncogene. 2004;23(50):8301–8309. doi: 10.1038/sj.onc.1208026. [DOI] [PubMed] [Google Scholar]
- Studzinski GP, Wang X, Ji Y, Wang Q, Zhang Y, Kutner A, Harrison JS. The rationale for deltanoids in therapy for myeloid leukemia: role of KSR-MAPK-C/EBP pathway. J Steroid Biochem Mol Biol. 2005;97(1–2):47–55. doi: 10.1016/j.jsbmb.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tatake RJ, O’Neill MM, Kennedy CA, Wayne AL, Jakes S, Wu D, Kugler SZ, Jr, Kashem MA, Kaplita P, Snow RJ. Identification of pharmacological inhibitors of the MEK5/ERK5 pathway. Biochem Biophys Res Commun. 2008;377(1):120–125. doi: 10.1016/j.bbrc.2008.09.087. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang X, Studzinski GP. Jun N-terminal kinase pathway enhances signaling of monocytic differentiation of human leukemia cells induced by 1,25-dihydroxyvitamin D3. J Cell Biochem. 2003;89(6):1087–1101. doi: 10.1002/jcb.10595. [DOI] [PubMed] [Google Scholar]
- Wang QF, Friedman AD. CCAAT/enhancer-binding proteins are required for granulopoiesis independent of their induction of the granulocyte colony-stimulating factor receptor. Blood. 2002;99(8):2776–2785. doi: 10.1182/blood.v99.8.2776. [DOI] [PubMed] [Google Scholar]
- Wang X, Gocek E, Novik V, Harrison JS, Danilenko M, Studzinski GP. Inhibition of Cot1/Tlp2 oncogene in AML cells reduces ERK5 activation and up-regulates p27Kip1 concomitant with enhancement of differentiation and cell cycle arrest induced by silibinin and 1,25-dihydroxyvitamin D3. Cell Cycle. 2010;9(22):4542–4551. doi: 10.4161/cc.9.22.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Studzinski GP. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvitamin D3-induced differentiation of HL60 cells. J Cell Biochem. 2001;80(4):471–482. doi: 10.1002/1097-4644(20010315)80:4<471::aid-jcb1001>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Wang X, Studzinski GP. The requirement for and changing composition of the activating protein-1 transcription factor during differentiation of human leukemia HL60 cells induced by 1,25-dihydroxyvitamin D3. Cancer Res. 2006;66(8):4402–4409. doi: 10.1158/0008-5472.CAN-05-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tournier C. Regulation of cellular functions by the ERK5 signalling pathway. Cell Signal. 2006;18(6):753–760. doi: 10.1016/j.cellsig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood. 2008;111(5):2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- Yan C, Luo H, Lee JD, Abe J, Berk BC. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J Biol Chem. 2001;276(14):10870–10878. doi: 10.1074/jbc.M009286200. [DOI] [PubMed] [Google Scholar]
- Yan L, Carr J, Ashby PR, Murry-Tait V, Thompson C, Arthur JS. Knockout of ERK5 causes multiple defects in placental and embryonic development. BMC Dev Biol. 2003;3:11. doi: 10.1186/1471-213X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, Yates JR, 3rd, Gray NS, Lee JD. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18(3):258–267. doi: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J, Harrison JS, Uskokovic M, Danilenko M, Studzinski GP. Silibinin can induce differentiation as well as enhance vitamin D3-induced differentiation of human AML cells ex vivo and regulates the levels of differentiation-related transcription factors. Hematol Oncol. 2010;28(3):124–132. doi: 10.1002/hon.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Bao ZQ, Dixon JE. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270(21):12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]