Abstract
Intracellular signaling pathways present targets for pharmacological agents with potential for treatment of neoplastic diseases, with some disease remissions already recorded. However, cellular compensatory mechanisms usually negate the initial success. For instance, attempts to interrupt aberrant signaling downstream of the frequently mutated ras by inhibiting ERK1/2 has shown only limited usefulness for cancer therapy. Here, we examined how ERK5, that overlaps the functions of ERK1/2 in cell proliferation and survival, functions in a manner distinct from ERK1/2 in human AML cells induced to differentiate by 1,25D-dihydroxyvitamin D3 (1,25D). Using inhibitors of ERK1/2 and of MEK5/ERK5 at concentrations specific for each kinase in HL60 and U937 cells, we observed that selective inhibition of the kinase activity of ERK5, but not of ERK1/2, in the presence of 1,25D resulted in macrophage-like cell morphology and enhancement of phagocytic activity. Importantly, this was associated with increased expression of the macrophage colony stimulating factor receptor (M-CSFR), but was not seen when M-CSFR expression was knocked down. Interestingly, inhibition of ERK1/2 led to activation of ERK5 in these cells. Our results support the hypothesis that ERK5 negatively regulates the expression of M-CSFR, and thus has a restraining function on macrophage differentiation. The addition of pharmacological inhibitors of ERK5 may influence trials of differentiation therapy of AML.
Keywords: MAPK, ERK5, ERK1/2, M-CSFR, vitamin D, AML
INTRODUCTION
Differentiation and lineage selection of hematopoietic stem cells present multiple points at which errors can occur and, thus, lead to hematopoietic malignancies. For instance, point mutations in the c-fms proto-oncogene, which encodes the receptor for Macrophage-Colony Stimulating Growth Factor (M-CSF or CSF1), known as M-CSFR or CSF1R, are not infrequently found in Myelodysplastic Syndrome (MDS) and Acute Myeloid Leukemia (AML), particularly of the monocytic subtypes, M4 and M5 [1]. Also, the loss of the c-fms (M-CSFR) has been reported to play a role in microglial (brain macrophage) proliferation and differentiation [2] These findings suggest that the differentiation of bone marrow promonocytes to macrophages is a potential control point which requires an intact M-CSFR, and its loss or malfunction can lead to neoplastic differentiation arrest.
M-CSFR, as well as receptors for the granulocyte (G-CSF or CSF3) and granulocyte-macrophage (GM-CSF or CSF2) colony simulating factors, are individually or collectively responsible for mediating the effects of cell environment on proliferation, survival and differentiation of progenitor cells of the corresponding lineage (see [3, 4] for reviews). The downstream signaling from these plasma membranespanning receptors, which function as protein tyrosine kinases [5–8], are usually transmitted by several phosphorylation cascades (e.g. [7, 9–11]).
In the case of M-CSFR the reported signaling includes JAK/STAT, PI3K/AKT and MAPK pathways [10, 12–14]. The latter pathway consists of a family of related protein kinases, of which ERK1/2 (MAPK3/MAPK1) has received most attention (e.g., [15–18]). However, ERK5 (MAPK7) shares a number of properties and some functions with ERK1/2, yet the overlap is often overlooked in the analysis of MAPK role in carcinogenesis and the therapeutic approaches to malignancies. On the other hand, considerable attention has been recently given to the role of ERK5 in organ development and cell differentiation. For instance, ERK5 has an important role during cardiovascular development [19]. In neural tissues, ERK5 is required for neural outgrowth [20], and Z Xia group made extensive studies of the role of ERK5 in neurogenesis in several regions of the brain (e.g., [21–23]). At cellular level, ERK5 pathway is required for Colony-Stimulating Factor-1(CSF-1)-induced proliferation of macrophages [24], and is linked to cell metabolism in this cell type [25].
We have previously reported that the MAP3K8 known as COT1 is activated during differentiation of cultured AML cells induced by a combination of two differentiation agents, 1,25-dihydroxyvitamin D3 (1,25D) and the plant derived-polyphenol silibinin [26]. Interestingly, ERK5, a known downstream target of COT1 was also activated, and its inhibition appeared to alter the expression of conventional markers in 1,25D-induced differentiation of several types of cultured AML cell [27, 28]. Although the activation of ERK5 was paralleled by the expression of several markers of monocytic differentiation, there was a reciprocal modulation of the relative levels of these markers, with general myeloid marker CD11b being increased by the addition of inhibitors of the ERK5 pathway to either untreated or 1,25D-treated AML cells, while the specific monocytic marker CD14 was concurrently decreased. This suggested that this altered phenotype was due to the reduced ERK5 activity resulting in a change in differentiation state of the monocytes. Such possibility was explored here, and our data suggest that ERK5 functions to retard the progression of monocytes to the next functional stage of differentiation, the macrophage. The principal mechanism for this partial and transient arrest at the stage of monocyte is, at least in part, due to the ability of ERK5, but not of ERK1/2, to inhibit upregulation of M-CSFR levels, necessary for the macrophage phenotype.
MATERIALS AND METHODS
Reagents
1,25D was a kind gift from Dr. Milan Uskokovic (Bioxell, Nutley, NJ). The pharmacological inhibitors of MAP2K5/MEK5 kinase (BIX02189), and of ERK5 (XMD8-92) were purchased from Selleck Chemicals (Houston, TX) and Santa Cruz Biotechnology Inc., respectively. The MEK1/2 (MAP2K1/MAP2K2) inhibitors PD98059 and U0126 were obtained from Cell Signaling Technologies (Danvers, MA). 12-O-Tetradecanoylphorbol 13-acetate (TPA) was from Sigma-Aldrich (St. Louis, MO). Crk-L (#sc-319) antibody was obtained from Santa Cruz Biotechnology (Dallas, TX). Phospho-Erk1/2 (Thr202/Tyr204, #9101), Erk1/2 (#9102), phospho-ERK5 (Thr187/Tyr220, #3371), Erk5 (#3372), M-CSF Receptor (#3152), anti-rabbit (#7074) and anti-mouse (#7076) antibodies linked to HRP were purchased from Cell Signaling Technologies. Nitrocellulose membranes were purchased from GE Healthcare (Pittsburgh, PA). All kinase inhibitors were dissolved in DMSO.
Cells and culture
HL60-G cells were subcloned from HL60 cells derived from a patient with promyeloblastic leukemia [29], and U937 monoblastic cells were derived from a patient with histiocytic lymphoma [30]. These cell lines were cultured in suspension in RPMI-1640 medium supplemented with 10% bovine calf serum (Hyclone, Logan, UT), and incubated at 37°C in a humidified atmosphere with 5% CO2. Routine microbiology testing for Mycoplasma was conducted by selective culture techniques [31]. For experiments involving kinase inhibitors, cells (0.1 × 106/ml) were pretreated with experimental agents or 0.1% DMSO (vehicle control) for 1 h before the addition of 1,25D for additional 96 h. Cells were considered adherent where pre-incubation with EDTA was required to loosen the cells from the substratum. Cell number and viability were estimated on the basis of the trypan blue exclusion assay.
Ex vivo samples
AML blasts were obtained from three patients and from two individuals without malignant disease who provided normal bone marrow cells. All specimens were accrued in accordance with an IRB approved protocol and with informed consent. Mononuclear cells were isolated as described previously [27, 32]. The cells were placed in primary culture and treated in a manner described for the established cell lines, but because the experiments were limited by the quantity of cells available, the investigations were not able to be as extensive.
Morphological analysis of Wright-Giemsa-stained cells
Following incubations, cells were washed twice with PBS and once with bovine serum. Cells were then smeared on glass slides, air-dried, and subjected to Wright-Giemsa staining procedure in a Hema Tek 2000 automated slide staining apparatus (Siemens Healthcare Diagnostics, Tarrytown, NY, USA) in accordance with the manufacturer’s protocol. The slides were then air-dried and examined under an Eclipse E600W light microscope (Nikon, Tokyo, Japan) at 1000× magnification. Images were obtained by a DS-5M digital camera (Nikon).
Phagocytosis assay
After incubation with the indicated compounds for 96 h, 5 × 105 cells were washed twice with PBS, suspended in 495 µl RPMI-1640 containing 10% heat-inactivated FCS and incubated at 37°C for 15 min with 5 µl of opsonized zymosan (1 mg/ml). Subsequently, the cells were smeared on glass slides and stained with Wright-Giemsa, as described above. Phagocytosis was determined under light microscope (500× magnification) in at least 500 cells, and defined as the percent of cells containing two or more phagocytized particles of opsonized zymosan, as described previously [33].
Flow Cytometry
Aliquots of one million cells were harvested, washed twice with phosphate buffered saline (PBS) and incubated for 45 min at room temperature with 0.5 µl CD14-RD1, 0.5 µl CD11b-FITC or 5 µl human PE-conjugated CD115 (c-fms) antibodies (all from Beckman Coulter Inc., Brea, CA) to analyze the expression of the corresponding surface markers of differentiation. The cells were then washed three times with ice-cold PBS, resuspended in 1 ml PBS and analyzed using EPICS XL flow cytometer (Beckman Coulter). IgG-FITC, IgG2b-RD1 and IgG2a-PE isotype controls were used to set threshold parameters.
Transfection of siMCSFR oligonucleotides
The siRNA targeting M-CSFR and a generic scrambled control siRNA were purchased from Santa Cruz Biotechnology. Transfection was carried out by using Endo-Porter delivery reagent from Gene Tools Inc. (Philomath, OR). siRNAs were transfected at a final concentration of 20 nM for 48 hours, and the transfected cells were seeded at 2 × 105 cells/ml and exposed to the specified agents for the indicated times. The cells were examined for the efficiency of the knock down of M-CSFR expression at both mRNA and protein levels.
RT-PCR
Total cellular RNA was extracted by using Trizol (Invitrogen, CA) according to manufacturer’s protocol and reverse transcribed for quantification by ABI cDNA Archive Kit (Applied Biosystems, Foster City, CA) as previously described [26]. Quantitative RT-PCR for M-CSFR was carried out using an ABI SYBR Green master kit (Applied Biosystems, Foster City, CA). Fold changes of mRNA levels in target gene relative to the RNA polymerase II (RPII) control were calculated by relative quantification analysis. Primers used for M-CSFR were: upstream 5’- CTG CTG ACT GTT GAG ACC TTA G -3’, downstream 5’- GGT ACT TGG GCT TCT GCT TAT -3’; for RPII, upstream 5’-GCA CCA CGT CCA ATG ACAT-3’, downstream 5’-GTG CGG CTG CTT CCA TAA-3’. The quality of PCR products was monitored using post-PCR melting curve analysis.
Western blotting
Western blotting was performed using total cell extracts, as described before [26]. Briefly, membranes were incubated with different primary antibodies for 1–2 h, and then blotted with HRP-linked secondary antibodies for 1 h. The protein bands were visualized using a chemiluminescence assay system (Pierce Biotechnology, Rockford, IL), each membrane was stripped and reprobed for Crk-L, as the internal loading control. The optical density of each band was quantitated using ImageQuant 5.0 (Molecular Dynamics, Sunnyvale, CA), and normalized to the signal of Crk-L loading control.
Statistical analysis
Each cell line experiment was performed at least three times and the results are expressed as the mean ± S.D. Significance of the differences between mean values was assessed by a two-tailed Student’s t-test. A p value less than 0.05 was considered to be statistically significant. All computations were performed using Microsoft EXCEL or GraphPad Prism ver. 6.0 (GraphPad, La Jolla, CA).
RESULTS
Specificity of kinase inhibitors for ERK1/2 and ERK5 pathways
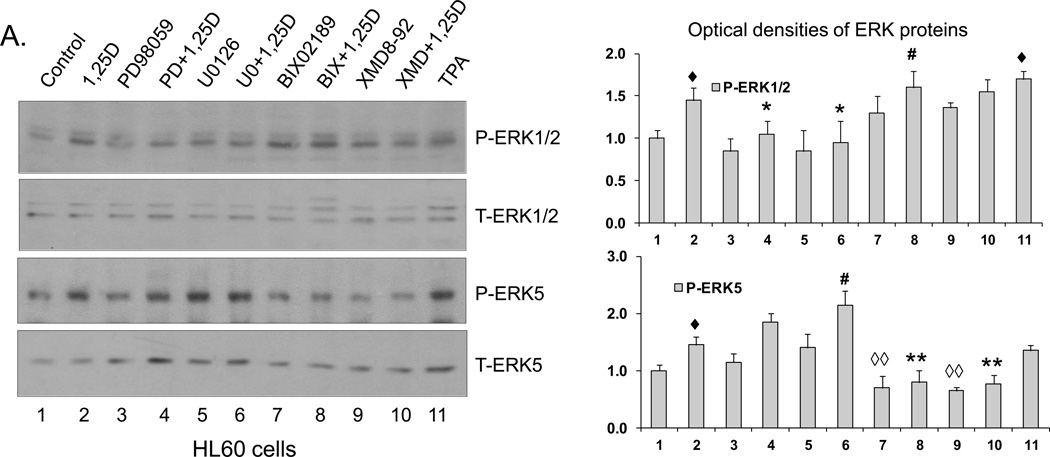
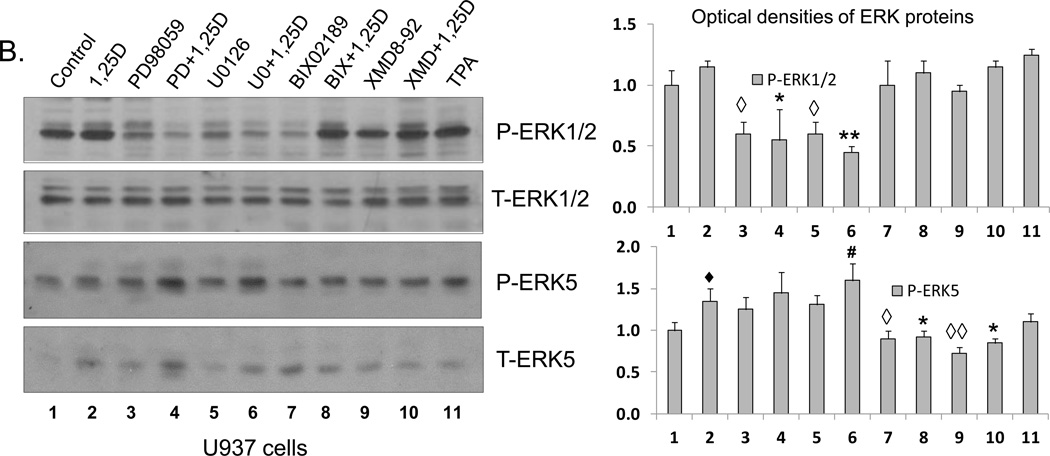
A number of synthetic inhibitors has been used in studies of MAPK function, and some ERK1/2 inhibitors have been and currently are in clinical trials of various solid tumor and leukemia therapy (e.g., [34–38]). However, there has been some discussion whether compounds such as PD98059 and U0126, the well-studied inhibitors of ERK1/2 [39], also inhibit the activation of ERK5 (e.g., [40, 41]). Since at high concentrations the specificity of pharmacological inhibitors is routinely lost (e.g., [42]), we used inhibitor concentrations near or at the lowest concentration reported in previous studies, as summarized in Supplemental Table 1. More importantly, in initial experiments we established that in the cell system studied here 20 µM PD98059 (lane 4) and 1 µM U0126 (lane 6) inhibit 1,25D-induced activation of ERK1/2, and the latter inhibitor actually increased the activation of ERK5, as shown by its phosphorylation levels (Fig.1).
Fig. 1. Western blots showing the specificity of ERK1/2 and ERK5 inhibitors.
(A) HL60 cells were pretreated with either MEK1/2 inhibitors, PD98059 (20 µM) and U0126 (1 µM), or MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), for 1 h, then 1,25D (1 nM) was added for an additional 96 h. TPA (10 nM) treated group was used as positive control. P-ERK1/2 and P-ERK5 protein levels were determined by Western blots. Normalized optical densities of each band are shown in the bar charts. The blots shown are representative of three experiments. CTL = Control. (B) U937 cells were treated in the same manner as HL60 cells. ◆ = p<0.05, significantly increased vs. control group; ◇ = p<0.05, ◇◇ = p<0.01, significantly decreased vs. control group; # = p<0.05, significantly increased vs. 1,25D-treated group; * = p<0.05, ** = p<0.01, significantly decreased vs. 1,25D-treated group; n=3.
Conversely, Fig.1 also shows that 10 µM BIX02189 (lanes 7 and 8) and 5 µM XMD8-92 (lanes 9 and 10) inhibit both basal and 1,25D-induced activation of ERK5 in both cell lines studied. Additionally, in HL60 cells there is increased phosphorylation of ERK1/2, also most likely a compensatory event. Thus, in human AML cells 20 µM PD98059 or 1 µM U0126 specifically inhibit 1,25D-induced activation of ERK1/2, while 10 µM BIX02189 and 5 µM XMD8-92 specifically inhibit the ERK5 pathway, and a compensatory activation of the parallel MAPK pathway can also occur, though these are inhibitorspecific events.
Inhibition of ERK5 pathway changes AML cell morphology and cell surface marker expression in a manner similar to changes seen with TPA, a recognized inducer of macrophage phenotype
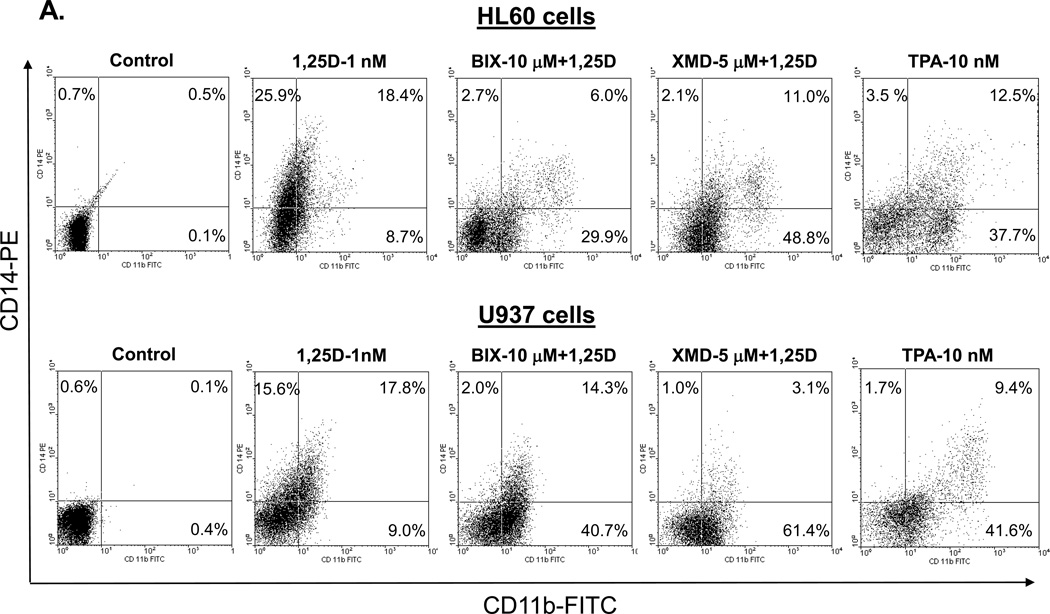
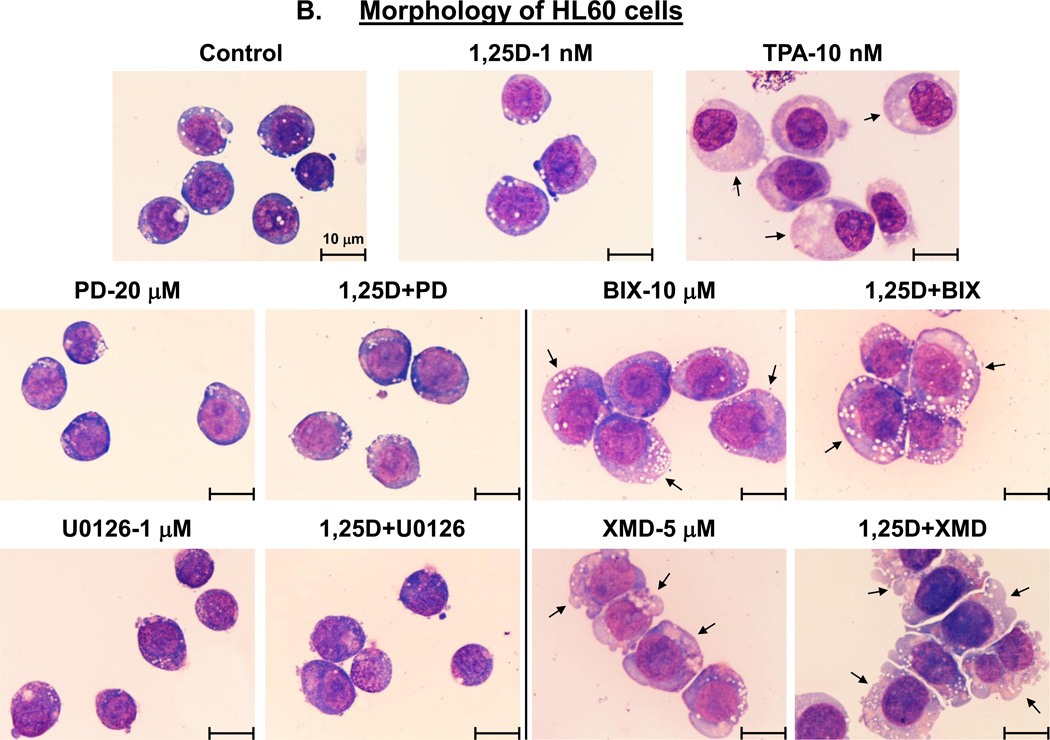
We have previously observed that the exposure of AML cells to 1,25D in the presence of BIX02189 or XMD8-92 markedly reduces the surface expression of CD14, the principal marker of monocytic differentiation [27, 28]. As mentioned above, this was accompanied by an increase in the expression of CD11b, the general myeloid marker [43], raising the possibility that the inhibition of the ERK5 pathway results in a switch in the differentiation lineage. Accordingly, we compared patterns of CD11b and CD14 expression in HL60 and U937 cells treated with 1,25D and its combinations with MEK5/ERK5 inhibitors to those in cells incubated with TPA, a well-known inducer of macrophage differentiation [44]. In addition, we performed microscopic examination of Wright-Giemsa stained cells following different treatments.
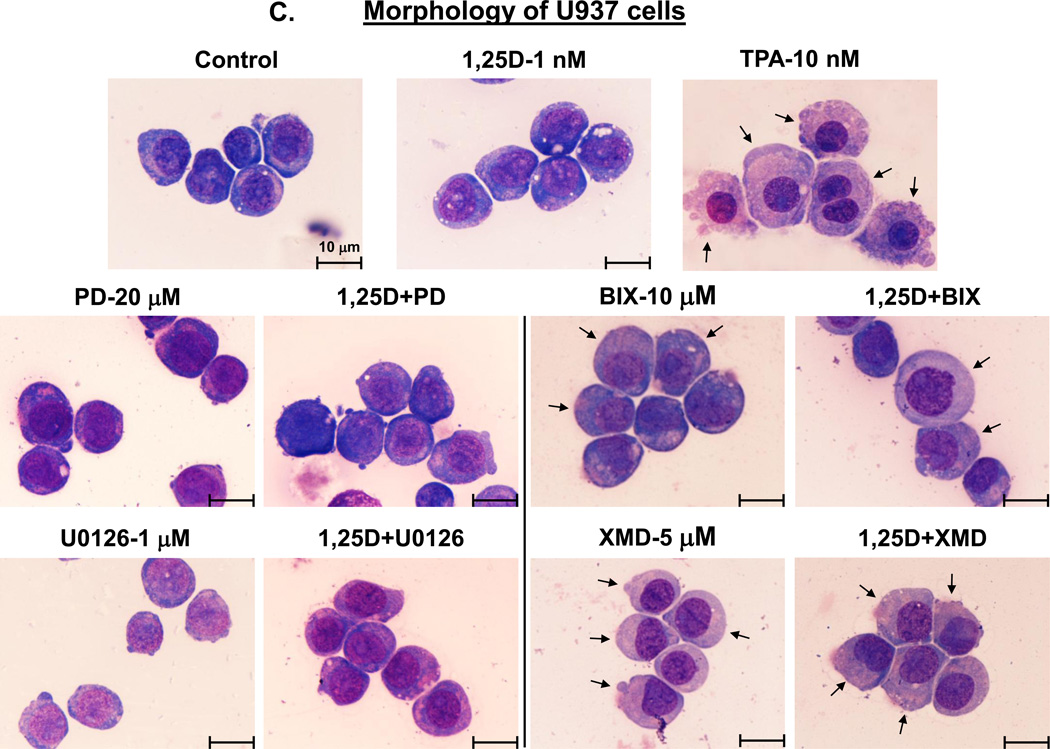
Fig. 2A demonstrates that, in a striking similarity to the samples treated with BIX02189+1,25D or XMD8-92+1,25D, TPA treatment resulted in a marked increase in surface CD11b levels accompanied by a much less pronounced elevation of CD14 levels. Further, as illustrated in Figs. 2B and C, control cells (treated with the vehicle only) displayed blast-like morphology (large nuclear/cytoplasmic ratio, strongly basophilic cytoplasm).Treatment with 1,25D alone induced moderate morphological changes (slightly less basophilic cytoplasm and smaller nuclear/cytoplasmic ratio). The morphological features of cells treated with PD98059 or U0126, in the absence or presence of 1,25D, showed almost no difference compared with untreated control cells. However, cells treated with BIX02189+1,25D and, particularly, XMD8-92+1,25D acquired the morphological features consistent with macrophage-like cells, including enlarged cells with diffuse or ameboid shape, abundant and less basophilic cytoplasm, ruffled cell membrane, and adherence to the substratum. In some cells ameboid protrusions were seen, generally considered to be specific for macrophage morphology [45]. BIX02189 and, more clearly, XMD8-92 alone, had similar, but less pronounced effects. TPA treatment, used as a positive control for macrophage-like phenotype induced in AML cells [44], showed substantial similarity to the treatment with BIX02189+1,25D or XMD8-92+1,25D with regard to morphological features (Fig. 2B and C), Thus, both cell morphology and cell surface marker expression changes strongly suggest that inhibition of ERK5 in HL60 and U937 cells results in macrophage-like phenotype.
Fig. 2. Inhibition of the MEK1/2-ERK1/2 and MEK5/ERK5 pathways differentially affects the cell surface marker expression and morphology of AML cells.
(A) An example of primary flow cytometry data. These frames show the similarity of changes in cell surface expression of differentiation markers induced by treatment with TPA or MEK5/ERK5 inhibitors together with 1,25D. Note that TPA, like the inhibitors and 1,25D combinations, increases the expression of CD11b (horizontal scale), but reduces the expression of CD14 (vertical scale). HL60 and U937 cells were pretreated with either MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), or TPA 10 (nM) for 1 h, then 1,25D (1 nM) was added for an additional 96 h. N=3. (B) HL60 cells were pretreated with either MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), or MEK1/2 inhibitors, PD98059 (20 µM) and U0126 (1 µM) for 1 h, then 1,25D (1 nM) was added for an additional 96 h. (C) U937 cells were subjected to identical treatments. Following incubations, cell smears were stained with Wright-Giemsa and photographed at 1000× magnification. TPA (10 nM) was used as the positive control for the induction of macrophage differentiation. Arrows indicate the more abundant cytoplasm in cells treated with MEK5/ERK5 inhibitors, alone and in combination with 1,25D, and TPA, typical of macrophages. Other typical features evident in this group are nuclear morphology, ameboid pseudopodia, and abundant cytoplasmic inclusions, better seen in HL60 cells. Scale lines within the panels indicate 10 µm.
Changes in the expression of M-CSFR, a molecular marker of macrophage phenotype
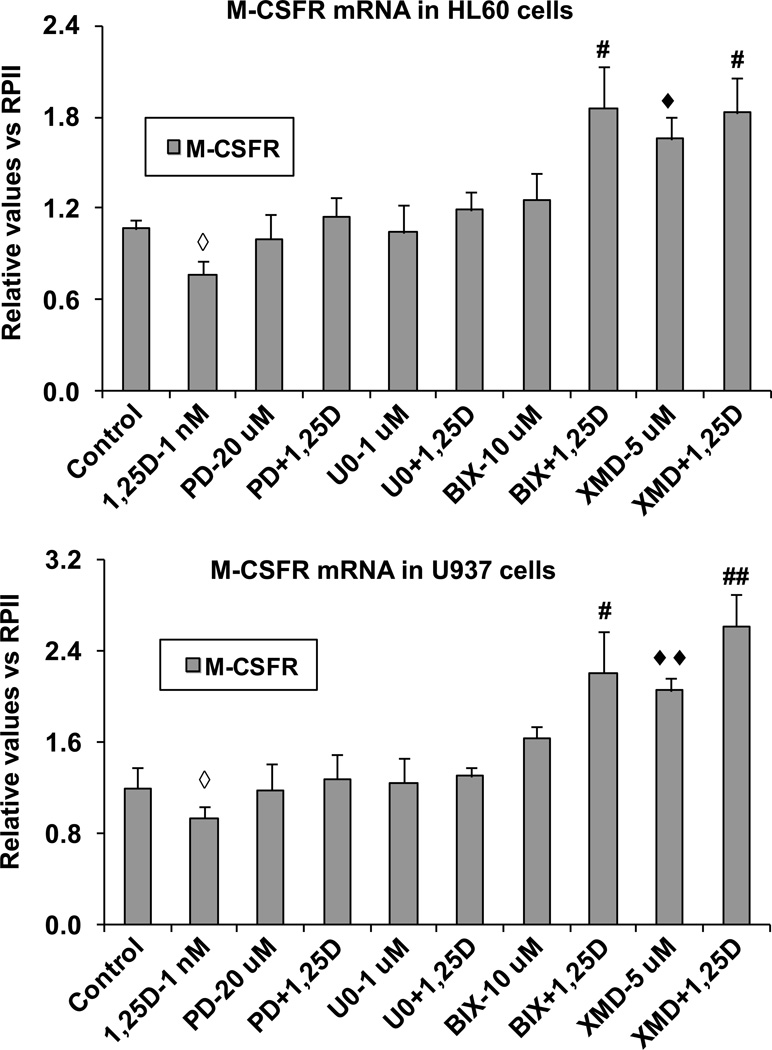
Since cell morphology clearly excluded granulocyte phenotype in AML cells treated with 1,25D and inhibitors of the ERK5 pathway, we have focused on a definitive documentation of the macrophage-like phenotype of the treated cells. First, we examined the expression of mRNA for M-CSFR in HL60 and U937 in basal state and after 96 h exposure to 1,25D. As a control we used cells treated with 10 nM TPA, known to upregulate M-CSFR expression in AML cells [46]. Interestingly, while we detected small but consistent decreases in M-CSFR mRNA levels after 1,25D exposure which were statistically significant (p < 0.05) in both cell lines studied, addition of ERK1/2 inhibitors abrogated these decreases, whereas inhibitors of ERK5 increased M-CSFR mRNA expression in cells in basal state, and even more when combined with 1,25D treatment (Fig. 3). As in most respects, XMD8-92 was somewhat a more potent upregulator of M-CSFR than BIX02189 (Fig. 3). These data suggest that while 1,25D promotes differentiation of AML cells, it tends to inhibit the progression of the monocytes to macrophages, and that ERK5, which is upregulated by 1,25D (Fig. 1 and ref. [27]), is the principal mediator of this effect by inhibiting the expression of M-CSFR. In contrast, ERK1/2 MAPKs do not appear to be involved in this aspect of differentiation control (Fig. 3).
Fig. 3. ERK5 inhibition selectively increases the expression of M-CSFR, a molecular marker of macrophage differentiation, at mRNA level.
HL60 or U937 cells were pretreated with either MEK1/2 inhibitors, PD98059 (20 µM) and U0126 (1 µM) or MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), for 1 h, then 1,25D (1 nM) was added for an additional 96 h. The levels of M-CSFR mRNA were determined by SYBRGreen RT-qPCR in both HL60 and U937 cells. ◆ = p<0.05, ◆◆ = p<0.01, significantly increased vs. control group; ◇ = p<0.05, significantly decreased vs. control group; # = p<0.05, ## = p<0.01, significantly increased vs. 1,25D-treated group.
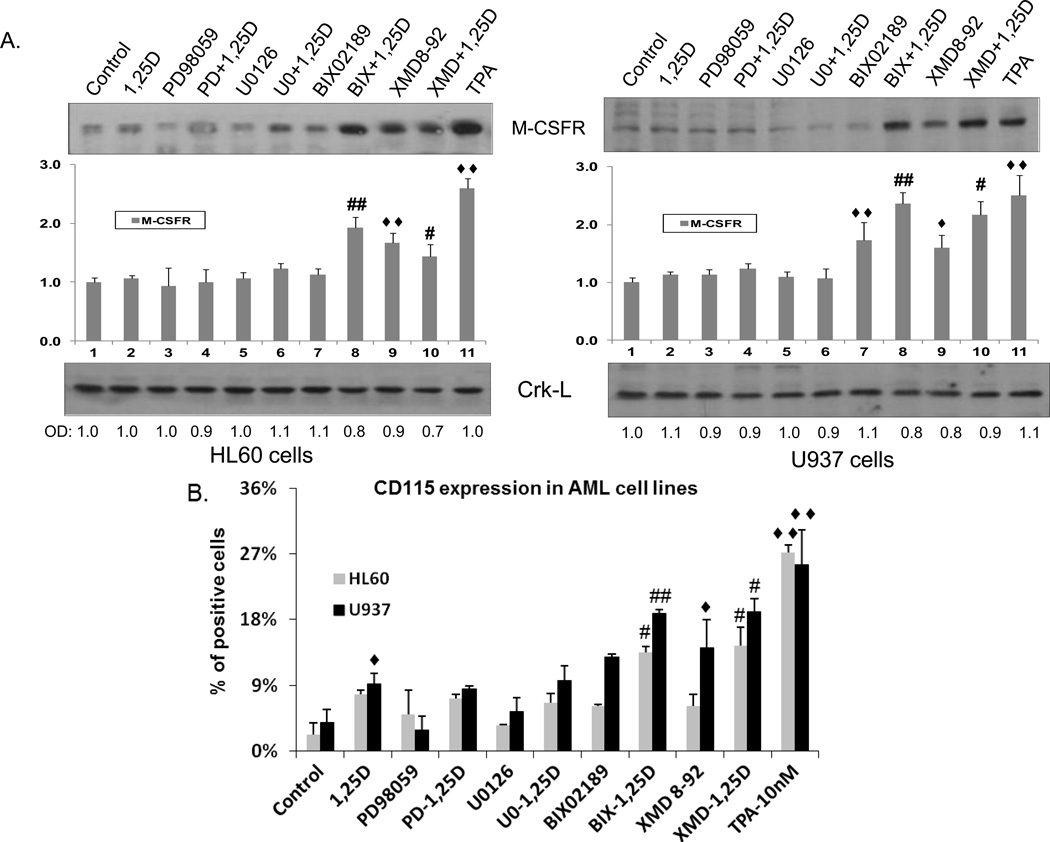
We determined cellular protein levels of M-CSFR by two methods, Western blotting for total protein, and by the expression of M-CSFR on the cell surface, known here as the CD115 antigen. In these experiments we also used as a control cells treated with 10 nM TPA, the inducer of macrophage-like differentiation. The Western blots, shown in Fig. 4, are largely consistent with the mRNA results seen following exposure to ERK inhibitors, indicating that the effects of these kinase inhibitors take place at the transcriptional level. However, the decrease in M-CSFR mRNA in 1,25D-treated cells was not seen at protein level, suggesting that 1,25D may stabilize this protein. Indeed, the surface detectable CD115 showed an increase in 1,25D-treated cells which reached statistical significance in U937 cells (Fig. 4B). These flow cytometric determinations confirmed that inhibitors of ERK1/2 had no discernible effect on the expression of M-CSFR in AML cells, while inhibitors of ERK5 can significantly (p< 0.05 to p< 0.01) increase the level of this receptor, approaching the levels seen in TPA-treated AML cells, an established in vitro model of macrophages.
Fig. 4. Expression of M-CSFR, a molecular marker of macrophage differentiation, at protein level.
AML cells were pretreated with either MEK1/2 inhibitors, PD98059 (20 µM) and U0126 (1 µM) or MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), for 1 h, then 1,25D (1 nM) was added for an additional 96 h. TPA (10 nM) treated cells were used as the positive control. (A) M-CSFR (also known as CD115) total protein levels as determined by Western blotting. Normalized optical densities of each band are shown in the bar charts. Crk-L was used as a loading control. The blots shown are representative of three experiments. CTL = Control. PD = PD98059, U0 = U0126, BIX = BIX02189, XMD = XMD8-92.
(B) Expression of surface M-CSFR (CD115) was determined by flow cytometry. ◆ = p<0.05, ◆◆ = p<0.01, significantly increased vs. control group; # = p<0.05, ## = p<0.01, significantly increased vs. 1,25D-treated group, n=3.
M-CSFR protein levels are critical for the enhancement of macrophage phenotype by inhibition of ERK5 activity
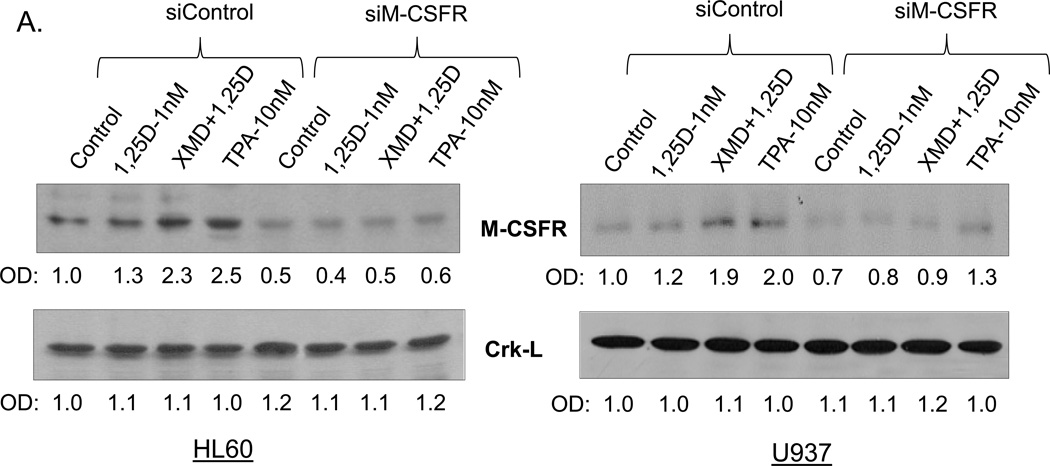
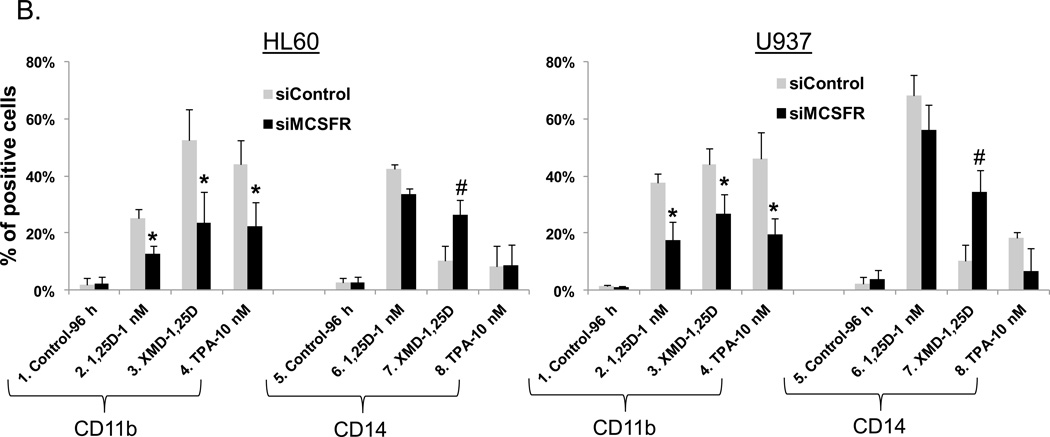
Further confirmation of the importance of M-CSFR for the modulation by ERK5 of differentiation was obtained by knock down (KD) of M-CSFR expression using silencing oligonucleotides to M-CSFR. The KD greatly reduced the expression of M-CSFR in all groups tested, but this was most evident when M-CSFR levels were enhanced by XMD8-92+1,25D or TPA (Fig. 5A). Concurrently, KD of M-CSFR markedly diminished the effect of XMD8-92 on increasing CD11b expression (Fig. 5B, lanes 3 in both HL60 and U937 cells), and on the inhibition of CD14 expression (Fig. 5B, lanes 7). Thus, the inhibition of ERK5 activity by pharmacological agents BIX02189 and XMD8-92 requires high M-CSFR levels to have an effect on macrophage differentiation.
Fig. 5. High levels of M-CSFR are required for ERK5 inhibition of macrophage differentiation.
AML cells were transfected with silencing oligonucleotides to M-CSFR before addition of 1,25D and ERK5 inhibitor XMD8-92. (A) siM-CSFR abrogated the XMD+1,25D-induced increase in M-CSFR protein levels in HL60 and U937 cells. The protein levels of a loading control, Crk-L, were not significantly altered. (B) The knock down of M-CSFR markedly diminished the effect of XMD8-92 on increasing CD11b expression and inhibition of CD14 expression in both HL60 and U937 cells. Asterisks (*) show a significant (p<0.05) decrease in cells subjected to M-CSFR knockdown, while # denotes a significant (p<0.05) increase.
Ex vivo AML cell studies
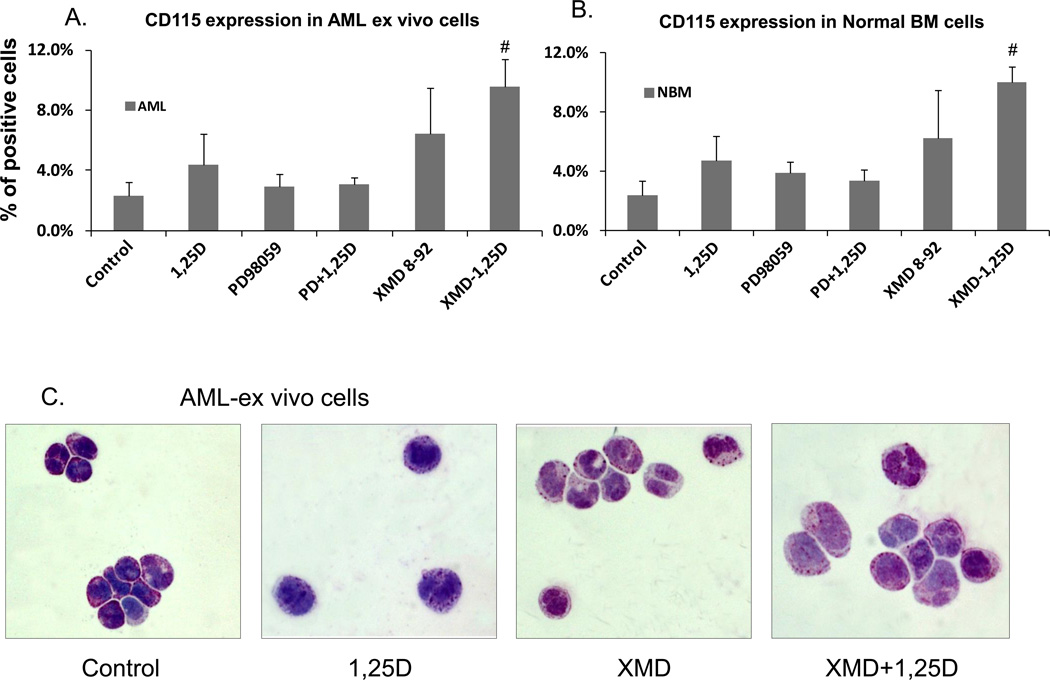
We also examined the effect of the addition of the potent ERK5 inhibitor XMD8-92 on cells obtained from the bone marrow of patients without hematologic disease (“Normal BM”), and patients with AML. The accrual of these specimens was limited by patient availability during these studies. The AML and normal human BM cells responded to 1,25D, XMD8-92, and to their combination in a manner similar to AML cells in established culture. Namely, 1,25D increased surface expression of the monocytic marker CD14 more than of CD11b, and, as previously reported [27, 28], this ratio was reversed by an exposure to the ERK5 inhibitor XMD8-92, but not to the ERK1/2 inhibitor PD98059 (data not shown). Importantly, XMD8-92, but not PD98059, enhanced the expression of M-CSFR (CD115) in both AML cells ex vivo, and in normal BM cells (Fig. 6A and B).These data imply that ERK5 plays a role in normal human as well as neoplastic hematopoiesis by regulating the expression of M-CSFR. The examination of morphological changes induced in the experiments shown in Fig. 5B confirmed the macrophage-like morphology of AML cells exposed to XMD8-92+1,25D (Fig. 6C).
Fig. 6. Macrophage-like phenotype induced by inhibition of ERK5 activity in human AML cells ex vivo.
(A) Flow cytometric determination of surface expression of M-CSFR of AML cells in primary culture. Mononuclear cells were separated from the total cells in specimens of bone marrow, and then subjected to the same treatment as AML cell lines. The expression of M-CSFR (CD115) in primary cultures was determined by flow cytometry. (B) Normal bone marrow cells in primary culture were treated as described for AML cells. # = p<0.05, significantly increased vs. 1,25D-treated group, n=3. (C) Effect of MEK5/ERK5 inhibitors on cell morphology of human AML cells ex vivo. The cells were stained and photographed similarly to those shown in Fig. 2A and B. The cells shown here were from the FAB subtype M2 sample.
AML cells in which the ERK5 pathway is inhibited show macrophage function
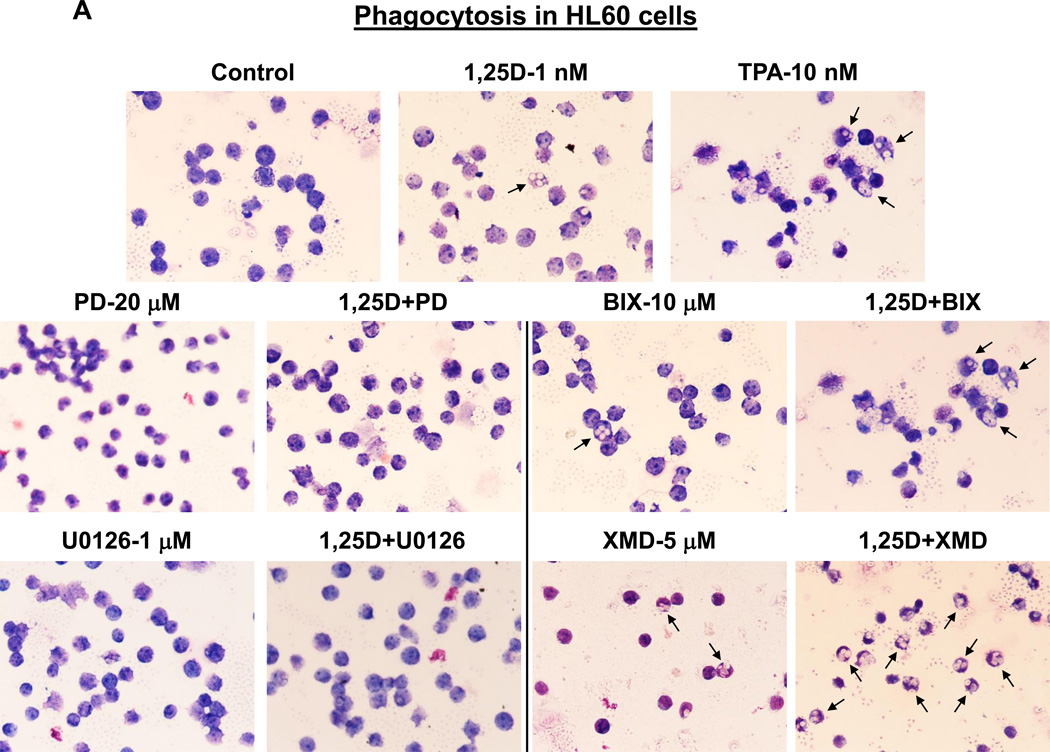
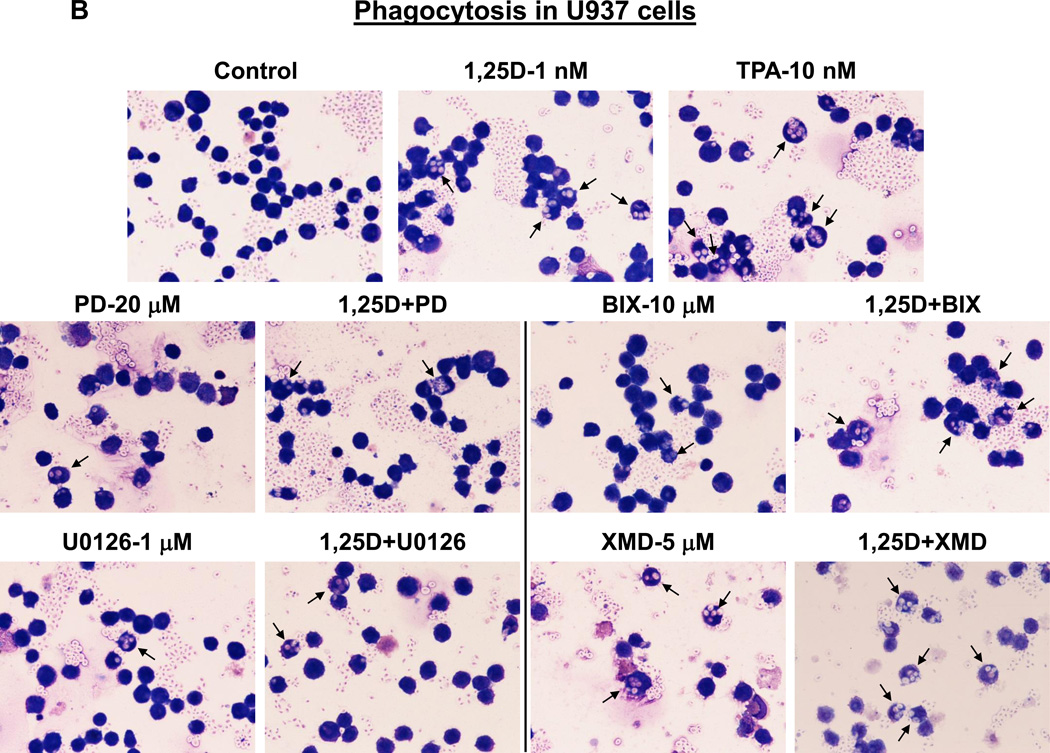
One of the principal functions of macrophages is phagocytosis, and this function is shared with only a few other cell types, such as granulocytes and dendritic cells (e.g., [47, 48]). To further confirm that inhibition of ERK5, but not of ERK1/2, kinase activity confers macrophage characteristics, we determined the ability of HL60 and U937 cells to phagocytose opsonized zymosan particles in response to treatment with 1,25D and kinase inhibitors, alone and in combination. The results illustrated in Fig. 6 and summarized in Table 1 indicate that untreated control HL60 and U937 cells had a marginal phagocytic activity which was moderately but significantly (p< 0.01) increased by 1 nM 1,25D in both cell lines. The ability of cells to phagocytose was minimally influenced by kinase inhibitors alone, except for XMD8-92, whose effect was comparable with that of 1,25D. Interestingly, combined treatment with 1,25D and ERK1/2 inhibitors ether did not significantly affect 1,25D-induced phagocytic activity (in U937 cells) or even reduced it (in HL60 cells). On the other hand, ERK5 inhibitors strongly cooperated with 1,25D to stimulate phagocytosis in both cell lines, XMD8-92 being the more effective, synergistically acting potentiating agent (p< 0.001 in U937 cells; p< 0.0001 in HL60 cells). Importantly, the effects of the 1,25D+XMD8-92 combination were superior to those induced by 10 nM TPA, the known inducer of macrophage-like phenotype [44, 49]. Thus, taken together, our results provide evidence that the reason for the well documented action of the near-physiological concentrations of 1,25D to induce differentiation of AML cells at monocyte, rather than macrophage stage [50, 51], is the activity of ERK5 upregulated by 1,25D [26], which retards the completion of the maturation process.
Table 1. Quantitation of phagocytic activity following induction of macrophage phenotype.
Following treatments and phagocytosis assay (illustrated in Fig. 6, A and B) at least 500 cells were counted on each slide. The percentage of cells containing two or more phagocytized particles of opsonized zymosan was calculated and expressed as the mean ± S.D. (n=3).
| Level of significance | ||||
|---|---|---|---|---|
| Phagocytosis (%) | vs. Control | vs. 1,25D alone | vs. Inhibitor alone | |
| HL60 cells | ||||
| Control | 0.2 ± 0.4 | |||
| D3-1 nM | 6.1 ± 2.2 | ** | ||
| PD-20 µM | 0.9 ± 0.1 | * | ||
| D3+PD | 2.8 ± 0.6 | ** | ns | ** |
| U0126-1 µM | 0.6 ± 0.6 | ns | ||
| D3+U0126 | 4.9 ± 2.2 | * | ns | * |
| BIX-10 µM | 1.7 ± 0.8 | * | ||
| D3+BIX | 10.2 ± 1.4 | *** | ns | *** |
| XMD-5 µM | 6.2 ± 1.4 | ** | ||
| D3+XMD | 48.0 ± 3.8 | **** | **** | **** |
| TPA 10 nM | 22.0 ± 4.9 | ** | ||
| U937 cells | ||||
| Control | 0.8 ± 1.0 | |||
| D3-1 nM | 6.0 ± 1.1 | ** | ||
| PD-20 µM | 2.6 ± 1.0 | ns | ||
| D3+PD | 6.8 ± 2.8 | * | ns | ns |
| U0126-1 µM | 1.0 ± 0.8 | ns | ||
| D3+U0126 | 6.5 ± 2.8 | * | ns | ** |
| BIX-10 µM | 1.4 ± 0.6 | ns | ||
| D3+BIX | 9.2 ± 1.8 | *** | * | *** |
| XMD-5 µM | 4.4 ± 0.7 | * | ||
| D3+XMD | 22.0 ± 4.8 | *** | *** | ** |
| TPA 10 nM | 14.5 ± 3.5 | *** | ||
= p<0.05;
= p<0.01;
= p<0.001;
= p<0.0001.
DISCUSSION
One of the fundamental questions in the biology of neoplasia is how differentiating cells restrain the unnecessarily rapid progression of the successive steps in this process. This restraint, although necessary and beneficial under normal conditions, when due to mutations or epigenetic errors can lead to inappropriately arrested, immature cells, and thus to unrestrained proliferation. In this study we elucidated several novel aspects of this process. Most importantly, we found that two seemingly parallel signaling pathways, while overlapping to some extent, can also antagonize each other’s functions. Specifically, we demonstrate that ERK5 retards the progression of differentiation of macrophage precursors that can be driven by ERK1/2 [52, 53].
Another intriguing finding is that treatment with MEK5 inhibitor BIX02189 can produce a compensatory increase in ERK1/2 activation. This is evident in HL60 cells, and although some of the increases did not reach statistical significance, BIX02189 clearly enhanced the activation of 1,25D-treated cells (Fig. 1A). Perhaps due to the more advanced maturation state of U937 cells, this was not observed in these cells. However, the reverse situation, the compensatory increases in ERK5 levels following treatment with ERK1/2 inhibitors, was seen in both cell types treated with the inhibitor of ERK5 autophosphorylation XMD8-92 and 1,25D, and was previously noted to occur in HeLa cells in which the ERK1/2 pathway stimulated by EGF was inhibited by PD184352 [42]. We therefore extend the previous finding to show that ERK1/2 and ERK5 pathways can provide a negative feedback to each other.
The conclusions that we have reached regarding the effects of ERK5 inhibition by BIX02189 and XMD8-92 can be considered well founded, since they are based on the use two inhibitors of the ERK5 pathway with entirely different mode of ERK5 inhibition - BIX02189 inhibits MEK5 phosphorylation and thus the activation of ERK5, while XMD8-92 inhibits ERK5 auto-phosphorylation. Since both have essentially the same effects on M-CSFR expression in 1,25D-treated cells, using two cell lines and examining M-CSFR expression at both transcriptional (Fig. 3) and protein (Fig. 4) levels, we felt confident that this is a specific effect on ERK5 activity. This conclusion was reinforced by the absence of such effects of two ERK1/2 inhibitors, PD988059 and U0126 (Figs. 3 and 4). This is important, as in many cell systems ERK5 and ERK1/2 have overlapping effects, yet Figs. 3 and 4 show starkly contrasting effects of their inhibitors on M-CSFR expression. Further, the results obtained with these pharmacological inhibitors were confirmed by the knock-down of M-CSFR expression which also resulted in inhibited macrophage-like differentiation (Fig. 4B). Further, our findings in AML cells in primary culture support the view that our observations are not due to artifacts of prolonged culture, but reflect to a large extent the situation in vivo.
However, some caveats need to be mentioned. Detailed immunophenotyping is unlikely to help to identify the monocyte-macrophage phenotype, as various subtypes of these cells have been described, with different functions and immunophenotypes (e.g., [54–57]). Therefore, our conclusion that treatment of AML cells with ERK5 inhibitors combined with 1,25D results in macrophage-like cells is based on cell morphology, induction of M-CSFR, and most importantly, on vigorous phagocytosis by these cells. The latter finding is supported by previous studies which demonstrated that increased levels of M-CSFR results in enhanced phagocytosis (e.g., [58, 59]), while downregulation of M-CSFR leads to impaired phagocytosis (e.g., [60, 61]) by tissue macrophages. The similarity in all these respects to TPA-treated THP-1 cells, generally accepted to exhibit macrophage phenotype [62] provides supporting evidence for this conclusion. To date, our experiments do not show if the ERK5 inhibitors act directly on M-CSFR expression, or are mediated by some intermediaries, which are still unknown. In any case, ERK5 inhibition requires M-CSFR for optimal induction of macrophage phenotype, as shown by the knock down of M-CSFR expression (Fig. 5A and B).
Some differences in the effects of BIX02189 and XMD8-82 are to be expected, as the former inhibits activating phosphorylation of ERK5 by MEK5, and the latter inhibits the auto phosphorylation of activated ERK5. Thus, the more pronounced effects of XMD8-82 are likely due to its more direct effect on the ERK5 pathway, as auto-phosphorylated ERK5 enters the cell nucleus, and is thus closer to the interacting transcription factors that transmit the ERK5 pathway signals to the gene promoters (e.g., [63]). Therefore, the main significance of this study is that the recognition of events altered by these pharmacological agents should allow them to be included in appropriate modifications of regimens currently not totally effective in the treatment of AML, and perhaps other neoplastic diseases.
Supplementary Material
Fig. 7. MEK5/ERK5 inhibitors, but not MEK1/2-ERK1/2 inhibitors, promote phagocytic activity of AML cells.
(A) HL60 cells were pretreated with either MEK5/ERK5 inhibitors, BIX02189 (10 µM) and XMD8-92 (5 µM), or MEK1/2 inhibitors, PD98059 (20 µM) and U0126 (1 µM) for 1 h, then 1,25D (1 nM) was added for an additional 96 h. Cells (5 × 105) were then incubated with opsonized zymosan, smeared on glass slides and stained with Wright-Giemsa. Phagocytosis was determined microscopically at 500× magnification. Arrows indicate examples of cells containing phagocytized zymosan particles. TPA (10 nM) was used here as the positive control for the induction of phagocytosis. (B) U937 cells treated as described above.
Highlights.
ERK5 has at least some functions in AML cells which are distinct from those of ERK1/2
ERK5 activity negatively controls the expression of M-CSFR
ERK5 retards the progression of differentiation from monocyte to functional macrophage
Acknowledgements
Supported by the NIH grant R01-CA 044722-23 from the National Cancer Institute to GPS, and by the Israel Science Foundation grant 635/11 to MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest to disclose.
References
- 1.Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA. FMS mutations in myelodysplastic, leukemic, and normal subjects. Proc Natl Acad Sci U S A. 1990;87:1377–1380. doi: 10.1073/pnas.87.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rademakers R, Baker M, Nicholson AM, Rutherford NJ, Finch N, Soto-Ortolaza A, Lash J, Wider C, Wojtas A, DeJesus-Hernandez M, Adamson J, Kouri N, Sundal C, Shuster EA, Aasly J, MacKenzie J, Roeber S, Kretzschmar HA, Boeve BF, Knopman DS, Petersen RC, Cairns NJ, Ghetti B, Spina S, Garbern J, Tselis AC, Uitti R, Das P, Van Gerpen JA, Meschia JF, Levy S, Broderick DF, Graff-Radford N, Ross OA, Miller BB, Swerdlow RH, Dickson DW, Wszolek ZK. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nature genetics. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreda DR, Hanington PC, Belosevic M. Regulation of myeloid development and function by colony stimulating factors. Developmental and comparative immunology. 2004;28:509–554. doi: 10.1016/j.dci.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton JA, Achuthan A. Colony stimulating factors and myeloid cell biology in health and disease. Trends in immunology. 2013;34:81–89. doi: 10.1016/j.it.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Broughton SE, Hercus TR, Lopez AF, Parker MW. Cytokine receptor activation at the cell surface. Current opinion in structural biology. 2012;22:350–359. doi: 10.1016/j.sbi.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Mouchemore KA, Pixley FJ. CSF-1 signaling in macrophages: pleiotrophy through phosphotyrosine-based signaling pathways. Crit Rev Clin Lab Sci. 2012;49:49–61. doi: 10.3109/10408363.2012.666845. [DOI] [PubMed] [Google Scholar]
- 7.Hercus TR, Broughton SE, Ekert PG, Ramshaw HS, Perugini M, Grimbaldeston M, Woodcock JM, Thomas D, Pitson S, Hughes T, D'Andrea RJ, Parker MW, Lopez AF. The GM-CSF receptor family: mechanism of activation and implications for disease. Growth factors. 2012;30:63–75. doi: 10.3109/08977194.2011.649919. [DOI] [PubMed] [Google Scholar]
- 8.Liongue C, Wright C, Russell AP, Ward AC. Granulocyte colony-stimulating factor receptor: stimulating granulopoiesis and much more. Int J Biochem Cell Biol. 2009;41:2372–2375. doi: 10.1016/j.biocel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Broughton SE, Dhagat U, Hercus TR, Nero TL, Grimbaldeston MA, Bonder CS, Lopez AF, Parker MW. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunological reviews. 2012;250:277–302. doi: 10.1111/j.1600-065X.2012.01164.x. [DOI] [PubMed] [Google Scholar]
- 10.Bourette RP, Rohrschneider LR. Early events in M-CSF receptor signaling. Growth factors. 2000;17:155–166. doi: 10.3109/08977190009001065. [DOI] [PubMed] [Google Scholar]
- 11.Suzu S, Hiyoshi M, Yoshidomi Y, Harada H, Takeya M, Kimura F, Motoyoshi K, Okada S. M-CSF-mediated macrophage differentiation but not proliferation is correlated with increased and prolonged ERK activation. J Cell Physiol. 2007;212:519–525. doi: 10.1002/jcp.21045. [DOI] [PubMed] [Google Scholar]
- 12.Douglass TG, Driggers L, Zhang JG, Hoa N, Delgado C, Williams CC, Dan Q, Sanchez R, Jeffes EW, Wepsic HT, Myers MP, Koths K, Jadus MR. Macrophage colony stimulating factor: not just for macrophages anymore! A gateway into complex biologies. International immunopharmacology. 2008;8:1354–1376. doi: 10.1016/j.intimp.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Jenkins B, Shin JL, Rohrschneider LR. Scaffolding protein Gab2 mediates differentiation signaling downstream of Fms receptor tyrosine kinase. Mol Cell Biol. 2001;21:3047–3056. doi: 10.1128/MCB.21.9.3047-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novak U, Harpur AG, Paradiso L, Kanagasundaram V, Jaworowski A, Wilks AF, Hamilton JA. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948–2956. [PubMed] [Google Scholar]
- 15.Balmanno K, Cook SJ. Tumour cell survival signalling by the ERK1/2 pathway. Cell Death Differ. 2009;16:368–377. doi: 10.1038/cdd.2008.148. [DOI] [PubMed] [Google Scholar]
- 16.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 17.Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO reports. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deschenes-Simard X, Kottakis F, Meloche S, Ferbeyre G. ERKs in cancer: friends or foes? Cancer Res. 2014;74:412–419. doi: 10.1158/0008-5472.CAN-13-2381. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Merritt AJ, Seyfried J, Guo C, Papadakis ES, Finegan KG, Kayahara M, Dixon J, Boot-Handford RP, Cartwright EJ, Mayer U, Tournier C. Targeted deletion of mek5 causes early embryonic death and defects in the extracellular signal-regulated kinase 5/myocyte enhancer factor 2 cell survival pathway. Mol Cell Biol. 2005;25:336–345. doi: 10.1128/MCB.25.1.336-345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obara Y, Yamauchi A, Takehara S, Nemoto W, Takahashi M, Stork PJ, Nakahata N. ERK5 activity is required for nerve growth factor-induced neurite outgrowth and stabilization of tyrosine hydroxylase in PC12 cells. J Biol Chem. 2009;284:23564–23573. doi: 10.1074/jbc.M109.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan YW, Zou J, Wang W, Sakagami H, Garelick MG, Abel G, Kuo CT, Storm DR, Xia Z. Inducible and conditional deletion of extracellular signal-regulated kinase 5 disrupts adult hippocampal neurogenesis. J Biol Chem. 2012;287:23306–23317. doi: 10.1074/jbc.M112.344762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Pan YW, Wietecha T, Zou J, Abel GM, Kuo CT, Xia Z. Extracellular signal-regulated kinase 5 (ERK5) mediates prolactin-stimulated adult neurogenesis in the subventricular zone and olfactory bulb. J Biol Chem. 2013;288:2623–2631. doi: 10.1074/jbc.M112.401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Pan YW, Zou J, Li T, Abel GM, Palmiter RD, Storm DR, Xia Z. Genetic activation of ERK5 MAP kinase enhances adult neurogenesis and extends hippocampus-dependent long-term memory. J Neurosci. 2014;34:2130–2147. doi: 10.1523/JNEUROSCI.3324-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rovida E, Spinelli E, Sdelci S, Barbetti V, Morandi A, Giuntoli S, Dello Sbarba P. ERK5/BMK1 is indispensable for optimal colony-stimulating factor 1 (CSF-1)-induced proliferation in macrophages in a Src-dependent fashion. J Immunol. 2008;180:4166–4172. doi: 10.4049/jimmunol.180.6.4166. [DOI] [PubMed] [Google Scholar]
- 25.Charni S, de Bettignies G, Rathore MG, Aguilo JI, van den Elsen PJ, Haouzi D, Hipskind RA, Enriquez JA, Sanchez-Beato M, Pardo J, Anel A, Villalba M. Oxidative phosphorylation induces de novo expression of the MHC class I in tumor cells through the ERK5 pathway. J Immunol. 2010;185:3498–3503. doi: 10.4049/jimmunol.1001250. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Gocek E, Novik V, Harrison JS, Danilenko M, Studzinski GP. Inhibition of Cot1/Tlp2 oncogene in AML cells reduces ERK5 activation and up-regulates p27Kip1 concomitant with enhancement of differentiation and cell cycle arrest induced by silibinin and 1,25-dihydroxyvitamin D3. Cell Cycle. 2010;9:4542–4551. doi: 10.4161/cc.9.22.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Pesakhov S, Harrison JS, Danilenko M, Studzinski GP. ERK5 pathway regulates transcription factors important for monocytic differentiation of human myeloid leukemia cells. J Cell Physiol. 2014;229:856–867. doi: 10.1002/jcp.24513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Pesakhov S, Weng A, Kafka M, Gocek E, Nguyen M, Harrison JS, Danilenko M, Studzinski GP. ERK 5/MAPK pathway has a major role in 1a,25-(OH)2 vitamin D-induced terminal differentiation of myeloid leukemia cells. J Steroid Biochem Mol Biol. 2013 doi: 10.1016/j.jsbmb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher R, Collins S, Trujillo J, McCredie K, Ahearn M, Tsai S, Metzgar R, Aulakh G, Ting R, Ruscetti F, Gallo R. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 30.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 31.Garner CM, Hubbold LM, Chakraborti PR. Mycoplasma detection in cell cultures: a comparison of four methods. British journal of biomedical science. 2000;57:295–301. [PubMed] [Google Scholar]
- 32.Zhang J, Harrison JS, Uskokovic M, Danilenko M, Studzinski GP. Silibinin can induce differentiation as well as enhance vitamin D3-induced differentiation of human AML cells ex vivo and regulates the levels of differentiation-related transcription factors. Hematol Oncol. 2010;28:124–132. doi: 10.1002/hon.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liel Y, Rudich A, Nagauker-Shriker O, Yermiyahu T, Levy R. Monocyte dysfunction in patients with Gaucher disease: evidence for interference of glucocerebroside with superoxide generation. Blood. 1994;83:2646–2653. [PubMed] [Google Scholar]
- 34.Kim KB, Kefford R, Pavlick AC, Infante JR, Ribas A, Sosman JA, Fecher LA, Millward M, McArthur GA, Hwu P, Gonzalez R, Ott PA, Long GV, Gardner OS, Ouellet D, Xu Y, DeMarini DJ, Le NT, Patel K, Lewis KD. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J Clin Oncol. 2013;31:482–489. doi: 10.1200/JCO.2012.43.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain N, Curran E, Iyengar NM, Diaz-Flores E, Kunnavakkam R, Popplewell L, Kirschbaum MH, Karrison T, Erba HP, Green M, Poire X, Koval G, Shannon K, Reddy PL, Joseph L, Atallah EL, Dy P, Thomas SP, Smith SE, Doyle LA, Stadler WM, Larson RA, Stock W, Odenike O. Phase II study of the oral MEK inhibitor selumetinib in advanced acute myelogenous leukemia: a University of Chicago phase II consortium trial. Clin Cancer Res. 2014;20:490–498. doi: 10.1158/1078-0432.CCR-13-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akinleye A, Furqan M, Mukhi N, Ravella P, Liu D. MEK and the inhibitors: from bench to bedside. Journal of hematology & oncology. 2013;6:27. doi: 10.1186/1756-8722-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daver N, Cortes J. Molecular targeted therapy in acute myeloid leukemia. Hematology. 2012;17(Suppl 1):S59–S62. doi: 10.1179/102453312X13336169155619. [DOI] [PubMed] [Google Scholar]
- 38.Bachegowda L, Gligich O, Mantzaris I, Schinke C, Wyville D, Carrillo T, Braunschweig I, Steidl U, Verma A. Signal transduction inhibitors in treatment of myelodysplastic syndromes. Journal of hematology & oncology. 2013;6:50. doi: 10.1186/1756-8722-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fremin C, Meloche S. From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. Journal of hematology & oncology. 2010;3:8. doi: 10.1186/1756-8722-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamakura S, Moriguchi T, Nishida E. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J Biol Chem. 1999;274:26563–26571. doi: 10.1074/jbc.274.37.26563. [DOI] [PubMed] [Google Scholar]
- 41.Razumovskaya E, Sun J, Ronnstrand L. Inhibition of MEK5 by BIX02188 induces apoptosis in cells expressing the oncogenic mutant FLT3-ITD. Biochem Biophys Res Commun. 2011;412:307–312. doi: 10.1016/j.bbrc.2011.07.089. [DOI] [PubMed] [Google Scholar]
- 42.Mody N, Leitch J, Armstrong C, Dixon J, Cohen P. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 43.Mazzone A, Ricevuti G. Leukocyte CD11/CD18 integrins: biological and clinical relevance. Haematologica. 1995;80:161–175. [PubMed] [Google Scholar]
- 44.Rovera G, O'Brien TG, Diamond L. Induction of differentiation in human promyelocytic leukemia cells by tumor promoters. Science. 1979;204:868–870. doi: 10.1126/science.286421. [DOI] [PubMed] [Google Scholar]
- 45.Gutsch R, Kandemir JD, Pietsch D, Cappello C, Meyer J, Simanowski K, Huber R, Brand K. CCAAT/enhancer-binding protein beta inhibits proliferation in monocytic cells by affecting the retinoblastoma protein/E2F/cyclin E pathway but is not directly required for macrophage morphology. J Biol Chem. 2011;286:22716–22729. doi: 10.1074/jbc.M110.152538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hass R, Prudovsky I, Kruhoffer M. Differential effects of phorbol ester on signaling and gene expression in human leukemia cells. Leuk Res. 1997;21:589–594. doi: 10.1016/s0145-2126(97)00010-6. [DOI] [PubMed] [Google Scholar]
- 47.Nagl M, Kacani L, Mullauer B, Lemberger EM, Stoiber H, Sprinzl GM, Schennach H, Dierich MP. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clinical and diagnostic laboratory immunology. 2002;9:1165–1168. doi: 10.1128/CDLI.9.6.1165-1168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fossati-Jimack L, Ling GS, Cortini A, Szajna M, Malik TH, McDonald JU, Pickering MC, Cook HT, Taylor PR, Botto M. Phagocytosis is the main CR3-mediated function affected by the lupus-associated variant of CD11b in human myeloid cells. PLoS One. 2013;8:e57082. doi: 10.1371/journal.pone.0057082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwende H, Fitzke E, Ambs P, Dieter P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J Leukoc Biol. 1996;59:555–561. [PubMed] [Google Scholar]
- 50.Collins SJ. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood. 1987;70:1233–1244. [PubMed] [Google Scholar]
- 51.Studzinski GP, Bhandal AK, Brelvi ZS. A system for monocytic differentiation of leukemic cells HL 60 by a short exposure to 1,25-dihydroxycholecalciferol. Proc Soc Exp Biol Med. 1985;179:288–295. doi: 10.3181/00379727-179-42098. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Studzinski GP. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvitamin D3- induced differentiation of HL60 cells. J Cell Biochem. 2001;80:471–482. doi: 10.1002/1097-4644(20010315)80:4<471::aid-jcb1001>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 53.Marcinkowska E. Evidence that activation of MEK1,2/erk1,2 signal transduction pathway is necessary for calcitriol-induced differentiation of HL-60 cells. Anticancer Res. 2001;21:499–504. [PubMed] [Google Scholar]
- 54.Grieser C, Heine G, Stelter L, Steffen IG, Rothe JH, Walter TC, Fischer C, Bahra M, Denecke T. Morphological analysis and differentiation of benign cystic neoplasms of the pancreas using computed tomography and magnetic resonance imaging. RoFo : Fortschritte auf dem Gebiete der Rontgenstrahlen und der Nuklearmedizin. 2013;185:219–227. doi: 10.1055/s-0032-1325551. [DOI] [PubMed] [Google Scholar]
- 55.Grage-Griebenow E, Lorenzen D, Fetting R, Flad HD, Ernst M. Phenotypical and functional characterization of Fc gamma receptor I (CD64)-negative monocytes, a minor human monocyte subpopulation with high accessory and antiviral activity. European journal of immunology. 1993;23:3126–3135. doi: 10.1002/eji.1830231213. [DOI] [PubMed] [Google Scholar]
- 56.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews. Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon S. The macrophage: past, present and future. European journal of immunology. 2007;37(Suppl 1):S9–S17. doi: 10.1002/eji.200737638. [DOI] [PubMed] [Google Scholar]
- 58.Mitrasinovic OM, Murphy GM., Jr Accelerated phagocytosis of amyloidbeta by mouse and human microglia overexpressing the macrophage colony-stimulating factor receptor. J Biol Chem. 2002;277:29889–29896. doi: 10.1074/jbc.M200868200. [DOI] [PubMed] [Google Scholar]
- 59.Mitrasinovic OM, Murphy GM., Jr Microglial overexpression of the M-CSF receptor augments phagocytosis of opsonized Abeta. Neurobiology of aging. 2003;24:807–815. doi: 10.1016/s0197-4580(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 60.Hoebe EK, Le Large TY, Tarbouriech N, Oosterhoff D, De Gruijl TD, Middeldorp JM, Greijer AE. Epstein-Barr virus-encoded BARF1 protein is a decoy receptor for macrophage colony stimulating factor and interferes with macrophage differentiation and activation. Viral immunology. 2012;25:461–470. doi: 10.1089/vim.2012.0034. [DOI] [PubMed] [Google Scholar]
- 61.Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ, Sharma A, Kaplan D, Greaves DR, Wang Y, Simon DI. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112:4699–4711. doi: 10.1182/blood-2008-01-137018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erazo T, Moreno A, Ruiz-Babot G, Rodriguez-Asiain A, Morrice NA, Espadamala J, Bayascas JR, Gomez N, Lizcano JM. Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol Cell Biol. 2013;33:1671–1686. doi: 10.1128/MCB.01246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.