Abstract
Plasmodium falciparum, the causative agent of severe human malaria, employs antigenic variation to avoid host immunity. Antigenic variation is achieved by transcriptional switching amongst polymorphic var genes, enforced by epigenetic modification of chromatin. The histone-modifying ‘sirtuin’ enzymes PfSir2a and PfSir2b have been implicated in this process. Disparate patterns of var expression have been reported in patient isolates as well as in cultured strains. We examined var expression in three commonly used laboratory strains (3D7, NF54 and FCR-3) in parallel. NF54 parasites express significantly lower levels of var genes compared to 3D7, despite the fact that 3D7 was originally a clone of the NF54 strain. To investigate whether this was linked to the expression of sirtuins, genetic disruption of both sirtuins was attempted in all three strains. No dramatic changes in var gene expression occurred in NF54 or FCR-3 following PfSir2b disruption, contrasting with previous observations in 3D7. In 3D7, complementation of the PfSir2a genetic disruption resulted in a significant decrease in previously-elevated var gene expression levels, but with the continued expression of multiple var genes. Finally, rearranged chromosomes were observed in the 3D7 PfSir2a knockout line. Our results focus on the potential for parasite genetic background to contribute to sirtuin function in regulating virulence gene expression and suggest a potential role for sirtuins in maintaining genome integrity.
Introduction
Malaria caused by Plasmodium falciparum gives rise to widespread morbidity and approximately a million deaths each year. During its asexual replicative lifecycle, the parasite lives inside human erythrocytes and is spread between hosts via mosquito bite. The parasite can maximize transmission by avoiding the human immune system and sustaining chronic infections; consequently, P. falciparum has evolved a complex system of antigenic variation, allowing infections to persist for months or years.
Antigenic variation is best characterized for the major surface antigen expressed on infected erythrocytes, PfEMP1 [1–4]. This large transmembrane protein mediates adhesion to a variety of host endothelial molecules, sequestering infected cells out of the circulation to avoid splenic clearance. The exposed PfEMP1 protein is thus a target for host immunity [5] and a large family of variant ‘var’ genes encoding PfEMP1 has evolved. There are 63 var genes in the sequenced 3D7 strain, with the majority located sub-telomerically and a subset arranged in tandem arrays at chromosome-internal locations [6]. The sub-telomeric genes are divided into groups termed upsA and upsB according to similarities in upstream sequence and gene orientation. There is also a unique sub-telomeric ‘upsE’ gene. Chromosome-internal genes, meanwhile, belong to a separate ‘upsC’ group. Several studies have linked high expression of sub-telomeric var genes to severe forms of malaria, showing that the upsA and/or upsB genes are expressed in children with severe disease [7–11] and the upsE gene, in pregnancy related malaria [12].
The majority of var genes are silenced at any one time, with periodic switching of active gene(s). Although var genes appear to be expressed in a mutually exclusive fashion [13–16], a more relaxed transcriptional pattern has been described in the 3D7 parasite strain. 3D7 expresses several var genes at significant levels [17–20] and represents these simultaneously as protein on the cell surface [20], whereas NF54 shows tight allelic exclusion [21].
The mechanism that controls var silencing, activation and switching appears to be epigenetic because expression patterns are maintained semi-stably for multiple generations [22] and because var family silencing is disrupted by mutation of either of the two P. falciparum sirtuin genes [4,23]. Sirtuins are NAD+-dependent histone deacetylases that facilitate chromatin condensation and heterochromatic gene silencing in model systems. The larger of the two P. falciparum sirtuins, PfSir2b, remains biochemically uncharacterized but the smaller, PfSir2a, is a histone deacetylase with a role in chromatin condensation and var gene silencing [24,25]. Initial studies conducted in 3D7 showed that the loss of PfSir2a causes de-silencing of multiple var genes, particularly the sub-telomeric upsA, BA and E groups [23], while the loss of PfSir2b causes a modest loss of silencing amongst upsB genes [4].
Sirtuins in model organisms often act specifically at telomeres. The term ‘telomere position effect’ (TPE) refers to a gradient of sirtuin-dependent heterochromatic silencing that extends into chromosomes from telomeres in budding yeast [26]. The PfSir2a and PfSir2b mutant phenotypes suggest that TPE also exists in P. falciparum. Furthermore, PfSir2a binds to sub-telomeric chromatin [25] and may have a direct effect on telomere maintenance, since telomeres are dramatically elongated in the absence of PfSir2a [4]. Consistently, a mammalian sirtuin, SIRT1, has been shown to affect telomere maintenance in mouse cells [27], as well as having complex effects upon DNA repair and genomic integrity [28].
Despite accumulating evidence that the sirtuins affect both virulence gene expression and telomere maintenance in P. falciparum, some important questions remain unanswered. Firstly, TPE may not account for the entire mechanism of var gene silencing. Histone modification (according to genome-wide chromatin immunoprecipitation) changes evenly throughout sub-telomeres when PfSir2a is mutated [29], yet not every sub-telomeric var gene is activated and some chromosome-central genes are also affected [30]. Therefore, the present study investigated whether PfSir2a may affect gene expression via molecular mechanisms besides TPE. Secondly, we wished to establish whether all sirtuin-related phenotypes are conserved across strain backgrounds and to evaluate whether the transcription of different var gene sub-types may be differently controlled by PfSir2a and PfSir2b. Thirdly, we investigated the effect of complementing PfSir2a in the 3D7 mutant. Our results reveal significant strain-to-strain differences in Sir2 phenotypes in long-term culture-adapted parasite strains, and point to a plethora of roles for sirtuins in controlling virulence gene expression, telomere maintenance and chromosomal stability.
Materials and Methods
Parasites and parasite culture
3D7 parasite isolates were obtained from three different sources and clonal parasites were generated from bulk culture by limiting dilution. The three sources included the MR4 strain (clones 4,5,11,14,18), the HR1.2 strain (clones 2, 10) and strain KW (gift obtained from Dr. Kim Williamson, Loyola University) (clones G10 and G11). NF54 clones were generated and analyzed from the MR4 strain (clones 21 and 23). NF54 clones were compared with data from previously generated clones of NF54 (a3, c3, g6, b3) as processed in Frank et al. [31]. Clones from FCR-3 were generated as described in Janes et al. [11].
All P. falciparum lines were cultured in human O+ erythrocytes obtained from Research Blood Components (Boston, MA) at 4% haematocrit in RPMI 1640 supplemented with 0.5% albumax (Invitrogen) and 0.25% sodium bicarbonate, using standard procedures [32]. Donor consent was waived by the institutional review board due to deidentification of the samples. Cloning was achieved by limiting dilution [33] and var expression profiles were assessed as soon as possible after cloning (15–20 generations). ~2x108 ring-stage parasites were collected for all ring-stage RNA preparations: synchronized rings were produced by treatment with 5% sorbitol. These parasites were then kept in culture until schizonts were observed to collect late-stage RNA samples.
Plasmid construction and generation of knockout parasite lines
Knockout parasites were generated using PfSir2a and PfSir2b targeting plasmids previously used and described in [4,23]. An adapted schematic for generation of these lines is presented in S3 Fig. Transfection of the PfSir2a and PfSir2b targeting plasmids into the FCR-3 and NF54 backgrounds was carried out as previously described [34]. For complementation studies, a plasmid containing the coding sequence for PfSir2a (PF3D7_1328800) under 1.75kb of its own upstream sequence from 3D7 was constructed in the plasmid vector pLNENRGFP [35]. The PfSir2a coding sequence was amplified from 3D7 genomic DNA (Sir2aF/Sir2aR) and cloned into pLNENRGFP as an AvrII/AflII fragment. The construct was verified by sequencing. The PfSir2a upstream sequence was then amplified (Sir2proF/Sir2proR) and cloned into this plasmid as an Apa1/AvrII fragment, replacing the calmodulin promoter sequence. Primers are described in S7 Table. Transfection of the PfSir2a complementation plasmid was performed as above, with selection on 2.5mg/ml Blasticidin HCl (Sigma).
Southern blotting and pulsed field gel electrophoresis
Telomere Restriction Fragment Southern blots were performed as previously described [36]; briefly, genomic DNA was digested with AluI, DdeI, MboII and RsaI, and blotted using a probe amplified from the P. falciparum telomeric sequence. The median telomere length in each genome was measured using Telometric software. (Note that one replicate blot for 3D7, 3D7ΔSir2a and 3D7ΔSir2b was measured from Tonkin et al. [4].) Southern blots to confirm successful disruptions of the PfSir2a and PfSir2b genes were carried out as previously described [4,23], using the 5’ ends of the Sir2a and Sir2b genes respectively as probes. Acc1/Nde1 were used for digestion of FCR-3Δsir2a and NF54Δsir2a and ScaI/HpaII for FCR-3Δsir2b and NF54Δsir2b digestion. Southern blots to assess rearrangement or amplification of sub-telomeric repeat sequences were carried out by digesting genomic DNA with Taq1 and using the probes ‘TARE1’ and ‘ATS’ (kind gift from Dr. Ingrid Felger, Swiss Tropical Institute). Pulsed Field Gel Electrophoresis was carried out as previously described [37].
RT-PCR for var gene expression
cDNAs were prepared and assessed essentially as described in [38]. Briefly, RNA was extracted from parasites using Trizol Reagent (Invitrogen), purified on PureLink columns (Invitrogen), treated with DNaseI (Invitrogen) and then reverse-transcribed using the Superscript II reverse transcriptase kit (Invitrogen). Each cDNA was checked for gDNA contamination using PCR across the intron of the gene PFD1155w, as described by Frank et al. [31]. RT-PCR for var gene expression in the 3D7 and NF54 backgrounds was carried out using the primer set designed by Salanti et al. [12], with modifications as in [31,38]. Primers and PCR conditions for the var family in FCR-3 are previously described [39].
RT-PCR was carried out using ITAQ SYBR SUPERMIX (Bio-Rad) in an ABI Prism 7500 thermocycler. Conditions for 3D7 and NF54 cDNAs are described in [38]; for FCR-3, 20 μl reactions were carried out using Power-SYBR Green Master Mix at final primer concentrations of 0.05 μM-0.5 μM under the following PCR conditions: 50°C for 1 min, 95°C for 10 min, then 40 cycles of dissociation, annealing, and extension at 95°C for 15 sec, 52°C for 15 sec, and 60°C for 45 sec, respectively.
Slight variations in primer efficiency were accounted for by the method of Dzikowski et al. [38] with correction factors listed in S8 Table; RT-PCR was carried out in duplicate on gDNA and a ΔCt for each primer set was then calculated relative to a housekeeping gene: PF07_0073 (seryl-tRNA synthetase) for 3D7 or adenylosuccinate lyase (ASL, PFB0295w) for FCR-3. This ‘correction factor’ was applied to all experimental ΔCt values. The gDNA analysis also confirmed NF54 clones analysed here have a 3D7-like genome not an E5-like genome [44]. Relative Copy Number (RCN) were calculated as 2ΔCt, where ΔCt was relative to control genes. For var subset analysis between strains, control genes were PF07_0073 (seryl-tRNA synthetase) for 3D7/NF54 or adenylosuccinate lyase (ASL, PFB0295w) for FCR-3. For the individual var gene RCN of sirtuin knockout strains, the average Ct of the 3 control genes: PF07_0073 (seryl-tRNA synthetase), PFL0900c (arginyl-tRNA synthetase), PF13_0170 (glutaminyl-tRNA synthetase) was used for all strains.
RT-PCR for sirtuin expression
cDNAs were prepared from schizont-stage samples as described for ring-stage samples, and quantitative PCR was carried out using the primers listed in S7 Table for PfSir2a, PfSir2b and the control genes MAL13P1.148 (myosin) and PF07_0073 (seryl-tRNA synthetase). ΔΔCt analysis was carried out to yield the Relative Copy Number (RCN) of the sirtuin genes calculated as 2ΔCt, where ΔCt was relative to the average Ct of the 2 control genes. Experiments were carried out in both biological and technical duplicate.
Comparative genome hybridization
A custom 385 k NimbleGen array designed for the P. falciparum 3D7 reference genome (PlasmoDB, 2006) using the standard CGH probe design protocol [40] was used [41]. The array comprises 385,585 probes semi-tiled across the genome at a 4 bp interval spacing with a minimum probe length of 45 bp, and a maximum length of 85 bp. Labeling and hybridization was carried out according to the standard NimbleGen CGH protocol [40]. Genomic DNAs (gDNA) from the NF54 and the 3D7ΔPfSir2a parasite lines were co-hybridized to CGH microarrays with reference 3D7. DNA fragmentation, labeling, hybridization, washing and scanning were carried out using the standard NimbleGen CGH protocol at the Genomics Core Facility (University of Notre Dame, Notre Dame, IN). The microarrays were hybridized and washed in a NimbleGen Hybridization System 4 (NimbleGen Systems, Inc.). Images were acquired by using a The NimbleGen MS 200 Microarray Scanner (NimbleGen Systems, Inc.) at a 5 μm resolution. Probe intensity values were extracted from scanned images using NimbleScan extraction software (NimbleGen Systems, Inc.). The Cy3 and Cy5 signal intensities were normalized according to standard Nimblegen protocol (http://www.nimblegen.com/products/lit/cgh_userguide_v6p0.pdf). The normalized values were used for calculation of Log2ratio values and used for CNV detection. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [42] and are accessible through the GEO series accession number GSE61940 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61940).
CNV detection criteria
Each probe was blasted (NCBI BLAST 2.1.1, without low complexity filtering) against the 3D7 reference genome (PlasmoDB v5.4) and non-unique probes were discarded. A total of 383,333 probes were used for CNV analysis. Segmentation analysis for identification of CNV regions was performed with CGHSeg algorithm [43] using CGHweb (http://compbio.med.harvard.edu/CGHweb). As P. falciparum is a haploid organism, relatively low single value cutoffs of log2ratio of normalized Cy5/Cy3 values, 0.5 and -0.5, were used for the primary level identification of CNVs. Additionally for a region to be considered a CNV, the lists of candidate CNV regions were filtered using 2 different stringency criteria. 1) CNV regions were required to carry 10 or more probes; 2) the average log2ratio value of the normalized Cy5/Cy3 values of all the probes spanning a CNV region was required to be ≤ -0.8 to be considered a loss, or ≥0.8 to be considered a gain.
Results
Polarized expression and quantitative variation in expression of var gene subsets in common lab strains
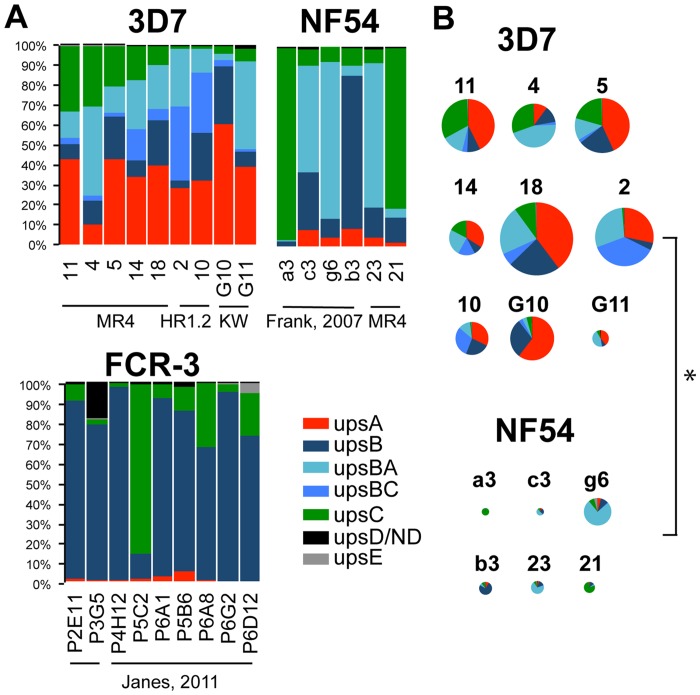
To understand the expression patterns of var subsets in different parasite lines, we obtained three common lab strains for comparison. Since var expression patterns are only semi-stable with periodic switching, all parasites were cloned by limiting dilution to obtain homogenous populations prior to assessment. For each strain, clones were obtained from lines that had been separately maintained for months or years. Nine 3D7 clones were obtained from three backgrounds, including five derived from the MR4 strain, two from ‘3D7 HR1.2’, a derivative of the MR4 strain with a drug-resistance gene integrated at a non-coding sub-telomeric locus [23], and two clones from ‘3D7 KW’, a derivative selected for the inability to produce gametocytes. Similarly, clones from different sources of NF54 and FCR-3 were compared. Two NF54 clones were freshly derived from the MR4 strain and compared to previous data obtained from independently-derived NF54 clones [31]. Data from nine FCR-3 clones, previously derived from two different backgrounds, were also included in the analysis [11].
Var transcription patterns in these three lab strains were compared via quantitative reverse-transcriptase PCR (q-RTPCR) using primers for almost every member of the fully-sequenced var gene family of the strains 3D7 and NF54 [12], and of FCR-3. This assessed the specific subsets of var genes expressed in each strain. Strikingly, the clones from the different strains expressed very different var subsets (Fig. 1A). Statistical analyses demonstrated that var subsets expressed by 3D7 were significantly different from both NF54 and FCR-3 (Fig. 1A, S1 Table and S2A-C Tables). 3D7 expressed predominantly upsA and upsB types (B, BC and BA)). In contrast, neither NF54 nor FCR-3 highly expressed upsA transcripts (S2B and S2C Tables).
Fig 1. Polarized expression of var genes subsets in different strains.
A: Analysis of var subset expression within clones of 3D7, NF54 and FCR-3 from disparate sources, demonstrating skews in var sub-class expression. 3D7 clones were derived from three different sources (MR4 3D7, 3D7 HR1.2 and 3D7 KW). NF54 clones were derived from the MR4 strain and var gene expression was compared to that previously obtained from clones of a distinct source of NF54 [31]. Var expression data for FCR-3 was obtained from clones previously examined from two parental parasite sources [11]. Var expression patterns are group representations that were determined by qRT-PCR using gene-specific primers for the entire var family in 3D7/NF54, normalized to seryl-tRNA-synthetase and sorted by ups-group. Var expression patterns were measured by qRT-PCR using gene-specific primers for the majority of the family in FCR-3, sorted by ups-group and normalized to the average abundance of the transcript of adenylosuccinate lyase. Independent clones are shown for each strain (different sources are denoted by underline). B: Relative copy numbers (RCN) for all of the genes within each 3D7 and NF54 strain were summed, and the proportions of expression of each subset within each genetic background are indicated. RCN was calculated by comparison to the abundance of transcripts encoding seryl-tRNA-synthetase (results were also similar if compared to the ring-stage-specific genes encoding SBP1 (PF3D7_0501300) and MAHRP (PF3D7_1370300)). Pie charts show proportions of each var transcript and the size of the chart is proportional to the total RCN. * denotes significant difference of p < 0.01.
The difference between the strains was particularly striking considering that 3D7 was originally considered to be a clone derived from the NF54 isolate, although the NF54 isolate has since been reported to contain two closely-related genotypes, 3D7 and ‘E5’ [44]. In fact, we found that in all cases, the total level of var transcription was about ten times higher in 3D7 than in NF54 (Fig. 1B). Furthermore, NF54 clones tended to express single dominant var transcripts, whereas all 3D7 clones, prepared identically and cultured for a similar number of cycles, expressed several transcripts (S1 Fig.).
Sirtuin expression is variable between P. falciparum strains
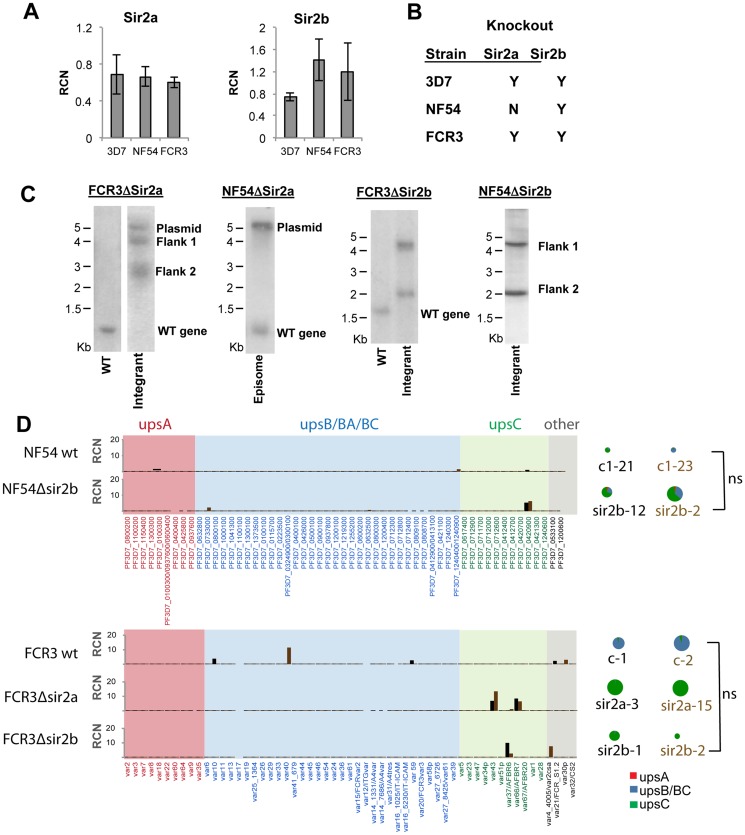
The only proteins known thus far to have a direct effect on var expression patterns are chromatin-modifying proteins, including heterochromatin protein 1 [45]; histone methyltransferases [46] and the histone deacetylases PfHda2 [47], PfSir2a and PfSir2b [4,23,30]. Focusing on the sirtuins, both PfSir2a and PfSir2b are encoded in the genomes of 3D7, NF54 and FCR-3 and their expression was therefore measured to see if variation at the transcriptional level might correlate with the observed differences in var expression. All three strains expressed both sirtuins, although the precise balance of expression levels varied, with a trend (albeit non-significant) toward lower expression of PfSir2b in 3D7 than in NF54 and FCR-3 (Fig. 2A).
Fig 2. Sirtuin expression and genetic targeting in NF54 and FCR-3.
A: Quantitative RT-PCR analysis of PfSir2a and PfSir2b expression in schizont-stage 3D7, NF54 and FCR-3. RCN is calculated by comparison to the average abundance of transcripts encoding two control genes: seryl-tRNA-synthetase and myosin. Plot shows average of 2 biological replicates, measured in technical duplicate. B: Table showing sirtuin gene disruptions attempted and achieved in 3D7, NF54 and FCR-3. C: Southern blots showing disruption of PfSir2b in NF54, PfSir2a and PfSir2b in FCR-3, and attempted but unsuccessful disruption of PfSir2a in NF54. The expected sizes for the FCR-3Δsir2a and NF54Δsir2a endogenous genomic locus, integrated flank 1, integrated flank 2 and concatamerized plasmid following AccI/NdeI digestion are 1.1 kb, 3.9 kb, 2.4 kb and 5.2 kb, respectively. The expected sizes for the FCR-3Δsir2b and NF54Δsir2b endogenous genomic locus, integrated flank 1, integrated flank 2 and concatamerized plasmid following ScaI/HpaII digestion are 1.7 kb, 4.2 kb, 2.1 kb and 4.5 kb. D: Var expression patterns in two clones each of NF54 wt, NF54Δsir2b, FCR-3 wt, FCR-3Δsir2a and FCR-3Δsir2b measured by qRT-PCR using gene-specific primers for the majority of the var gene family, sorted by ups-group. RCN was calculated by comparison to the abundance of transcripts encoding the average of the three control genes seryl-tRNA synthetase, arginyl-tRNA synthetase and glutaminyl-tRNA synthetase. Individual clones denoted in black and brown bars. Pie charts define percentage of each ups class expressed, and are sized proportional to the total RCN. ns = no significant difference between total var gene expression levels in the individual strains.
Disruption of PfSir2a in FCR-3 and disruption of PfSir2b in FCR-3 and NF54 does not alter var gene expression as dramatically as in 3D7
To investigate the var expression phenotypes caused by sirtuin knockouts in various strain backgrounds, gene disruption by homologous recombination was attempted for both PfSir2a and PfSir2b in 3D7, FCR-3 and NF54 (Fig. 2B and 2C, S2 Fig.). Both knockouts are previously published for 3D7 [4,23] (as with any gene disruption strategy, a low level of disrupted or unstable transcripts may still be produced in these lines, but Duraisingh et al. detected no stable full-length transcript by Northern blot, and previously-published immunofluorescence data has also shown the absence of Sir2a protein in this mutant line [48]). Both PfSir2a and b disruptions were successfully obtained in FCR-3 but it was only possible to disrupt PfSir2b in NF54; drug selection and cycling for over a year failed to disrupt PfSir2a in the NF54 strain (Fig. 2C), providing some evidence that it might be essential in this parasite genetic background.
The disruption of PfSir2a has previously been shown to cause extensive relaxation of var gene silencing in 3D7, with clonal parasites expressing a fixed set of ~10 var genes and also having an elevated total transcript level [30]. Here, by contrast, FCR-3ΔPfSir2a showed no significant change in overall transcript abundance or in the number of genes expressed in a single clone, albeit both clones switched to an upsC var transcript (Fig. 2D and S3A Table).
Previously, changes in var gene expression were also observed following disruption of PfSir2b in 3D7 [4]. The NF54 and FCR-3 PfSir2b knockouts were likewise assessed for var expression (Fig. 2D). No gross alterations in the dominant var expression patterns were observed (S3A and S3B Tables) although there were some subtle changes: some NF54ΔPfSir2b clones expressed more than one gene at detectable levels and there was also a slight increase in the level of total var transcripts compared to WT NF54, whereas FCR-3ΔPfSir2b showed a slight decrease in total transcripts. All of the Sir2b gene-disruption clones in NF54 and FCR-3 switched to predominantly upsC transcripts.
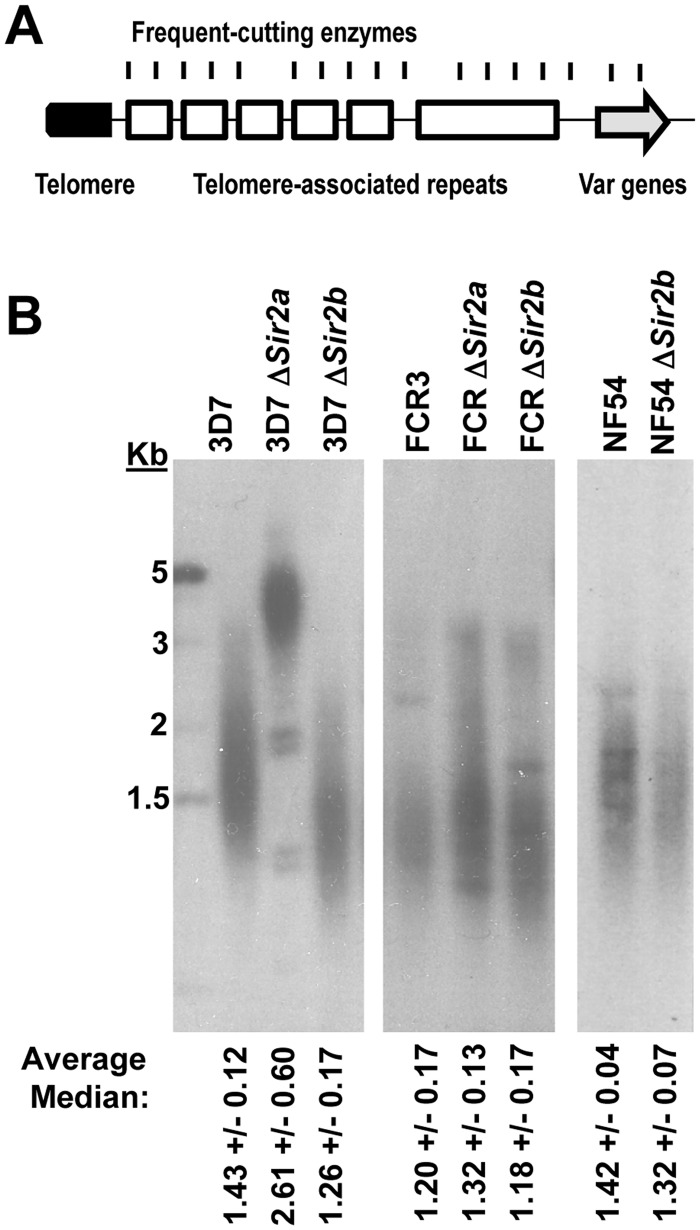
Telomere maintenance is affected by loss of either sirtuin
Telomere elongation has previously been reported in 3D7ΔPfSir2a [4], therefore, telomere lengths were examined in all sirtuin mutant strains using Telomere Restriction Fragment (TRF) Southern blotting (Fig. 3A). Comparing densitometry measurements from at least three replicate blots, 3D7ΔPfSir2a did indeed show highly elongated telomeres, as well as several distinct bands that may possibly represent telomeric repeats that have recombined into chromosome-internal locations and are now ‘fixed’, whereas FCR-3ΔPfSir2a showed only a slight elongation (Fig. 3B). The phenotypes of elongated telomeres was stable after many months of 3D7 growth in culture (S3A Fig.). In 3D7, loss of PfSir2b had the opposite effect to loss of PfSir2a, shortening the median telomere length by ~0.2 kb. A marginal telomere decrease (0.1 kb) was observed in NF54ΔPfSir2b. Although many of the telomeres in FCR-3ΔPfSir2b did clearly decrease in size, the bulk population remained unchanged due to the presence of a small but distinct population of long telomeres (Fig. 3B).
Fig 3. Telomere length is slightly altered in NF54 and FCR-3 sirtuin mutants.
A: Schematic showing structure of a P. falciparum telomere. The genome is digested with frequent-cutting restriction enzymes which do not cut within the telomere repeat to make a Telomere Restriction Fragment (TRF) Southern blot. B: TRF Southern blots of sirtuin-mutant lines compared to parental lines. Representative blots are shown and represent more than 3 experiments for each strain. The average median telomere lengths +/- S.D. from replicate blots are listed below each smear.
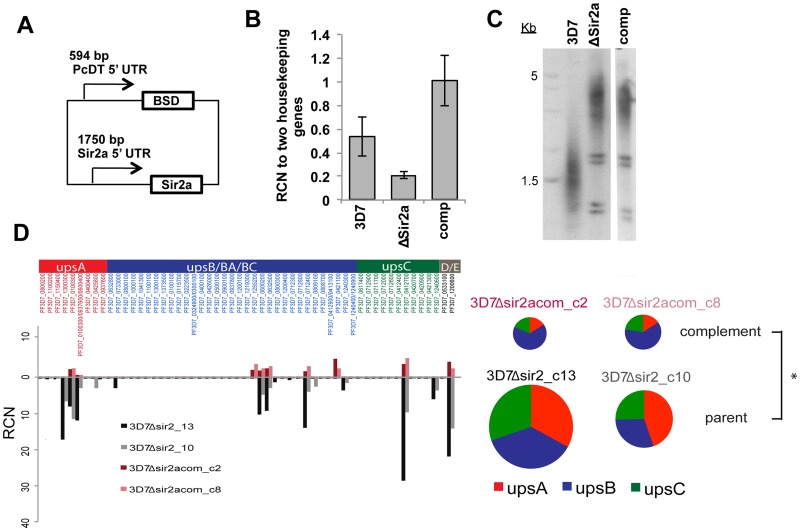
Complementation of 3D7ΔSir2a with wildtype Sir2a reverses the level of var gene expression
Since 3D7ΔPfSir2a showed the most dramatic phenotypes in both var expression and telomere length, complementation of this mutant was attempted. PfSir2a was cloned under the endogenous upstream sequence from the PfSir2a gene and transfected episomally into 3D7ΔPfSir2a (Fig. 4A). PfSir2a was expressed at about twice its original level in ‘3D7ΔPfSir2a _comp’, while only a fragment of the disrupted gene was expressed in the mutant, at about half of wildtype levels (likely due to low-level detection of disrupted transcripts in the mutant) (Fig. 4B & [23]). After more than 30 generations post-transfection, neither telomere elongation nor the deregulated pattern of var gene expression was reversed in 3D7ΔPfSir2a_comp (Fig. 4C and 4D) but the overall level of var transcription was significantly lowered (S3C Table). The total var transcript levels in the complemented knockout line were comparable to wildtype 3D7.
Fig 4. Complementation of 3D7ΔSir2a results in reduced var gene expression.
A: Schematic of PfSir2a complementation plasmid. B: PfSir2a expression levels measured by qRT-PCR in schizonts of 3D7, 3D7ΔSir2a and 3D7ΔSir2a_comp. RCN is calculated by comparison to the average abundance of transcripts encoding two control genes: seryl-tRNA-synthetase and myosin. Plot shows average of 2 biological replicates measured in technical duplicate. C: TRF Southern blot of 3D7, 3D7ΔSir2a and 3D7ΔSir2a_comp. D: Var expression patterns in 2 clones each of 3D7ΔSir2a (red, pink) and 3D7ΔSir2a_comp (black, grey), measured as in Fig. 2D. 3D7ΔSir2a_comp shows significant reduction in var expression levels as compared to 3D7ΔSir2a. Pie charts represent var expression by ups class. * denotes significant difference of p < 0.01.
Disruption of PfSir2a is associated with extensive genome rearrangement
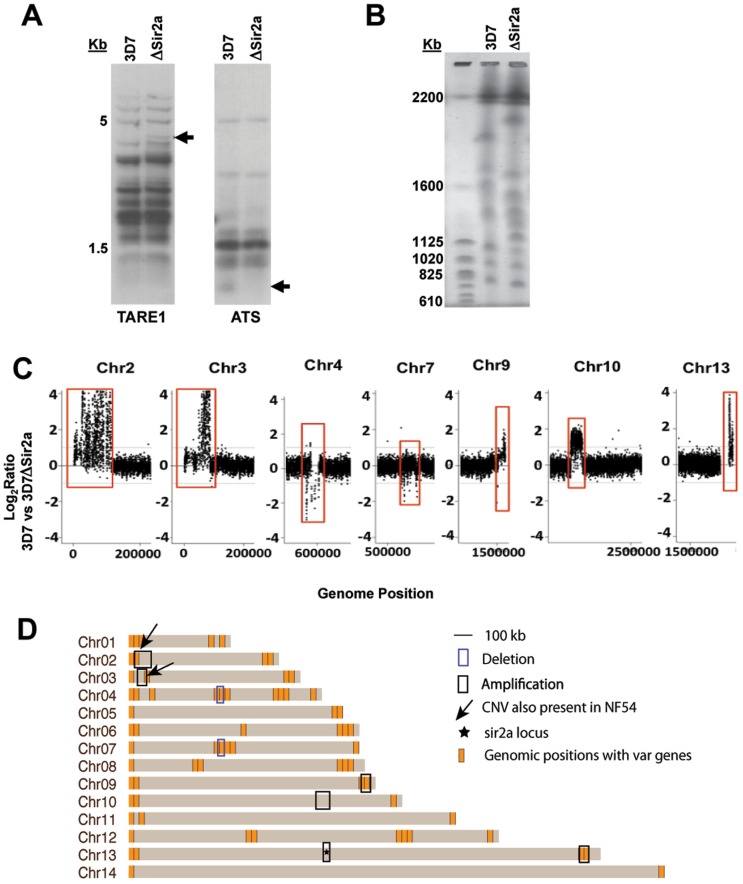
To further understand the genetic impact of gene disruption in 3D7ΔPfSir2a parasites, we examined chromosomal changes that occurred in the knockout strain. Since PfSir2a binds to sub-telomeric DNA [25], Southern blots were performed using genomic DNA from 3D7 and 3D7ΔPfSir2a to establish whether changes in sub-telomeric repeat sequences and/or sub-telomeric genes were observed, These Southern blots produced similar, although not identical, profiles in 3D7 and 3D7ΔPfSir2a for both the sub-telomeric repeat TARE1 and the conserved ‘ATS’ region of the var gene family (Fig. 5A). Pulsed field gel electrophoresis, however, revealed that the karyotype of 3D7ΔPfSir2a was extensively altered, with large changes in chromosome sizes encompassing tens or hundreds of kilobases: too large to be accounted for by ~2kb of extra telomere repeat sequence (Fig. 5B). The 3D7ΔPfSir2a line is karyotypically identical in a recent clone and in a bulk population (S3B Fig.) suggesting that these chromosomal changes may have occurred due to a major disruption following gene knockout, and were then fixed into a homogenous population of surviving parasites,
Fig 5. Dramatic chromosomal rearrangements in 3D7ΔSir2a gene disruption mutants.
A: Southern blots of genomic DNA from 3D7 and 3D7ΔSir2a, digested with Taq1 and probed for the first sub-telomeric repeat, TARE1, or the conserved ‘ATS’ region of var genes. Arrows indicate bands that differ between the two lines. B: Pulsed-field gel electrophoresis showing altered karyotype in 3D7ΔSir2a compared to 3D7. C: Comparative genomic hybridization plots of 3D7 and 3D7ΔSir2a. Only those chromosomes containing significant CNVs are shown. The log2ratio of Cy3/Cy5 value is plotted against chromosomal position. D: Chromosomal rearrangements in 3D7ΔSir2a detected by array CGH. The location of each var gene is shown in orange in the schematic chromosomes. The two locations that show CNV in NF54 are indicated with arrows. Deletions and amplifications are highlighted with boxes.
To further analyze the changes causing the altered karyotypes, comparative genomic hybridization (CGH) was carried out to pinpoint genetic differences between 3D7 and 3D7ΔPfSir2a that could account for their differing var transcription patterns. Although no major rearrangement or amplification of var-encoding regions had apparently occurred, CGH revealed widespread change throughout many chromosomes in 3D7ΔPfSir2a (Fig. 5C and S4 Table). Amplifications and deletions were observed on seven chromosomes. Interestingly, all of the changes were associated with var gene dense regions of the chromosome, except one on chromosome 10 (Fig. 5D and S5 Table). It should be noted that the PfSir2a knockout was created by a single-crossover insertion resulting in CGH detection as a genetic amplification, even though no protein product is produced.
CGH comparison was also carried out between 3D7 and NF54. Several copy number variations (CNVs) were detected, mostly as losses in 3D7 or amplifications in NF54. These were primarily in sub-telomeric regions, including the previously-noted chromosome 2 truncation which encompasses Knob-associated Histidine-Rich Protein and Erythrocyte Membrane Protein 3 and which is well-tolerated in many isolates [49] (S4 Fig. and S6 Table). The changes in chromosome 2 and 3 of both 3D7ΔPfSir2a and NF54 likely occur spontaneously, suggesting that these regions are unstable and therefore may not have a connection to the PfSir2a knockout. Additional changes detected in the 3D7 versus 3D7ΔPfSir2a analysis were, however, much more extensive than in NF54 versus 3D7, although the mutant strain had been separated from its parent for only a few months, compared to many years for NF54. No clear candidate emerged with a strong potential link to the regulation of var transcription but the roles of the genes affected by PfSir2a knockout will be pursued in further work.
Discussion
The expression and silencing of adhesin-encoding var genes is a key virulence factor in P. falciparum and the sirtuins were until very recently the only proteins reported to directly affect this process. This study evaluated and compared the var expression patterns and phenotypes of sirtuin mutants in three laboratory strains. Var expression patterns were found to be highly variable, both qualitatively and quantitatively, while the sirtuins had pleiotropic effects upon var gene expression, telomere maintenance and genome stability. The sensitivity of different strains to altered sirtuin levels, however, varied significantly with PfSir2a knockout in 3D7 having the most severe impact and NF54 and FCR-3 knockouts demonstrating a more minimal effect.
This side-by-side comparison of var gene expression in several laboratory strains helps to resolve what has been an ongoing conflict between reports of mutual exclusion and poly-genic var expression patterns. Both modes apparently exist in different strains; NF54 and FCR-3 show tighter mutually exclusive expression than 3D7, which transcribes several var genes quite abundantly at a similar age post-cloning. The concept that var expression is strictly mutually exclusive was initially derived from reverse transcription and cloning of var cDNAs from lines of FCR-3 selected for different adhesive phenotypes. This previous analysis detected only a single var gene per adhesive phenotype [22] but the proposed model has since been debated. In particular, single-cell RT-PCR experiments detected several var transcripts in single cells of FCR-3 [17,50]; while in 3D7, poly-allelic var expression has been demonstrated in single cells by quantitative PCR and RNA-FISH at the mRNA level, immunofluorescence at the protein level and cytoadherence assays at the phenotypic level [20]. Clones of 3D7 have, furthermore, been monitored for switching over many cycles, showing that the expression of multiple var genes is quite stable and hence not consistent simply with rapid switching [30] (although other models, such as very rapid on/off switching amongst largely the same subset of var genes, cannot be entirely excluded without further experimental validation). NF54 however, was independently reported to conform to the mutual exclusion model [21,31,51]. Since 3D7 was originally a clone of NF54, var expression patterns must have altered in one or both of these lines after lengthy in vitro culture. The original ex vivo phenotype is not recorded and it is not clear whether the 3D7 or NF54 type of allelic expression more closely represents that found in in vivo infections.
A second novel finding from this work is the dramatic disparity in var transcript levels between 3D7 and NF54. Again, it is not known whether either strain represents the ‘real’ level of var transcription in vivo, although it has been reported that var transcription falls rapidly after parasites are transferred to in vitro culture, suggesting that the higher level may be closer to reality [52]. In either case, the disparity suggests that a var-specific transcription factor or other regulators, such as chromatin proteins, may exist at very different levels in the two cultured strains. CGH revealed no obvious candidate at the genetic level, although there were several CNVs in these two strains. The majority of CNVs represented losses of sub-telomeric genes, which are probably dispensable in in vitro culture. The remaining genes require further investigation but exhibit no obvious link to var transcription.
Turning to the effects of sirtuins on var gene expression and genome stability, these could all stem mechanistically from histone deacetylase activity, although additional enzymatic targets or structural roles for sirtuins remain possible. Sirtuins in model organisms generate sub-telomeric heterochromatin via histone deacetylation, silencing sub-telomeric genes and probably also regulating access for telomerase. Telomere maintenance is, indeed, affected by sirtuin complexes in species from budding yeast to mammals, e.g. telomere shortening is seen after loss of human SIRT1 [27,53]. Thus, the relaxation of var gene silencing and the altered telomere lengths observed in sirtuin mutants could both be explained by altered acetylation and relaxation of sub-telomeric heterochromatin. (Telomere length between clones has recently been shown to vary in parasites grown under drug pressure and this may also have an influence on telomere maintenance [54].) Interestingly, truncated P. falciparum chromosomes which lack their sub-telomeric repeats also show abnormally elongated telomeres, perhaps because sirtuin binding is reduced on these chromosomes [36]. PfSir2a localizes to nucleoli as well as telomeres, where it was recently reported to act as a negative regulator of ribosomal RNA gene transcription in 3D7 [55]. Again, this effect was apparently due to altered histone acetylation upstream of rRNA genes.
The third phenotype of chromosomal rearrangement seen in 3D7ΔPfSir2a may result from a crisis in genome stability caused by altered sirtuin activity, although the exact timing of unfolding of this phenotype cannot be determined simply because genetic manipulation in P. falciparum is a very lengthy and unpredictable process. Again, such a ‘crisis’ could be due to altered histone acetylation, although other roles for PfSir2a in chromatin structure or the regulation of DNA replication remain possible. A human sirtuin, SirT6, targets histones in DNA sequences that are difficult to replicate, including telomeres, and the absence of SirT6 causes stochastic chromosome breaks and fusions [56]. This phenotype is significantly similar, although not identical, to that observed in 3D7ΔPfSir2a. If a genome ‘crisis’ did occur when 3D7ΔPfSir2a was generated, it must have been resolved, resulting in a stable karyotype that permits in vitro survival (and yet that cannot directly account for the dysregulated var gene expression phenotype simply in terms of missing or amplified genes). However, the failure to disrupt PfSir2a in NF54 suggests that this may be a rare and difficult event. Nevertheless, remarkable levels of genome variation are apparently tolerated in P. falciparum [57], certain large deletions are commonly observed in in vitro culture [49] and it is notable that the karyotypes of different strains, including 3D7 and FCR-3, are not identical ([58,59] and data not shown). Pressures to retain a particular karyotype may be greater in vivo than in vitro, yet it has been observed that other pathogens such as Candida glabrata also have surprisingly flexible genomes even in vivo, perhaps as an adaptive strategy [60].
All phenotypes relating to PfSir2a were seen more dramatically in 3D7 than in FCR-3, suggesting that 3D7 is significantly more sensitive to changes in PfSir2a activity. The difference could reflect a spectrum of sirtuin-sensitivity existing in the wild, or changes following lab adaptation. Indeed, a strong effect of genetic background is seen when SIRT1 is mutated in the mouse [28]. One explanation is that PfSir2a and PfSir2b may act more redundantly in FCR-3 than in 3D7; another is that putative PfSir2a-interacting factors may be transcribed at different levels, or may differ in their sirtuin-interacting domains. In S. cerevisiae, alterations in sirtuin partner proteins can certainly modulate function [61] but no such partners have yet been identified for P. falciparum sirtuins.
Regarding PfSir2b, its mutation caused distinct but related phenotypes to PfSir2a mutation and these phenotypes were similar in 3D7, NF54 and FCR-3. Firstly, telomeres were slightly shortened in all three strains, suggesting that the two sirtuins play opposing roles in controlling telomere maintenance. Secondly, in contrast with the dramatic effect on var transcription reported in 3D7ΔPfSir2a, there was no major change in dominant var transcripts, although the silencing of sub-telomeric vars was previously reported to be slightly altered in 3D7ΔPfSir2b [4], while the clones of FCR-3ΔPfSir2b and NF54ΔPfSir2b analyzed here had all switched to predominantly upsC var genes. PfSir2b remains uncharacterized as an enzyme but its possible roles include deacetylating a distinct set of histone residues to PfSir2a, acting as a structural chromatin element, or perhaps performing both functions in partial overlap with PfSir2a.
Finally, with respect to complementation of 3D7ΔPfSir2a, there was a dramatic reversion in the level of var gene expression, but mutually exclusive expression of var genes was not restored. This effect on var gene expression levels was also unlinked from effects on telomere length and genome integrity. The absence of full phenotypic reversion probably results from the extensive genome rearrangements seen in this line. The massive relaxation of var gene silencing in 3D7ΔPfSir2a may in fact be a complex result of altered TPE and altered histone modification together with altered chromatin context, explaining why sub-telomeric var genes are not uniformly turned on and also why the transcriptional pattern is not reversible. Alternatively, the complete ‘re-setting’ of epigenetic control over var transcription from a deregulated state may require passage through meiosis. Nevertheless, the striking quantitative reversal of var over-expression in the complemented line suggests that PfSir2a has a distinct—and reversible—effect upon the level of var transcription. It also argues against the hypothesis that another var gene regulator was deleted during chromosome rearrangement and that the phenotypes of the mutant are unrelated to PfSir2a itself, although the nature of this genetic experiment makes it impossible to completely exclude this. It is not clear why the elongated telomeres in 3D7ΔPfSir2a could not be re-shortened; however, ‘set-point’ control in telomere maintenance is a subject of ongoing investigation in model systems. For example, aberrantly shortened telomeres in telomerase-mutant human cells cannot be ‘reset’ by simply restoring telomerase [62]. Much remains to be discovered about telomere maintenance in P. falciparum, particularly since it may form a target for novel anti-malarial drugs.
In conclusion, var expression patterns in some of the most commonly-used laboratory strains are shown here to be highly variable, while the sirtuins have pleiotropic—and likewise variable—effects upon var expression, telomere maintenance and genome stability. It will now be important to characterize sirtuin-interacting proteins and other chromatin-modifying enzymes in order to better understand the mechanisms underlying antigenic variation in in vivo infections with this important human pathogen.
Supporting Information
Var gene expression was assessed using a 3D7-specific primer subset [12] for 3D7 and NF54 or an FCR-3-specific subset for the var family in FCR-3 [63], which contains similar, but not identical, numbers of upsA, B and C type var genes.
(PDF)
A: Strategy for single crossover disruption of PfSir2a locus adapted from Duraisingh et al., 2005. Probes used for Southern blot confirmation of integration are shown in red. Dashed lines represent plasmid sequence. A = AccI sites, N = NdeI sites. B: Strategy for single crossover disruption of PfSir2b locus adapted from Tonkin et al., 2009. Probes used for Southern blot confirmation of integration are shown in red. Dashed lines represent plasmid sequence. S = ScaI sites, H = HpaII sites.
(PDF)
A: TRF blots showing telomere lengths for parasites in the bulk population versus a recent clone for 3D7 WT and 3D7Δsir2a. B: PFGE showing karyotypes of the bulk population versus a recent clone for 3D7 WT and 3D7Δsir2a (markers = S. cerevisiae chromosomes).
(PDF)
Only those chromosomes containing significant CNVs are shown. The log2ratio of Cy3/Cy5 value is plotted against chromosomal position.
(PDF)
A: RCN values for var gene expression levels in 3D7. B: RCN values for var gene expression levels in NF54. C: RCN values for var gene expression levels in FCR-3.
(XLSX)
A: Results of non-parametric tests (unpaired Wilcoxon test) performed between the total var gene expression levels of 3D7 and NF54. B: Results of pairwise Chi-squared tests performed between the averaged expression levels of (upsA, upsB and upsC) between 3D7, NF54 and FCR-3. C: Results of pairwise Chi-squared tests of the differences in var gene expression patterns (composition of upsA, upsB and upsC) between clones of 3 strains.
(PDF)
A: Paired Wilcoxon tests were performed between the individual var gene expression levels of different FCR-3, FCR-3Δsir2a and FCR-3Δsir2b strains. No significant var gene expression differences were detected. B: Paired Wilcoxon tests were performed between the individual var gene expression levels of NF54 and NF54Δsir2b strains. No significant var gene expression differences were detected. C: Paired Wilcoxon tests were performed between the individual var gene expression levels of different strains. Significant reduction in var gene expression occurs between 3D7Δsir2a-comp and 3D7Δsir2a.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful to Dasein Howell and Joseph Smith (Seattle BioMed) for help with var gene expression analysis, Kim Williamson (Loyola University) for the 3D7KW strain, John Tan and the Genomics Core Facility at University of Notre Dame for technical support and Richard Pinapati (University of Notre Dame) for assistance with database submission.
Funding Statement
This work was supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases award to MTD, a Charles H. Hood Foundation fellowship to CJM, and by NIH Grant AI071121 to MTF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995; 82: 77–87. [DOI] [PubMed] [Google Scholar]
- 2. Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995; 82: 89–100. [DOI] [PubMed] [Google Scholar]
- 3. Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995; 82: 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009; 7: e84 10.1371/journal.pbio.1000084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bull PC, Marsh K. The role of antibodies to Plasmodium falciparum-infected-erythrocyte surface antigens in naturally acquired immunity to malaria. Trends Microbiol. 2002; 10: 55–58. [DOI] [PubMed] [Google Scholar]
- 6. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419: 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006; 150: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis. 2006; 193: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004; 199: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun. 2006; 74: 3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janes JH, Wang CP, Levin-Edens E, Vigan-Womas I, Guillotte M, Melcher M, et al. Investigating the host binding signature on the Plasmodium falciparum PfEMP1 protein family. PLoS Pathog. 2011; 7: e1002032 10.1371/journal.ppat.1002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003; 49: 179–191. [DOI] [PubMed] [Google Scholar]
- 13. Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, et al. Developmental selection of var gene expression in Plasmodium falciparum. Nature. 1998; 394: 392–395. [DOI] [PubMed] [Google Scholar]
- 14. Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. Embo J. 1998; 17: 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, et al. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007; 8: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006; 439: 1004–1008. [DOI] [PubMed] [Google Scholar]
- 17. Duffy MF, Brown GV, Basuki W, Krejany EO, Noviyanti R, Cowman AF, et al. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin sulphate A binding phenotype. Mol Microbiol. 2002; 43: 1285–1293. [DOI] [PubMed] [Google Scholar]
- 18. Mok BW, Ribacke U, Rasti N, Kironde F, Chen Q, Nilsson P, et al. Default Pathway of var2csa switching and translational repression in Plasmodium falciparum. PLoS One. 2008; 3: e1982 10.1371/journal.pone.0001982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mok BW, Ribacke U, Winter G, Yip BH, Tan CS, Fernandez V, et al. Comparative transcriptomal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. Mol Biochem Parasitol. 2007; 151: 184–192. [DOI] [PubMed] [Google Scholar]
- 20. Joergensen L, Bengtsson DC, Bengtsson A, Ronander E, Berger SS, Turner L, et al. Surface co-expression of two different PfEMP1 antigens on single plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathog.6: e1001083 10.1371/journal.ppat.1001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank M, Deitsch K. Activation, silencing and mutually exclusive expression within the var gene family of Plasmodium falciparum. Int J Parasitol. 2006; 36: 975–985. [DOI] [PubMed] [Google Scholar]
- 22. Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. Embo J. 1998; 17: 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, et al. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005; 121: 13–24. [DOI] [PubMed] [Google Scholar]
- 24. Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot Cell. 2007; 6: 2081–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005; 121: 25–36. [DOI] [PubMed] [Google Scholar]
- 26. Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990; 63: 751–762. [DOI] [PubMed] [Google Scholar]
- 27. Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010; 191: 1299–1313. 10.1083/jcb.201005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008; 14: 312–323. 10.1016/j.ccr.2008.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009; 5: 179–190. 10.1016/j.chom.2008.12.012 [DOI] [PubMed] [Google Scholar]
- 30. Merrick CJ, Dzikowski R, Imamura H, Chuang J, Deitsch K, Duraisingh MT. The effect of Plasmodium falciparum Sir2a histone deacetylase on clonal and longitudinal variation in expression of the var family of virulence genes. Int J Parasitol. 2010; 40: 35–43. 10.1016/j.ijpara.2009.06.012 [DOI] [PubMed] [Google Scholar]
- 31. Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol. 2007; 64: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trager W, Jenson JB. Cultivation of malarial parasites. Nature. 1978; 273: 621–622. [DOI] [PubMed] [Google Scholar]
- 33. Kirkman LA, Su XZ, Wellems TE. Plasmodium falciparum: isolation of large numbers of parasite clones from infected blood samples. Exp Parasitol. 1996; 83: 147–149. [DOI] [PubMed] [Google Scholar]
- 34. Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A. 1997; 94: 10931–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR Jr., et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat Methods. 2006; 3: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. Embo J. 2002; 21: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hernandez-Rivas R, Scherf A. Separation and mapping of chromosomes of parasitic protozoa. Mem Inst Oswaldo Cruz. 1997; 92: 815–819. [DOI] [PubMed] [Google Scholar]
- 38. Dzikowski R, Frank M, Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006; 2: e22 Epub 2006 Mar 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Viebig NK, Levin E, Dechavanne S, Rogerson SJ, Gysin J, Smith JD, et al. Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One. 2007; 2: e910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005; 44: 305–319. [DOI] [PubMed] [Google Scholar]
- 41. Tan JC, Patel JJ, Tan A, Blain JC, Albert TJ, Lobo NF, et al. Optimizing comparative genomic hybridization probes for genotyping and SNP detection in Plasmodium falciparum. Genomics. 2009; 93: 543–550. 10.1016/j.ygeno.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Picard F, Robin S, Lavielle M, Vaisse C, Daudin JJ. A statistical approach for array CGH data analysis. BMC Bioinformatics. 2005; 6: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frank M, Kirkman L, Costantini D, Sanyal S, Lavazec C, Templeton TJ, et al. Frequent recombination events generate diversity within the multi-copy variant antigen gene families of Plasmodium falciparum. Int J Parasitol. 2008; 38: 1099–1109. 10.1016/j.ijpara.2008.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brancucci NM, Bertschi NL, Zhu L, Niederwieser I, Chin WH, Wampfler R, et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe.16: 165–176. 10.1016/j.chom.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 46. Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013; 499: 223–227. 10.1038/nature12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coleman BI, Skillman KM, Jiang RH, Childs LM, Altenhofen LM, Ganter M, et al. A Plasmodium falciparum Histone Deacetylase Regulates Antigenic Variation and Gametocyte Conversion. Cell Host Microbe.16: 177–186. 10.1016/j.chom.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deshmukh AS, Srivastava S, Herrmann S, Gupta A, Mitra P, Gilberger TW, et al. The role of N-terminus of Plasmodium falciparum ORC1 in telomeric localization and var gene silencing. Nucleic Acids Res. 2012; 40: 5313–5331. 10.1093/nar/gks202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Biggs BA, Kemp DJ, Brown GV. Subtelomeric chromosome deletions in field isolates of Plasmodium falciparum and their relationship to loss of cytoadherence in vitro. Proc Natl Acad Sci U S A. 1989; 86: 2428–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fernandez V, Chen Q, Sundstrom A, Scherf A, Hagblom P, Wahlgren M. Mosaic-like transcription of var genes in single Plasmodium falciparum parasites. Mol Biochem Parasitol. 2002; 121: 195–203. [DOI] [PubMed] [Google Scholar]
- 51. Fastman Y, Noble R, Recker M, Dzikowski R. Erasing the epigenetic memory and beginning to switch—the onset of antigenic switching of var genes in Plasmodium falciparum. PLoS One.7: e34168 10.1371/journal.pone.0034168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peters JM, Fowler EV, Krause DR, Cheng Q, Gatton ML. Differential changes in Plasmodium falciparum var transcription during adaptation to culture. J Infect Dis. 2007; 195: 748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Narala SR, Allsopp RC, Wells TB, Zhang G, Prasad P, Coussens MJ, et al. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity. Mol Biol Cell. 2008; 19: 1210–1219. 10.1091/mbc.E07-09-0965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet.9: e1003293 10.1371/journal.pgen.1003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A. Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat Commun.4: 1530 10.1038/ncomms2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008; 452: 492–496. 10.1038/nature06736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009; 5: e1000644 10.1371/journal.ppat.1000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kemp DJ, Corcoran LM, Coppel RL, Stahl HD, Bianco AE, Brown GV, et al. Size variation in chromosomes from independent cultured isolates of Plasmodium falciparum. Nature. 1985; 315: 347–350. [DOI] [PubMed] [Google Scholar]
- 59. Corcoran LM, Forsyth KP, Bianco AE, Brown GV, Kemp DJ. Chromosome size polymorphisms in Plasmodium falciparum can involve deletions and are frequent in natural parasite populations. Cell. 1986; 44: 87–95. [DOI] [PubMed] [Google Scholar]
- 60. Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, Stenderup J, et al. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci U S A. 2009; 106: 2688–2693. 10.1073/pnas.0809793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005; 25: 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chiang YJ, Calado RT, Hathcock KS, Lansdorp PM, Young NS, Hodes RJ. Telomere length is inherited with resetting of the telomere set-point. Proc Natl Acad Sci U S A. 2010; 107: 10148–10153. 10.1073/pnas.0913125107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Viebig NK, Gamain B, Scheidig C, Lepolard C, Przyborski J, Lanzer M, et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 2005; 6: 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Var gene expression was assessed using a 3D7-specific primer subset [12] for 3D7 and NF54 or an FCR-3-specific subset for the var family in FCR-3 [63], which contains similar, but not identical, numbers of upsA, B and C type var genes.
(PDF)
A: Strategy for single crossover disruption of PfSir2a locus adapted from Duraisingh et al., 2005. Probes used for Southern blot confirmation of integration are shown in red. Dashed lines represent plasmid sequence. A = AccI sites, N = NdeI sites. B: Strategy for single crossover disruption of PfSir2b locus adapted from Tonkin et al., 2009. Probes used for Southern blot confirmation of integration are shown in red. Dashed lines represent plasmid sequence. S = ScaI sites, H = HpaII sites.
(PDF)
A: TRF blots showing telomere lengths for parasites in the bulk population versus a recent clone for 3D7 WT and 3D7Δsir2a. B: PFGE showing karyotypes of the bulk population versus a recent clone for 3D7 WT and 3D7Δsir2a (markers = S. cerevisiae chromosomes).
(PDF)
Only those chromosomes containing significant CNVs are shown. The log2ratio of Cy3/Cy5 value is plotted against chromosomal position.
(PDF)
A: RCN values for var gene expression levels in 3D7. B: RCN values for var gene expression levels in NF54. C: RCN values for var gene expression levels in FCR-3.
(XLSX)
A: Results of non-parametric tests (unpaired Wilcoxon test) performed between the total var gene expression levels of 3D7 and NF54. B: Results of pairwise Chi-squared tests performed between the averaged expression levels of (upsA, upsB and upsC) between 3D7, NF54 and FCR-3. C: Results of pairwise Chi-squared tests of the differences in var gene expression patterns (composition of upsA, upsB and upsC) between clones of 3 strains.
(PDF)
A: Paired Wilcoxon tests were performed between the individual var gene expression levels of different FCR-3, FCR-3Δsir2a and FCR-3Δsir2b strains. No significant var gene expression differences were detected. B: Paired Wilcoxon tests were performed between the individual var gene expression levels of NF54 and NF54Δsir2b strains. No significant var gene expression differences were detected. C: Paired Wilcoxon tests were performed between the individual var gene expression levels of different strains. Significant reduction in var gene expression occurs between 3D7Δsir2a-comp and 3D7Δsir2a.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)