SUMMARY
Cells continuously adjust their behavior in response to changing environmental conditions. Both intensity and duration of external signals are critical factors in determining what response is initiated. To understand how intracellular signaling networks process such multidimensional information, we studied the AtRGS1-mediated glucose response system of Arabidopsis. By combining experiments with mathematical modeling, we discovered a reciprocal dose and duration response relying on the orchestrated action of three kinases (AtWNK1, AtWNK8, AtWNK10) acting on distinct time scales and activation thresholds. Specifically, we find that high concentrations of D-glucose rapidly signal through AtWNK8 and AtWNK10, whereas low, sustained sugar concentration slowly activate the pathway through AtWNK1, allowing the cells to respond similarly to transient, high-intensity signals, and sustained low-intensity signals. This “dose-duration reciprocity” allows encoding of both the intensity and persistence of glucose as an important energy resource and signaling molecule.
INTRODUCTION
Cells constantly sense and respond to changes in their extracellular environment (Rué et al., 2012; Waltermann and Klipp, 2011). The heterogeneous and fluctuating nature of the micro-environment requires that cells possess complex signaling pathways to interpret and respond to a broad range of environmental conditions. Therefore, an important problem in cell biology is to understand the design principles of the intracellular signaling networks that allow cells to process multi-dimensional information, such as the amplitude and duration of external signals, and respond appropriately. There are numerous examples in which both signal amplitude and duration play critical roles in cellular decision making (Purvis and Lahav, 2013). These include: 1) The discovery that an incoherent feed-forward loop between three transcriptional regulators enhances the inflammatory response of immune cells during short-term bacterial infection, but dampens inflammation if the infection becomes protracted (Litvak et al., 2009). 2) The observation that both the intensity and duration of MAP kinase activity are critical factors in regulating the differentiation of thymocytes into mature T lymphocytes (Mariathasan et al., 2001). 3) The demonstration that in B cells, an acute spike of intracellular calcium induces activation of the transcriptional activator NFκB, whereas low sustained calcium levels signal through another transcriptional activator NFAT (nuclear factor of activated T-cells) (Dolmetsch et al., 1997). These examples clearly demonstrate the importance of both signal intensity and duration in determining cell fate. However, the regulatory mechanisms that allow cells to interpret and respond to these properties are for the most part unknown.
Plants also must be able to respond to continuously changing environmental conditions. For example, sugars produced from photosynthesis vary not only under regular night/day cycles but also in the presence of unpredictable weather conditions and other random changes in the light environment. Thus, the micro-environment experienced by plant cells contains sugar levels that are constantly fluctuating from micromolar to high millimolar concentrations over time scales that can range from minutes to hours (Deuschle et al., 2006). Therefore, it seems likely that the sugar response system of plants evolved mechanisms to filter out small transient fluctuations while at the same time responding to sustained low sugar concentrations. To investigate this possibility, we studied signaling through the D-glucose response pathway in the model plant Arabidopsis thaliana. Our initial investigation suggested that plants possess dose-duration reciprocity in sugar sensing that allows them to respond similarly to transient, high-intensity and sustained low-intensity sugar signals. Therefore, we set out to determine the regulatory mechanisms that underlie dose-duration reciprocity.
There are at least three main glucose-sensing and signaling pathways in Arabidopsis, including a plasma membrane heterotrimeric G-protein system pathway, a hexokinase (HXK) enzymatic activity independent pathway, and a glycolysis-dependent SnRK1/TOR pathway (Smeekens et al., 2010; Urano et al., 2012b). Here, we focused on the G protein mediated D-glucose pathway. Heterotrimeric G proteins regulate various physiological processes including growth and development, host defense, and hormonal responses in mammals, fungi and plants (Gilman, 1987; Moriyama et al., 2006). The Arabidopsis G protein senses and responds to sugar levels to evaluate available energy resources and to determine proper developmental strategies (Booker et al., 2010; Chen et al., 2003; Chen and Jones, 2004; Johnston et al., 2007; Phan, 2013; Urano et al., 2012b). Distinct from mammalian G proteins, which require ligand-bound G protein-coupled receptors (GPCRs), the Arabidopsis Gα (AtGPA1) readily exchanges GDP for GTP without a GPCR (Johnston et al., 2007; Urano et al., 2012a). Instead, the D-glucose-induced G protein activation is mediated primarily through endocytosis of the Regulator of G protein Signaling 1 (AtRGS1) (Urano et al., 2012b), a seven-transmembrane GTPase-activating protein (GAP) that keeps AtGPA1 in its inactive state (Chen et al., 2003; Johnston et al., 2007; Jones et al., 2011). Under D-glucose treatment, AtRGS1 and the heterotrimeric G-protein temporarily favor formation of the AtRGS1-AtGPA1 complex over the AtRGS1-heterotrimer complex (Johnston et al., 2007), resulting in the accumulation of free Gβγ dimers. The Gβγ heterodimer then recruits WNK (With No Lysine) kinases that phosphorylate AtRGS1 at its C-terminal region (amino acid 400–459). The phosphorylated AtRGS1 undergoes endocytosis, allowing AtGPA1 to self-activate and initiate, along with Gβγ dimers, the downstream signaling (Urano et al., 2012b). This effect of D-glucose on AtRGS1 endocytosis is stereospecific to D-glucose and is not an osmotic stress (Urano et al., 2012b).
By integrating experimental investigations and mathematical modeling, we determined that dose-duration reciprocity is achieved through the orchestrated action of three kinases (AtWNK1, AtWNK8, AtWNK10) acting on distinct time scales and activation thresholds. Specifically, we found that high concentrations of D-glucose rapidly signal through AtWNK8 and AtWNK10, whereas low, sustained sugar concentrations slowly activate the pathway through AtWNK1. Our results demonstrate how plant cells encode both the intensity and the temporal information of extracellular sugar concentrations to initiate appropriate responses.
RESULTS
Dose-duration reciprocity in AtRGS1 endocytosis
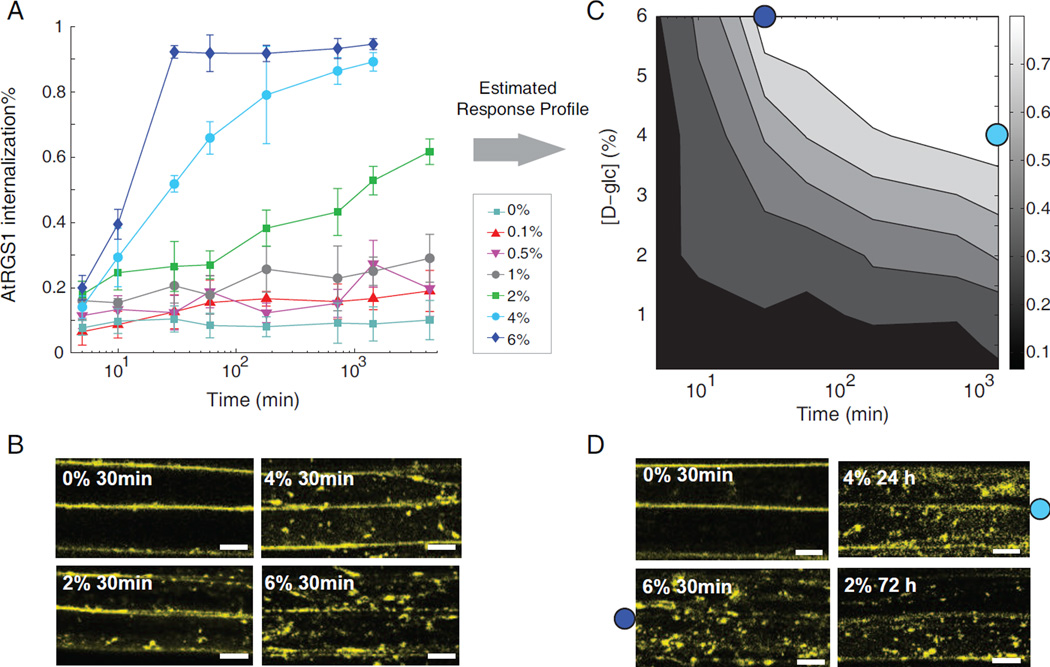
To investigate the relationship between sugar-induced dose response and duration response, the dynamic profiles of AtRGS1-YFP endocytosis in Arabidopsis hypocotyl epidermal cells were measured using confocal laser scanning microscopy (see Experimental Procedures). In terms of the maximum AtRGS1 internalization level, only 9% internalization was observed with treatment of water, whereas approximately 95% internalization was obtained under 6% D-glucose. Increasing D-glucose concentration increased the rate (Figure 1A) and the amount of internalization of AtRGS1 (Figure 1B).
Figure 1. The reciprocal dose-and-duration response in the AtRGS1 endocytosis.
(A) Transgenic AtRGS1-YFP Arabidopsis seedlings are treated with D-glucose at the indicated concentrations and monitored over time at the hypocotyl regions. Fluorescence was detected using a confocal laser scanning microscopy. Measurements of the internalized fluorescence percentage were used to evaluate AtRGS1 endocytosis, error bar = 2 × S.E.M (See Experimental Procedures) (B) Representative snap shots taken at 30 min for the AtRGS1 internalization with varying D-glucose (D-glc) concentrations. Scale bar = 10 μm (C) Experimentally determined 2D contour plot of AtRGS1 internalization as a function of D-glucose concentration and treatment time. (D) Dose-duration reciprocity. Snap shot of AtRGS1 endocytosis under D-glc concentration and time as indicated. (C, D) Typical images of AtRGS1 endocytosis shown in panel D at conditions shown in panel C are 6% D-glucose for 30 min (labeled as dark blue dot) and 4% D-glucose for 24 h (labeled as light blue dot). Scale bar = 10 μm.
The AtRGS1 internalization time courses reveal a reciprocal relationship between sugar dose and treatment duration required to reach maximum internalization. Specifically, the data demonstrate that near saturating internalization is achieved by either high or low doses of sugar, however drastically different exposure times are required to reach steady state. For example, 6% D-glucose induced a maximum steady-state internalization of AtRGS1 within 30 minutes of treatment, whereas with 4% D-glucose, a similar steady-state level was reached after 24 hours (Figure 1C and 1D). Treatment with 2% D-glucose led to over 60% of AtRGS1 being internalized within 3 days (Figure 1D) and this percentage increased to 70% after 4 days (Figure 2B). Note that treatment with 1% D-glucose for 1 d reached maximum internalization around 25%, and further extension of the sugar duration to 3 d did not significantly boost AtRGS1 internalization (30%) (p>0.1, student t-test), therefore it is likely that the dose-duration reciprocity operates above 1% of D-glucose. These data demonstrate millimolar levels are the minimum concentration of D-glucose required for pathway activation (1% D-glucose=55mM).
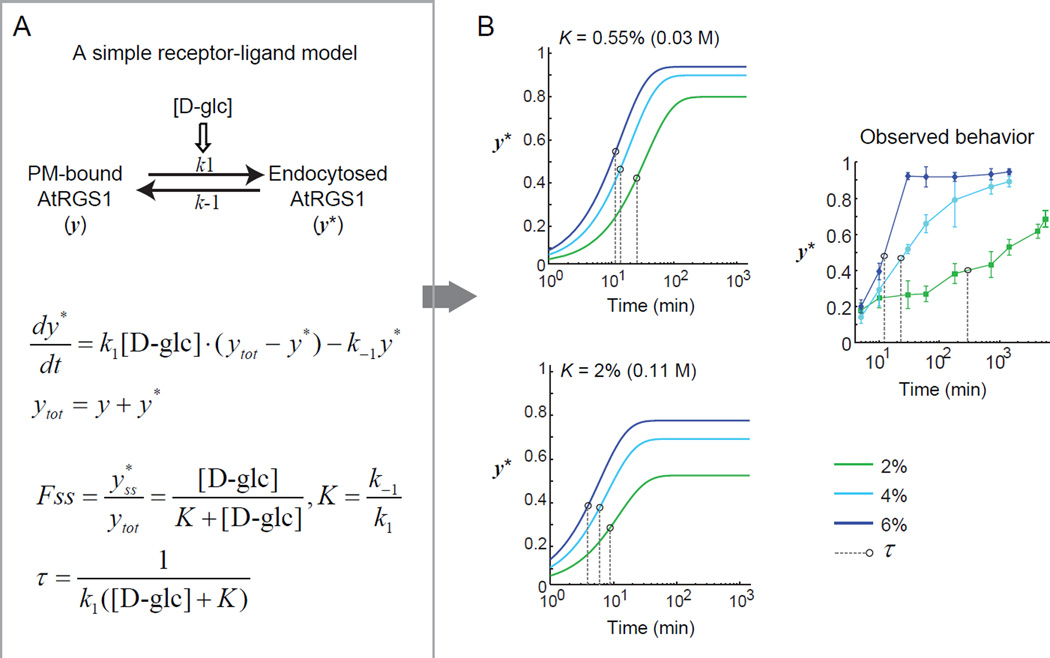
Figure 2. A simple kinetic model for AtRGS1 deactivation.
(A) A simple “receptor-ligand” model with a deactivation rate of AtRGS1 being proportional to the D-glucose concentration. The steady-state fraction of internalized AtRGS1 Fss and the characteristic time scale τ to reach steady state is analytically calculated. (B) Full pathway activation requires [D-glc] ≫ K. In this limit τ ≈ 1/(k1[D-glc]) and the time to reach steady state is relatively insensitive to [D-glc] (top panel). For the time to reach steady state to change significantly with [D-glc] requires [D-glc] ≈ K (bottom panel). However, in this scenario the Fss changes significantly with [D-glc]. For comparison the experimental data are shown (right panel). Black circles indicate the characteristic time scale τ of each curve. Refer to Figure S1 for the results of a thorough parameter search for this simple kinetic model.
Simple activation kinetics cannot generate dose-duration reciprocity
The rate at which AtRGS1 is removed from the G protein complex depends on the D-glucose concentration. However, a single glucose-dependent rate is insufficient to explain the observed dose-duration reciprocity. To illustrate this point, consider a simple two-state model in which the internalization rate of AtRGS1 is directly proportional to the concentration of D-glucose (Figure 2A). In this model, the steady-state fraction of internalized AtRGS1 is Fss = [D-glc]/(K+[D-glc]), where K = k−1/k1, and the characteristic time scale for the system to reach steady state is τ=1/(k1([D-glc]+K)). If Fss ≈ 1, then this implies [D-glc] ≫ K. In this situation τ ≈ 1/(k1[D-glc]). Therefore, with this model a 10-fold increase in the D-glucose concentration leads to a 10-fold decrease in the time to reach steady state. However, as noted above, an increase from 4% to 6% D-glucose reduced the time to reach steady state from around 24 hours to 30 min, a greater than 50 fold change. Therefore this simple “ligand-receptor” model cannot account for both the near full activation of the pathway over a wide range of D-glucose concentrations and the dependence of the time to reach steady state on the D-glucose concentration (Figure. 2B). To further verify this conclusion, we attempted to fit this simple model to the internalization time course data using an evolutionary algorithm (see Experimental Procedures). However, none of the 900,000 parameter sets generated by the algorithm produced a satisfactory fit to the data (Figure S1).
Development of a quantitative mathematical model of G protein activation in Arabidopsis
To investigate potential mechanisms for dose-duration reciprocity, we developed a detailed mathematical model to quantitatively describe the dynamics of AtRGS1-mediated G protein signaling. The model consists of 14 key signaling components in the G protein activation pathway (Figure 3A, Table S1 and Table S2), capturing five different G protein states: GαGDPβγ, GαGTPβγ, GαGDP, GαGTP, and Gβγ. Each of these subunits (except perhaps the last one, Gβγ) forms a complex with AtRGS1.
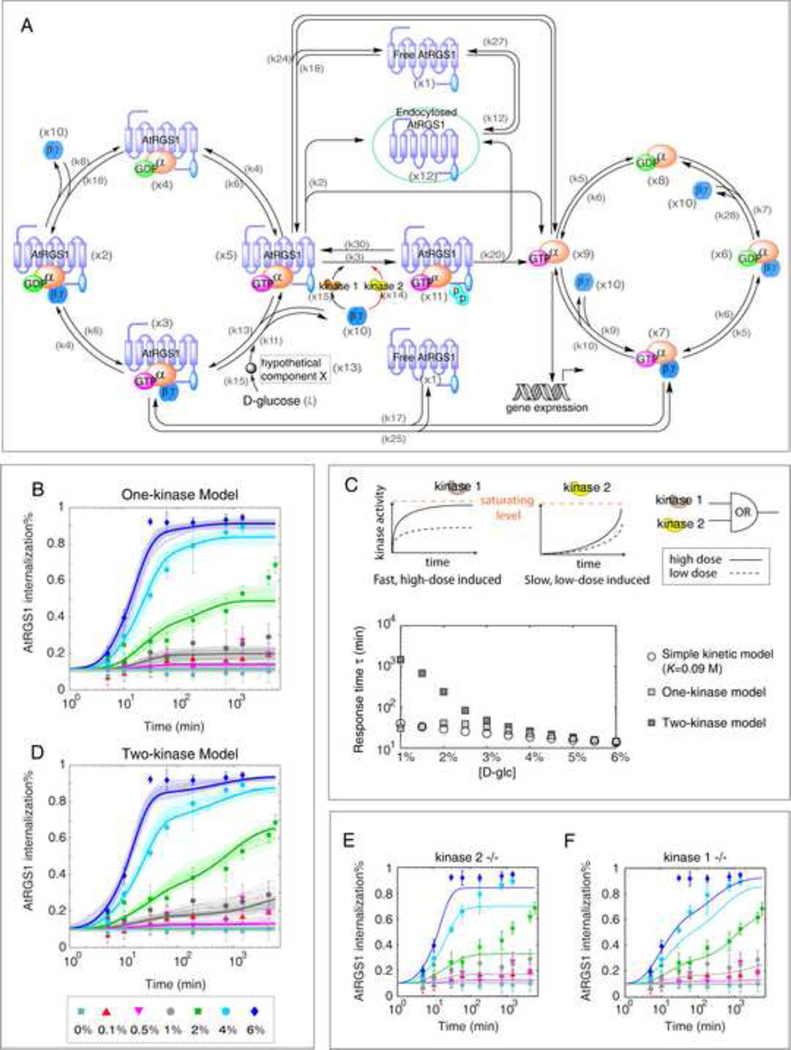
Figure 3. Modeling the dose-duration reciprocity in AtRGS1 endocytosis.
(A) Molecular interactions in the Arabidopsis G signaling network included in the models. The initial model includes a single kinase (one-kinase model) that phosphorylates AtRGS1. In the two-kinase model the second kinase acting on a different time scale is added (center red arrows). Refer to the main text for a description of the model.(B) The one-kinase model fails to reproduce the observed dose-duration reciprocity. Fitting results for the top 50 scored parameter sets are shown in thin lines with light tone. Thick lines represent the prediction calculated from the median of the top 50 scored parameter sets (, values shown in Table S3). (C) Top panels: In the two kinase model, the two kinases are activated with different kinetics (top panels). AtRGS1 phosphorylation follows “OR” logic with either kinase capable of phosphorylating the protein. Bottom panel: Response time (τ) to reach half of the steady state under differential sugar concentrations based on the simple model (with parameter values from Figure S1D), one-kinase model (,Table S3) and two-kinase model (, Table S3). (D) The two-kinase model reproduces the AtRGS1 internalization data. Fitting results for the top 50 parameter sets are plotted with thin lines in light tone. Thick lines are calculated from the median of the top 50 scored parameter sets (, values shown in Table S3). The modeling prediction in (E) kinase 2 deleted mutant and (F) kinase 1 deleted mutant. The prediction is given based on with the action from one kinase blocked. Western blot showing stable AtGPA1 and AGB1 levels independent of D-glucose treatment is provided in Figure S2, as evidence supporting assumption on conserved G protein level upon glucose treatment. Top parameter sets and time courses for each signaling component using the two-kinase model with parameter values are shown in Figure S3. In addition, the model predicts that the accumulation of Gα in the active G protein cycle is proportional to AtRGS1 endocytosis over the 7 different D-glucose concentrations tested as shown in Figure S5. Variables of the quantitative model are provided in Table S1, and the ODEs of the one- and two- kinase model provided in Table S2.. The median of the top scored parameter sets based on the one-kinase () and the two-kinase () are listed in Table S3.
There are two major chemical cycles in the model: one containing AtRGS1-bound G proteins (“inactive cycle”, the left cycle in Figure 3A) and one containing free G proteins (“active cycle”, the right cycle in Figure 3A). In the absence of D-glucose, the majority of G proteins participate in the inactive cycle, i.e. in complexes with AtRGS1. In the presence of D-glucose, Gβγ is released from the AtRGS1:GαGTPβγ complex. Free Gβγ then recruits a kinase (e.g. AtWNK8) to phosphorylate AtRGS1 which triggers the trafficking of AtRGS1 from the plasma membrane to endosomes, leaving GαGTP at the plasma membrane to initiate signaling. In this way, AtRGS1 trafficking redistributes the G protein pool from the inactive cycle into the active cycle (Figure 3A). Because the free GαGTP and perhaps internalized AtRGS1 are responsible for downstream signaling and sugar-induced gene expression (Chen et al., 2003), the variables are the key outputs of the model.
We made the following assumptions in developing the model:
Because no evidence supports direct binding of D-glucose to AtRGS1, the model incorporates a hypothetical protein X that binds D-glucose and initiates signaling.
AtRGS1 endocytosis is triggered by phosphorylation. AtRGS1 phosphorylation is necessary for its endocytosis (Urano et al., 2012c).
In addition to the kinase-dependent AtRGS1 endocytosis pathway, the model also includes a basal level of endocytosis.
The model allows tonic cycling (a D-glucose-independent background cycling) of AtRGS1 in which endocytosed AtRGS1 trafficks back to the plasma membrane as free AtRGS1.
Finally, the model is based on the total amount of G protein subunits remaining the same following sugar stimulation as supported by immunoblot analysis (Figure S2). The model assumes that AtRGS1 steady-state level does not change following D-glucose stimulation.
We refer to this model as the one-kinase model, because it assumes a single kinase is responsible for phosphorylating AtRGS1. The model parameters were either taken from the literature (Table S3) or estimated by fitting the model to the AtRGS1 internalization data in Figure 1A. Parameter estimation was again performed using our evolutionary algorithm (Experimental Procedures). The top 50 estimated parameter sets were chosen out of a total 9 million trials generated by the algorithm. Interestingly, despite the intensive parameter search, these top scored parameter sets (Figure 3B) only generated a dose-duration behavior that is qualitatively similar to that of the simple model discussed in Figure 2 and Figure S1. The median of the top 50 parameter sets () is shown in Table S3. This result indicates that there are additional elements required to obtain the observed dose-duration reciprocity.
A two-kinase model is capable of dose and duration reciprocity
Our previous study showed that sugar-induced endocytosis of AtRGS1 requires kinase phosphorylation at its C-terminal region (amino acid 400–459) (Urano et al., 2012b). D-glucose favors the AtRGS1-AtGPA1 complex over AtRGS1-heterotrimer complex, resulting in accumulation of freed Gβγ dimers. The latter then recruits AtWNK kinases, including AtWNK8 and perhaps AtWNK10, to phosphorylate AtRGS1 for endocytosis (Urano et al., 2012b). Thus, one potential strategy to generate dose-duration reciprocity is through an integrated action of multiple kinases that are differentially activated by Gβγ and/or phosphorylate AtRGS1 on different time scales. This regulatory mechanism represents a Gβγ-mediated positive feedback loop (Figure 3A).
To test this hypothesis, we modified the detailed model to include two kinase groups, kinase 1 and kinase 2, which are activated by freed Gβγ and mediate AtRGS1 internalization (Figure 3A, red lines present kinase 2). The combined activity of both kinases determines the phosphorylation status of AtRGS1. We again used our evolutionary algorithm to fit the two kinase model to our AtRGS1 internalization data. The model successfully generated the observed dose-duration reciprocity (Figure 3D and Figure S3A). The median values of the top 50 scored parameter sets () was taken as the final parameter set for the two-kinase model (Table S3). Time courses for signaling components in the network were calculated (Figure S3B). The simulations revealed that there are two critical requirements for the model to capture the data: (1) the two kinases are activated on different time scales (kinase 1 is quickly activated by Gβγ whereas the activation of kinase 2 is slow) and (2) they have different activation thresholds (Km) with kinase 1 activated by high sugar concentrations (large Km) and kinase 2 activated under lower concentration (small Km) (Figure 3C, top panels). Note that plotting the response time τ as a function of D-glucose concentration demonstrates the qualitatively different behavior of the two-kinase model as compared to the one-kinase model and the simple kinetic model (Figure 3C, bottom panel).
The model predicted that blocking kinase 2 (the slow kinase) would result in the dose and duration relationship predicted by the simple kinetic model (Figure 3E). On the other hand, the model predicts blocking kinase 1 (the fast kinase) would result in a delay in AtRGS1 endocytosis under acute sugar stimulation (Figure 3F). Consequently, the internalization under high dose D-glucose(e.g. 6%) would be reduced at an early phase of glucose stimulation (e.g. 30 minutes) (Figure 3F). Note that this prediction is consistent with decreased AtRGS1 internalization shown with mutants lacking either AtWNK8 or AtWNK10 following 6% D-glucose for 30 minutes (Urano et al., 2012b). Therefore, AtWNK8 and AtWNK10 are good candidates for kinase 1 in mediating the dose-duration reciprocity.
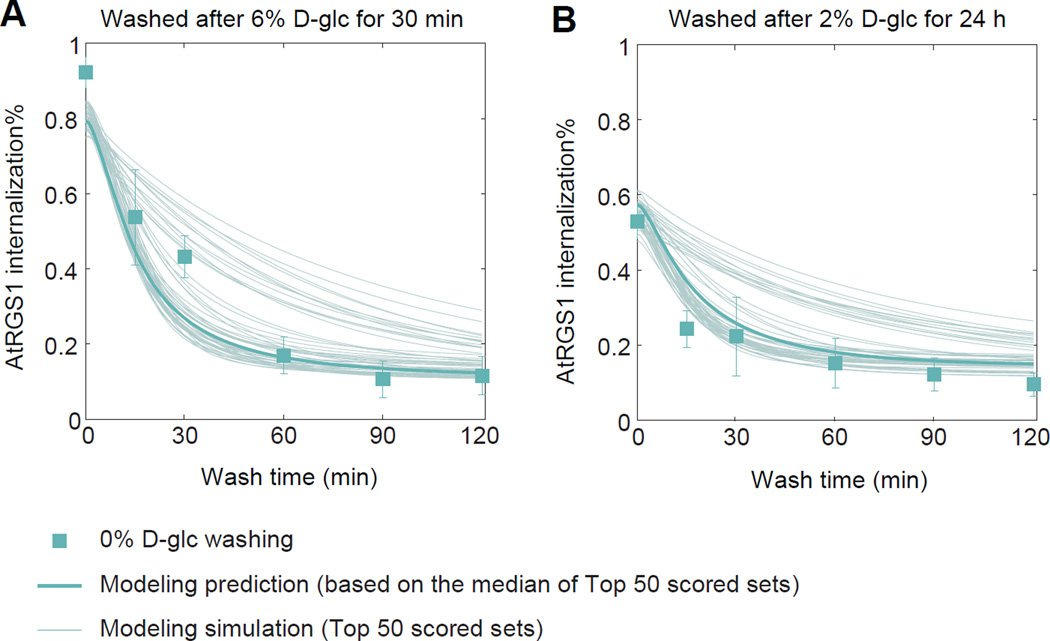
We validated the quantitative two-kinase model using wash-out experiments. Arabidopsis seedlings were exposed to D-glucose (either 6% for 30 min or 2% for 24 h), and then the seedlings were washed with water. AtRGS1 internalization was then monitored for 2 hours following washout (Figure 4). Following both concentrations, the model predicted fast relaxation dynamics in which AtRGS1 internalization returns to basal levels within 1 hour. This prediction is in good qualitative agreement with the wash-out data, demonstrating the ability of the model to AtRGS1 endocytosis dynamics under novel experimental conditions.
Figure 4. Testing the model on relaxation experiment.
Arabidopsis seedlings that were treated with either (A) 6% D-glucose for 30 min or (B) 2% for 24 h were quickly washed 3 times with water before transferred into water medium. AtRGS1-YFP internalization was monitored using confocal microscope at specific times in water (data presented in green squares, error bar = 2 × S.E.M.). The predicted internalization (fine lines) from the model based on the top 50 scored parameter sets. Prediction based on the median of the top 50 scored parameter sets () was plotted in thick.
Experimental validation of WNK kinases role in AtRGS1 endocytosis
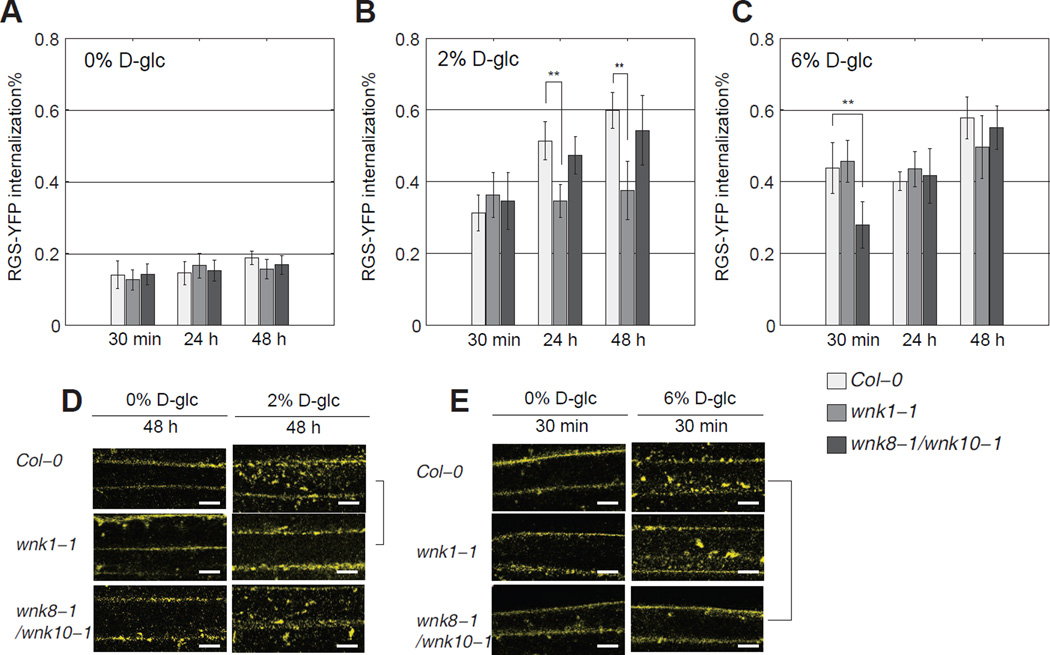
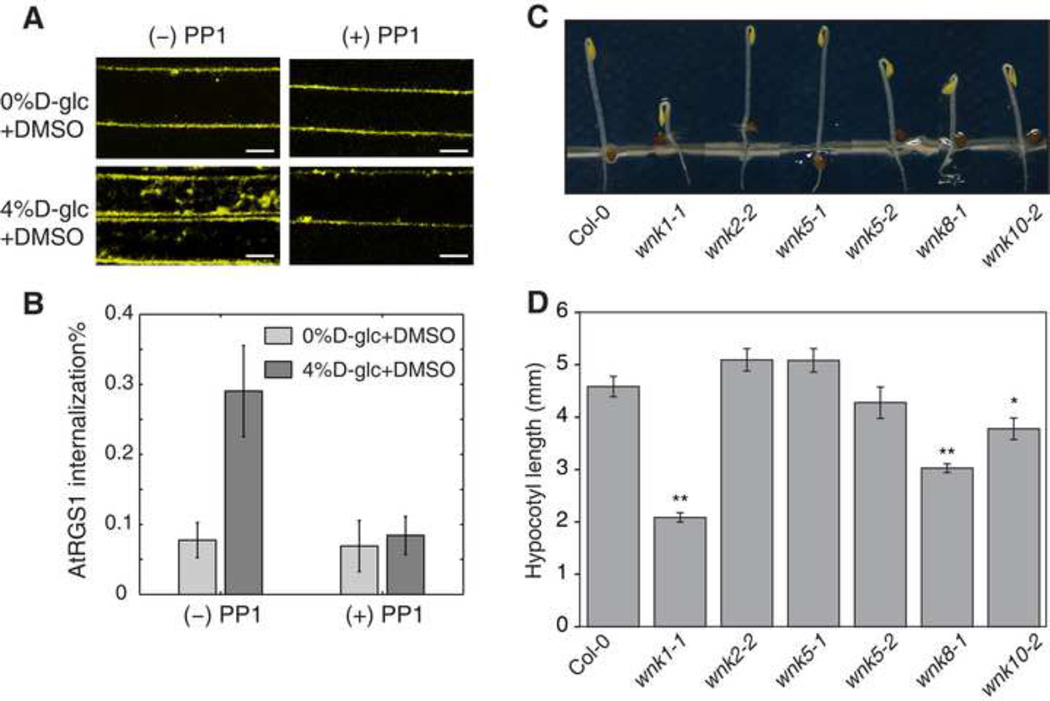
AtWNK8 and AtWNK10 phosphorylates AtRGS1 at its C terminus and this phosphorylation is critical for its endocytosis (Urano et al., 2012b). However the double mutant wnk8-1/wnk10-1 still showed partial internalization of AtRGS1 by sugar (Figure 6C). This suggests other kinases are involved in the process of AtRGS1 endocytosis. To test whether kinases outside the WNK family are involved in the D-glucose-induced AtRGS1 endocytosis, we blocked total WNK kinase activity with the inhibitor PP1 (Yagi et al., 2009). PP1 almost completely blocked AtRGS1 endocytosis (Figure 5A–B) indicating that one or more other WNK kinases are required for AtRGS1 endocytosis. Thus, it follows that a set of WNK kinases may be activated in a coordinate fashion to mediate the dose-duration reciprocity. This hypothesis was tested genetically. The Arabidopsis genome encodes ten AtWNK kinases (Wang et al., 2008). However five of these were excluded from the test due to their incongruent expression pattern, leaving AtWNK1, AtWNK2, AtWNK5 along with AtWNK8 and AtWNK10 as candidate kinases responsible for the dose-duration reciprocity property. Genetic ablation of AtWNK2 or AtWNK5 had no effect on D-glucose-induced AtRGS1 internalization (Figure S4). Both dose and duration of D-glucose was manipulated with cells transiently expressing AtRGS1-YFP in either the wnk1 single or wnk8/10 double mutants (Figure 6) (see Experimental Procedures for transient transfection). A significant decrease of internalized fluorescence was observed in wnk1-1, but not in wnk8-1/wnk10-1 cells, under a relatively low and long-term sugar treatment (2% for 24 hours and for 48 hours, with p<0.001) (Figures 6B and 6D). The difference between Col-0 and wnk1-1 increased from 24 hours to 48 hours. Note that no significant difference was detected between Col-0 and wnk1-1 within 30 minutes over low dose treatment (Figure 6B). This suggests that AtWNK1 functions as kinase 2 (the slow kinase with small Km) in the model, because loss-of-function mutations confer decreased internalization under low doses of sugar at later times (Figure 3E). Genetically ablating AtWNK8 and AtWNK10 (wnk8-1/wnk10-1) resulted in the loss of AtRGS1 internalization at a high dose sugar treatment for 30 minutes (p<0.001), compared to Col-0, whereas no difference was detected between Col-0 and wnk1-1 (Figure 6C and 6E). This suggests AtWNK8 and AtWNK10 function as kinase 1 (the fast kinase with large Km) in the model and account for the predicted decrease in internalization under high dose sugar at early phase (Figure 3F). Loss of either AtWNK1 or AtWNK8 and AtWNK10 had no effect on the response to a long duration of high D-glucose amount (Figure 6C). Overall, in either wnk1-1 or wnk8-1/wnk10-1, but not Col-0, wnk2-2 or wnk5-1, the dose-duration reciprocity was broken. The wnk1-1 mutant lost sensitivity to low concentration of D-glucose for a long duration, whereas the wnk8-1/wnk10-1 mutant lost the ability to detect a high concentration of D-glucose for a short duration.
Figure 6. Coordinated kinase activity between AtWNK1, AtWNK8 and AtWNK10 under different sugar conditions.
(A–C) Wild type Col-0, mutant wnk1-1 and wnk8-1/wnk10-1 seedlings transiently transfected with AtRGS1-YFP (see Materials and Methods) are treated with D-glucose at the indicated concentration and time, and imaged with confocal microscopy for AtRGS1 internalization amount (see Materials and Methods). Error bar = 2 × S.E.M. (D, E) Representative microscope images. Scale bar = 10 μm.
Figure 5. Physiological phenotypes of wnk mutants.
(A, B) Four-day-old seedlings that express AtRGS1-YFP were pre-treated with a kinase inhibitor, 50 μM PP1 for 3 h followed by D-glucose treatment for 1 h as shown in the panel. The control seedlings were treated with DMSO as the solvent control. PP1 is commonly used as a Src-family kinase inhibitor, but also suppressed human WNK kinase activity in vitro (Yagi et al., 2009). Among 42 kinase inhibitors comprehensively examined, the specificity of PP1 was relatively specific, although PP1 still inhibited 4 of 28 tested mammalian kinases (Bain et al., 2003; Davies et al., 2000). The Arabidopsis genome encodes no homologous genes to Src-family kinase genes. (B) Quantified data are shown as mean ± 2 × S.E.M. (C, D) Seedlings were grown on ½ × MS plates with 1% sucrose for 2 d in darkness. Length of hypocotyls was measured and quantified. Data are mean ± SEM. Student’s t-test was used to compare values to the Col-0. Sample size = 23–49,*, p < 0.01; **, p < 0.001. Representative seedlings of Columbia (Col-0) and wnk mutants are shown in C.
The physiological genetic linkages between G protein and WNK kinase mutants were also examined. G proteins regulate various developmental processes including growth in leaves, fruits, and hypocotyls (Urano et al., 2013). We compared early hypocotyl growth of the wnk and G protein mutants because this development is regulated in part by G protein-mediated sugar signaling (Gupta et al., 2012; Lilley et al., 2012; Ullah et al., 2001; Zhang et al., 2010). Sugars derived from carbohydrate reserves in the cotyledons drive, in part, elongation of the subtending epidermal cells. The wnk1-1 single, wnk8-1 and wnk10-2 alleles, but not wnk2-2, wnk5-1, wnk5-2 alleles conferred short hypocotyls (Figure 5C–D) like in G protein mutants (Chen et al., 2006; Ullah et al., 2001). This observation suggests that these three AtWNK kinases operate as a functional complex and loss of any one of the three compromises G protein-mediated cell expansion.
WNK kinase recruitment to AtRGS1 is sugar dose and duration dependent
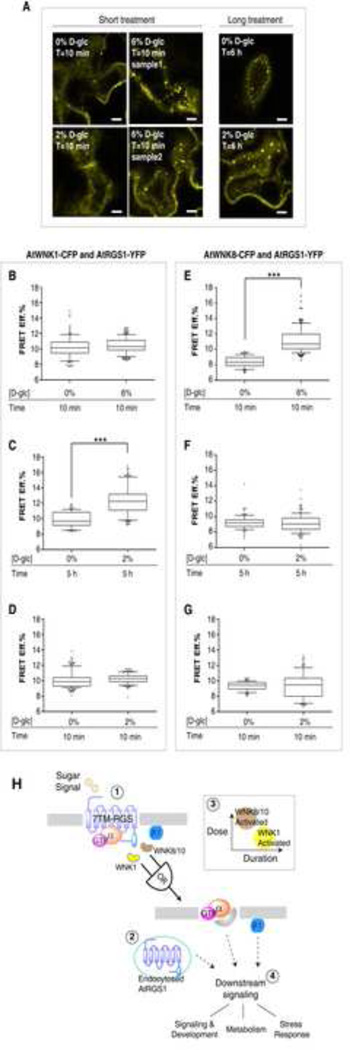
The mechanism for dose-duration reciprocity is, in part, by physical recruitment of the WNK kinases to AtRGS1. We showed this using FRET analysis of tobacco epidermal cells expressing AtRGS1-YFP and AtWNK-CFP genes (Experimental Procedures). AtWNK1-CFP becomes closer to AtRGS1-YFP when a lower concentration of sugar was applied however this increase in proximity required a long duration (e.g. 2% for 5 h, Figure 7C). An acute pulse of D-glucose regardless of concentration showed no increase in FRET efficiency between AtWNK1-CFP and AtRGS1-YFP (Figure 7B and 7D). On the other hand, AtWNK8-CFP became closer to AtRGS1-YFP within a short pulse of a high concentration of sugar (Figure 7E). For example, as shown in Figure 7F & 7G, 2% glucose was below the threshold for AtWNK8 in AtRGS1 endocytosis, whereas at 6% glucose, AtWNK8 was recruited to AtRGS1 within 10 minutes (Figure 7E). Thus, the FRET data supports the “OR” gate linking two kinases with differential working thresholds and kinetics is one of the potential mechanisms that lead to the observed dose-duration reciprocity in the sugar-induced AtRGS1 endocytosis (Figure 7H).
Figure 7. Specific WNK kinases are recruited to interact with AtRGS1 on differential sugar dose-and-duration regime.
(A) Paradermal optical section of AtRGS1 endocytosis in tobacco epidermal cells transiently transfected by AtRGS1-YFP with D-glucose treatment as indicated. Scale bar = 10 μm. Refer to Figure S6 for top view and middle view of the AtRGS1 endocytosis along Z-stack in tobacco epidermal cells. Sensitized emission FRET was carried out between AtWNK1-CFP and AtRGS1-YFP or between AtWNK8-CFP and AtRGS1-YFP in tobacco epidermal cells at the indicated dose and duration (see Experimental Procedures). Quantification is represented as the FRET Efficiency (FRET Eff.%) between AtWNK1-CFP and AtRGS1-YFP (B–D) and between AtWNK8-CFP and AtRGS1-YFP (E–G) as described in Experimental Procedures. ***: p-value < 0.0001, unpaired t-test, sampling size (ROIs) = 95 – 176 (the AtWNK1 case), 89–121 (the AtWNK8 case). The whiskers represent 10–90 percentile of the data. Outliers (<10% or >90%) are presented in black dots surrounding the whiskers. (H) Proposed model of dual-kinase coordination on specific sugar condition. Through recruiting or activating kinases at different dose and duration regime, the plant cells may endow the ability to decode environmental clue of the sugar availability, through which specific downstream signal may be propagated. (1) Arabidopsis G protein is kept in its inactive state by the action of a 7TM-RGS1. The presence of D-glucose, in the mM range, releases inhibition of the G protein complex by physically uncoupling and thus allowing the G protein to self-activate. (2) Physical uncoupling is achieved by AtRGS1 endocytosis. (3) This physical uncoupling requires the action of three kinases designated AtWNK1, AtWNK8 and AtWNK10 operating with the indicated OR-gate. We speculate that WNK1 is recruited to AtRGS1 by low D-glucose for a long duration and WNK8/10 are recruited to AtRGS1 by a high dose of D-glucose for a short duration (inset, dose-duration relationship). (4) Arrows indicate that downstream signaling is propagated by the action of both G proteins on the plasma membrane and AtRGS1 on the endosome.
AtRGS1 endocytosis and Gα activation are stoichiometrically related
In addition, the model predicts that the accumulation of Gα in the active G protein cycle is proportional to AtRGS1 endocytosis over the 7 different D-glucose concentrations tested (Figure S5). This suggests that the stoichiometric relationship between internalized AtRGS1 and the G-protein is independent of D-glucose concentration. The model also predicted that within 24 h treatment of D-glucose, the fraction of free Gα:GTP in the total Gα pool would peak around the D-glucose concentration that generated 50% AtRGS1 internalization, as observed. One explanation why the active pool of Gα:GTP does not increase above 50% AtRGS1 endocytosis is due to Gα captured by newly released Gβγ in the active cycle (Figure S5).
DISCUSSION
A reciprocal dose and duration response was found in AtRGS1 endocytosis where sensitivity to the sustained low concentration of sugar was observed. Interestingly, similar reciprocal dose-duration behavior was also observed in the photosynthetic leaf cells transiently expressing AtRGS1-YFP (described in Experimental Procedures), where either treatment with 6% D-glucose for 10 min or treatment with 2% for 6 h induced abundant AtRGS1 endocytosis (typical images shown in Figure 7A). It is likely that similar dose-duration compensation also present in photosynthetic cells.
We speculate that dose-duration reciprocity in sugar sensing provides a mechanism for plants to evaluate the availability of energy based on both the concentration and persistence of sugar in the environment. Glucose levels derived from photosynthesis dramatically vary in plants within a single day from micromolar to millimolar levels (Deuschle et al., 2006). Moreover, variations in sunlight throughout the day are expected to cause rapid fluctuations in the local sugar concentration. Our results suggest that a simple dual kinase system, consistent with this time frame, is sufficient to distinguish short term changes in glucose caused by intermittent fluctuations, such as cloud movement, from slower changes occurring from day night cycles. Though D-glucose induced AtRGS1-YFP internalization in wnk mutants showed clear differences in threshold and time for recruiting specific kinases, it remains an open question as to whether additional factors are required to implement the specific response to D-glucose dose and duration.
One goal of systems biology is to identify signaling motifs, sets of molecular interactions, which perform specific information processing functions. We demonstrated how a dual kinase system forms a signaling motif for dose-duration reciprocity (Figure 7). We speculate this motif occurs in other biological processes where both the concentration and persistence of an external signal are important factors in determining cell fate. For example, photosynthetic efficiency is controlled by a pair of kinases that phosphorylate photosystem II core proteins. While it is not understood how, detection of light intensity and duration may be a property of this dual kinase regulatory system (Bonardi et al., 2005). Clearly, plants cells evolved regulatory mechanisms that allow them to respond to a wide range of fluctuating sugar concentrations. Glucose and other sugars regulate photosynthesis efficiency in part through regulation of the genes encoding rate-limiting steps in photosynthesis, including carbon fixation (Koch, 1996). Our understanding of information processing by cells is far from complete, and the two-kinase motif considered here is only one mechanism used by cells to survive in noisy environments.
Finally, our work presents a new concept in light of the body of knowledge of G protein signaling in yeast. While yeast and Arabidopsis both use the same repertoire of G protein signaling components, the network architectures in these two cell types are significantly different demonstrating how the same building blocks can be used to perform different network functions. This last point profoundly impacts the way we envisage evolution of signal perception/transduction pathways. Network architecture, per se, is responsible for emergent behavior that presumably is under positive and negative selection.
EXPERIMENTAL PROCEDURES
Microscopy and quantification of AtRGS1 internalization fraction
The method is as described in details in (Urano et al., 2012b). Briefly, stable Arabidopsis expressing AtRGS1-YFP (Col-0) were stratified on plates with ½ Murashige and Skoog (MS) for 3 d, followed by 2 h light and then grown in darkness for 5 d to germinate. The seedlings were treated with sterile water or D-glucose solutions before imaging as indicated. Hypocotyl epidermal cells located 2–4 mm below the cotyledons were imaged using vertical optical sectioning (Z-stack acquisition) by a Zeiss LSM710 confocal laser scanning microscope with C-Apochromat × 40/1.2 water immersion objective. An argon laser (514 nm excitation) was used to excite YFP and a photomultiplier detector set between 526 nm – 569 nm acquired the quantification. For each D-glucose condition, at least two (typically three or four) seedlings were imaged to reduce bias.
For transient expression of AtRGS1-YFP in the indicated genotypes, seeds were stratified for 3 d then grown in darkness in ½ MS liquid medium with no sugar for 4 d. Agrobacterium tumifaciens carrying RGS-YFP were grown in 3 ml LB medium with 50 μg ml−1 rifampicin, 50 μg ml−1 gentamycin and 100 μg ml−1 kanamycin, for overnight in 28°C incubator with 220 rpm. A second growth cycle was then performed by diluting 1 ml of the overnight growth to 1:10 with LB medium and growing in the 28°C incubator with 220 rpm for 6 h. Bacteria cells were then harvested (centrifuged at full speed for 3 min) and re-suspended with ½ MS medium with targeted volume to reduce OD600 to 0.3. To the culture was added acetosyringone and Sylwet-77 (final concentration of 150 μM and 0.003%, respectively). The Arabidopsis seedlings were then transferred and cultured in the Agrobacterium solution at 25°C chamber for additional 2 d. Before microscopy imaging, the seedlings were taken out from the agro solution, washed twice in sterile water and treated with sugar with specified concentration and time.
For each condition of D-glucose concentration and treatment duration, the top layer of cells on the Z-stack were used to quantify the AtRGS1 endocytosis fraction. Images were converted to 8-bit and a threshold for fluorescence signal chosen. Regions of cells were determined based on the outlines of cells in corresponding bright-field image. The ratio between the integrated fluorescence signal intensity detected within cytosolic compartments (Ic) and the whole cell (Iw) is defined as the fraction of internalized AtRGS1.
For each condition (one combination of a proposed dose and duration), at least 6 images were sampled randomly from the Z-stacks to calculate the AtRGS1 internalization percentage. All the percentages of internalization were with standard error less than 0.1, thereby excluding potential bias.
Transient transformation of tobacco epidermal cells
Agrobacteria carrying a binary plasmid encoding either AtRGS1-YFP or P19 (to reduce gene silencing) were grown for the first cycle in 3 ml fresh LB medium with 50 μg ml−1 rifampicin, 50 μg ml−1 gentamycin and 100 μg ml−1 kanamycin overnight in 28°C incubator with 220 rpm. In the second cycle, 500 μl of the first cycle culture were diluted in 50 ml fresh LB with 50 μg ml−1 rifampicin, 50 μg ml−1 gentamycin, 100 μg ml−1 kanamycin and 20 μM acetosyringone. The culture was grown overnight in the 28°C incubator with 220 rpm. Bacteria cells were then harvested by centrifuge at 7,000 rpm for 15 min (temperature kept at 20°C – 24°C). The supernatant was discarded and cells were re-suspended and diluted in buffer (0.2% MES and 0.2% MgCl2 dissolved in sterile water with 200 μM acetosyringone) to reach OD600=1. On the day of infiltration, the Agrobacteria with either AtRGS1-YFP and P19 were mixed at 1:1 ratio and gently infiltrate on the back of the tobacco leaves. On the 4th d post infiltration, D-glucose was infiltrated at the same spots where Agrobacterium were infiltrated. Leave disks from these infiltration sites were taken and incubated in the same sugar concentration for the indicated times before imaging by confocal microscopy.
Immunoblot analysis and relative protein quantitation
Arabidopsis seedlings of wild type Col-0 were grown in ½ MS liquid medium with 1% sucrose under continuous light (40 μE m−2 s−1) at 23°C for 4 d. The seedlings were starved of added sugars for 3 days under darkness, then stimulated with 6% D-glucose for 6 hs, and then harvested and ground in liquid nitrogen. The protein extracts suspended in buffer A (50 mM Tris-HCl (pH 8.0), 10 mM 2-mercaptoethanol, 10% glycerol and protease inhibitors) were centrifuged at 18,000 rpm in a Beckman SM-24 rotor. The pellet was solubilized in buffer A with 1% NP-40 and 2% ASB-14. Protein levels of AtGPA1 and AGB1 of the solubilized fractions were evaluated by immunoblot using anti-AtGPA1 and anti-AGB1 antibodies.
Early development of etiolated seedlings
Sterilized seeds were placed in a row 1 cm from an edge on 10 × 10 cm square plates containing ½ × Murashige and Skoog (MS) media with 1% sucrose and stratified. The plates were placed for 2 h in a light chamber, and covered in foil and held vertically at 25°C for 48 h in a dark chamber. Hypocotyl images were scanned, and the lengths were measured with the image analyzing software, Image J.
Accessions
The mutant alleles for the wnk mutants are described in [10]. The accession numbers are shown as follows: AtWNK1, At3g04910; AtWNK2, At3g22420; AtWNK5, At3g51630; AtWNK8, At5g41990; AtWNK10, At1g64630.
Mathematical modeling and computer simulation of G protein activation dynamics
An ordinary differential equation (ODE)-based model was constructed to simulate the dynamics of G protein activation (Table S1). The model used mass action kinetics and Hill function typed equations to quantitatively capture the dynamical flows within and between the inactive and the active G protein cycles. The quantitative model consisted of 14/15 ODEs (Table S1 & S2) with 30 kinetic rate parameters, with the values taken from existing biochemical studies or estimated based on the AtRGS1 internalization profiles presented in this work (Table S3). Parameter estimation and optimization was implemented by a large-scale evolution algorithm with least squares regression as described below. The median values of the top 50 best fitted parameter sets from 9 million trials were set as the final parameter values of the model (Table S3).
Evolutionary algorithm for parameter estimation
To perform parameter estimation, we used a modified evolutionary algorithm (Singhania, 2011). The algorithm iterates M generations of parameter values, with each generation containing P parents with C offspring. In each generation, a percentage, μ, of parents are randomly selected to undergo “macro-mutations” in which new parameter values a randomly selected from uniform distribution within a range of 0–100 fold of the initial values. Macro-mutations allow the algorithm to escape local minima and sample new regions of parameter space. Each parent, whether it undergoes macro-mutation or not, produces C children that contain small random variations in the parameter values. This process is designated as “micro-mutation” and is computed as follows: cj,i = pj ∘ (1⃗+λ(δ − 0.5·1⃗)), where pj is the vector of parameter values for the jth parent and cj,i is the ith child of the jth parent, λ is the maximum possible percent change of a parameter, δ is a random vector whose elements are chosen from a uniform distribution on the unit interval, and 1⃗ is a vector whose components are all 1. The operator ∘ denotes component-wise multiplication. In each mutation step, all parameters (except the ones being fixed on the known values, see below) are mutated at the same time.
All P*C progenies are scored based on a weighted sum of the squared differences between the model output and experimental data, the score is denoted as Ωji. Children are then subject to selection to become a parent in the next generation based on the criterion: θ = e−βΩji > ε, where β is an effective energy and ε is a random number uniformly distributed between 0 and 1. For example, a perfect fit implies the score Ωji = 0 and this child will be selected with probability θ = e−β·0 = 1. The closer the parameter set fits the data, the smaller its score and the larger the chance it will be selected as a parent in the next generation. P children from these “good” parent candidates are randomly selected and the algorithm goes to the next generation. The algorithm continues until the maximum generation number M is reached. For each model, we ran X instances of the algorithm in parallel, each starting from a set of randomly chosen values. In our implementation of the simple “ligand-receptor” model, μ = 30%, λ = 20%, β = 2, P = C =6, M = 1000, X = 25. These values generated 0.9 million trials in the parameter search. For the detailed models, k4, k5, k6, k7 and k14 were fixed (see Table 3 for their values), and the other 22 in the one-kinase model and 25 parameters in the two-kinase model were searched by the algorithm. To avoid slow random diffusion in the high-dimensional parameter space, a moderately good parameter set was found before initiating the program and set as the initial condition for half of the parents in the first generation. The parameter values for the other parents were chosen at random value. Note, all of the parents were still subject to the macro-mutation process. For these models, μ = 30%, λ = 20%, β = 2, P = C = 6, M = 5000, X = 50, which generated 9 million trials in the parameter search. Comparison of the top 50 scored parameter sets from one-kinase model and two-kinase model are shown in Figure 3B and 3D, respectively, with the corresponding scores Ω compared in Figure S3A.
Förster Resonance Energy Transfer Analyses
Sensitized emission FRET was performed with an Zeiss LSM 710 instrument on tobacco cells co-expressing eCFP- or eYFP-tagged proteins. eCFP and eYFP were excited using 405 nm and 489 nm diode laser lines, respectively, and 460–520 and 526–569nm band pass filters to collect emission images, respectively. In brief, 4–5 week old tobacco leaves were injected with Agrobacterium GV3101 harboring binary plasmids for expression of AtWNK-CFP and AtRGS1-YFP pairs. Imaging was performed on the 4th–7th day post-infiltration during which time, leaves disks were taken of leaves injected with D-glucose at different concentration for the designated times. The FRET was calculated by the Fiji pFRET plugin based on the method used by Chen and co-workers (Chen and Periasamy, 2005). Using this algorithm, FRET images were corrected for spectral bleed through by analyzing images of control cells expressing donor proteins alone or acceptor proteins alone with the same intensity distributions as the sample.
Supplementary Material
Highlights.
An atypical G signaling pathway architecture confers dose-duration reciprocity.
Three unusual kinases provide the kinetic drivers for the dose-duration mechanism.
Endocytosis of AtRGS1 from its cognate Gα subunit is sufficient for G activation.
Acknowledgement
We would like to thank Ariko Urano, Jing Yang and Tony Purdue for their technical assistance, and Drs. Joshua B Kelley, Matthew Pena, Alejandro Colaneri, Susanne Wolfenstetter and Jianping Huang for useful discussions. This work was supported by grants from the NIGMS (R01GM065989) and NSF (MCB-0718202) to A.M.J and NIGMS (R01GM079271) to T.C.E. The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through the grant DE-FG02-05er15671 to A.M.J. funded technical support in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Booker KS, Schwarz J, Garrett MB, Jones AM. Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Gao Y, Jones AM. Differential Roles of Arabidopsis Heterotrimeric G-Protein Subunits in Modulating Cell Division in Roots. Plant Physiology. 2006;141:887–897. doi: 10.1104/pp.106.079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A Seven-Transmembrane RGS Protein That Modulates Plant Cell Proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- Chen JG, Jones AM. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 2004;389:338–350. doi: 10.1016/S0076-6879(04)89020-7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Periasamy A. FRET Data Analysis: The Algorithm. New York: Oxford University Press; 2005. [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell. 2006;18:2314–2325. doi: 10.1105/tpc.106.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R, Lewis R, Goodnow C, Healy J. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Gupta A, Singh M, Jones AM, Laxmi A. Hypocotyl directional growth in Arabidopsis: A complex trait. Plant Physiology. 2012;159:1463–1476. doi: 10.1104/pp.112.195776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci U S A. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Temple BR, Jones AM, Dohlman HG. Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem. 2011;286:13143–13150. doi: 10.1074/jbc.M110.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated Gene Expression in Plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Lilley JLS, Gee CW, Sairanen I, Ljung K, Nemhauser JL. An Endogenous Carbon-Sensing Pathway Triggers Increased Auxin Flux and Hypocotyl Elongation. Plant Physiology. 2012;160:2261–2270. doi: 10.1104/pp.112.205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBP delta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Zakarian A, Bouchard D, Michie AM, Zúñiga-Pflücker JC, Ohashi PS. Duration and Strength of Extracellular Signal-Regulated Kinase Signals Are Altered During Positive Versus Negative Thymocyte Selection. The Journal of Immunology. 2001;167:4966–4973. doi: 10.4049/jimmunol.167.9.4966. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Strope PK, Opiyo SO, Chen Z, Jones AM. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:R96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan N, Urano Daisuke, Srba Miroslav, Fischer Lukas, Jones Alan. Sugar-induced endocytosis of plant 7TM-RGS proteins. Plant Signaling & Behavior. 2013;8:0–1. doi: 10.4161/psb.22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis Jeremy E, Lahav G. Encoding and Decoding Cellular Information through Signaling Dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rué P, Domedel-Puig N, Garcia-Ojalvo J, Pons AJ. Integration of cellular signals in chattering environments. Progress in Biophysics and Molecular Biology. 2012;110:106–112. doi: 10.1016/j.pbiomolbio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Singhania R. Modeling Protein Regulatory Networks that Control Mammalian Cell Cycle Progression and that Exhibit Near-Perfect Adaptive Responses. 2011 [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology. 2010;13:273–278. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen J-G, Young JC, Im K-H, Sussman MR, Jones AM. Modulation of Cell Proliferation by Heterotrimeric G Protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- Urano D, Chen J-G, Botella JR, Jones AM. Heterotrimeric G protein signalling in the plant kingdom. Open Biology. 2013;3 doi: 10.1098/rsob.120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones JC, Wang H, Matthews M, Bradford W, Bennetzen JL, Jones AM. G Protein Activation without a GEF in the Plant Kingdom. PLoS Genet. 2012a;8:e1002756. doi: 10.1371/journal.pgen.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Philip Taylor J, Jones AM. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012b doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Phan N, Jones JC, Yang J, Huang J, Grigston J, Philip Taylor J, Jones AM. Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat Cell Biol. 2012c;14:1079–1088. doi: 10.1038/ncb2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltermann C, Klipp E. Information theory based approaches to cellular signaling. Biochimica et Biophysica Acta (BBA) - General Subjects. 2011;1810:924–932. doi: 10.1016/j.bbagen.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biology. 2008;10:548–562. doi: 10.1111/j.1438-8677.2008.00072.x. [DOI] [PubMed] [Google Scholar]
- Yagi YI, Abe K, Ikebukuro K, Sode K. Kinetic mechanism and inhibitor characterization of WNK1 kinase. Biochemistry. 2009;48:10255–10266. doi: 10.1021/bi900666n. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Wang L, Zheng S, Xie J, Bi Y. Sucrose-induced hypocotyl elongation of Arabidopsis seedlings in darkness depends on the presence of gibberellins. Journal of Plant Physiology. 2010;167:1130–1136. doi: 10.1016/j.jplph.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.