Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is a positive-sense ssRNA virus whose envelope contains four glycoproteins and three nonglycosylated proteins. Glycans of major envelope glycoprotein 5 (GP5) are proposed as important for virus assembly and entry into permissive cells. Structural characterization of GP5 glycans would facilitate the mechanistic understanding of these processes. Thus, we purified the PRRSV type 2 prototype strain, VR2332, and analyzed the virion-associated glycans by both biochemical and mass spectrometric methods. Endoglycosidase digestion showed that GP5 was the primary protein substrate, and that the carbohydrate moieties were primarily complex-type N-glycans. Mass spectrometric analysis (HPLC-ESI-MS/MS) of GP5 N-glycans revealed an abundance of N-acetylglucosamine (GlcNAc) and N-acetyllactosamine (LacNAc) oligomers in addition to sialic acids. GlcNAc and LacNAc accessibility to ligands was confirmed by lectin co-precipitation. Our findings help to explain PRRSV infection of cells lacking sialoadhesin and provide a glycan database to facilitate molecular structural studies of PRRSV.
Keywords: PRRSV, GP5, N-glycan, lectin, mass spectrometry, veterinary virology, virion structure, GlcNAc, LacNAc
Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV), first isolated as the type 1 European strain, Lelystad virus (LV), in the Netherlands (Wensvoort, 1993) and shortly after as the type 2 North American strain, VR-2332, in the USA (Benfield et al., 1992, Collins et al., 1992), is the etiologic agent of “mystery swine disease” that emerged in 1980s and spread worldwide thereafter. The representative syndromes of the disease include reproductive failure in sows and respiratory distress in growing pigs. Based on similar genomic organization and transcription strategy, PRRSV, together with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV) and simian hemorrhagic fever virus (SHFV), belongs to the order Nidovirales, family Arteriviridae, genus Arterivirus (Cavanagh, 1997).
Mature PRRS virions are composed of a nucleocapsid core enclosing a positive-sense, single-stranded RNA genome of ~15 kb, and an envelope harboring critical transmembrane proteins (Conzelmann et al., 1993, Dea et al., 1995, Dea et al., 2000, Mardassi et al., 1996). The major envelope proteins GP5 and matrix (M) form heterodimeric complexes linked by N-terminal ectodomain disulfide bonds and together comprise at least half of the viral proteins (Dea et al., 2000, Mardassi et al., 1996, Meulenberg et al., 1995, Wissink et al., 2005). PRRSV particles display a smooth outline of the envelope with few protruding features, consistent with predicted small ectodomains of GP5 and M (30 residues for GP5 and 16 for M) (Dokland, 2010, Spilman et al., 2009). GP5 contains 3 putative N-glycosylation sites at residues 33, 44 and 51 in VR-2332 and 2 putative N-glycosylation sites at residues 46 and 53 in LV. Lack of the oligosaccharides linked to N44 (type 2 PRRSV) and N46 (LV) in GP5 impairs the production of infectious progeny virus and significantly reduces viral infectivity (Ansari et al., 2006, Wissink et al., 2004). Minor proteins GP2a, E, GP3 and GP4 are incorporated as multimeric complexes in the envelope, with the glycoproteins containing conserved N-glycosylation sites in both strains (Wissink et al., 2005). Therefore, the broadly distributed viral glycans likely cover the virion surface and stretch out as antennae, thus interacting with host cells and contributing to viral biology. Removal of complex-type N-glycans from PRRSV reduced infectivity in porcine macrophages, suggesting an important role of viral glycans in infection (Delputte and Nauwynck, 2004). In particular, sialic acids on GP5 bind sialoadhesin on macrophages, mediating virus attachment and internalization (Delputte and Nauwynck, 2004, Van Breedam et al., 2010, Van Gorp et al., 2008). An N-acetylglucosamine (GlcNAc)-specific ligand also binds and reduces viral infectivity in MARC-145 cells (Keirstead et al., 2008).
Significant roles for PRRSV-associated glycans have been postulated in virus assembly, virus attachment to target cells, virus neutralization and immunological protection (Ansari et al., 2006, Das et al., 2011, Delputte and Nauwynck, 2004, Wissink et al., 2004). However, detailed knowledge of glycan structural information and distribution in viral envelope glycoproteins is essential to further evaluate the contributions of viral glycans to PRRSV pathogenesis and immune protection.
Therefore, we digested highly purified PRRSV with endoglycosidases and showed that GP5 is the major source of predominantly complex-type N-glycans. Mass spectrometric analysis confirmed this finding, and further revealed that the characteristic glycan structures contain N-acetylglucosamine (GlcNAc) and N-acetyllactosamine (LacNAc) oligomers and terminal sialic acids, whose accessibility was confirmed by lectin co-precipitation.
Results
GP5 contains complex-type N-glycans
There are four glycoproteins in the PRRSV envelope, the major protein GP5 and minor proteins GP2a, GP3 and GP4. According to the glycosylation prediction programs NetNGlyc 1.0 and NetOGlyc 3.1 (Center for Biological Sequence Analysis, Technical University of Denmark), all the envelope glycoproteins have exclusively N-linked glycosylation sites, but no O-linked glycosylation sites. Thus we focused our study on N-linked glycans.
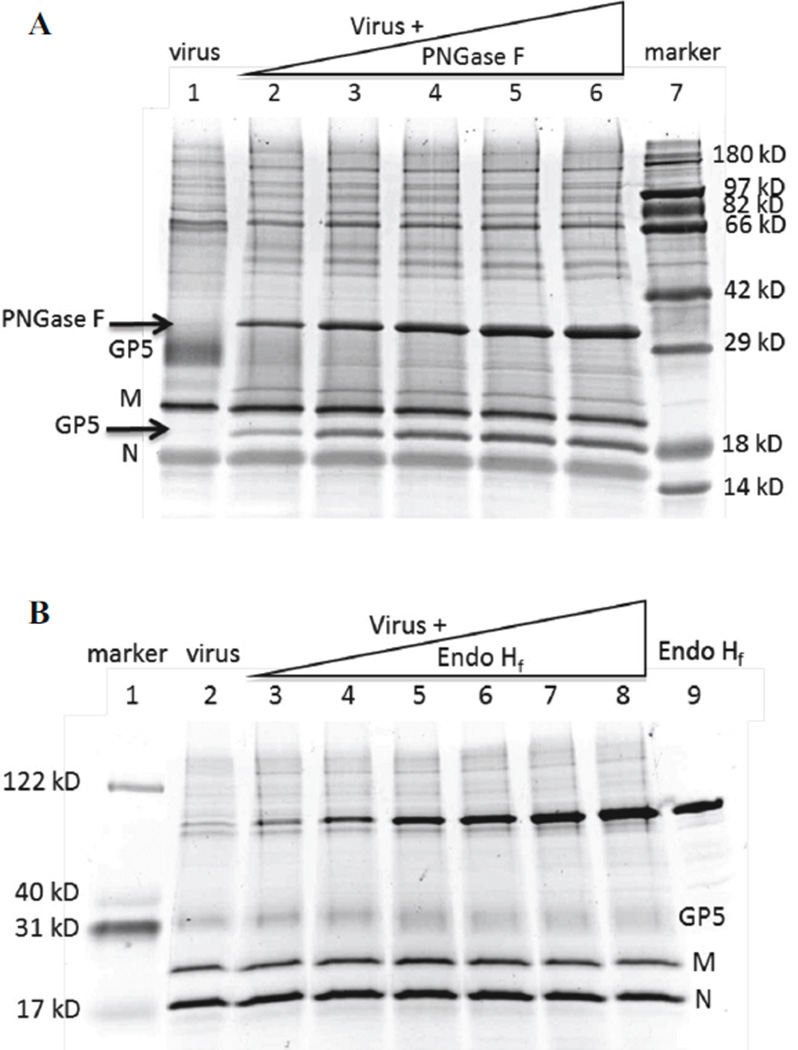
In reducing SDS-PAGE, purified PRRSV showed 3 major protein bands, GP5 (~25 kD), M (19 kD) and N (14 kD), the three major structural proteins of PRRSV (Fig. 1A). The minor envelope glycoproteins, GP2a, GP3 and GP4, were not visible due to low abundance. Incubation of purified virus with increasing amounts of PNGase F (36 kD, Fig. 1A arrow) caused a disappearance of GP5 at 25 kD and the appearance with increasing intensity of a new band between M and N (Fig. 1A). Mass spectrometric analysis identified this new band to be GP5 (arrow labeled GP5). Deglycosylated GP5 is about 19 kD, similar to M, but has a lower isoelectric point (pI 8.87) than M (pI 10.03), accounting for its appearance below M in the gel. Therefore, GP5 from VR-2332 contains exclusively PNGase F-sensitive N-glycans. No other bands shifted in the gel after PNGase F treatment, showing that GP5 contains the vast majority of viral N-glycans. A faint contaminating band between GP5 and M in purified virus was identified as trypsin by mass spectrometry, and stayed at the same position after PNGase F treatment.
Figure 1. Endoglycosidase digestion of purified PRRSV.
(A) Purified virus was incubated with PNGase F, electrophoresed in reducing SDS-PAGE, and the gel was stained with Ruby Protein Gel Stain. Lane 1, virus only. Lanes 2–6, virus treated with 100 to 500 units of PNGase F at 37° for 1 h. Lane 7, protein markers. (B) Purified virus was incubated with Endo Hf, samples were electrophoresed as above, and the gel was stained with Deep Purple. Lane 1, protein markers. Lane 2, virus only. Lanes 3–8, virus treated with 100 to 400 units of Endo Hf at 37° for 1 h. Lane 9, Endo Hf only. Bands containing GP5, M, and N are indicated to the right.
Treatment of purified virus with Endo Hf (70 kD), which cleaves high-mannose and hybrid-type N-glycans, caused the GP5 band to broaden but did not shift the protein to a new location (Fig. 1B). Thus, enzymatic digestion predicted that GP5 is predominantly composed of Endo Hf-resistant complex-type N-glycans with small amounts of high-mannose and/or hybrid-type N-glycans.
GP5-linked N-glycans contain GlcNAc and LacNAc oligomers and terminal sialic acids
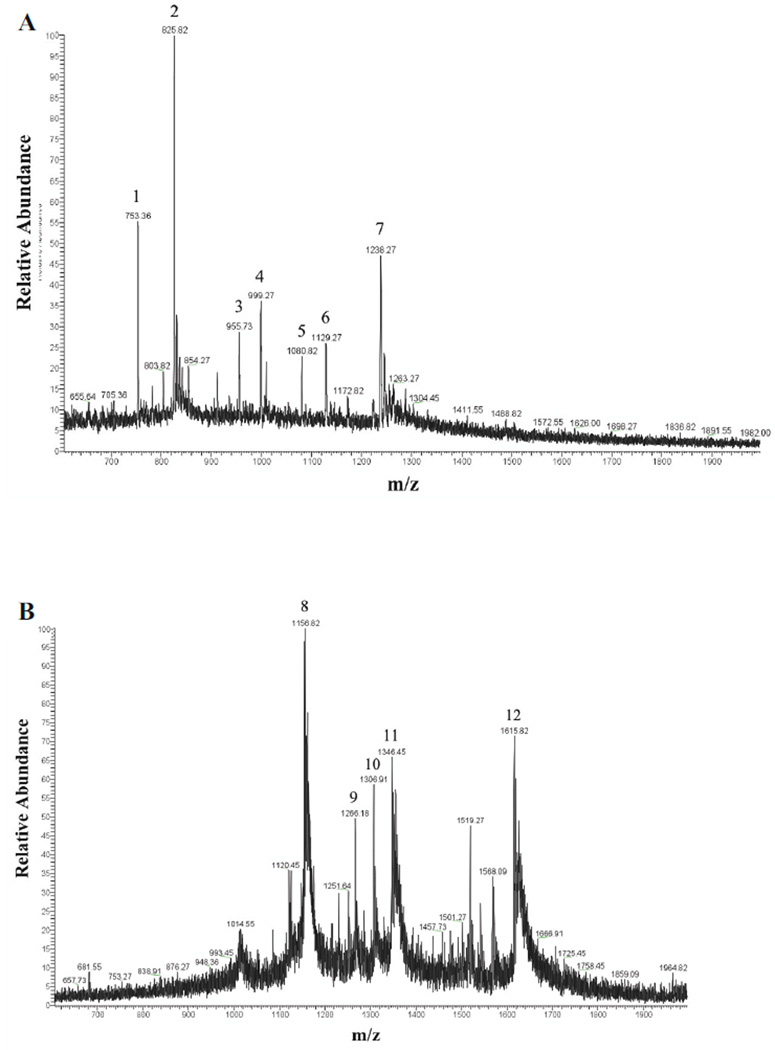
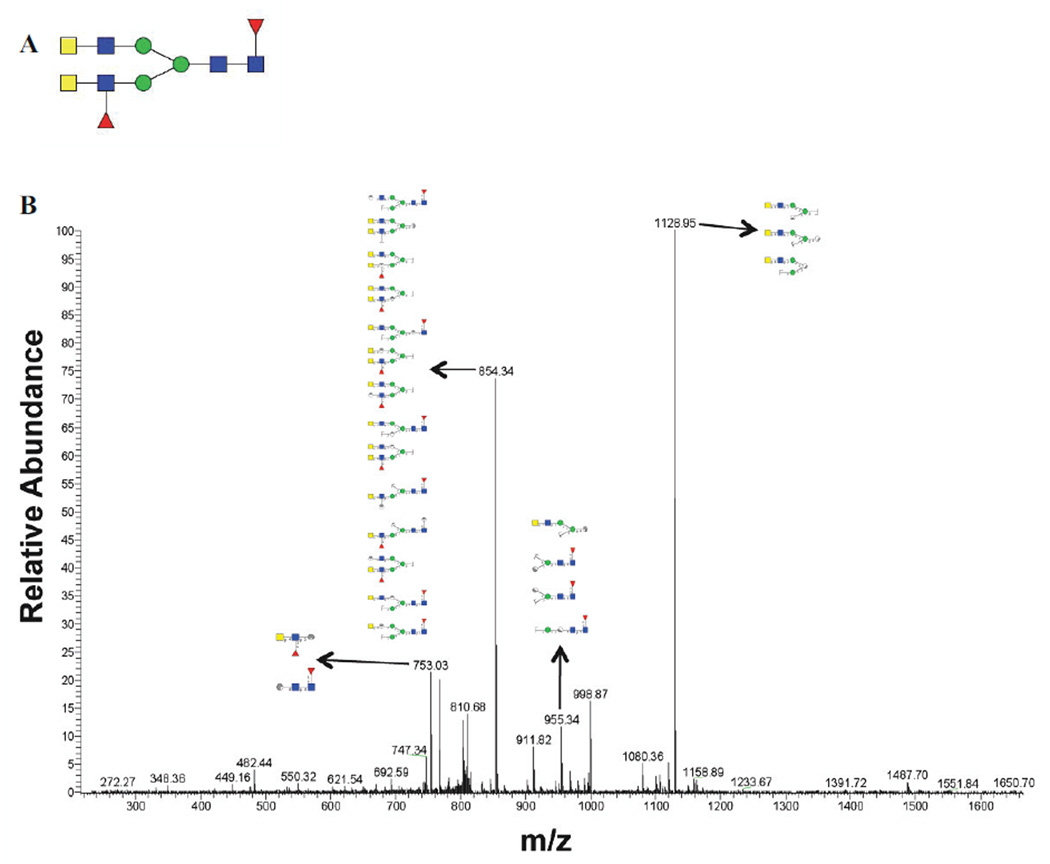
Mass spectrometric analysis of GP5-derived N-glycans was performed to characterize specific glycan compositions and structures. Figure 2 shows the positive ion electrospray spectra with 12 mass/charge (m/z) peaks identified as candidate N-linked glycans. Candidate glycan structures from Carbbank (Doubet and Albersheim, 1992) were assigned by GlycoWorkbench (Ceroni et al., 2008) according to the m/z values with a tolerance of ±1 Da. MS/MS fragmentation spectra were used to further reduce the candidate glycan structures by comparing the masses of the computed fragment ions to those observed experimentally. Glycan structures were considered to be reasonable candidate glycans on GP5 if their calculated precursor mass-to-charge ratios and fragmentation mass values were consistent with the experimental data. As an example of structural elucidation, the most abundant glycan (m/z 825.82) was assigned a structure with the monosaccharide composition Hex3HexNAc6Fuc2 (Fig. 2A). The MS/MS fragmentation spectrum showed ions in agreement with computed fragments of the proposed structure (Fig. 3). This approach was followed to determine the compositions and expected structures that would correspond to all 12 observed glycan peaks. The findings are summarized in Table 1. Thus, the major m/z 825.82 peak, for example, would result from any of 5 possible glycan compositions with 7 possible structures. Five of the seven possible structures do not contain sialic acid.
Figure 2. Electrospray spectra of PRRSV GP5 N-glycans.
The 12 numbered peaks were identified as N-glycans. (A) LC Retention time 53:47–54:28 min. (B) LC Retention time 55:52–56:14 min.
Figure 3. One of the candidate glycan structures of peak 2 (m/z 825.82) and its corresponding MS/MS spectrum.
(A) A candidate glycan structure of the major peak 2. Monosaccharide symbols follow the Consortium for Functional Glycomics guidelines and are detailed in Supplementary Data Table 1. (B) MS/MS fragmentation spectrum of peak 2 with matching substructures matching the predicted structure in panel A.
Table 1.
Monoisotopic masses, compositions and putative structures of GP5-linked N-glycans.
| Peak | Observed m/z |
Theory m/z |
Mass | Charge | Composition | Proposed structure | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | Xyl | NeuAc | NeuGc | Kdn | ||||||
| 1 | 753.36 | 753.36 | 2191.12 | 3 | 4 | 4 | 2 | 0 | 0 | 0 | 0 | 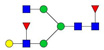 |
| 753.38 | 1482.75 | 2 | 3 | 2 | 1 | 1 | 0 | 0 | 0 | |||
| 752.82 | 1459.66 | 2 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | |||
| 752.37 | 2210.12 | 3 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | |||
| 2 | 825.82 | 825.73 | 2408.22 | 3 | 5 | 4 | 0 | 0 | 1 | 0 | 0 | 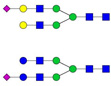 |
| 825.73 | 2408.22 | 3 | 4 | 4 | 1 | 0 | 0 | 1 | 0 | |||
| 826.73 | 2411.22 | 3 | 6 | 4 | 1 | 1 | 0 | 0 | 0 | |||
| 826.75 | 2455.24 | 3 | 7 | 4 | 0 | 0 | 0 | 0 | 0 | 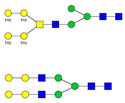 |
||
| 826.77 | 2477.27 | 3 | 3 | 6 | 2 | 0 | 0 | 0 | 0 | 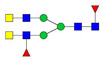 |
||
| 3 | 955.73 | 955.83 | 2864.47 | 3 | 3 | 9 | 0 | 0 | 0 | 0 | 0 | 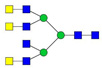 |
| 955.46 | 2797.42 | 3 | 4 | 5 | 2 | 0 | 1 | 0 | 0 | 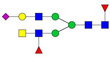 |
||
| 955.45 | 1908.88 | 2 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | |||
| 956.12 | 2799.40 | 3 | 5 | 4 | 0 | 0 | 1 | 1 | 0 | |||
| 956.14 | 2799.44 | 3 | 5 | 5 | 1 | 0 | 0 | 1 | 0 | |||
| 956.49 | 2844.45 | 3 | 6 | 5 | 2 | 0 | 0 | 0 | 0 |
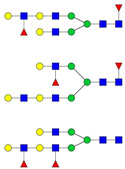 
|
||
| 4 | 999.27 | 999.50 | 2973.49 | 3 | 5 | 4 | 1 | 0 | 1 | 1 | 0 | |
| 999.50 | 2973.49 | 3 | 6 | 4 | 0 | 0 | 2 | 0 | 0 | |||
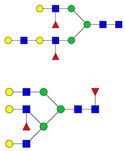
| 999.82 | 2930.49 | 3 | 5 | 4 | 3 | 0 | 1 | 0 | 0 | 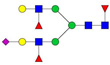 |
||
| 1000.00 | 1976.01 | 2 | 5 | 3 | 1 | 0 | 0 | 0 | 0 | 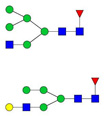 |
||
| 998.50 | 1973.01 | 2 | 3 | 4 | 1 | 1 | 0 | 0 | 0 | |||
| 998.49 | 2926.50 | 2 | 4 | 7 | 2 | 0 | 0 | 0 | 0 | 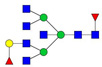 |
||
| 5 | 1080.82 | 1081.21 | 3218.61 | 3 | 6 | 5 | 0 | 0 | 2 | 0 | 0 | 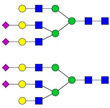 |
| 1081.54 | 2139.08 | 2 | 7 | 2 | 1 | 0 | 0 | 0 | 0 | 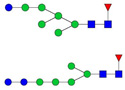 |
||
| 1081.56 | 2161.11 | 2 | 3 | 4 | 3 | 0 | 0 | 0 | 0 | 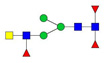 |
||
| 1079.87 | 3214.61 | 3 | 6 | 5 | 4 | 0 | 0 | 0 | 0 | 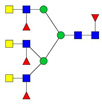 |
||
| 6 | 1129.27 | 1129.23 | 3362.69 | 3 | 5 | 5 | 2 | 0 | 2 | 0 | 0 | 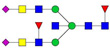 |
| 1129.91 | 3364.71 | 3 | 7 | 7 | 1 | 0 | 0 | 0 | 0 | 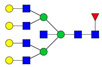 |
||
| 7 | 1238.27 | 1238.28 | 3667.84 | 3 | 7 | 6 | 0 | 0 | 2 | 0 | 0 | 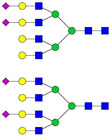 |
| 1238.09 | 2474.16 | 2 | 5 | 4 | 0 | 0 | 1 | 0 | 0 | |||
| 1238.63 | 3712.85 | 3 | 7 | 4 | 0 | 0 | 3 | 0 | 0 | 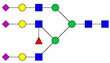 |
||
| 1238.64 | 3712.88 | 3 | 7 | 7 | 3 | 0 | 0 | 0 | 0 | 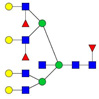 |
||
| 1238.96 | 3669.89 | 3 | 6 | 5 | 1 | 0 | 0 | 3 | 0 | |||
| 1237.62 | 3687.86 | 3 | 9 | 6 | 1 | 1 | 0 | 0 | 0 |  |
||
| 1237.61 | 3665.83 | 3 | 5 | 4 | 1 | 0 | 4 | 0 | 0 |  |
||
| 8 | 1156.82 | 1156.93 | 3467.76 | 3 | 7 | 6 | 3 | 0 | 0 | 0 | 0 | 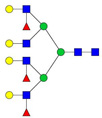 |
| 9 | 1266.18 | 1265.98 | 3794.90 | 3 | 5 | 6 | 1 | 0 | 3 | 0 | 0 | 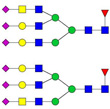 |
| 1265.97 | 3772.91 | 3 | 9 | 7 | 1 | 0 | 0 | 0 | 0 | |||
| 1265.64 | 2507.29 | 2 | 4 | 6 | 1 | 0 | 0 | 0 | 0 | 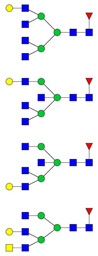 |
||
| 1266.95 | 3731.89 | 3 | 10 | 6 | 1 | 0 | 0 | 0 | 0 | 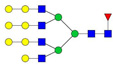 |
||
| 1266.96 | 3753.88 | 3 | 6 | 5 | 1 | 0 | 3 | 0 | 0 | 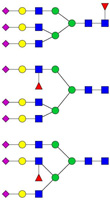 |
||
| 10 | 1306.91 | 1307.17 | 2612.31 | 2 | 6 | 4 | 0 | 0 | 1 | 0 | 0 | 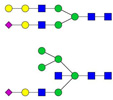 |
| 1307.17 | 2612.31 | 2 | 5 | 4 | 1 | 0 | 0 | 1 | 0 | |||
| 1307.64 | 3875.93 | 3 | 6 | 6 | 1 | 0 | 0 | 0 | 3 | 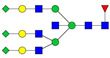 |
||
| 1307.65 | 2569.31 | 2 | 5 | 4 | 3 | 0 | 0 | 0 | 0 | 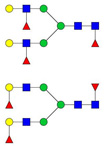 |
||
| 1306.18 | 2610.34 | 2 | 4 | 5 | 3 | 0 | 0 | 0 | 0 | 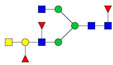 |
||
| 11 | 1346.45 | 1347.11 | 2648.25 | 2 | 5 | 4 | 1 | 0 | 1 | 0 | 0 | 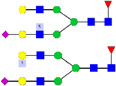 |
| 1347.18 | 2670.36 | 2 | 6 | 5 | 1 | 0 | 0 | 0 | 0 | 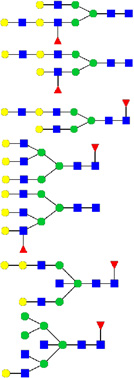 |
||
| 1345.67 | 1322.68 | 1 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | |||
| 1345.65 | 2689.28 | 2 | 4 | 5 | 1 | 0 | 1 | 0 | 0 | |||
| 12 | 1615.82 | 1614.81 | 1590.81 | 2 | 6 | 5 | 2 | 0 | 1 | 0 | 0 | 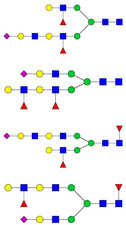 |
Candidate GP5-linked N-glycan structures were a mixture of bi-, tri-, and tetra-antennary complex (n=63), high-mannose (n=5), and hybrid (n=11) carbohydrates with or without core fucose (Table 1). Consistent with the endoglycosidase digestion results, GP5 contained primarily complex-type N-glycans. GlcNAc oligomers were found in all the candidate hybrid and complex-type structures. Bisecting GlcNAc was detected in 2 hybrid and 9 complex candidate structures. Type 2 LacNAc (Galβ1-4GlcNAc) was present in 4 hybrid and 55 complex glycan structures, with 12 containing 2–4 tandem repeats of LacNAc. Antennae were mainly terminated with galactose (Gal), N-acetylglucosamine (GlcNAc), N-acetyllactosamine (LacNAc) and various forms of sialic acid (N-acetylneuraminic acid, NeuAc; N-glycolylneuraminic acid, NeuGc; and 2-keto-3-deoxynononic acid, Kdn). NeuAc was the major sialic acid terminating the candidate glycan structures.
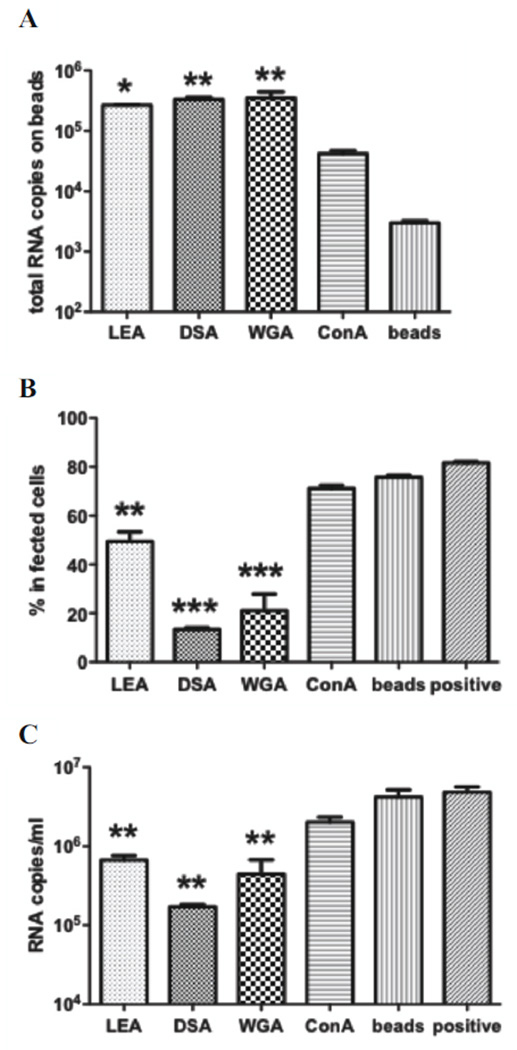
Lectins specific for GlcNAc and LacNAc oligomers and sialic acids bind PRRSV
To verify the presence of GlcNAc and LacNAc oligomers and terminal sialic acids in the viral glycan structures, lectins of varied specificities were coupled to Sepharose beads and incubated with PRRSV to evaluate their virus-binding capabilities. Based on the amount of precipitated virus, the order of the bead treatments was wheat germ agglutinin (WGA) ≈ Datura stramonium agglutinin (DSA) > Lycopersicon esculentum agglutinin (LEA) > concanavalin A (ConA) > blank beads (Fig. 4A). WGA, DSA and LEA, which bind GlcNA oligomers, significantly depleted PRRSV, and significantly reduced cell-culture infectivity of the resulting mixture, compared to ConA, a mannose-specific lectin, blank beads, and the untreated virus preparation (Fig. 4B). Also, viral growth, as measured by progeny virus in culture supernatant, was significantly reduced in the groups treated with WGA, DSA and LEA (Fig. 4C). These results demonstrate that GlcNAc and LacNAc oligomers, in addition to terminal sialic acids, are accessible on PRRSV virions for specific ligand binding, whereas high mannose glycans do not contribute substantially to ligand binding.
Figure 4. Lectin co-precipitation of PRRSV.
LEA, DSA, WGA and ConA-coupled beads and control beads were incubated with PRRSV, pelleted, and washed. Effect of treatment was assessed by quantitation of virus amount bound to beads, by qRT-PCR (A), depletion of infectious virus from supernatant determined by inoculation of supernatant onto MARC-145 cells and assessment of infection 1 day later by flow cytometry (B), and reduction in progeny virus from MARC-145 culture. Data were analyzed by one-way ANOVA with Tukey’s multiple comparison test. Significant differences shown were compared to blank beads. *, p <0.05; **, p <0.01 ***, p <0.001.
Although the lectins, WGA, DSA and LEA, all recognize GlcNAc oligomers, there are subtle differences in carbohydrate specificity. WGA reacts strongly with the chitobiose core of bisected hybrid-type N-glycans, specifically with the oligosaccharide GlcNAcβ1-4Manβ1-4GlcNAcβ1-4GlcNAc (Yamamoto et al., 1981, Yodoshi et al., 2011), which was proposed in the candidate glycan structure of peak 10 (Fig. 2B, m/z 1306.91). To a lesser extent, WGA has an affinity for broadly linked sialic acids (Neu5Ac), which were found in the termini of 1 hybrid and 27 complex-type candidate structures. DSA and LEA recognize GlcNAc oligomers that may not be consecutive (Kawashima et al., 1990) and were found in most candidate hybrid and complex-type N-glycan structures. In addition, DSA reacts with the complex-type N-glycans containing the structure Galβ1-4GlcNAcβ1-6(Galβ1-4GlcNAcβ1-2)Man or at least one LacNAc (Galβ1-4GlcNAc) unit in an outer chain, and the interaction is not affected by the presence of a bisecting GlcNAc residue (Yamashita et al., 1987). There were 55 candidate glycan structures that satisfy the LacNAc binding properties of DSA. In contrast, LEA requires 2 consecutive LacNAc residues (Kawashima et al., 1990), which were found in only 12 candidate structures. The difference in LacNAc-binding properties explains why DSA bound more virus than LEA. ConA is specific for terminal α-D-mannose and α-D-glucose (Maupin et al., 2011), which were present in 5 high-mannose and 11 hybrid-type candidate structures. The lack of significant ConA-binding is consistent with endoglycosidase digestion results showing that high-mannose and hybrid-type N-glycans are minor components of the PRRSV virion.
Discussion
PNGase F sensitivity and Endo H resistance, indicating the predominance of complex-type N-glycans, is a general characteristic of PRRSV grown on MA-104 cells, including type 1 Lelystad virus and type 2 IAF-Klop strain (Mardassi et al., 1996, Meulenberg et al., 1995). These glycans reside primarily on GP5 and, as we now show, have characteristic features of GlcNAc and LacNAc oligomers and terminal sialic acids. Viral protein glycans are obtained from host cells, and the glycan patterns are dependent on the specific host-cell modifying enzymes (Dalpathado et al., 2006, Liedtke et al., 1994). The main pathway of N-glycan biosynthesis is conserved among eukaryotic species, beginning as a dolichol-linked precursor that is transferred to an Asn-X-Ser/Thr sequon in a protein and modified to a high-mannose type in the endoplasmic reticulum (Varki et al., 2009). The protein-bound N-glycan is then translocated to the Golgi and further processed to hybrid and complex-type N-glycans. Thus, presence of high-mannose glycans frequently implies minimal processing and a more protected local conformational structure. Thus, the presence of fully processed (hybrid and complex) glycans in PRRSV indicates that the glycosylation site is readily accessible, and thus reflects a more flexible and exposed protein structure (Go et al., 2008, Kong et al., 2010, Leonard et al., 1990, Yeh et al., 1993).
PRRSV has a restricted cell tropism, infecting primarily porcine macrophages, the natural host cell, and a green monkey kidney epithelial-like cell line (MA-104, also known as CL2621, and a subclone, MARC-145) that has been routinely used for in vitro propagation of PRRSV isolates and modified live vaccines. Structural glycan characterization of the virus grown in simian cells will facilitate a better understanding of PRRSV molecular and cellular biology, and provide a reference for interpretation of glycan features of PRRSV grown in macrophages. Interestingly, the external envelope glycoprotein of human immunodeficiency virus type 2 (HIV-2) grown in macrophages displays a complex-type N-glycan pattern with LacNAc repeats as is observed in PRRSV (Liedtke et al., 1994).
Among the characteristic features of PRRSV glycans, sialic acid binding to sialoadhesin on porcine macrophages is reported to mediate virus attachment and internalization (Delputte and Nauwynck, 2004, Van Breedam et al., 2010, Van Gorp et al., 2008). Our data show that GlcNAc and LacNAc oligomers on the surface of PRRSV also can bind protein ligands. Since macrophages express galectin-3, which binds the poly-LacNAc-containing N-glycans, these oligomers may provide an alternative route mediating viral attachment and internalization (Dumic et al., 2006). Indeed, lectin binding to viral GlcNAc and LacNAc oligomers may be the principal route for glycan-assisted PRRSV infection of MA-104 cells and CD163-transfected PK-15 cells, neither of which express sialoadhesin, (Calvert et al., 2007). Similarly, in vivo ablation of sialoadhesin had no measurable effect on PRRSV infection of susceptible pigs (Prather et al., 2013). Alternatively, it also is possible that viral glycans are not involved in permissive cell infection, but play a role in other host-cell interactions such as immune avoidance.
The VR2332 strain GP5 contains 3 N-glycosylation sites but it is not certain if all are glycosylated. The total mass of glycans on GP5 is estimated by SDS gel electrophoresis to be 6–7 kD. Thus, it was postulated that all 3 sites contain oligosaccharides of ~2.5 kD (Ansari et al., 2006). Since the glycan structures identified here are in the range of 2 to 4 kD, either 2 or 3 glycosylation sites may be utilized. The VR2332 GP5 N33 in the sequon NDS, in particular, may be nonglycosylated since the site is not highly conserved (Murtaugh et al., 2010), it contains D34, which is linked to poor glycosylation efficiency (Kasturi et al., 1997), and it contains S35, which is less preferred in the third position than is T that is present in the N44 and N51 sequons (Kasturi et al., 1997, Rao and Bernd, 2010). We attempted to determine the prevalence of glycosylation at N33, N44 and N51 by PNGase F digestion of glycopeptides in 18O-labeled or normal water. Product peptides with an “occupied” glycosylation site, when digested in 18O water, would show a 3-Dalton molecular weight increase compared to the peptides digested in normal water. Since all the three GP5 glycosylation sites reside in the same tryptic peptide, endoproteinases Glu-C and Asp-N were used to isolate each glycosylation site on an individual peptide. Unfortunately, in-gel digestion of GP5 with these enzymes was not successful for unknown reasons (data not shown).
The discovery that GlcNAc and LacNAc oligomers as well as terminal sialic acids are on the PRRSV envelope and readily accessible for specific binding may explain the ability of PRRSV to infect permissive cells in the absence of sialoadhesin and it helps provide a mechanistic basis for variation in efficiency of cell culture of PRRSV isolates (de Abin et al., 2009). The potential for structural variation in viral glycan composition differences due to growth in porcine macrophages versus simian cells might also contribute to attenuation of vaccine strains or other biological characteristics.
Materials and Methods
Virus and cells
The North American prototype PRRSV, ATCC VR-2332, GenBank ID: PRU87392 (American Type Culture Collection, Manassas VA) (Benfield et al., 1992, Nelsen et al., 1999), was propagated in MARC-145 cells (Kim et al., 1993) using MEM (Mediatech, Herndon VA) containing 10% FBS (Mediatech), 1 mg/ml sodium bicarbonate (Sigma-Aldrich, St. Louis MO), 1% (v/v) 100× nonessential amino acid solution (Mediatech), 10 mM HEPES solution (Mediatech) and 20 µg/ml gentamicin (Invitrogen, Carlsbad CA) at 37°C in a 5% CO2 incubator. After 2 days of infection, virus was collected from culture supernatant, clarified by centrifugation, titered on MARC-145 cells, and stored in aliquots at −80°C.
Virus purification
VR-2332 was propagated in MARC-145 cells for two days. Debris was removed from spent media by centrifugation at 17,000 × g for 1 h, and supernatant was mixed with 10% polyethyleneglycol-8000 (Fisher Scientific, Fair Lawn NJ) overnight at 4°C. The virus was pelleted at 22,000 × g for 2 h, resuspended in Tris-buffered saline (TBS), and pelleted twice through a 0.5 M sucrose (Fisher Scientific) cushion at 110,000 × g for 3 h. The final pellet was re-suspended in 20% iodixanol (Sigma) in TBS and banded twice in a self-generating gradient by ultracentrifugation at 250,000 × g for 9 h. The purified virus band was removed with a sterile needle and stored in aliquots at −80°C.
Endoglycosidase digestion and SDS-PAGE
In non-reducing conditions, 2 µg of purified PRRSV was incubated with 100~500 units of peptide-N-glycosidase F (PNGase F, New England Biolabs, Ipswich MA) in 50 mM sodium phosphate, pH 7.5, or 100~400 units of endoglycosidase Hf (Endo Hf, New England Biolabs) in 50 mM sodium citrate, pH 5.5, at 37°C for 1 h. The mixture of virus and endoglycosidase was added to gel loading buffer with 5% β-mercaptoethanol, boiled for 10 min and electrophoresed in 10–20% gradient Tris-HCl Ready Gels (Bio-Rad Laboratories, Hercules CA). For PNGase F samples, CandyCane Glycoprotein Molecular Weight Standards (Invitrogen) were used to estimate protein size and protein bands were visualized with RubyProtein Gel Stain (Invitrogen). For Endo Hf samples, Kaleidoscope Prestained Standards (Bio-Rad) were used to estimate protein size and protein bands were visualized with Deep Purple Total Protein Stain (GE Healthcare, Buckinghamshire, UK). Finally, proteins were analyzed in EpiChemi3 Darkroom (UVP, Upland CA) using LabWorks 4.5 software (UVP).
Mass spectrometry identification of structural proteins
The identity of glycosylated and nonglycosylated GP5, M and N bands visualized in polyacrylamide gels was confirmed in excised gel pieces by micro liquid chromatorgraphy-pulsed Q collision induced dissociaton (µLC-PDQ) tandem mass spectrometry (MS/MS) at the Center for Mass Spectrometry and Proteomics, University of Minnesota. Data were analyzed as described with the SEAQUEST program (Bioworks version 3.2, Thermo Fisher) to search against a composite database of all PRRSV VR2332 ORFs and a subset of monkey, pig and random contaminants from the NCBI nonredundant protein database (Johnson et al. 2011).
N-glycan release from GP5
Approximately 150 µg of purified VR-2332 was run with 5% β-mercaptoethanol in SDS-PAGE as above and visualized with Imperial Protein Stain (Thermo Scientific, Rockford IL). In-gel trypsin digestion was performed to extract GP5 glycopeptides (adapted from Shevchenko et al., 1996). Briefly, GP5 bands were excised from the acrylamide gel and Coomassie blue dye was extracted with a mixture of 50% acetonitrile (Sigma) and 50% 100 mM ammonium bicarbonate (Sigma). The protein bands were then treated with 10 mM dithiothreitol to reduce cysteines, alkylated in 55 mM iodoacetamide and thereafter incubated with 12.5 ng/µl trypsin (Invitrogen) at 37°C overnight. GP5 glycopeptides were then collected and lyophilized in a Speed-Vac refrigerated concentrator (Thermo Scientific). To deactivate trypsin, sample digests were reconstituted in 50 mM ammonium bicarbonate and heated at 100°C for 10 min. After cooling, 1500 units of PNGase F was added and incubated at 37°C overnight to release N-glycans.
N-glycan purification and modification
Lyophilized PNGase F digests were reconstituted in 5% acetic acid and passed through a Sep-Pak C18 cartridge (J.T. Baker, Phillipsburg NJ) to separate peptides and glycans. Glycans were eluted with 5% acetic acid and lyophilized overnight. Purified glycans were resuspended in a slurry of NaOH in DMSO and permethylated with iodomethane. After chloroform extraction, permethylated glycans were dried in a stream of N2. Permethylated N-glycans were dissolved in 50% methanol and applied to a C18 column to remove salts and other contaminants before MS analysis. The column was washed with Milli-Q water and followed by 15% acetonitrile. N-glycans were eluted with 85% acetonitrile, and dried in a stream of N2.
Liquid chromatography and electrospray mass spectrometry
Reversed phase HPLC-ESI-MS/MS analysis of permethylated N-glycans was performed on a hybrid linear ion-trap Fourier Transform ion cyclotron resonance (FTICR) mass spectrometer (LTQ-FT, Thermo Finnigan). The separation column was a C18 reversed phase capillary column (Magic C18 AQ, 0.2×50mm). Mobile phases consisted of solvent A, 99.9% deionized H2O and 0.1% formic acid, and solvent B, 99.9% acetonitrile and 0.1% formic acid, which were pumped at a flow rate of 5 µl/min. For LC separation, the gradient went from 20% to 85% of solvent B in 65 min. MS/MS analysis was operated in a data-dependent scanning mode; a full MS scan within the m/z range 600–2000 followed by data-dependent MS/MS scans of the nine most intense glycan ions from the full MS scan. Glycan precursor ions were isolated for MS/MS using an isolation width of 3.0 m/z, and a normalized collision energy of 35% was applied for fragmentation. Manual interpretation of all glycan structures was carried out with in-house fragmentation rules and GlycoWorkbench (http://www.glycoworkbench.org/). Cartoon representations of the glycan structures were constructed using standard Consortium for Functional Glycomics (CFG) nomenclature for monosaccharides.
Lectin-virus co-precipitation
Lycopersicon esculentum agglutinin (LEA), Datura stramonium agglutinin (DSA), wheat germ agglutinin (WGA) and concanavalin A (ConA) were purchased from Sigma. Approximately 15 nmoles of lectin was coupled to CNBr-activated Sepharose 4B beads. Coupled and blank beads were then incubated with virus solution at 4°C for 3 h. After washing with PBS three times, the amount of virus bound to beads was determined by quantitative RT-PCR.
PRRSV quantitative reverse transcriptase (RT)-PCR
Total RNA isolation was performed using a Viral RNA Mini Kit following the manufacturer’s protocol (Qiagen, Valencia CA). RNA was eluted in 50 µl water and 10 µl was used to prepare cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City CA). Quantitative real time PCR was carried out with 5 µl of 1:10 diluted cDNA, 10 µl of PerfeCta SYBR Green Fast Mix (Quanta Biosciences, Gaithersburg MD), and 5 µl of 1 µM primers with forward primer GATAACCACGCATTTGTCGTC, and reverse primer TGCCGTTGTTATTTGGCATA. To quantify viral RNA copies, RNA extracted from a titered VR-2332 stock was used as standards. The final results were expressed as total RNA copies according to the standard curve.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software, San Diego CA). Lectin co-precipitation data were analyzed by one-way ANOVA with Tukey’s multiple comparison test. A p-value <0.05 was considered statistically significant.
Supplementary Material
Highlights.
PRRSV glycans are primarily N-acetylglucosamine and N-acetyllactosamine oligomers
Lectin binding to virions is strongest to GlcNAc and LacNAc moieties
PRRSV can bind cells via multiple glycan structures including GlcNAc and LacNAc
The findings help explain PRRSV infection of permissive cells lacking sialoadhesin
Acknowledgements
The work was funded in part by grant number 09-227 from the U.S. National Pork Board, the National Institutes of Health/NCRR-funded Integrated Technology Resource for Biomedical Glycomics (P41 RR018502), and USDA NIFA multistate project MIN 63-084.
Abbreviations
- PRRSV
porcine reproductive and respiratory syndrome virus
- N-glycan
asparagine-linked glycan
- PNGase F
peptide-N-glycosidase F
- Endo Hf
endoglycosidase Hf
- GlcNAc
N-acetylglucosamine
- LacNAc
N-acetyllactosamine
- Gal
galactose
- NeuAc
N-acetylneuraminic acid
- NeuGc
N-glycolylneuraminic acid
- Kdn
2-keto-3-deoxynononic acid
- Fuc
fucose
- Sia
sialic acid
- LEA
Lycopersicon esculentum agglutinin
- DSA
Datura stramonium agglutinin
- WGA
wheat germ agglutinin
- ConA
concanavalin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 2006;80:3994–4004. doi: 10.1128/JVI.80.8.3994-4004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J. Vet. Diagn. Invest. 1992;4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- Calvert JG, Slade DE, Shields SL, Jolie R, Mannan RM, Ankenbauer RG, Welch SK. CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. J. Virol. 2007;81:7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpathado DS, Irungu J, Go EP, Butnev VY, Norton K, Bousfield GR, Desaire H. Comparative glycomics of the glycoprotein follicle stimulating hormone: glycopeptide analysis of isolates from two mammalian species. Biochemistry. 2006;45:8665–8673. doi: 10.1021/bi060435k. [DOI] [PubMed] [Google Scholar]
- Das PB, Vu HL, Dinh PX, Cooney JL, Kwon B, Osorio FA, Pattnaik AK. Glycosylation of minor envelope glycoproteins of porcine reproductive and respiratory syndrome virus in infectious virus recovery, receptor interaction, and immune response. Virology. 2011;410:385–394. doi: 10.1016/j.virol.2010.12.002. [DOI] [PubMed] [Google Scholar]
- de Abin MF, Spronk G, Wagner M, Fitzsimmons M, Abrahante JE, Murtaugh MP. Comparative infection efficiency of Porcine reproductive and respiratory syndrome virus field isolates on MA104 cells and porcine alveolar macrophages. Can. J. Vet. Res. 2009;73:200–204. [PMC free article] [PubMed] [Google Scholar]
- Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates. Arch. Virol. 2000;145:659–688. doi: 10.1007/s007050050662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dea S, Sawyer N, Alain R, Athanassious R. Ultrastructural characteristics and morphogenesis of porcine reproductive and respiratory syndrome virus propagated in the highly permissive MARC-145 cell clone. Adv. Exp. Med. Biol. 1995;380:95–98. doi: 10.1007/978-1-4615-1899-0_13. [DOI] [PubMed] [Google Scholar]
- Delputte PL, Nauwynck HJ. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 2004;78:8094–8101. doi: 10.1128/JVI.78.15.8094-8101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T. The structural biology of PRRSV. Virus Res. 2010;154:86–97. doi: 10.1016/j.virusres.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubet S, Albersheim P. CarbBank. Glycobiology. 1992;2:505. doi: 10.1093/glycob/2.6.505. [DOI] [PubMed] [Google Scholar]
- Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim. Biophys. Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 2011;92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi L, Chen H, Shakin-Eshleman SH. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J. 1997;323(Pt 2):415–419. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H, Sueyoshi S, Li H, Yamamoto K, Osawa T. Carbohydrate binding specificities of several poly-N-acetyllactosamine-binding lectins. Glycoconj. J. 1990;7:323–334. doi: 10.1007/BF01073376. [DOI] [PubMed] [Google Scholar]
- Keirstead ND, Lee C, Yoo D, Brooks AS, Hayes MA. Porcine plasma ficolin binds and reduces infectivity of porcine reproductive and respiratory syndrome virus (PRRSV) in vitro. Antiviral Res. 2008;77:28–38. doi: 10.1016/j.antiviral.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kwang J, Yoon IJ, Joo HS, Frey ML. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- Kong L, Sheppard NC, Stewart-Jones GB, Robson CL, Chen H, Xu X, Krashias G, Bonomelli C, Scanlan CN, Kwong PD, Jeffs SA, Jones IM, Sattentau QJ. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 2010;403:131–147. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- Liedtke S, Adamski M, Geyer R, Pfutzner A, Rubsamen-Waigmann H, Geyer H. Oligosaccharide profiles of HIV-2 external envelope glycoprotein: dependence on host cells and virus isolates. Glycobiology. 1994;4:477–484. doi: 10.1093/glycob/4.4.477. [DOI] [PubMed] [Google Scholar]
- Mardassi H, Massie B, Dea S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology. 1996;221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- Maupin KA, Liden D, Haab BB. The fine-specificity of mannose-binding and galactose-binding lectins revealed using outlier-motif analysis of glycan array data. Glycobiology. 2011;22:160–169. doi: 10.1093/glycob/cwr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg JJ, Petersen-den Besten A, De Kluyver EP, Moormann RJ, Schaaper WM, Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh MP, Stadejek T, Abrahante JE, Lam TT, Leung FC. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 2010;154:18–30. doi: 10.1016/j.virusres.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather RS, Rowland RR, Ewen C, Trible B, Kerrigan M, Bawa B, Teson JM, Mao J, Lee K, Samuel MS, Whitworth KM, Murphy CN, Egen T, Green JA. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J. Virol. 2013;87:9538–9546. doi: 10.1128/JVI.00177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Spilman MS, Welbon C, Nelson E, Dokland T. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J. Gen. Virol. 2009;90:527–535. doi: 10.1099/vir.0.007674-0. [DOI] [PubMed] [Google Scholar]
- Van Breedam W, Van Gorp H, Zhang JQ, Crocker PR, Delputte PL, Nauwynck HJ. The M/GP(5) glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 2010;6:e1000730. doi: 10.1371/journal.ppat.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gorp H, Van Breedam W, Delputte PL, Nauwynck HJ. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2008;89:2943–2953. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Wensvoort G. Lelystad virus and the porcine epidemic abortion and respiratory syndrome. Vet. Res. 1993;24:117–124. [PubMed] [Google Scholar]
- Wissink EH, Kroese MV, Maneschijn-Bonsing JG, Meulenberg JJ, van Rijn PA, Rijsewijk FA, Rottier PJ. Significance of the oligosaccharides of the porcine reproductive and respiratory syndrome virus glycoproteins GP2a and GP5 for infectious virus production. J. Gen. Virol. 2004;85:3715–3723. doi: 10.1099/vir.0.80402-0. [DOI] [PubMed] [Google Scholar]
- Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 2005;79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Tsuji T, Matsumoto I, Osawa T. Structural requirements for the binding of oligosaccharides and glycopeptides to immobilized wheat germ agglutinin. Biochemistry. 1981;20:5894–5899. doi: 10.1021/bi00523a037. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Totani K, Ohkura T, Takasaki S, Goldstein IJ, Kobata A. Carbohydrate binding properties of complex-type oligosaccharides on immobilized Datura stramonium lectin. J. Biol. Chem. 1987;262:1602–1607. [PubMed] [Google Scholar]
- Yeh JC, Seals JR, Murphy CI, van Halbeek H, Cummings RD. Site-specific N-glycosylation and oligosaccharide structures of recombinant HIV-1 gp120 derived from a baculovirus expression system. Biochemistry. 1993;32:11087–11099. doi: 10.1021/bi00092a019. [DOI] [PubMed] [Google Scholar]
- Yodoshi M, Oyama T, Masaki K, Kakehi K, Hayakawa T, Suzuki S. Affinity entrapment of oligosaccharides and glycopeptides using free lectin solution. Anal. Sci. 2011;27:395. doi: 10.2116/analsci.27.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.