Abstract
Linkage between the adherens junction (AJ) and the actin cytoskeleton is required for tissue development and homeostasis. In vivo findings indicated that the AJ proteins E-cadherin, β-catenin, and the filamentous (F)-actin binding protein αE-catenin form a minimal cadherin-catenin complex that binds directly to F-actin. Biochemical studies challenged this model since the purified cadherin-catenin complex does not bind F-actin in solution. Here we reconciled this difference. Using an optical trap-based assay, we showed that the minimal cadherin-catenin complex formed stable bonds with an actin filament under force. Bond dissociation kinetics can be explained by a catch bond model in which force shifts the bond from a weakly to a strongly bound state. These results may explain how the cadherin-catenin complex transduces mechanical forces at cell-cell junctions.
Introduction
Epithelia serve as barriers between the organism and its environment. A defining feature of these tissues is adhesion between cells at specialized intercellular junctions. The mechanical connection at these junctions imparts shape, organization and structural integrity to the tissue, and enables morphogenetic changes such as the movement of epithelial sheets and the formation of tubes during development (1, 2). Dysregulation of cell-cell junctions is common in cancer metastasis, which is characterized by loss of contact inhibition, epithelial-to-mesenchymal transitions, and abnormal cell invasiveness (3).
Classical cadherins and their cytoplasmic binding partners play a central role in intercellular adhesion in many tissues (4). In epithelial tissues, the extracellular domain of cadherin forms adhesive contacts between neighboring cells, and its cytoplasmic domain binds β-catenin, which in turn binds the F-actin binding protein αE-catenin (5), the most widely expressed of the three α-catenin family members (6). αE-catenin binds strongly to the E-cadherin/β-catenin complex (KD ~1 nM) (7, 8), but more weakly to F-actin (KD ~1 μM) (9-11). Cell biological studies led to the hypothesis that αE-catenin directly links the E-cadherin/β-catenin complex to F-actin, consistent with its role in force transmission between cadherins and the actin cytoskeleton (12). However, in vitro binding of αE-catenin to the cadherin cytoplasmic domain/β-catenin complex further weakens the affinity of αE-catenin for F-actin by at least 20-fold, to a level that would not be useful for transmitting force between E-cadherin and the actin cytoskeleton (8, 10, 13).
These biochemical studies, performed with proteins from Mus musculus (Mm; mouse) and Danio rerio (Dr; zebrafish), cast doubt on the simple model that αE-catenin directly links the cadherin/catenin complex to the actin cytoskeleton. Other proteins, including vinculin (14-18), EPLIN (19), α-actinin (20), and afadin (21, 22) bind to both αE-catenin and F-actin, and could be a link between the cadherin-catenin complex and F-actin. Notably, force exerted on the cadherin-catenin complex appears to recruit vinculin to cell junctions (15, 23-26). However, it is unclear how the changes in αE-catenin conformation required for vinculin binding (14, 27) can be induced by force if αE-catenin does not link the E-cadherin/β-catenin complex to F-actin.
Given the importance of actomyosin-generated tension in cell-cell adhesion (12, 15, 25), we posited that tension stabilizes a direct link between the minimal cadherin-catenin complex and F-actin. Therefore, we developed a single-molecule optical trap-based assay that replicated the geometry of the AJ and mechanical forces between cadherin-catenin complexes and actin filaments. We found that the application of physiological, pN-level forces increased the lifetime of normally transient bonds between cadherin-catenin complexes and an actin filament. This behavior is indicative of a catch bond, in which the dissociation rate decreases with applied force (28, 29), rather than the more typical slip bond in which the dissociation rate increases exponentially with increasing applied force. We show that a two-state catch bond model (30) fits the distribution of lifetimes of cadherin-catenin complex/F-actin bonds under force. In this model, the cadherin-catenin complex and an actin filament interact in a short-lived, weakly bound state under low forces, and transition to a stable, strongly bound state at higher forces. Thus our data reveal that the cadherin-catenin complex is a force-sensitive, direct linker to the actin cytoskeleton, and our model offers a kinetic basis for understanding mechanotransduction at AJs.
Experimental Approach
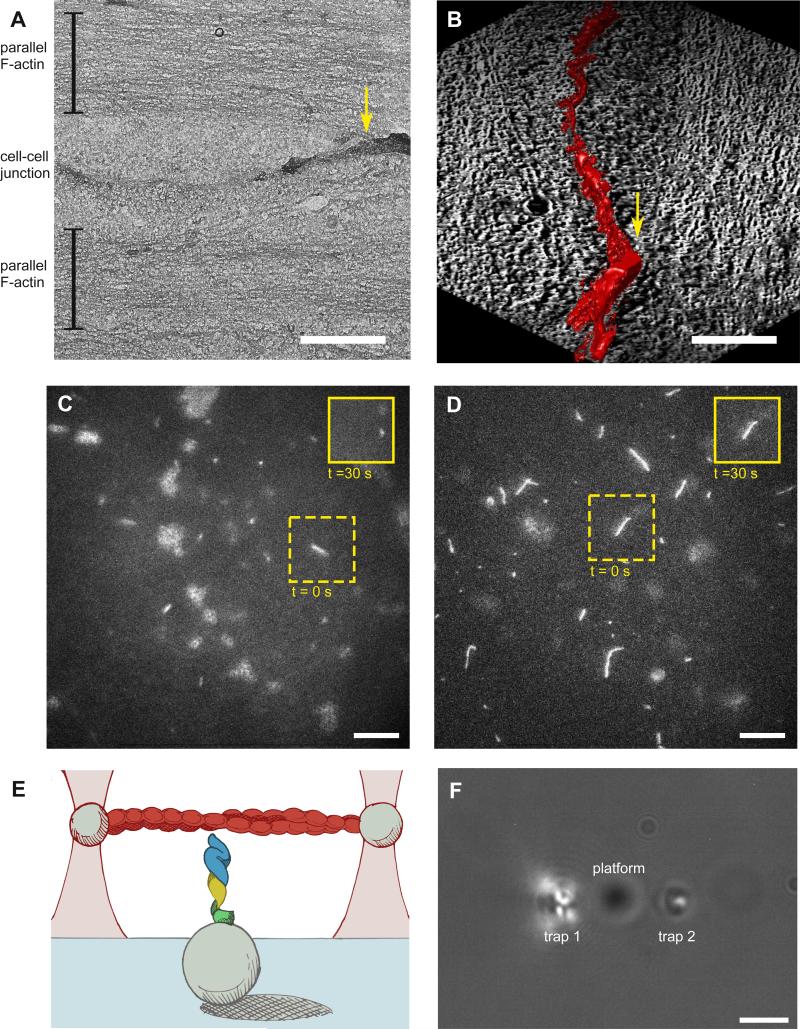
To replicate in vitro the spatial organization of the cadherin-catenin complex and F-actin, we examined electron tomographic reconstructions of cell-cell contacts in Caco-2 cells (31), which are derived from human intestinal epithelia. These images showed dense arrays of actin filaments parallel to the plasma membrane (Fig. 1, A and B, and fig. S1). Although the centerline distance between cell-cell contacts and the F-actin arrays averaged between 0.5 and 1.5 μm, there were regions where the edges of the parallel F-actin arrays were in close proximity to the cell-cell contacts (supplementary online text), and in other areas a less dense organization of actin filaments was present between the actin arrays and plasma membrane. Note that cell-cell contacts are dynamic (8, 32), and therefore these images represent only a temporal snapshot of the proximity of junctions and the underlying actin filament bundles. Parallel actin arrays tended to appear at cell-cell contacts tens of nm above the extracellular matrix (ECM) interface, whereas non-parallel actin networks were spatially correlated with the basal membrane near the ECM contact. These observations are consistent with previous super-resolution microscopy data, which revealed actin filaments parallel to intercellular junctions in simple epithelia (9).
Fig. 1. Electron microscopy of cell-cell junctions and optical trap-based assay setup.
(A) Transmission electron microscopy (TEM) image of a cell-cell junction between Caco-2 epithelial cells cultured on EM-amenable substrates. Scale bar is 1 μm. Brackets label actin filament arrays parallel to the junction, and the yellow arrow marks where the actin arrays were in close proximity to cell-cell contacts. (B) Three-dimensional electron tomography reconstruction of the same region shown in (A) rotated 90° clockwise and then tilted 45° around the horizontal axis. The cell-cell junction is highlighted in red. Yellow arrow marks the same region as in (A). Scale bar is 1 μm. (C, D) Fluorescence time-lapse micrographs of tetramethylrhodamine (TMR)-labeled F-actin (white lines) at the surface of a coverslip coated with either 1 μM Mm cadherin-catenin complex (C) or 2 μM Mm αE-catenin homodimer (D). Insets in solid yellow lines show the corresponding images for regions bounded in the dashed yellow lines after 30 s had elapsed; actin filaments remained stably bound to coverslips coated with Mm αE-catenin homodimer, whereas actin filaments diffused away from coverslips coated with the Mm cadherin-catenin complex within seconds. Out-of-focus features are glass microsphere platforms (see below). Scale bars are 10 μm. (E) Illustration of a cadherin-catenin complex and actin filament reconstituted in the optical trap assay (not to scale). GFP-E-cadherin cytoplasmic tail (green), β-catenin (yellow) and αE-catenin (blue) are immobilized on a coverslip. Glass microspheres (1.5 μm-diameter) on the coverslip act as platforms such that cadherin-catenin complexes can contact the actin filament. A single biotinylated, TMR-phalloidin-coated actin filament (red) extends between two 1 μm-diameter streptavidin-coated beads held in optical traps (pink). (F) Top view of the assay in bright field. Beads attached to the actin filament (not visible) are held in traps 1 and 2, and the platform bead is positioned between the traps and below the filament. Scale bar is 2 μm.
Without applied tension, cadherin-catenin complex/F-actin bonds are much weaker than αE-catenin homodimer/F-actin bonds (8, 10, 13). This difference was apparent in time-lapse movies of tetramethylrhodamine-labeled actin filaments diffusing in solution near the surface of coverslips coated with either 1 μM Mm GFP-E-cadherin/β-catenin/αE-catenin or 2 μM Mm GFP-αE-catenin homodimer (KD for F-actin is ~1 μM) (Fig. 1, C and D). Actin filaments bound stably to Mm GFP-αE-catenin homodimers for at least thirty seconds, whereas actin filaments did not bind stably to the reconstituted Mm cadherin-catenin complex. Thus these results replicate previous bulk sedimentation binding assays (13, 19).
To apply tension on transient cadherin-catenin complex/F-actin bonds we replicated the orientation of actin filaments and cadherin-catenin complexes observed at intercellular junctions in cells (Fig. 1). A single actin filament was optically trapped and extended above Mm or Dr cadherin-catenin complexes immobilized on a coverslip surface pre-coated with glass microspheres (31) (Fig. 1, E and F). The glass microspheres acted as spacers to prevent the surface of the coverslip from interfering with force measurements. The coverslip was mounted on a stage that moved back and forth parallel to the actin filament. Upon formation of cadherin-catenin complex/F-actin bonds, the motion of the stage was transmitted to the trapped beads, and the displacement of beads within the optical trap caused a restoring force that was applied to the cadherin-catenin complex/F-actin bonds (Fig. 2A). The amount of applied force and the rate of its application were controlled by adjusting the frequency and amplitude of the stage oscillation. Since an optical trap works like a simple spring, the magnitude of this force was calculated from the stiffness of the trap and the displacement of the trapped beads caused by this force (31).
Fig. 2. Mm cadherin-catenin complexes bound actin filaments in oscillating-stage experiments.
(A) The illustrations show that upon binding, the motion of the stage was transmitted to the trapped beads, and the force exerted on them was correlated with the motion of the stage, as shown in the time series in (B). The trapped beads stopped moving along with the stage when the surface-bound cadherin-catenin complex detached from the actin filament. (B) Part of a representative time series of force exerted on one of the two optically trapped beads attached to an actin filament (blue). ~1 μM Mm cadherin-catenin complex was purified and added to the flow cell. The stage was driven by a 150 nm-amplitude, 50 Hz-frequency sine wave (gray). (C) Full force time series from which the binding events in (B) came (shaded in teal). Peaks in the series are individual binding events, most of which lasted for approximately one half period of the sine wave used to drive the stage. (D) Force time series from a negative control experiment in which ~1 μM Mm E-cadherin/β-catenin complex was purified without Mm αE-catenin and added to the flow cell. Oscillation amplitude and frequency of the sine wave used to drive stage were the same as in (B). (E) Frequency of observed Mm cadherin-catenin complex/F-actin binding as a function of maximum stage speed (amplitude × angular frequency); n = 297, bin width is 104 nm/s. Event frequency is the number of binding events divided by the total time sampled. (F) Sine histogram of the number of Mm cadherin-catenin complex/F-actin binding events which started in each angle bin (legend shows counts; n = 235, bin width is 36°, binding events from trap 1 are in blue and those from trap 2 are in green). All events were from the 4×104 nm/s-bin shown in (E). The glass microsphere platform (A) was farthest from trap 1 when it was at +1, and from trap 2 when the stage position was at -1. Stage position was normalized by the maximum amplitude of the wave.
The cadherin-catenin complex binds to F-actin under tension
Mm cadherin-catenin complexes, added at a concentration of 1 μM to the flow cell, bound an actin filament when the stage was driven back and forth by sine waves with 150 nm amplitudes and frequencies of up to 150 Hz (6.7-ms period, Fig. 2B). The stage motion was transmitted to the optically trapped beads whenever cadherin-catenin complexes bound to the suspended actin filament (Fig. 2A). We observed many changes in the force experienced by the trapped beads as a function of time, indicating the formation of robust Mm cadherin-catenin complex/F-actin bonds (Fig. 2, B and C).
Binding of the cadherin-catenin complex to F-actin required αE-catenin (Fig. 2D, supplementary online text). The observed unbinding events were most likely caused by the dissociation of the intact cadherin-catenin complex from the suspended actin filament, rather than dissociation of αE-catenin from the E-cadherin/β-catenin heterodimer. In solution (without applied tension), the αE-catenin monomer binds strongly to E-cadherin/β-catenin (KD = 1 nM), but binds at least 1000x more weakly to F-actin (7-9, 13). At the concentrations in our experiments, any αE-catenin molecules that detached from the surface-bound cadherin-catenin complex would occupy a negligible number of binding sites on the actin filament (31). Thus, it is unlikely that αE-catenin molecules attached to the actin filament could generate reversible binding events with surface-bound E-cadherin/β-catenin heterodimers. In contrast, the actin filament provided a very large number of binding sites for αE-catenin in the platform-bound cadherin-catenin complexes. Moreover, if αE-catenin separated from the E-cadherin/β-catenin complex during every actin filament-binding event, αE-catenin would most likely dissociate quickly from the actin filament and be irreversibly lost (this assumes a reasonably high off rate, consistent with the weak affinity of monomeric αE-catenin for F-actin). This scenario would rapidly depopulate a platform of active cadherin-catenin complexes, whereas we observe tens to hundreds of binding events per glass microsphere platform (for example Fig. 2C). In any case, the key finding remains that the cadherin-catenin complex bound the actin filament under tension.
Cadherin-catenin complex/F-actin binding was observed more frequently at intermediate stage oscillation speeds (150-nm amplitude, 50-Hz frequency) than at either lower or higher speeds (Fig. 2E). At these intermediate oscillation speeds, the majority of binding events started shortly after the stage had changed direction and was in the slowest part of the sine cycle (Fig. 2F). This indicates that some minimum contact time was necessary to establish Mm cadherin-catenin complex/F-actin bonds before they were subjected to increasing tension. The observation that more events occurred at intermediate rather than low loading rates further indicated that load stabilized the transient initial bonds. These results motivated us to examine the mechanism by which force might modulate the lifetime of the cadherin-catenin complex bond to F-actin.
Force regulates cadherin-catenin complex dissociation from F-actin
To investigate how force modulated the dissociation kinetics of individual cadherin-catenin complex/F-actin bonds, we modified our optical trap-based assay to observe the detachment of cadherin-catenin complexes from the actin filament under constant force. In these experiments, a signal drove the stage 100 nm back and forth at a constant rate of 1×104 nm/s (1.5×103 pN/s). Before reversing direction, the stage paused for 150 ms and the forces exerted on the trapped beads were measured. If these forces surpassed a user-defined threshold that indicated binding of cadherin-catenin complexes to the trapped actin filament, then the stage paused until complete detachment of all cadherin-catenin complex/F-actin bonds returned the trapped beads to their zero-force baselines. In these experiments, we used αE-catenin and β-catenin from Danio rerio (Dr) rather than Mus musculus (Mm). Mm αE-catenin forms homodimers (13) whose potential presence during the preparation of the flow cell or during the assay could complicate the interpretation of cadherin-catenin complex/F-actin binding events. Importantly, Dr αE-catenin is a monomer and, like Mm αE-catenin, its affinity for F-actin decreases 20-fold upon binding Dr β-catenin (10).
When we reconstituted cadherin-catenin complexes with 10 nM Dr αE-catenin (in these experiments Dr β-catenin and E-cadherin were pre-absorbed onto the coverslip and glass microspheres (31)), we observed stepwise changes in the forces exerted on the trapped beads (Fig. 3, A and B), indicating that several cadherin-catenin complexes were initially bound to the actin filament, and that they unbound sequentially. The number of stepwise changes scaled with the concentration of Dr αE-catenin used to reconstitute the complexes, but the binding frequency decreased abruptly when less than 10 nM Dr αE-catenin was added to the flow cell (table S1). The lowest concentration of Dr αE-catenin that still resulted in binding activity was ~5 nM. Even at this minimal concentration, however, unbinding events still comprised a few stepwise changes (similar to Fig. 3A), indicating that binding by several complexes was favored over binding by a single complex (table S1).
Fig. 3. Measurement of Dr cadherin-catenin complex/F-actin bond lifetimes in constant-stage-position experiments.
(A) An actin filament binding event representative of those observed in the presence of cadherin-catenin complexes reconstituted with 10 nM of added Dr αE-catenin (see text). The stage was driven using a wave in the shape of a trapezoid with a 100-nm height and 1×104 nm/s slope (top: force time series in blue; bottom: stage position time series in black). Piecewise-constant fit of the force signal is shown in red, and the black arrow points to the segment whose duration and magnitude are the lifetime of the last Dr cadherin-catenin/F-actin bond and the force exerted on it, respectively. The black bar underneath the trace represents the total bound time of the entire event. (B) A representative force time series with the event in (A) shaded in teal. (C) Representative single-molecule force time series showing binding between a surface-bound Dr cadherin-catenin complex and an actin filament in the presence of 100 nM Mm αE-catenin ABD. Flow cell was prepared using 1 nM added Dr αE-catenin. (D) Force time series showing an actin-binding event measured for multiple surface-bound Dr cadherin-catenin complexes and 100 nM Mm ABD. The black bar above the trace represents the total bound time of the entire event. Flow cell was prepared using 5 nM added Dr αE-catenin. (E) Survival frequency of total bound times, as marked by the black bar in (A) and (D), measured in experiments using flow cells prepared with 5 nM added Dr cadherin-catenin complex (red: no Mm ABD present, and n = 412; blue: 100 nM Mm ABD, and n = 107). The survival frequency at time t is the fraction of complexes that remain bound for durations greater than t.
To test whether multiple cadherin-catenin complexes might bind F-actin more readily than a single, isolated complex, we introduced the actin-binding domain (ABD) of αE-catenin into the reaction buffer. Since ABD binds cooperatively to F-actin but does not bind to β-catenin (9), we reasoned that the presence of ABD would mimic the effect of having many αE-catenin molecules bound to F-actin. When we prepared flow cells with 1 nM Dr αE-catenin to reconstitute the cadherin-catenin complex, none of the microsphere platforms interacted with the actin filament. However, when we included 100 nM ABD in the assay buffer, we observed many binding interactions that dissociated in a single step (Fig. 3C), indicative of the interaction of a single cadherin-catenin complex with the actin filament. Under these conditions, approximately 1 in 10 platforms interacted with the actin filament, providing further evidence that the large majority of the platforms contained at most one active cadherin-catenin complex (supplementary online text). These observations indicate that addition of ABD was sufficient to replicate the presence of multiple cadherin-catenin complexes interacting with the actin filament. Additionally, since we could observe unbinding of single cadherin-catenin complexes reliably only in the presence of ABD, we conclude that multiple cadherin-catenin complexes may be required for actin filament binding to occur at an observable rate. The increased on-rates for individual complexes in the presence of ABD may be due to changes in the actin filament induced by cooperative binding of ABD as reported previously (9), although further experiments are needed to show this unequivocally.
We next asked how the presence of ABD might alter the interaction of the actin filament with many surface-bound cadherin-catenin complexes. Remarkably, in experiments using 5 nM Dr αE-catenin to reconstitute the cadherin-catenin complex, addition of ABD greatly increased the total bound times of the cadherin-catenin complex/F-actin bonds (Fig. 3, D and E). This observation indicates that cooperative interactions between neighboring cadherin-catenin complexes and the actin filament enhanced the load-bearing capacity of cadherin-catenin/F-actin bonds by significantly extending their total bound time (Fig. 3D).
Two bound states in force-lifetime distributions
The duration of the last segment of stepwise unbinding events (black arrow in Fig. 3A) represented the lifetime of a single cadherin-catenin complex/F-actin bond, and the displacement from baseline represented the load experienced by the bond. The distribution of lifetimes of the last segment in multi-step unbinding events revealed the existence of at least two bound states: a subpopulation of short-lived events at all forces, and a subpopulation of long-lived events (up to 25 s) at forces between 5 pN and 10 pN (Fig. 4A).
Fig. 4. Force-lifetime distribution of Dr cadherin-catenin complex/F-actin bonds and lifetime survival analysis.
(A) Two-dimensional histogram of Dr cadherin-catenin complex/F-actin bond force-lifetime values measured from the last piecewise constant segment in multi-step unbinding events, as shown in Fig. 3A. Tick labels on color bar are bin counts (17 force bins, 32 lifetime bins, and n = 803). (B) Kinetic schemes representing dissociation from either one bound state (blue) or two bound states (red). (C) Survival frequencies of Dr cadherin-catenin complex/F-actin bond lifetimes from three force bins indicated in Fig. 4A (black arrows, n = 188 in bin F1, 185 in F2, and 129 in F3; errors are SEM). Red lines are least-square fits of a bi-exponential function (two bound states), and blue lines are those of a single exponential function (one bound state). R2 > 0.90 for the bi-exponential function in all force bins, and the additional parameters of the bi-exponential function are justified (p ~0 in F-test). Additional force bins are shown in fig. S2A.
These results formed the basis for testing several models to explain the distribution of cadherin-catenin complex/F-actin bond lifetimes. Models in which dissociation occurs from a single bound state (state 1) result in a bond survival probability that is described by a single exponential function. In contrast, dissociation from two distinct bound states (states 1 and 2) results in a survival probability that is described by a bi-exponential function, with a separate exponential decay rate corresponding to each bound state (Fig. 4B). In both of these models, state 0 represents the unbound state.
Bond survival probabilities over a broad range of forces were better fit by a bi-exponential function than a single exponential function; the improved fit was not due to chance (p ~0 in F-test; Fig. 4C and fig. S2). Furthermore, the bi-exponential function fits identified 24% of the lifetimes in the 4-pN bin as long-lived, 43% in the 6.8-pN bin, and 45% in the 13.7-pN bin. Based on this analysis, we conclude that: 1) a model with a single bound state did not explain the distribution of cadherin-catenin complex/F-actin bond lifetimes, and 2) models with two-bound states must recapitulate how the ratio of short-lived to long-lived lifetimes depended on force.
Two-state catch bond model
Several quantitative models have been developed to account for the effect of force on how fast a bond dissociates. Most models are based on the Bell equation (33-35):
In this equation, kij (F) is the rate of transition from state i to state j at force F, the rate constant, xij the distance to the transition state, kB Boltzmann's constant, and T the absolute temperature. As formulated, the Bell equation fits the behavior of slip bonds, in which dissociation rates increase exponentially with respect to applied tension. On the other hand, the equation with a negative exponential argument has been used in catch bond models that include transitions in which rates decrease exponentially with respect to applied tension (29).
Based on the requirements determined by survival analysis of cadherin-catenin complex/F-actin bond lifetimes, we considered a two-state catch bond model, which has been proposed to explain FimH adhesion (30, 36), and a two independently bound states model (Fig. 5A). In the two-state catch bond model, bonds at low force prefer a weakly bound state (state 1) and dissociate quickly (k10) to an unbound state (state 0). As force increases to an intermediate value, bonds transition (k12) into a strongly bound state (state 2) and remain in that state because force opposes transitions (k21) back to the weakly bound state (state 1). In this intermediate force regime, dissociation from the strongly bound state (k20) is not accelerated significantly resulting in long bond lifetimes. As force increases further, unbinding occurs directly from the strongly bound state. In a two independently bound states model (Fig. 5A), transitions between states 1 and 2 are not allowed and bonds remain in the weakly (state 1) or strongly (state 2) bound state in which they originally formed until the cadherin-catenin complex dissociates (k10 or k20) from the actin filament to an unbound state (state 0).
Fig. 5. Two-state catch bond model.
(A) Kinetic schematics of the 2-bound state models that were considered. In a two-state catch bond model (left), states 1 and 2 are weakly and strongly bound states, respectively. State 0 is the unbound state. Unbinding rates k10 and k20 increase exponentially with respect to force. Transitions between states 1 and 2 occur at rates k12 and k21. The transition rate k12 increases exponentially with force, while k21 does the opposite. These transitions do not occur in the independently bound states model (right). (B) Averages of Dr cadherin-catenin complex/F-actin bond lifetimes binned by force (black dots, error bars are SEM, and n = 803). The red curves show the mean lifetime distributions predicted by the two-state catch bond model (solid red) and a two independently bound states model (dashed red). Model parameters were computed using maximum likelihood estimation. (C) Survival frequencies of Dr cadherin-catenin complex/F-actin bond lifetimes from Fig. 4C, compared with survival probability curves predicted by the two-state catch bond model (red lines, R2 > 0.90 for all bins except for the 4 pN bin, R2 = 0.67). (D) Two-state catch bond model dissociation rates as functions of force. (E) Table of maximum likelihood-estimated parameters of the two-state catch bond model and their 95% confidence intervals determined by parametric bootstrapping.
We found that the two-state catch bond model was consistent with mean lifetime distributions and bond survival frequencies from our data (Fig. 5, B and C). We used maximum likelihood estimation to calculate the parameters of the two-state catch bond model most likely to generate our un-binned, bond force-lifetime distributions (n = 803; supplementary online text; Fig. 5, D and E, fig. S3, and table S2). The estimated parameters predicted a mean bond lifetime curve that agreed well with the mean lifetime distribution (R2 = 0.86, Fig. 5B). Predicted survival probabilities also agreed well with survival frequencies of bond lifetimes in all force bins (R2 > 0.90), except for the lowest force bin (F = 4 pN, R2 = 0.61) in which the model underestimated the fraction of short-lived bonds (Fig. 5C). The most likely explanation for this discrepancy is that bond lifetimes at low forces were under-sampled, as our assay cannot detect events that were both short lived (less than ~5 ms) and low force (less than ~1 pN). The two independently bound states model, in which the bond is not allowed to transition between bound states, did not fit the mean lifetime distribution (Fig. 5, B), emphasizing the importance of force-dependent transitions in the bound states in the two-state catch bond model.
Discussion
Our data and kinetic model reconcile previous in vitro and in vivo studies of the interaction between the cadherin-catenin complex and the actin cytoskeleton. The cadherin-catenin complex/F-actin linkage could not be reconstituted in solution using purified proteins (8, 13, 19) because, as shown here, force needed to be applied to the αE-catenin/F-actin interface to form a stable bond between F-actin and the cadherin-catenin complex.
In our experiments, the duration of direct bonds between the cadherin-catenin complex and an actin filament was sensitive to load, such that at moderate loads (~8 pN) the lifetime of the bonds dramatically increased relative to those observed at lower forces. These data were explained better by a two-state catch bond model than any of the alternative models that we tested (supplementary online text). In the two-state catch bond model, force shifts the equilibrium from a weakly to a strongly bound state. Our model predicts that cadherin-catenin complex/F-actin bonds formed in the weakly bound state ~90% of the time. This probability was derived from the assumption that bound state transitions reached equilibrium before the application of force, which resulted in a model that was more consistent with our results than those that assumed different initial conditions, including those that incorporated force-loading history ((30, 37-39); supplementary online text; figs. S4, S5, and S6).
In the two-state catch bond model, cadherin-catenin complexes in the weakly bound state rapidly dissociate from actin filaments (k10). Force accelerates the transition into the strongly bound state (k12), from which cadherin-catenin complexes dissociate from actin filaments at a rate of ~0.1 s−1 at zero force (k20), a ~70-fold reduction compared to the dissociation rate from the weakly bound state (k10). Importantly, the transition from the strongly bound to weakly bound state is greatly decreased by tension (k21): in effect, tension locks the complex in the strongly bound state. Finally, forces greater than 10 pN are sufficient to accelerate the dissociation of cadherin-catenin complexes from the strongly bound state (k20), leading to a decrease in bond lifetimes at high forces.
The force-dependent distance parameters obtained from the model may be related to changes in protein structure that accompany the transition between the weakly and strongly bound states. In particular, the large value of x21, 4 nm, suggests that the αE-catenin/F-actin linkage undergoes a large decrease in length between the strongly and weakly bound states. This putative conformational change underpins catch bond behavior, since force that opposes this change maintains the bond in the strongly bound state. Previous work indicated that αE-catenin has multiple flexible domains that could be capable of mediating such a transition (14, 16, 21, 27, 40-43). At present, however, the precise structural changes in αE-catenin that accompany the transition between the weakly and strongly F-actin bound states are unknown.
Catch bond models have been used to describe the bonds formed by integrins, selectins, FimH, and myosin (28, 30, 44-47). Interestingly, the homophilic contacts between cadherin extracellular domains have also been shown to exhibit catch bond behavior (48, 49) indicating that the cadherin complex may be regulated by force on multiple levels. While a two-state catch bond model is a kinetic model that fits our data quantitatively, alternative models considering molecular details, such as the sliding-rebinding, deformation, and hydrogen bond network models that have been applied previously to selectins (50-52), may also describe the interaction between the cadherin-catenin complex and F-actin. Structural information about how αE-catenin, in a complex with β-catenin, binds F-actin will help establish the molecular basis of the kinetics observed in our experiments.
In summary, our study demonstrates that a strong interaction between the reconstituted cadherin-catenin complex and F-actin requires force, and is best described by a two-state catch bond model. Catch bond behavior provides a possible explanation for how cells transduce mechanical signals through cadherin-based adhesions, as observed in cell culture and in vivo studies (12, 15, 23-26, 53). Given the evidence for force-dependent conformational changes in αE-catenin (15, 16), the two-state catch bond model may also correspond to distinct conformational states of αE-catenin. In addition, changes in the structure of actin protomers within filaments observed in the presence of αE-catenin ABD (9) indicate that structural changes in the actin filament may also contribute to enhancing αE-catenin/F-actin interactions. Tension-stabilized states have been shown to regulate αE-catenin binding to vinculin, another actin-binding protein (15, 16). Thus, tension may not only shift the cadherin-catenin complex into a strongly bound state but also promote binding of vinculin, thereby creating a self-reinforcing system for strong linkage of the complex to the actin cytoskeleton. The experiments described here provide an approach to address this possibility directly.
Supplementary Material
One sentence summary.
The minimal cadherin-catenin complex forms a two-state catch bond with actin filaments under mechanical load.
Acknowledgements
The authors thank Sabine Pokutta, Julie Bianchini, and Phil Miller for help with protein purification and biochemistry. C.D.B. thanks Jongmin Sung and Suman Nag for assistance with optical trapping instrumentation. D.H. and N.V. thank Wendy Ochoa, Mariam Rodriguez Lee and Chris Page for their contribution to the study, and Hudson Freeze and Fred Levine for advice and discussion. The data reported in this paper are further detailed in the Supporting Online Material. This work was supported by NSF Pre-doctoral Fellowships (C.D.B., J.T.), Stanford Bio-X Pre-doctoral Fellowships (C.D.B., J.T.), a Burroughs Wellcome Career Award at the Scientific Interface (A.R.D.), the NSF (EFRI 1136790 to W.I.W., W.J.N., and A.R.D.), and the NIH (New Innovator Award 1DP2OD007078 to A.R.D., GM35527 to W.J.N., GM56169 to W.I.W., P01GM098412 to D.H. and N.V, and UO1GM094663 to W.J.N., W.I.W., D.H., N.V.).
References and Notes
- 1.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 2.Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasioukhin V. Adherens junctions and cancer. Subcell Biochem. 2012;60:379–414. doi: 10.1007/978-94-007-4186-7_16. [DOI] [PubMed] [Google Scholar]
- 4.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 5.Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssens B, et al. αT-catenin: a novel tissue-specific β-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114:3177–3188. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- 7.Pokutta S, Choi HJ, Ahlsen G, Hansen SD, Weis WI. Structural and thermodynamic characterization of cadherin·β-catenin·α-catenin complex formation. J Biol Chem. 2014;289:13589–13601. doi: 10.1074/jbc.M114.554709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen SD, et al. αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol Biol Cell. 2013;24:3710–3720. doi: 10.1091/mbc.E13-07-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller PW, et al. Danio rerio αE-catenin is a monomeric F-actin binding protein with distinct properties from Mus musculus αE-catenin. J Biol Chem. 2013;288:22324–22332. doi: 10.1074/jbc.M113.458406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rimm DL, Koslov ER, Kebriaei P, Cianci CD, Morrow JS. α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc Natl Acad Sci U S A. 1995;92:8813–8817. doi: 10.1073/pnas.92.19.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi HJ, et al. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci U S A. 2012;109:8576–8581. doi: 10.1073/pnas.1203906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. α-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 16.Yao M, et al. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- 17.Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL. Vinculin is associated with the E-cadherin adhesion complex. J Biol Chem. 1997;272:32448–32453. doi: 10.1074/jbc.272.51.32448. [DOI] [PubMed] [Google Scholar]
- 18.Weiss EE, Kroemker M, Rüdiger AH, Jockusch BM, Rüdiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe K, Takeichi M. EPLIN mediates linkage of the cadherin-catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen KA, Soler AP, Johnson KR, Wheelock MJ. Interaction of α-actinin with the cadherin/catenin cell-cell adhesion complex via α-catenin. J Cell Biol. 1995;130:67–77. doi: 10.1083/jcb.130.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J Biol Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tachibana K, et al. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol. 2000;150:1161–1175. doi: 10.1083/jcb.150.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twiss F, et al. Vinculin-dependent Cadherin mechanosensing regulates efficient epithelial barrier formation. Biol Open. 2012;1:1128–1140. doi: 10.1242/bio.20122428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huveneers S, et al. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J Cell Biol. 2012;196:641–652. doi: 10.1083/jcb.201108120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barry AK, et al. α-Catenin cytomechanics: role in cadherin-dependent adhesion and mechanotransduction. J Cell Sci. 2014;127:1779–1791. doi: 10.1242/jcs.139014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangarajan ES, Izard T. The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J Biol Chem. 2012;287:18492–18499. doi: 10.1074/jbc.M112.351023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall BT, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 29.Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 30.Thomas W, et al. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Materials and methods are available as supplementary material on Science Online.
- 32.Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J Cell Biol. 2007;178:517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 34.Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 35.Kramers HA. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7:284–304. [Google Scholar]
- 36.Bartolo D, Derenyi I, Ajdari A. Dynamic response of adhesion complexes: beyond the single-path picture. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:051910. doi: 10.1103/PhysRevE.65.051910. [DOI] [PubMed] [Google Scholar]
- 37.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci U S A. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barsegov V, Thirumalai D. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc Natl Acad Sci U S A. 2005;102:1835–1839. doi: 10.1073/pnas.0406938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall BT, Sarangapani KK, Lou J, McEver RP, Zhu C. Force history dependence of receptor-ligand dissociation. Biophys J. 2005;88:1458–1466. doi: 10.1529/biophysj.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rangarajan ES, Izard T. Dimer asymmetry defines α-catenin interactions. Nat Struct Mol Biol. 2013;20:188–193. doi: 10.1038/nsmb.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pokutta S, Weis WI. Structure of the dimerization and β-catenin-binding region of α-catenin. Mol Cell. 2000;5:533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 43.Ishiyama N, et al. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J Biol Chem. 2013;288:15913–15925. doi: 10.1074/jbc.M113.453928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarangapani KK, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 46.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci U S A. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clobes AM, Guilford WH. Loop 2 of myosin is a force-dependent inhibitor of the rigor bond. J Muscle Res Cell Motil. 2014;35:143–152. doi: 10.1007/s10974-014-9375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci U S A. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manibog K, Li H, Rakshit S, Sivasankar S. Resolving the molecular mechanism of cadherin catch bond formation. Nat Commun. 2014;5:3941. doi: 10.1038/ncomms4941. [DOI] [PubMed] [Google Scholar]
- 50.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J. 2007;92:1471–1485. doi: 10.1529/biophysj.106.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer TA. Structural basis for selectin mechanochemistry. Proc Natl Acad Sci U S A. 2009;106:91–96. doi: 10.1073/pnas.0810784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chakrabarti S, Hinczewski M, Thirumalai D. Plasticity of hydrogen bond networks regulates mechanochemistry of cell adhesion complexes. Proc Natl Acad Sci U S A. 2014;111:9048–9053. doi: 10.1073/pnas.1405384111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai D, et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delorme-Walker V, et al. Toxofilin upregulates the host cortical actin cytoskeleton dynamics, facilitating Toxoplasma invasion. J Cell Sci. 2012;125:4333–4342. doi: 10.1242/jcs.103648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanein D, Volkmann N. Correlative light-electron microscopy. Adv Protein Chem Struct Biol. 2011;82:91–99. doi: 10.1016/B978-0-12-386507-6.00004-X. [DOI] [PubMed] [Google Scholar]
- 56.Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struct Biol. 1999;184:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- 57.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 61.Sommese RF, et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc Natl Acad Sci U S A. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sung J, Sivaramakrishnan S, Dunn AR, Spudich JA. Single-molecule dual-beam optical trap analysis of protein structure and function. Methods Enzymol. 2010;475:321–375. doi: 10.1016/S0076-6879(10)75014-X. [DOI] [PubMed] [Google Scholar]
- 64.Berg-Sørensen K, Flyvbjerg H. Power spectrum analysis for optical tweezers. Rev. Sci. Instrum. 2004;75:594. [Google Scholar]
- 65.Hansen PM, Tolic-Norrelykke IM, Flyvbjerg H, Berg-Sorensen K. tweezercalib 2.1: Faster version of MatLab package for precise calibration of optical tweezers. Comput Phys Commun. 2006;175:572–573. [Google Scholar]
- 66.Little MA, Jones NS. Generalized methods and solvers for noise removal from piecewise constant signals. I. Background theory. Proc Math Phys Eng Sci. 2011;467:3088–3114. doi: 10.1098/rspa.2010.0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peacock JA. Two-dimensional goodness-of-fit testing in astronomy. Mon Not R Astron Soc. 1983;202:615–627. [Google Scholar]
- 68.Efron B, Tibshirani RJ. An introduction to the bootstrap. Vol. 57. CRC press; 1994. [Google Scholar]
- 69.Delorme V, et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13:646–662. doi: 10.1016/j.devcel.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of β-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–12309. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, et al. Quantifying the effects of molecular orientation and length on two-dimensional receptor-ligand binding kinetics. J Biol Chem. 2004;279:44915–44923. doi: 10.1074/jbc.M407039200. [DOI] [PubMed] [Google Scholar]
- 72.Pereverzev YV, Prezhdo OV, Forero M, Sokurenko EV, Thomas WE. The two-pathway model for the catch-slip transition in biological adhesion. Biophys J. 2005;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.