Abstract
Actin filaments (F-actin) are protein polymers that undergo rapid assembly and disassembly and control an enormous variety of cellular processes ranging from force production to regulation of signal transduction. Consequently, imaging of F-actin has become an increasingly important goal for biologists seeking to understand how cells and tissues function. However, most of the available means for imaging F-actin in living cells suffer from one or more biological or experimental shortcomings. Here we describe fluorescent F-actin probes based on the calponin homology domain of utrophin (Utr-CH), which binds F-actin without stabilizing it in vitro. We show that these probes faithfully report the distribution of F-actin in living and fixed cells, distinguish between stable and dynamic F-actin, and have no obvious effects on processes that depend critically on the balance of actin assembly and disassembly.
Keywords: wound healing, cytokinesis, actomyosin, F-actin imaging
INTRODUCTION
Actin is one of the most abundant and important proteins in eukaryotes. Actin monomers (G-actin) assemble into actin filaments (F-actin) and thereby drive force production either directly, by pushing, or indirectly, via interaction with myosin motor proteins. In the typical cell populations of both dynamic (rapidly assembling and disassembling) and stable (slowly assembling and disassembling) F-actin are found [Stossel, 1993; Small and Resch, 2005]. The former are associated with transient force production on membranes, while the latter are associated with more sustained, myosin-dependent contraction. In addition to force production, actin plays a number of other roles, including maintenance of cell shape and regulation of intracellular signaling [Stossel, 1993; Schoenwaelder and Burridge, 1999; Small and Resch, 2005]. Due to its general importance, considerable effort has been expended to visualize F-actin distribution and dynamics in living cells and tissues [Merrifield, 2004; Dormann and Weijer, 2006].
Three general approaches have been employed for this purpose: microinjection or expression of fluorescent G-actin [Kreis et al., 1982; Westphal et al., 1997]; microinjection of fluorescent phalloidin, a mushroom toxin that binds specifically to F-actin [Wulf et al., 1979; Wang, 1987; Planques et al., 1991] and; expression of fluorescent F-actin binding proteins or domains [Gerisch et al., 1995; Edwards et al., 1997; Kost et al., 1998]. While each of these approaches have provided essential information about the distribution and dynamics of actin in living cells, each has one or more important disadvantages. Fluorescent G-actin, which is generated either via attachment of a chemical fluorophore or by fluorescent protein fusions, can alter actin assembly and processes dependent on actin assembly [Hird, 1996; Aizawa et al., 1997; Wu and Pollard, 2005] and, moreover, may not incorporate into all pools of F-actin [Kovar et al., 2006]. Further, because only a fraction of actin is in polymer form at any given time [Korn, 1982], imaging with fluorescent G-actin entails high background signal, unless specialized techniques such as speckling are employed [Waterman-Storer et al., 1998]. Phalloidin must be microinjected, and stabilizes actin filaments [Dancker et al., 1975], and thus changes the balance of actin assembly/disassembly in vivo [Planques et al., 1991; Dancker et al., 1975; Wehland et al., 1977]. Moreover, fluorescent phalloidin poorly labels dynamic actin in vivo [Mandato and Bement, 2003]. Expression of fluorescent F-actin binding proteins or domains has the potential to reduce the perturbation of actin assembly entailed by the use of fluorescent G-actin or fluorescent phalloidin. However, the localization of a fluorescent, full length F-actin binding protein is likely to be influenced by any regulatory interactions that the protein in question experiences. The use of isolated F-actin binding domains from proteins such as moesin or fimbrim [Edwards et al., 1997; Sheahan et al., 2004] has the potential to overcome this problem, but in general, the properties of a given F-actin binding protein or domain used for in vivo F-actin labeling have not been characterized with respect to their effects on F-actin stability. Further, in some cases, such constructs have been reported to alter actin-dependent processes in vivo [Hozumi et al., 2006].
To obtain F-actin probes that have the potential to overcome the problems described above, we generated fusion constructs based on the calponin homology domain (CH) of utrophin, an actin binding protein best known as the non-muscle counterpart of dystrophin [Winder et al., 1995]. We chose this domain (UtrCH) based on the demonstration that it binds to, without stabilizing, F-actin in in vitro dilution assays [Rybakova and Ervasti, 2005]. Three fusion constructs based on UtrCH were generated: Green fluorescent protein-UtrCH (GFP-UtrCH), Red fluorescent protein-UtrCH (RFP-UtrCH) and photoactivatable GFP-UtrCH [PaGFP-Utr-CH; Patterson and Lippincot-Schwartz, 2002]. We show that these probes faithfully report the distribution of F-actin in fixed and living samples without significantly altering the dynamics of actin-based processes. We also show that these probes report on F-actin populations that label poorly with fluorescent G-actin. Finally, we demonstrate that these probes, when used in conjunction with various photoactivation techniques can distinguish between highly dynamic and stable F-actin.
MATERIALS AND METHODS
Preparation of Probes
The UtrCH probes were cloned from the first 783 base pairs of the human Utrophin (NM_007124) (provided by J. Ervasti, The University of Minnesota) using the primers 5′-TAGATCCGGAACCATGGCCAAGT(s) and 5′-GGCGCTCGAGTTAGTCTATGGTGAC (as). The amplified fragment was digested with BspE1 and XhoI and inserted into a custom pcs2-eGFP or pcs2-mRFP vector as described in [Sokac et al., 2003]. The photoactivated variant was created according to [Patterson and Lippincott-Schwartz, 2002]. In all UtrCH variants, the fluorescent tag was n terminal to the UtrCH sequence. All clones were verified by DNA sequencing. All RNA was transcribed in vitro using the mMessage mMachine SP6 kit (Ambion). Plasmid DNA was purified using the Wizard Plus SV Minipreps DNA purification system (Promega).
Microinjection and Perturbation of the F-actin Cytoskeleton
Xenopus oocytes were obtained from adult females, defolliculated, and stored in 1X Barth’s Solution (88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, 10 mM Hepes, pH 7.4) containing 5 mg/ml BSA. UtrCH RNA and UtrCH plasmid DNA were pressure injected using glass microneedles. About 40 ng of UtrCH RNA was injected equatorially or 4 ng of UtrCH plasmid DNA was injected into the geminal vesicle and allowed to incubate for 18–20 h. Expression of DNA was comparable to the mRNA except that its expression level was slightly more variable. Alexa-568 G-actin was injected to a final concentration of 6 μg/ml. To perturb the actin cytoskeleton, oocytes were incubated in 10 μM Latrunculin A for 1 h or 5 μM Jasplakinolide for 30 min. Incubation of Xenopus oocytes in a 50% D2O Barth’s solution for 1 h prior to wounding was sufficient for F-actin comet formation during the healing process. Echinoderm embryos were obtained, cultured and microinjected as previously described [Bement et al., 2005]. About 4D imaging was performed as described in [Bement et al., 2003; Bement et al., 2005].
Fixation and Immunoflouresence
Prior to fixation, Xenopus oocytes were microinjected, wounded with a nitrogen pumped dye laser (Laser Science, Inc.), and allowed to heal for 90s. These oocytes were then fixed overnight with an aldehyde-based fixative (4% paraformaldehyde, 0.1% gluteraldehyde, 0.1% triton X-100, 10 mM EGTA, 100 mM KCl, 3 mM MgCl2, 150 mM sucrose, 10 mM HEPES, pH 7.6). Fixed cells were washed twice with PBS, quenched for 4 h with 100 mM NaBH4, and bisected. To label total actin distributions, JLA-20 anti-actin antibody (Calbiochem) was used at 1:200. Alexa-568 phalloidin (Molecular Probes) was used at a final concentration of 60 nM. For microtubule staining, DM1A anti-tubulin (Sigma) was used at 1:200. The methanol extraction step to improve the visualization of microtubules was adapted from Danilchik et al. [1998].
Cosedimentation and Immunoblotting
Oocytes were microinjected with 40 ng UtrCH RNA, 160 ng UtrCH RNA, or water and allowed to express overnight. Oocyte were washed twice with ice cold homogenization buffer (50 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 0.5 μM phalloidin, 20 mM PIPES, pH 7.0), and then homogenized in a 10 fold volume of the homogenization buffer supplemented with a cocktail of protease inhibitors (4 mM pefablock, 60 μg/ml chymostatin, 6 μg/ml leupeptin, 30 μg/ml pepstatin, 0.1 μg/ ml aprotinin, 0.5 μM phalloidin). Centrifuge homogenate at 15,000g for 15 min to remove yolk. The resulting supernatant was transferred to an ultracentrifuge tube and centrifuge at 100,000g for 1 h. The pellet was resuspended in 40 μl homogenization buffer without phalloidin. For actin blotting, JLA-20 anti-actin antibody (Calbiochem) was used at 1:1250. JL-8 anti-GFP (Clontech) was used at 1:2000.
Imaging, Photoactivation, and Image Analysis
Images were collected using a Zeiss Axiovert 100 M microscope (Carl Zeiss, Thornwood, NY) with Bio-Rad 1024 times 1024 Lasersharp Confocal software (Bio-Rad, Hercules, CA). This microscope was equipped with a 63× objective with a numerical aperture of 1.4. The 4D images and movies were collected using confocal microscopy as described in Bement et al. [2003, 2005]. Images of actin comets were captured with dual-wavelength, simultaneous confocal microscopy. Photoactivation of large squares of the cell cortex were accomplished by increasing the zoom setting in the Bio-Rad Lasersharp software to reduce the field of view. The reduced field of view was then scanned seven to nine times on the slowest setting with the power of imaging laser set to 30% of maximum. Immediately after the last high intensity scan, the zoom was reduced and image acquistion started. For point activation of the PaGFP-UtrCH probe, a 435 nm micropoint pulsed nitrogen pumped dye laser (Laser Science, Inc.) was used at the highest filter setting. Image rendering was done using Volocity 3.1 (Improvision). Fluorescent intensities were measured using Image J 1.37v or Adobe Photoshop 7.0. Flow rates were extracted from the slopes of kymographs as previously described in Mandato and Bement [2001]. Quantifications of fluorescence intensities at the leading edge, trailing edge, or on the contractile ring itself were measured at multiple locations and subsequently averaged. All statistical analysis was completed using Microsoft Excel.
RESULTS
UtrCH-Based Probes Faithfully Report the Distribution of F-Actin in Samples Fixed With and Without Methanol
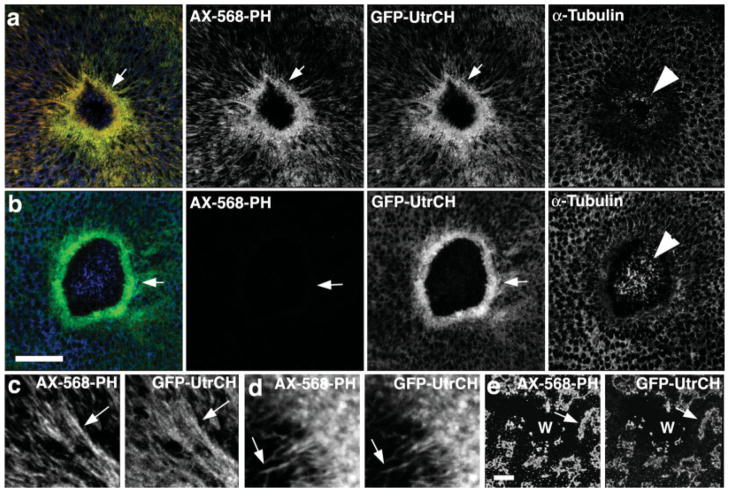
The most common means of monitoring F-actin distribution is staining fixed samples with fluorescent phalloidin. Accordingly, Xenopus oocytes were microinjected with an expression vector encoding GFP-UtrCH, and then wounded to trigger formation of an F-actin-rich contractile array [Mandato and Bement, 2001]. Samples were then fixed with an aldehyde-based fix (see methods), and processed for staining with AX-568 phalloidin and anti-tubulin antibodies (to detect microtubules). The UtrCH probe showed near complete colocalization with AX-568 phalloidin (Fig. 1a), implying that UtrCH detects endogenous actin filaments. This colocalization was not just evident on large structures, but also on much finer structures, such as the narrow cables of F-actin that extend away from the wound (Fig. 1c) and actin “fingers” that extend into the wound [Fig. 1d; Mandato and Bement, 2001]. Moreover, after disruption of the cortical F-actin network prior to wounding by latrunculin treatment, both the phalloidin and GFP-UtrCH colocalized in large aggregates (Fig. 1e), confirming that the normal GFP-UtrCH distribution is F-actin dependent.
Fig. 1.
Comparison of GFP-UtrCH and fluorescent phalloidin in wounded, fixed oocytes. (a) Low magnification images showing that in samples fixed with an aldehyde-based fixative, fluorescent phalloidin (AX-568-PH) and GFP-UtrCH colocalize, and reveal a striking concentration of F-actin around wounds (arrow). Immunolocalization of tubulin reveals modest preservation of microtubules in the interior of the wound (arrowhead). (b) In samples extracted with methanol after aldehyde fixation, phalloidin staining is completely eliminated, while staining with GFP-UtrCH is preserved. Microtubule preservation is superior under these conditions. (c) High magnification view showing colocalization of fluorescent phalloidin and GFP-UtrCH on cables of F-actin extending away from the wound. (d) High magnification view showing colocalization of fluorescent phalloidin and GFP-UtrCH on actin “fingers” that extend into the wound. (e) Images showing that in cells treated with latrunculin to disrupt F-actin, phalloidin and GFP-UtrCH colocalize in large, unorganized patches around wounds (W). Scale bars is 20 μm.
Methanol extraction is commonly employed to improve visualization of microtubules, but has the unfortunate consequence of rendering F-actin unable to bind phalloidin [Howard and Oresajo, 1985]. We therefore also compared the ability of GFP-UtrCH to label F-actin in samples extracted with methanol after an initial aldehyde fixation. Consistent with previous studies [Danilchik et al., 1998] methanol extraction did indeed improve microtubule visualization relative to aldehyde-only fixed samples (Fig. 1b). Further, whereas the methanol extraction completely eliminated phalloidin staining, the GFP-UtrCH was retained (Fig. 1b). Thus, the UtrCH probe works with fixed samples to detect F-actin.
UtrCH-Based Probes for Imaging F-Actin in Living Cells
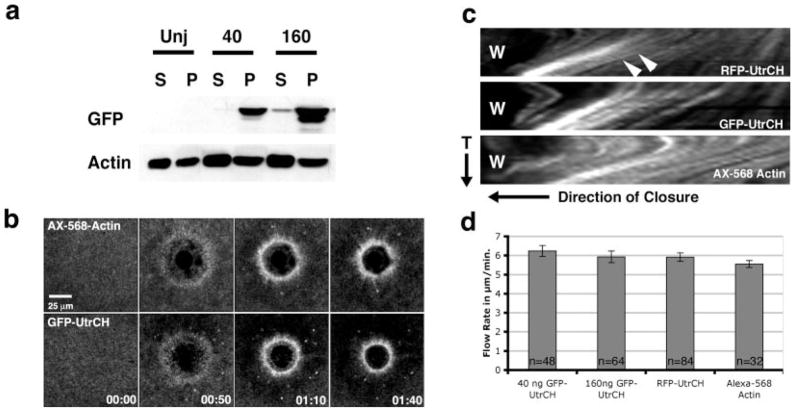
One of the most attractive features of the UtrCH was the report that it does not stabilize F-actin as determined by in vitro dilution assays [Rybakova and Ervasti, 2005], implying that unlike phalloidin, this domain could be employed in vivo without altering levels of endogenous F-actin. As a direct test of this possibility, we quantified F-actin levels by immunoblotting cytoskeletal fractions in control oocytes and oocytes injected with either 40 or 160 ng GFP-UtrCH mRNA (40 ng is approximately two to three times the amount required for imaging F-actin in living samples). High-speed centrifugation was then used to fractionate the oocytes into cytoskeletal (pellet) and soluble (supernatant) fractions, which were immunoblotted for actin and GFP. Injection of either 40 or 160 ng of GFP-UtrCH mRNA had no significant effect on actin pelleting in this assay (Fig. 2a; P > 0.05, n = 5).
Fig. 2.
The UtrCH probes do not perturb the F-actin cytoskeleton. (a) Microinjection of 40 or 160 ng of GFP-UtrCH mRNA into Xenopus oocytes does not increase the amount of pelletable (P) F-actin compared to uninjected (Unj) controls as indicated by Western blotting. (b) Oocytes microinjected with GFP-UtrCH appear to heal normally after wounding. Comparisons of several timepoints from 4D movies showed no physiological differences in overall appearance or structure, healing rate, or myosin-based cortical flow between GFP-UtrCH or AX-568 actin injected oocytes. (c) Kymographs of oocytes injected with GFP-UtrCH, RFP-UtrCH or AX-568 actin reveal similar rates of F-actin movement toward the wound site (W). Actin flow rates were determined by measuring the distance travelled by individual fluorescent points (arrowheads) over time (T). (d) Quantification of actin flow rates in cells injected with 40 or 160 ng GFP-UtrCH mRNA, 40 ng RFP-UtrCH mRNA, or AX-568 actin. There are no significant differences in the measured flow rates (P > 0.05).
To determine whether the UtrCH probes perturbed dynamic actin-dependent processes, we monitored the response to wound healing using 4D microscopy [Bement et al., 2003] in Xenopus oocytes microinjected with 40 ng GFP-UtrCH, 160 ng GFP-UtrCH, 40 ng RFP-UtrCH or, as a control, AX-568 actin. Comparison of 4D movies or kymographs derived therefrom revealed no apparent differences in the four groups, with the exception that the background signal (reflecting AX-568 actin not incorporated into filaments) was higher in the AX-568-actin sample (Figs. 2b and 2c). In addition, comparison of the rates of cortical flow (which is slowed when actin dynamics are reduced or myosin-2-dependent contraction is inhibited) [Mandato and Bement, 2001; Murthy and Wadsworth, 2005] revealed no significant differences between the four groups (Fig. 2d).
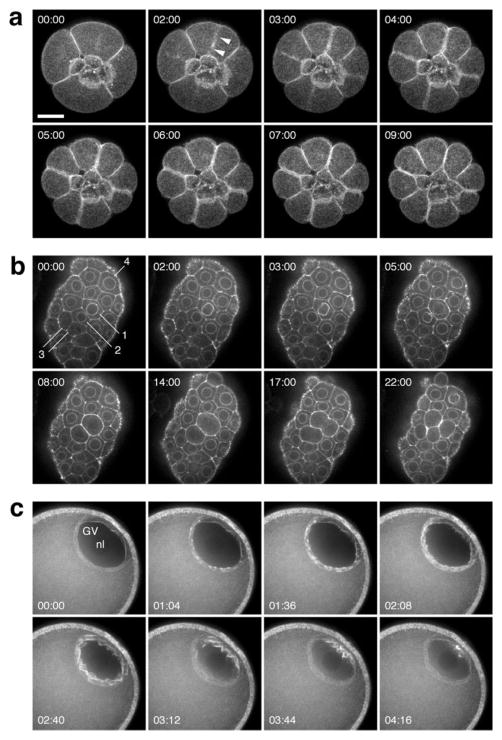
To extend the results obtained during Xenopus oocyte wound healing, GFP-UtrCH mRNA was injected into either sand dollar or sea urchin eggs shortly after fertilization. Within a few cell divisions, robust labeling of microvilli (data not shown), cortical F-actin, and the cytokinetic apparatus was seen (Fig. 3a) and the process of cytokinesis could be followed in exquisite detail (movie 1,2). This labeling persisted for at least 24 h and had no apparent effect on the rate of development, the pattern of cell division, or the organization of the blastula epithelium (Fig. 3b). Similar results were obtained with another species of echinoderm, the asteroid Pisaster ochraceus (Fig. 3c), Xenopus, and the nemertean worm Cerebratulus (data not shown). In several contexts we noted remarkably rapid recruitment of GFP-UtrCH to newly-assembled F-actin populations, and equally rapid disappearance: Fig. 3c illustrates germinal vesicle breakdown in a sea star oocyte, in which F-actin assembles transiently around the nuclear envelope [Lenart et al., 2005].
Fig. 3.
GFP-UtrCH labels F-actin structures in living echinoderm embryos and oocytes. (a) Frames from a 4D sequence of a 16-cell purple urchin embryo expressing GFP-UtrCH from injected mRNA; vegetal view. The four small cells in the center are the micromeres; their sisters, the macromeres, divide in this sequence. Each image is a brightest-point projection of ten 1-μm sections. Probe accumulation (arrowhead) in the equatorial cortex is approximately coincident with the onset of furrowing. (b) Frames from a time-lapse sequence at a single focal plane through the blastula epithelium in a sand dollar embryo. GFP-UtrCH accumulates on the nuclear membrane in inter-phase, brightening just before nuclear envelope breakdown (see cells labeled 1–4; the pair of cells labeled “3” has just divided at the beginning of the sequence, and by 3 min. have accumulated F-actin on the nuclear membrane). (c) Germinal vesicle (GV) breakdown in a sea star oocyte. Actin assembly proceeds in a wave starting at the interior side of the GV. Each timepoint is a projection of eight 1-μm sections. Scale bar is 25 μm.
Distinguishing Between Stable and Dynamic Actin in Living Cells
The forgoing results indicated that the UtrCH probes are generally useful in both fixed and living samples. We next sought to determine whether they could also be used to distinguish between stable and dynamic actin using the Xenopus oocyte wound healing model. This system is ideal for such an analysis in that it was previously found that the actin array that forms in response to wounding is comprised of pools of highly dynamic and relatively stable F-actin that are spatially distinct. That is, dynamic actin structures such as F-actin comets form around the outer portion of the ring, while relatively stable F-actin concentrates on the interior of the ring [Mandato and Bement, 2001]. Each of these populations has different labeling characteristics: the pool of dynamic actin is efficiently labeled by fluorescent actin and poorly labeled by fluorescent phalloidin in living cells, while the pool of stable actin is efficiently labeled by fluorescent phalloidin and poorly labeled by fluorescent actin [Mandato and Bement, 2001, 2003].
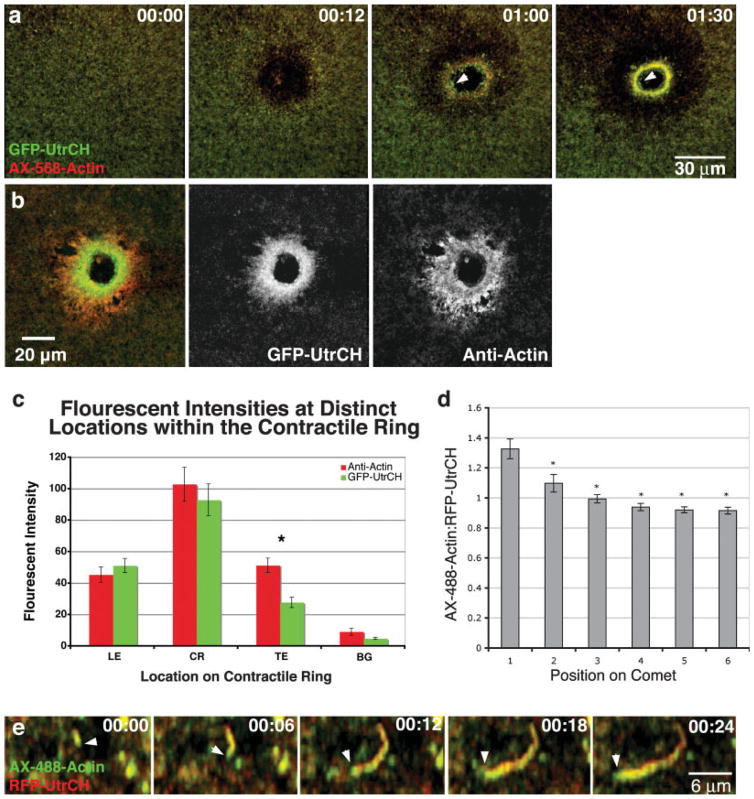
We first compared the distribution of GFP-UtrCH to AX-568 actin in wounded oocytes. Low magnification, 4D confocal movies showed that while the overall patterns were similar, GFP-UtrCH consistently labeled the interior of the actin array, where the stable actin is localized [Mandato and Bement, 2001], to a greater extent than the AX-568 actin (Fig. 4a). Similarly, when GFP-UtrCH expressing cells were wounded, fixed, and then stained with an anti-actin antibody, the GFP-UtrCH was relatively enriched on the interior of the wound array (Figs. 4b and 4c). Actin comets, another dynamic actin structure associated with oocyte wound healing, could also be utilized to gain further insight into UtrCH properties. While both the AX-488 actin and the UtrCH probes label actin comets, the ratio of AX-488 actin signal to RFP-UtrCH was consistently highest at the comet head (Figs. 4d and 4e). Because the comet head is the site of new actin assembly [Theriot et al., 1992], this indicates that the UtrCH probes bind the dynamic actin shortly after incorporation of actin itself.
Fig. 4.
Labeling and distinguishing between relatively more dynamic and stable actin structures in Xenopus oocytes. (a) Frames from a 4D movie show that GFP-UtrCH more effectively labels the stable actin that resides on the interior of contractile ring (arrowhead) than AX-568-actin. (b) Comparisons of GFP-UtrCH to total actin distributions by antibody staining in fixed oocytes show few differences at the leading or on the contractile ring but do exhibit differences at the trailing edge. (c) Quantifications of fluorescent intensities at the leading edge (LE), on the contractile ring (CR), and locations away from the contractile structure (BG) showed no differences (P > 0.05, n = 24) between antibody staining and the UtrCH probe. Significant differences in fluorescent intensities were observed at the trailing edge (TE, P < 0.05, n = 24). (d) Quantifications of actin comets from oocytes injected with AX-488 actin and RFP-UtrCH revealed that the ratio of AX-488 actin to RFP-UtrCH is greater at the newly assembled head of the comet than the older tail regions (Asterisks indicate P < 0.05, n = 24). (e) High magnification images showing the growing actin comet head (arrowhead) is labelled more intensely with AX-488 actin than RFP-UtrCH.
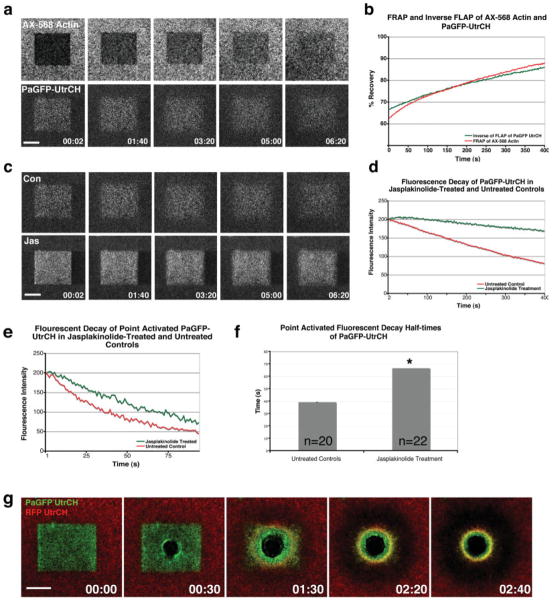
Based on the above, the UtrCH probes can be used in conjunction with fluorescent actin to distinguish between relatively stable and relatively dynamic actin. However, the differences between the two probes are not as striking as they might be. Moreover, one of the points of developing the UtrCH probes was to avoid problems associated with fluorescent actin (see Introduction). Thus, we sought another means to achieve the same result using only the UtrCH probes. Toward this end, we initially tested the ability of the PaGFP-UtrCH probe to monitor the stability of F-actin in one of two ways. First, in cells injected with both AX-568 actin and PaGFP-UtrCH, a region of the plasma membrane was illuminated with both 488 and 568 nm laser light to simultaneously photoactivate the PaGFP-UtrCH probe and photo-bleach the AX-568 actin. The rate of fluorescence loss in the former was compared to the rate of fluorescence recovery in the latter. The two closely matched (Figs. 5a and 5b), indicating that the photoactivated PaGFP-UtrCH probe monitors F-actin turnover as well as the photobleached fluorescent actin. Second, we compared the rate of PaGFP-UtrCH probe decay of either large (Figs. 5c and 5d) or small (Figs. 5e and 5f) regions in control oocytes vs. oocytes treated with jasplakinolide, which stabilizes F-actin [e.g. Murthy and Wadsworth, 2005]. In both cases, jasplakinolide greatly reduced the rate of PaGFP-UtrCH probe decay after photoactivation (Figs. 5c–5e) extending their fluorescence decay half-times from 39.3 to 66.7 s (Fig. 5f).
Fig. 5.
Using photobleaching and photoactivation with the UtrCH probes to monitor the contributions of the stable and dynamic F-actin populations. (a) Frames from a 4D sequence comparing the simultaneous fluorescence recovery of AX-568 actin and fluorescence loss of PaGFP-UtrCH in the same region of the oocyte cortex. (b) Measurements of fluorescence recovery of AX-568 actin parallels the inverse of fluorescence loss of PaGFP-UtrCH. (c) Frames from a 4D sequence contrasting the fluorescence loss of PaGFP-UtrCH in control and jasplakinolide treated oocytes. (d) Quantification of fluorescence loss of PaGFP-UtrCH in control and jasplakinolide treated cells reveals that fluorescence loss is reduced in jasplakinolide treated oocytes. (e) Measurements of PaGFP-UtrCH fluorescence loss in point-activated control and jasplakinolide treated oocytes also exhibited a reduced rate of fluorescence loss in jasplakinolide treated oocytes. (f) Fluorescence decay half-times of point-activated PaGFP-UtrCH in jasplakinolide-treated and untreated controls. Asterisks indicate P < 0.05. (g) Combined use of RFP-UtrCH and PaGFP-UtrCH reveals a clear subdivision in F-actin populations around wounds. The oldest (i.e. most stable; green around the wound) F-actin becomes enriched on the interior of the actin array. This population of F-actin becomes surrounded by more dynamic F-actin (red). Scale bars = 30 μm.
Because the above results indicated that the PaGFP-UtrCH probe could be used to monitor actin turnover, we next combined FRAP of the RFP-UtrCH probe with photoactivation of the PaGFP-UtrCH to monitor actin dynamics around wounds. Oocytes were injected with both RFP-UtrCH and PaGFP-UtrCH, and, after allowing for expression, were subjected to photoactivation and photobleaching of an 80 μm2 area as above, and then immediately wounded in the middle of the square. The 4D imaging revealed that the PaGFP-UtrCH probe accumulated on a region in the interior of the wound, while the RFP-UtrCH probe recovered in a circular region around the outer edge of the region occupied by the PaGFP-UtrCH probe (Fig. 5g). This presumably reflects retention of the limited amount of photoactivated GFP probe on the most stable filaments, combined with recruitment of the non-limiting RFP to newly-assembled filaments. The difference visible between these two regions was much more obvious than what was observed with the combination of AX-568 actin and GFP-UtrCH in Fig. 4a above, or in previous work with combinations of fluorescent actin and fluorescent phalloidin, demonstrating the power of this approach.
DISCUSSION
The UtrCH probes described here have a number of distinct advantages. First, they are experimentally flexible. That is, they function when expressed from mRNA or DNA and when used in living or fixed samples. Thus, they can be used not only for imaging the cytoskeleton in living cells, but also in fixed, transfected cells, one of the most common means of imaging F-actin. Furthermore, these constructs can be bacterially expressed and purified (unpublished results) to view early events where waiting for translation may be problematic. They also function in a variety of cell types and organisms: oocytes, embryos, and epithelial cells of amphibians and echinoderms, as shown here, but also in cultured neurons, zebrafish embryos, and cultured mammalian cells (unpublished results). Given the fact that the UtrCH we employed is from human Utrophin, and the relative conservation of actin, this is not surprising.
Second, the UtrCH probes have no apparent untoward effects on the actin cytoskeleton. This conclusion is based several findings: even excessive GFP-UtrCH does not increase the amount of pelletable F-actin, in keeping with the report showing that UtrCH does not reduce the rate of actin disassembly in vitro [Rybakova and Ervasti, 2005]. In addition, comparison of the wound healing process by both inspection and quantification revealed no differences between fluorescent actin and the UtrCH probes. Finally, The UtrCH probes provide robust labeling of F-actin in both echinoderm and amphibian embryos without any apparent effects on development. Biologically, this is the most rigorous test of potential artifacts, in that the rapid development of these organisms is critically dependent on the proper function of the actomyosin cytoskeleton not only during cytokinesis but also during morphogenetic processes such as formation of the blastular epithelium and gastrulation.
Third, the UtrCH probes label F-actin populations that are relatively poorly-labelled by fluorescent G-actin (e.g. the interior of the actin array around wounds). This is likely to be particularly important for studies in which formin-dependent processes are of interest, since fluorescent actins incorporate poorly into formin-nucleated actin filaments [Kovar et al., 2006]. Thus, efforts to visualize F-actin using fluorescent actin during processes such as cytokinesis, which is critically dependent on formins [Wu and Pollard, 2005], may underestimate local F-actin concentration.
Fourth, the UtrCH probes can reveal both stable and dynamic F-actin when used in combination. This is accomplished by the combined photobleaching of RFP-UtrCH and photoactivation of PaGFP-UtrCH. The utility of this approach is shown in Fig. 5 which clearly reveals that stable F-actin becomes concentrated on the inside of a zone of more dynamic F-actin around wounds. Because dynamic and stable actin make different contributions to actomyosin-based motility (see Introduction), this ability to distinguish between the two is critically important. For example, it has recently been reported that the cytokinetic apparatus in mammalian cells contains relatively dynamic actin [Guha et al., 2005; Murthy and Wadworth, 2005]. This was deduced from FRAP analysis of GFP-actin, which does not permit direct visualization of two different actin populations in the same area of the cell. In contrast, the combined photoactivation/photobleaching approach described here allows simultaneous visualization of stable and dynamic actin, and should therefore be of significant benefit.
Acknowledgments
Contract grant sponsor: National Institutes of Health; Contract grant numbers: GM52932, GM66050.
Thanks to J. Nahsti-Ervasti (University of Minnesota) for support and suggestions.
Footnotes
This article contains supplementary material available via the Internet at http://www.interscience.wiley.com/jpages/0886-1544/suppmat
References
- Aizawa H, Sameshima M, Yahara I. A green fluorescent protein–actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium. Cell Struct Funct. 1997;22:335–345. doi: 10.1247/csf.22.335. [DOI] [PubMed] [Google Scholar]
- Bement WM, Sokac AM, Mandato CA. Four-dimensional imaging of cytoskeletal dynamics in Xenopus oocytes and eggs. Differentiation. 2003;71:518–527. doi: 10.1111/j.1432-0436.2003.07109005.x. [DOI] [PubMed] [Google Scholar]
- Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancker P, Low I, Hasselbach W, Wieland T. Interaction of actin with phalloidin: Polymerization and stabilization of F-actin. Biochim Biophys Acta. 1975;400:407–414. doi: 10.1016/0005-2795(75)90196-8. [DOI] [PubMed] [Google Scholar]
- Danilchik MV, Funk WC, Brown EE, Larkin K. Requirement for microtubules in new membrane formation during cytokinesis of Xenopus embryos. Dev Biol. 1998;194:47–60. doi: 10.1006/dbio.1997.8815. [DOI] [PubMed] [Google Scholar]
- Dormann D, Weijer CJ. Imaging of cell migration. EMBO J. 2006;25:3480–3493. doi: 10.1038/sj.emboj.7601227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Albrecht R, Heizer C, Hodgkinson S, Maniak M. Chemoattractant-controlled accumulation of coronin at the leading edge of Dictyostelium cells monitored using a green fluorescent protein-coronin fusion protein. Curr Biol. 1995;5:1280–1285. doi: 10.1016/s0960-9822(95)00254-5. [DOI] [PubMed] [Google Scholar]
- Guha M, Zhou M, Wang YL. Cortical actin turnover during cytokinesis requires myosin II. Curr Biol. 2005;15:732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- Hird S. Cortical actin movements during the first cell cycle of the Caenorhabditis elegans embryo. J Cell Sci. 1996;109:525–533. doi: 10.1242/jcs.109.2.525. [DOI] [PubMed] [Google Scholar]
- Howard TH, Oresajo CO. A method for quantifying F-actin in chemotactic peptide activated neutrophils: Study of the effect of tBOC peptide. Cell Motil. 1985;5:545–557. doi: 10.1002/cm.970050609. [DOI] [PubMed] [Google Scholar]
- Hozumi S, Maeda R, Taniguchi K, Kanai M, Shirakabe S, Sasamura T, Speder P, Noselli S, Aigaki T, Murakami R, Matsuno K. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature. 2006;440:798–802. doi: 10.1038/nature04625. [DOI] [PubMed] [Google Scholar]
- Korn ED. Actin polymerization and its regulation by proteins from nonmuscle cells. Physiol Rev. 1982;62:672–737. doi: 10.1152/physrev.1982.62.2.672. [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;3:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kreis TE, Geiger B, Schlessinger J. Mobility of microinjected rhodamine actin within living chicken gizzard cells determined by fluorescence photobleaching recovery. Cell. 1982;29:835–845. doi: 10.1016/0092-8674(82)90445-7. [DOI] [PubMed] [Google Scholar]
- Lenart P, Bacher CP, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- Mandato CA, Bement WM. Contraction and polymerization cooperate to assemble and close actomyosin rings around Xenopus oocyte wounds. J Cell Biol. 2001;154:785–797. doi: 10.1083/jcb.200103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandato CA, Bement WM. Actomyosin transports microtubules and microtubules control actomyosin recruitment during Xenopus oocyte wound healing. Curr Biol. 2003;13:1096–1105. doi: 10.1016/s0960-9822(03)00420-2. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ. Seeing is believing: Imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Planques V, Warn A, Warn RM. The effects of microinjection of rhodamine-phalloidin on mitosis and cytokinesis in early stage Drosophila embryos. Exp Cell Res. 1991;192:557–566. doi: 10.1016/0014-4827(91)90076-7. [DOI] [PubMed] [Google Scholar]
- Rybakova IN, Ervasti JM. Identification of spectrin-like repeats required for high affinity utrophin-actin interaction. J Biol Chem. 2005;280:23018–23023. doi: 10.1074/jbc.M502530200. [DOI] [PubMed] [Google Scholar]
- Schoenwaelder SM, Burridge K. Bidirectional signaling between the cytoskeleton and integrins. Curr Opin Cell Biol. 1999;11:274–286. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 2004;136:3968–3978. doi: 10.1104/pp.104.049411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Resch GP. The comings and goings of actin: Coupling protrusion and retraction in cell motility. Curr Opin Cell Biol. 2005;17:517–523. doi: 10.1016/j.ceb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Sokac AM, Co C, Taunton J, Bement W. Cdc42-dependent actin polymerization during compensatory endocytosis in Xenopus eggs. Nat Cell Biol. 2003;5:727–732. doi: 10.1038/ncb1025. [DOI] [PubMed] [Google Scholar]
- Stossel TP. On the crawling of animal cells. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- Wang YL. Mobility of filamentous actin in living cytoplasm. J Cell Biol. 1987;105:2811–2816. doi: 10.1083/jcb.105.6.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- Wehland J, Osborn M, Weber K. Phalloidin-induced actin polymerization in the cytoplasm of cultured cells interferes with cell locomotion and growth. Proc Natl Acad Sci USA. 1977;74:5613–5617. doi: 10.1073/pnas.74.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Winder SJ, Hemmings L, Maciver SK, Bolton SJ, Tinsley JM, Davies KE, Critchley DR, Kendrick-Jones J. Utrophin actin binding domain: Analysis of actin binding and cellular targeting. J Cell Sci. 1995;108:63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wulf E, Deboben A, Bautz FA, Faulstich H, Wieland T. Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc Natl Acad Sci USA. 1979;76:4498–4502. doi: 10.1073/pnas.76.9.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]