Significance
Understanding dendritic cell (DC) migration during an immune response is fundamental to defining the rules that govern T cell-mediated immunity. We recently described mice deficient in the pattern recognition receptor NLRP10 (NLR family, pyrin domain containing 10) with a severe DC migration defect. Using whole-exome sequencing, we discovered that this defect was due to mutation of the guanine nucleotide exchange factor Dock8 (dedicator of cytokinesis 8). DOCK8 regulates cytoskeleton dynamics in leukocytes, and loss-of-function mutations cause an immunodeficiency syndrome. Mutations in other Dock genes have been reported in mice lacking innate immune pathways, and we now report two more lines with Dock8 mutations resulting in impaired DC migration. These results clarify the role of NLRP10 in DCs and confirm the essential function of DOCK8 in the immune system.
Keywords: dendritic cell, NLRP10, DOCK8, C3H/HeJ, CDC42
Abstract
Dendritic cells (DCs) are the primary leukocytes responsible for priming T cells. To find and activate naïve T cells, DCs must migrate to lymph nodes, yet the cellular programs responsible for this key step remain unclear. DC migration to lymph nodes and the subsequent T-cell response are disrupted in a mouse we recently described lacking the NOD-like receptor NLRP10 (NLR family, pyrin domain containing 10); however, the mechanism by which this pattern recognition receptor governs DC migration remained unknown. Using a proteomic approach, we discovered that DCs from Nlrp10 knockout mice lack the guanine nucleotide exchange factor DOCK8 (dedicator of cytokinesis 8), which regulates cytoskeleton dynamics in multiple leukocyte populations; in humans, loss-of-function mutations in Dock8 result in severe immunodeficiency. Surprisingly, Nlrp10 knockout mice crossed to other backgrounds had normal DOCK8 expression. This suggested that the original Nlrp10 knockout strain harbored an unexpected mutation in Dock8, which was confirmed using whole-exome sequencing. Consistent with our original report, NLRP3 inflammasome activation remained unaltered in NLRP10-deficient DCs even after restoring DOCK8 function; however, these DCs recovered the ability to migrate. Isolated loss of DOCK8 via targeted deletion confirmed its absolute requirement for DC migration. Because mutations in Dock genes have been discovered in other mouse lines, we analyzed the diversity of Dock8 across different murine strains and found that C3H/HeJ mice also harbor a Dock8 mutation that partially impairs DC migration. We conclude that DOCK8 is an important regulator of DC migration during an immune response and is prone to mutations that disrupt its crucial function.
Dendritic cells (DCs) are crucial for the initiation of an adaptive immune response. Upon acquiring antigens in the periphery, DCs undergo a maturation process that includes antigen processing, cytokine production, and up-regulation of costimulatory molecules. A mature DC must then migrate from peripheral tissues to draining lymph nodes (LNs) to fulfill its role as an antigen-presenting cell that primes naïve T cells (1). Although the signals that induce this maturation process are now well-established (1), relatively little is understood about DC migration aside from the primary chemotactic cue provided by CCR7 that guides DCs to the LN (2, 3).
We recently described a genetically modified NLRP10 (NLR family, pyrin domain containing 10) knockout strain in which this migration step was disrupted while leaving the remainder of the DC maturation program, including CCR7 expression, intact (4). NLRP10 is the only NOD-like receptor (NLR) without a leucine-rich repeat domain, the putative pathogen-associated molecular pattern (PAMP)–binding domain. It has been proposed to both positively and negatively regulate other NLRs, such as NOD1 and NLRP3, respectively (5, 6). Although we found that NLRP3 inflammasome activation was unaltered in the absence of NLRP10, we discovered that Nlrp10−/− mice could not mount a productive T- or B-cell immune response due to a DC-intrinsic failure to emigrate out of inflamed tissues (4, 7).
To understand the mechanism by which NLRP10 governs DC migration, we used an expression proteomic approach to identify molecules with altered expression in DCs generated from the Nlrp10−/− strain and discovered a profound reduction in DOCK8 (dedicator of cytokinesis 8). DOCK8 is a guanine nucleotide exchange factor (GEF) that has two functional domains, DOCK homology region (DHR) 1 and DHR2 (8). In murine DCs, the DHR2 domain has been implicated in regulating the Rho GTPase CDC42 (cell division control protein 42 homolog), which in turn maintains cell polarity of mature DCs during migration (9, 10). Furthermore, mice harboring inactivating mutations in Dock8 lack marginal zone B-cell development, long-term antibody production following immunization, and memory CD8+ T-cell responses to viral infections (11, 12). In humans, inactivating mutations in Dock8 were recently identified as the primary genetic cause underlying autosomal recessive hyper-IgE syndrome (13). This syndrome presents with eczema, recurrent infections of the skin and respiratory tract, increased serum IgE, eosinophilia, recurrent fungal and viral infections, extensive food and environmental allergies, and, in certain patients, squamous cell dysplasia and carcinomas (14).
Given that DOCK8 regulates a wide array of immunologic processes in mouse and human, we sought to understand how NLRP10 regulates DOCK8. To our surprise, we discovered that loss of DOCK8 in the Nlrp10−/− strain was secondary to a point mutation within the Dock8 gene itself. In this study, we demonstrate that restoring DOCK8 function in the Nlrp10−/− strain leads to normal DC migration in vivo. We further show that deletion of Dock8, as well as spontaneous mutation of Dock8 in another inbred strain of mice, results in defective DC migration and, depending on the degree of impaired migration, also abrogates CD4+ T-cell activation.
Results
Reduced Expression of the GEF DOCK8 in NLRP10-Deficient Mice.
Despite clear evidence demonstrating failed movement of adoptively transferred Nlrp10−/− bone marrow-derived dendritic cells (BMDCs) in vivo, classical assays to measure their migration in vitro, such as Transwell chambers, found no defect (4). Evidence exists that DCs behave differently when moving on a 2D surface versus through a 3D matrix (3). We reasoned that our previous in vitro migration assays might have failed to reveal a migration defect in DCs from the Nlrp10−/− strain because these systems failed to recapitulate the complex interactions between a DC and the 3D extracellular matrix of a tissue. Therefore, we studied BMDC migration using a 3D collagen gel assay (15).
In this system, BMDCs generated from WT, but not the Nlrp10−/− strain, could successfully traverse the matrix toward a CCL19 chemokine gradient (Fig. 1A). Although Nlrp10−/− BMDCs actively extended dendrites toward the chemokine gradient, their displacement through the matrix was severely impaired (Movie S1), mimicking the cellular phenotype we previously observed in vivo using two-photon microscopy (4). That the trapped Nlrp10−/− BMDCs could both sense the chemokine gradient and continuously move dendrites suggested that the basic chemotactic machinery was intact. However, the coordinated cell polarity needed for actual displacement through a complex 3D matrix was disrupted, arguing that a regulatory pathway needed for coordinated rearrangement of actin during this movement might be defective. An unbiased proteomic screen done in parallel confirmed that a component regulating actin dynamics was indeed disrupted in Nlrp10−/− BMDCs.
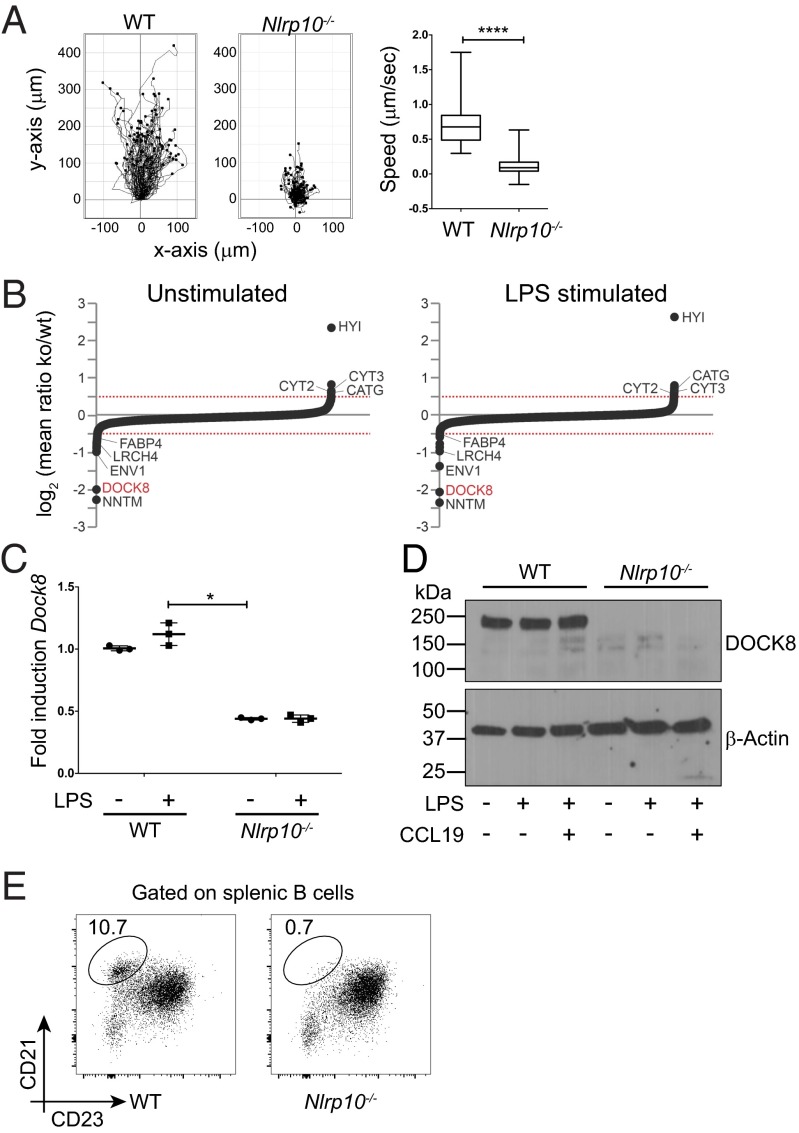
Fig. 1.
Loss of DOCK8 in NLRP10 knockout mice. (A) Individual-cell tracking showing the distance traveled by LPS-stimulated WT (n = 100 cells) and Nlrp10−/− (n = 100 cells) BMDCs toward a CCL19 source in 3D collagen gels. (Right) Average speed of WT and Nlrp10−/− BMDCs (****P < 0.0001; error bars indicate mean + SEM). BMDC migration was recorded by time-lapse microscopy over 4 h at 2 frames per min. (B) iTRAQ-based expression proteomics of WT and Nlrp10−/− BMDCs. Mean log2-transformed expression ratios (Nlrp10−/−/WT) of proteins from two biological replicates of unstimulated (Left) or LPS-stimulated (Right) BMDCs. Proteins that were significantly down- (log2 of mean ratio <−0.5) or up-regulated (log2 of mean ratio >0.5) under both conditions are indicated. (C) Semiquantitative PCR of Dock8 mRNA in unstimulated and LPS-stimulated (1 μg/mL) BMDCs from WT and Nlrp10−/− mice (n = 3 samples per group; *P < 0.05; error bars indicate mean + SEM). (D) Western blot analysis of DOCK8 from total cell lysates of WT and Nlrp10−/− BMDCs that were either unstimulated or treated with CCL19 (100 ng/mL) and/or LPS (1 μg/mL), as indicated. (E) Splenic marginal zone B cells in WT and Nlrp10−/− strains. The gating strategy is shown in Fig. S2A. One of three mice per group is shown. Figures shown are representative of two (B) or three (A and C–E) independent experiments.
To identify novel candidates differentially expressed in DCs lacking NLRP10, we performed a quantitative proteomic screen using iTRAQ (isobaric tag for relative and absolute quantitation) reagents (Fig. S1). Among the most significantly altered proteins detected, an important regulator of actin dynamics, DOCK8, was reduced in BMDCs generated from the Nlrp10−/− strain (Fig. 1B). We confirmed loss of DOCK8 at both the mRNA and protein level in Nlrp10−/− BMDCs (Fig. 1 C and D). DOCK8 is a GEF whose primary target is the Rho GTPase CDC42, which in turn regulates endocytosis of antigens in immature DCs (16) and migration of mature DCs to draining LNs (10). Nlrp10−/− BMDCs efficiently phagocytosed antigen in vitro (4), suggesting that CDC42 activity in immature DCs was intact. In contrast, migration of Nlrp10−/− DCs in the 3D collagen system was almost completely abrogated (Fig. 1A), similar to DOCK8- or CDC42-deficient DCs (9, 10). Further, the Nlrp10−/− strain exhibited other phenotypes that have been described in DOCK8-deficient mice, such as loss of marginal zone B cells (Fig. 1E and Fig. S2A).
The finding that NLRP10-deficient mice lack DOCK8 was intriguing but also surprising, as it suggests an unorthodox paradigm for how NLRs may function independent of pattern recognition. Other NLR family members have been suggested to regulate DC maturation or migration, such as NLRP3 and NLRP12, respectively (17, 18). The mechanisms by which these NLRs regulate DC function have not been determined. There is also precedent in other cell types for GEF-dependent NLR activation during bacterial cell invasion, although it is unknown whether this occurs via direct protein interaction (19). Therefore, it was possible that lack of DC migration in the NLRP10-deficient mice could result from positive regulation of DOCK8 by NLRP10; however, our subsequent studies in fact suggested that DOCK8 functions independent of NLRP10.
NLRP10-Deficient DCs Lack DOCK8 Due to a Point Mutation.
To study DC migration in vivo, we inject fluorescently labeled antigen s.c. along with a PAMP such as lipopolysaccharide (LPS). Eighteen hours later, antigen-carrying DCs that have migrated from the dermis can be found in the skin-draining LNs (conventional migratory DCs) (Fig. S2B). Consistent with our original observation (4), Nlrp10−/− mice showed a loss of antigen-positive DCs in draining LNs (Fig. 2A). However, when we crossed the Nlrp10−/− mice onto other backgrounds including BALB/c, DC migration was restored (Fig. 2A), indicating that the observed migration defect did not cosegregate with loss of NLRP10. Further, BALB/c Nlrp10−/− retained normal DOCK8 expression (Fig. 2 B and C), suggesting that a second genetic locus might regulate Dock8 expression.
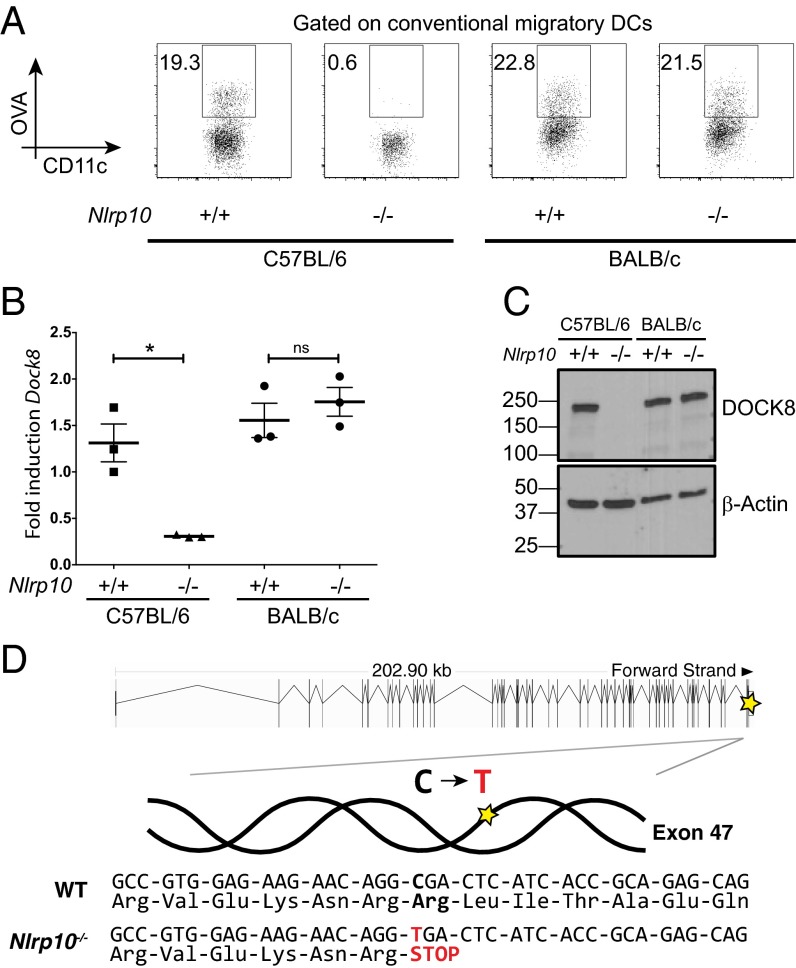
Fig. 2.
NLRP10 knockout dendritic cells lack DOCK8 due to a point mutation. (A) DC migration in WT and Nlrp10−/− strains on a C57BL/6 or BALB/c background following s.c. immunization with OVA-AF647 (10 μg per flank) and LPS (1 μg per flank). One of two to four mice per group is shown. (B and C) Semiquantitative PCR (B) and Western blot analysis (C) of DOCK8 in BMDCs from WT and Nlrp10−/− strains on a C57BL/6 or BALB/c background (n = 3 samples per group; *P < 0.05; ns, not significant; error bars indicate mean + SEM). (D) Whole-exome sequencing of the Nlrp10−/− strain revealed a C→T change in exon 47 of the Dock8 gene, causing a nonsense mutation (SNPEFF_AMINO_ACID_CHANGE=R2034*). Overview of the genomic structure of Dock8 adapted from Ensembl (36) highlighting the affected region (Top). Figures shown are representative of three independent experiments.
Initial single-nucleotide polymorphism (SNP) analysis across the whole genome of Nlrp10−/− mice revealed few differences from the reference C57BL/6 genotype (Fig. S3A). Because this methodology does not detect most single-point mutation differences, we screened for additional mutations in the Nlrp10−/− strain using whole-exome sequencing on a phenotype-positive mouse. We discovered a single-nucleotide change in exon 47 of Dock8 that results in a premature stop codon (Fig. 2D and Fig. S3B). We suspect that this base-pair change results in nonsense-mediated decay of the produced Dock8 mRNA in Nlrp10−/− mice (20). This is supported by our findings of reduced but detectable Dock8 mRNA levels (Fig. 1C) but a complete loss of protein (Fig. 1D).
From the sequencing data, we identified more than 500 other SNPs or indels in the Nlrp10−/− strain, 106 of which were homozygous. Of these, only 6 (including Dock8) were predicted to alter protein function (Table S1). Importantly, of these 6 genes, only Dock8 has a known or predicted role in actin dynamics or DC function. This high number of genetic lesions is perhaps not surprising, given that the estimated mutation rate in humans of 1.38 × 10−8 per bp per person results in potentially 30–50 mutations per generation (21). Although the mutation rate in inbred mice has been difficult to estimate, it is unlikely to be significantly different (22); in combination with the inbreeding that occurs in mouse colonies, numerous mutations could accumulate over time (22). Sequencing the region of interest in exon 47 of Dock8 in the original embryonic stem cells used to target Nlrp10 revealed that it was identical to a C57BL/6 reference sequence (AACAGGCGA; compare with Fig. 2D). Therefore, it appears that this mutation in Dock8 occurred spontaneously during the early intercrossing phase and became fixed in our colony before the initial phenotype analysis of these knockout mice.
NLRP10-Deficient Mice Have Normal DC Migration When DOCK8 Is Restored.
Through breeding, we restored functional DOCK8 in the original NLRP10 knockout strain (Nlrp10−/− Dock8+/+) and isolated the mutated Dock8 gene to an individual line (Nlrp10+/+ Dock8mu/mu). We confirmed that Nlrp10−/− mice in the absence of Dock8 mutation do not have a DC migration defect (Fig. 3A). However, they do retain some of the other phenotypes observed in our original study, including the aberrant up-regulation of Gdpd3 (Fig. 3B), a glycerophosphodiester phosphodiesterase family member with unknown function in the immune system. Related GDPD molecules have been shown to regulate actin cytoskeleton dynamics downstream of G protein-coupled receptor signaling (23). Although we have had limited success identifying the immunologic role of GDPD3 to date, it will be interesting to test whether this pathway could synergistically affect DOCK8-dependent cytoskeleton dynamics in an NLRP10-dependent manner. However, from the current studies, we conclude that NLRP10 is not required for efficient DC migration to draining LNs. Further consistent with our original report, we confirmed that activation of Nlrp10−/− DCs by NLRP3 inflammasome stimuli (LPS/ATP and LPS/Alum) was not enhanced, even when DOCK8 expression was restored (Fig. 3C) (4).
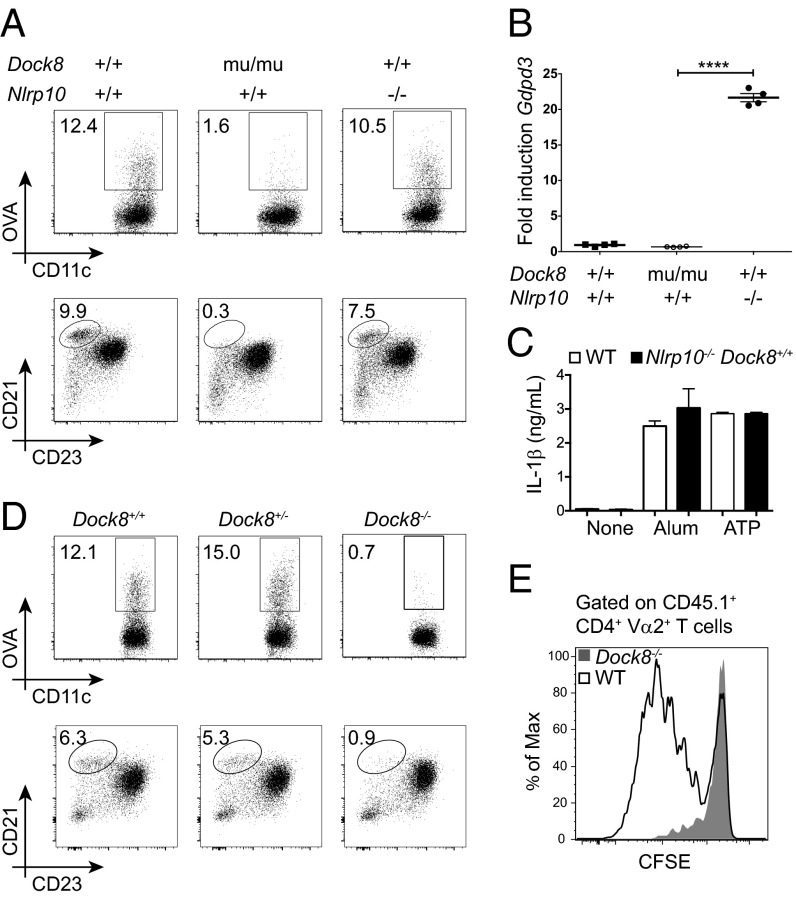
Fig. 3.
NLRP10-deficient and DOCK8-deficient mice have distinct phenotypes. (A) In vivo DC migration following s.c. immunization with OVA-AF647 (10 μg per flank) and LPS (1 μg per flank) (Top) and splenic marginal zone B cells (Bottom) in WT (Nlrp10+/+ Dock8+/+), DOCK8 mutant (Nlrp10+/+ Dock8mu/mu), and NLRP10-deficient (Nlrp10−/− Dock8+/+) mice. The gating strategy is shown in Fig. S2. One of two or three mice per group is shown. (B) Semiquantitative PCR of Gdpd3 from WT (Nlrp10+/+ Dock8+/+), DOCK8 mutant (Nlrp10+/+ Dock8mu/mu), and NLRP10-deficient (Nlrp10−/− Dock8+/+) BMDCs (n = 4 samples per group; ****P < 0.0001; error bars indicate mean + SEM). (C) Isolated NLRP10 deficiency in BMDCs does not impact inflammasome responsiveness. LPS-activated BMDCs from WT (Nlrp10+/+ Dock8+/+) and NLRP10-deficient (Nlrp10−/− Dock8+/+) mice were stimulated with NLRP3 inflammasome stimuli (ATP or Alum). IL-1β was measured from supernatants 8 h following the second stimulus (n = 3 samples per group). (D) In vivo DC migration following s.c. immunization with OVA-AF647 (10 μg per flank) and LPS (1 μg per flank) (Top) and splenic marginal zone B cells (Bottom) in newly generated DOCK8-deficient (Dock8−/−) mice. WT (Dock8+/+) and DOCK8 heterozygous littermates (Dock8+/−) were used as controls. The gating strategy is shown in Fig. S2. One of two to four mice per group is shown. (E) In vivo proliferation of OT-II cells by CFSE (carboxyfluorescein succinimidyl ester) dilution in WT (black histogram) and Dock8−/− (gray-filled histogram) mice 3 d after s.c. immunization with ovalbumin (0.5 μg per flank) and LPS (1 μg per flank). One of two or three mice per group is shown. Figures shown are representative of three independent experiments.
DOCK8 Knockout Mice Have Impaired DC Migration and CD4+ T-Cell Activation.
To confirm that the DC migration defect originally observed was solely due to loss of DOCK8, we used CRISPR/Cas9 technology to generate a DOCK8-deficient mouse (Fig. S4 A and B). Using these Dock8−/− mice, as well as the isolated Dock8 mutant mice (Nlrp10+/+ Dock8mu/mu), we confirmed that inefficient DC migration to draining LNs is associated with loss of DOCK8 (Fig. 3 A and D). Proliferation of adoptively transferred T-cell receptor (TCR) transgenic CD4+ T cells such as OT-II can be used to assess antigen presentation to naïve T cells in the LN. Consistent with failed DC migration to inguinal LNs following injected OVA antigen (Fig. 3D), OT-II T cells fail to significantly proliferate in immunized DOCK8-deficient hosts (Fig. 3E). As expected, marginal zone B-cell development was also disrupted in both mouse strains deficient in DOCK8 (Dock8mu/mu and Dock8−/−) but not in the mice only lacking NLRP10 (Fig. 3 A and D). Thus, the lack of DC migration in vivo and in vitro is due to DOCK8 deficiency, which is independent of NLRP10 and profoundly impacts the activation of naïve CD4+ T cells.
C3H/HeJ Mice Harbor a Point Mutation in Dock8.
The finding that NLRP10 knockout mice have a spontaneous mutation in Dock8 is striking for two reasons. The first is that a recent study of mice deficient in the NLR inflammasome adaptor ASC (apoptosis-associated speck-like protein containing a caspase activation and recruitment domain) identified a defect in DC antigen processing and movement but was subsequently found to be due to loss of another member of the DOCK family, DOCK2, and unrelated to loss of ASC (24, 25). The second is that more than 35 different mutations located throughout the Dock8 gene in humans are associated with immunodeficiency (ClinVar database) (13). The reason for this potential genetic instability that leads to functional changes of Dock8 is unclear and might relate to the sheer size of the gene (>200 kb in total with a >7 kb coding region in both mouse and human). Analysis of a wide array of standard and wild-derived inbred mouse strains found Dock8 to be highly polymorphic (Fig. S5 A–C and Dataset S1). Using the Jax MGI database, we found that of the 40 strains analyzed, 1 had a predicted deleterious polymorphism that would potentially result in loss of function: the C3H/HeJ strain (Fig. 4A and Fig. S5C).
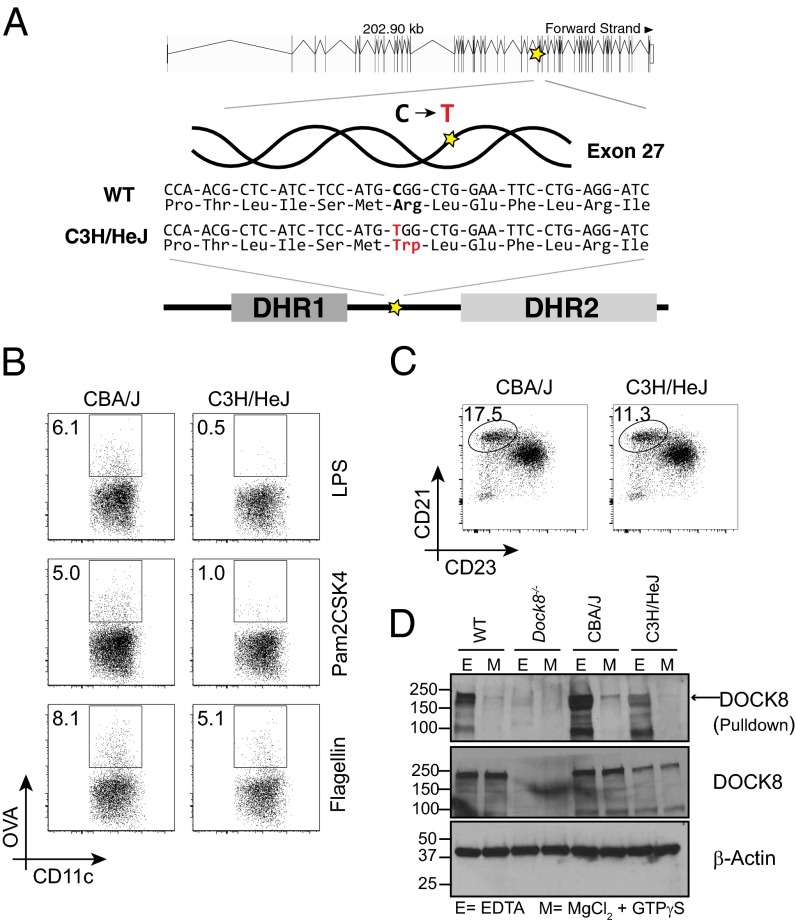
Fig. 4.
C3H/HeJ mice harbor a point mutation in Dock8 and have impaired DC migration. (A) Sequence confirmation of a Dock8 point mutation predicted in C3H/HeJ mice. The C→T mutation in exon 27 results in an amino acid change in the region between the DHR1 and DHR2 domains of DOCK8. (B) In vivo DC migration in CBA and C3H/HeJ mice following s.c. immunization with OVA-AF647 (10 μg per flank) and either LPS (1 μg per flank), Pam2CSK4 (1 μg per flank), or flagellin (1 μg per flank). The gating strategy is shown in Fig. S2B. One of three or four mice per group is shown. (C) Splenic marginal zone B cells in CBA and C3H/HeJ mice. The gating strategy is shown in Fig. S2A. One of five mice per group is shown. (D) Total cell lysates from C57BL/6N (WT), Dock8−/−, CBA/J, and C3H/HeJ BMDCs were pulled down with GST-tagged CDC42, followed by Western blot analysis for DOCK8 (Top). Assays were done in buffer containing either 20 mM EDTA (E buffer) or 100 mM MgCl2 and 200 μM GTPγS (M buffer). (Middle and Bottom) DOCK8 and β-actin levels in the total cell lysates used for pull-down experiments, respectively. Figures shown are representative of three independent experiments.
C3H/HeJ mice have traditionally been used to study the role of LPS in the innate immune response due to a mutation in the gene encoding TLR4 (Toll-like receptor 4) (26). We confirmed by sequencing that the closely related strains of mice included in the SNP analysis, CBA/J, C3H/HeOuJ, and C3HeB/FeJ (27), do not carry the same Dock8 polymorphism at this site (Fig. S5D). Both PROVEAN (Protein Variation Effect Analyzer; provean.jcvi.org) and SIFT (Sorting Intolerant From Tolerant; sift.jcvi.org) software predicted that this point mutation in C3H/HeJ mice would result in loss of DOCK8 function. Therefore, we tested in vivo DC migration in the C3H/HeJ mice in response to a non–TLR4-dependent innate immune stimulus and indeed found a partial loss of antigen-positive DCs in draining LNs (Fig. 4B). As expected, DC migration in C3H/HeJ mice is almost completely abrogated when the TLR4 ligand LPS is used as the adjuvant, whereas CBA/J mice demonstrate robust accumulation of antigen-positive DCs in the LN (Fig. 4B). DCs from C3H/HeJ mice should be able to respond to both the TLR2 agonist Pam2CSK4 and the TLR5 agonist flagellin. However, antigen-positive DC accumulation in the LN was reduced with both adjuvants in C3H/HeJ but robust in CBA/J mice (Fig. 4B). For unclear reasons, the degree of impairment in DC migration in the C3H/HeJ mice was consistently more pronounced when Pam2CSK4 rather than flagellin was used as the adjuvant and warrants further study. In contrast to the DOCK8-deficient mice (4) (Fig. 3), however, the DC migration defect in C3H/HeJ mice is mild enough to allow for adequate CD4+ T cell-dependent immune responses following immunization with an LPS-free adjuvant (Fig. S6A).
On average, the C3H/HeJ mice also demonstrated only a 30% reduction in splenic marginal zone B cells (Fig. 4C), suggesting that some degree of DOCK8 function is retained in these mice. Indeed, the Dock8 polymorphism in C3H/HeJ mice does not result in reduced mRNA levels (Fig. S6B), and only mildly reduced DOCK8 protein in BMDCs (Fig. 4D). Although we could not confirm whether activity of DOCK8 in C3H/HeJ mice is altered, we observed a reduction in binding to its main effector molecule CDC42 (Fig. 4D). Altogether, these data suggest that the Dock8 mutation in C3H/HeJ mice results in partially impaired activity and thus an incomplete DC migration and marginal zone B-cell development phenotype. It is intriguing that both of these responses might be analogous in nature and therefore “tunable,” that is, dependent on the degree of DOCK8 activity. Further work is necessary to identify how DOCK8 is actually affected in these mice to address this question.
Discussion
NLRP10-deficient DCs lack a crucial regulatory pathway for the coordinated rearrangement of the actin cytoskeleton during migration, but this is due to a coincidental genetic lesion in the gene Dock8. We discovered a point mutation in one of the terminal Dock8 exons resulting in a premature stop codon that likely results in nonsense-mediated decay. We confirmed that the DC migration phenotype originally reported (4) results from loss of DOCK8, both through the evaluation of a newly generated DOCK8 knockout mouse as well as by separating the Nlrp10 deletion from the Dock8 point mutation through breeding.
DOCK8 is an atypical GEF that regulates CDC42 (9). CDC42 is a member of the Rho family of small GTPases, which function as molecular switches by cycling between an active GTP-bound state and an inactive GDP-bound state (8). When CDC42 is GTP-bound and active, it can regulate a diverse set of cellular processes from cell migration to division. DCs require both CDC42 and DOCK8 to traverse a 3D matrix similar in nature to the one used in the current study. A seminal paper on the role of DOCK8 in DCs demonstrated colocalization of the two molecules at the membrane (9); a separate study characterized the necessity of CDC42 in leading-edge coordination during migration of mature DCs (10). Although how the DOCK8-CDC42 pathway directs cell polarity remains to be determined, loss of DOCK8 in the NLRP10 knockout mice is consistent with these previous observations. However, Nlrp10−/− BMDCs carrying the Dock8 mutation were fully capable of antigen phagocytosis and presentation to naïve T cells in vitro (4), suggesting CDC42 activity in immature DCs remains intact (16). We therefore propose that DOCK8-mediated CDC42 activity is limited to DC migration, which is essential for induction of CD4+ T-cell activation in mice (Fig. 3E) (4, 9). Dock8 mutations in humans are responsible for a rare form of immunodeficiency, autosomal recessive hyper-IgE syndrome, which affects both T-cell and B-cell function (13, 28, 29). The effect of Dock8 mutation on human DC migration and the subsequent impact on adaptive immune responses, however, remain unanswered questions.
It is interesting to note that Nlrp10−/− mice are not the first inbred knockout mouse line to develop a spontaneous mutation in a Dock gene. Ippagunta and colleagues first identified that certain lines of ASC-deficient mice acquired a DOCK2 deficiency, although the exact genetic alteration has not been characterized (24, 25). Even other genetically targeted mice, including IRF5-deficient mice, were discovered to harbor an unexpected frameshift mutation in Dock2 (30). Because the DOCK family of molecules appears to be frequently mutated in other inbred strains of mice, we analyzed the sequence diversity of Dock8. We found that another inbred strain, the commonly used C3H/HeJ mouse that lacks a functional TLR4, also has a predicted deleterious mutation in Dock8. These mice retain DOCK8 protein expression but demonstrate a mild impairment in DC migration as well as in marginal zone B-cell development (Fig. 4). It will be interesting to identify whether other lines of mice lacking specific pathways in the innate branch of the immune system harbor Dock mutations, which could suggest a possible link between the occurrence of genetic lesions and loss of innate immune defenses. All of the genes in the Dock family are large, with numerous exons and coding regions 6–10 kb in size; Dock8, in fact, has the smallest number of exons at 48. In comparison, the mean number of exons calculated for the human genome is 10.4, with an average coding region of 1.4 kb (31). This raises the alternative possibility that the identification of multiple point mutations in the Dock gene family might arise secondary to the sheer size of the genes involved in combination with induction of a severe phenotype.
Our proteomic analysis revealed other proteins that were also differentially expressed in Nlrp10−/− mice, including HYI (hydroxypyruvate isomerase) and NNTM (nicotinamide nucleotide transhydrogenase) (Fig. 1B). Hyi encodes for a putative hydroxypyruvate isomerase with no known mammalian function but potentially plays a role in carbohydrate transport and metabolism. HYI was highly up-regulated in Nlrp10−/− BMDCs, likely due to a point mutation, which was subsequently identified by exome sequencing (Table S1). NNTM, encoded by Nnt, catalyzes NADPH production for ATP synthesis and is highly reduced in Nlrp10−/− mice. Nnt lacks five exons from an in-frame deletion in C57BL/6 mice from The Jackson Laboratory (C57BL/6J) but not in C57BL/6 mice from the National Cancer Institute (NCI) (C57BL/6N) (32). The latter were used as WT controls for the proteomic screen. However, the exome sequencing results from the Nlrp10−/− mice aligned with the mm10 reference sequence derived from C57BL/6J mice (lacking these five exons), indicating that the Nlrp10−/− mice carried this region from the C57BL/6J background. Therefore, loss of the NNTM protein in Nlrp10−/− DCs (Fig. 1B) is consistent with this known gene deletion in C57BL/6J mice (32). These examples, as well as the Dock8 mutations identified in this paper, highlight the need for continual evaluation of the genetic stability of inbred mouse strains, especially those that are expanded by sibling matings beyond 10 generations. In addition, to limit genetic drift, periodically obtaining new breeding stock from founder lines or cryopreserved embryos from early generations has been recommended (22).
In a genome-wide association study of patients with atopic dermatitis, Nlrp10, which is highly expressed in the skin, was identified as one of eight susceptibility loci (33). Further suggesting NLRP10 is important to the immune response in the skin, Lautz et al. demonstrated that NLRP10 plays a crucial role in regulating dermal fibroblast immunity to Shigella flexneri by acting as a positive regulator of the NOD1 inflammasome (5). NLRP10 has also been shown to negatively regulate the NLRP3 inflammasome (6, 34). However, in contrast to this but consistent with our previous report, we and others failed to observe enhanced inflammasome function in the absence of NLRP10 (4, 5, 7). The reason for the differences in these results is unknown but could relate to different cell types, stimuli, and forms of NLRP10 modulation, such as overexpression systems versus knockout models. Understanding these differences and the various roles of NLRP10 in immunity, especially in the skin, will shed light on the function of this unusual NLR.
Materials and Methods
Mice.
To generate DOCK8-deficient mice, guide RNAs were designed against intronic sequences flanking exons 10–14. Cas9-mediated double-stranded DNA breaks resolved by nonhomologous end joining would ablate the intervening sequences containing exons 10–14, creating a frameshift and loss of DOCK8 protein expression. Cas9 targeting was performed as described in Yang et al. (35) with some modifications (SI Materials and Methods). Nlrp10−/− mice were generated by the targeted deletion of Nlrp10 exons 2 and 3 in C57BL/6 embryonic stem (ES) cell lines (Bruce4) as reported previously (4). Nlrp10−/− mice were also backcrossed for 10 generations to BALB/cJ mice (BALB/c Nlrp10−/−). Control mice used were C57BL/6N and BALB/cJ mice, purchased from the NCI and The Jackson Laboratory, respectively. CBA/J, C3H/HeJ, and OT-II [B6.Cg-Tg(TcraTcrb)425Cbn/J] mice were purchased from The Jackson Laboratory. OT-II mice were crossed onto the CD45.1 (B6Ly5.2Cr) background from the NCI. All protocols used in this study were approved by the Institutional Animal Care and Use Committee at the Yale University School of Medicine or the University of Iowa.
Collagen Gel Migration Assays.
In vitro migration of BMDCs in a 3D collagen gel system was performed as previously described (15). Briefly, a collagen gel matrix was prepared using PureCol (purified bovine collagen solution; Advanced BioMatrix), 1× Eagle’s MEM (Lonza), and 0.4% sodium bicarbonate (Sigma). This matrix was then mixed with LPS-matured BMDCs (3 × 106 per mL) in a 2:1 ratio, loaded into a glass chamber (2 × 2 × 0.1 cm), and allowed to polymerize at 37 °C. After 30 min, CCL19 (100 ng/mL) was added on top of the matrix and the chamber was sealed. BMDCs migrating in the direction of the chemokine gradient were imaged using a Leica 6000 microscope (DIC mode, 100× total magnification) for 4 h (2 frames per min). For data analysis, all images from a single run were imported into ImageJ software (NIH), and a cell-tracking tool (ibidi) was used to select and track single cells across all of the frames. For quantification, the distance migrated by 100 cells in each run was determined.
In Vivo DC Migration.
Mice were immunized s.c. in each flank with 10 μg OVA-Alexa Fluor 647 (Molecular Probes) and 1 μg LPS, 1 μg Pam2CSK4, or 1 μg flagellin from Bacillus subtilis (all from Invivogen), as indicated. Inguinal lymph nodes were harvested 18 h postimmunization and digested with collagenase IV (1 mg/mL; Sigma) for 40 min at 37 °C. Single-cell suspensions were prepared, stained, and then analyzed on an LSRII flow cytometer (BD Biosciences).
In Vivo T-Cell Activation.
OVA-specific CD4+ T cells were prepared from spleen and lymph nodes of OT-II TCR transgenic mice by negative selection using the CD4+ T Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions. OT-II CD4+ T cells were labeled with 2 μM CFSE, and 3 × 106 cells were transferred into mice by retroorbital injection. Mice were s.c. immunized 24 h later with 0.5 μg OVA (Sigma) and 1 μg LPS (Sigma) per flank. Inguinal lymph nodes were harvested 3 d postimmunization, and single-cell suspensions were prepared, stained, and then analyzed on an LSRII flow cytometer (BD Biosciences).
Supplementary Information.
Generation of Cas9 mRNA and guide RNAs, one-cell embryo injection, antibodies, BMDC culture, marginal zone B-cell analyses, relative gene expression analyses, Western blot analyses, CDC42 pull-down assay, quantitative proteomics, whole-exome sequencing, SNP analyses and hierarchical clustering, asthma model and intracellular cytokine staining, and statistical analyses are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mark Firla, Lucy Rowe, Mary Barter, and Katja Parapatics for technical assistance, Jennifer A. Doudna and Stephen Floor for guidance with establishing the CRISPR/Cas9 system, and Krishna Karuturi, Florian P. Breitwieser, and Jacques Colinges for support with bioinformatics analysis. This work was supported by NIH National Institute of Allergy and Infectious Diseases (NIAID) Grant K08 AI085038 (to S.C.E.), a Hartwell Foundation Individual Biomedical Research Award (to S.C.E.), NIH NIAID Grant R01 AI108829 (to S.C.E.), Austrian Academy of Sciences and European Research Council Advanced Grant i-FIVE 250179 (to G.S.-F.), and NIH NIAID Grant R01 AI087630 (to F.S.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRR1792904).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501554112/-/DCSupplemental.
References
- 1.Krishnaswamy JK, Chu T, Eisenbarth SC. Beyond pattern recognition: NOD-like receptors in dendritic cells. Trends Immunol. 2013;34(5):224–233. doi: 10.1016/j.it.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohl L, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21(2):279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Lämmermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 4.Eisenbarth SC, et al. NLRP10 is a NOD-like receptor essential to initiate adaptive immunity by dendritic cells. Nature. 2012;484(7395):510–513. doi: 10.1038/nature11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lautz K, et al. NLRP10 enhances Shigella-induced pro-inflammatory responses. Cell Microbiol. 2012;14(10):1568–1583. doi: 10.1111/j.1462-5822.2012.01822.x. [DOI] [PubMed] [Google Scholar]
- 6.Imamura R, et al. Anti-inflammatory activity of PYNOD and its mechanism in humans and mice. J Immunol. 2010;184(10):5874–5884. doi: 10.4049/jimmunol.0900779. [DOI] [PubMed] [Google Scholar]
- 7.Joly S, et al. Cutting edge: Nlrp10 is essential for protective antifungal adaptive immunity against Candida albicans. J Immunol. 2012;189(10):4713–4717. doi: 10.4049/jimmunol.1201715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikimi A, Kukimoto-Niino M, Yokoyama S, Fukui Y. Immune regulatory functions of DOCK family proteins in health and disease. Exp Cell Res. 2013;319(15):2343–2349. doi: 10.1016/j.yexcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Harada Y, et al. DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood. 2012;119(19):4451–4461. doi: 10.1182/blood-2012-01-407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lämmermann T, et al. Cdc42-dependent leading edge coordination is essential for interstitial dendritic cell migration. Blood. 2009;113(23):5703–5710. doi: 10.1182/blood-2008-11-191882. [DOI] [PubMed] [Google Scholar]
- 11.Randall KL, et al. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J Exp Med. 2011;208(11):2305–2320. doi: 10.1084/jem.20110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall KL, et al. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat Immunol. 2009;10(12):1283–1291. doi: 10.1038/ni.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361(21):2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Davis JC, Dove CG, Su HC. Genetic, clinical, and laboratory markers for DOCK8 immunodeficiency syndrome. Dis Markers. 2010;29(3-4):131–139. doi: 10.3233/DMA-2010-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sixt M, Lämmermann T. In vitro analysis of chemotactic leukocyte migration in 3D environments. Methods Mol Biol. 2011;769:149–165. doi: 10.1007/978-1-61779-207-6_11. [DOI] [PubMed] [Google Scholar]
- 16.Garrett WS, et al. Developmental control of endocytosis in dendritic cells by Cdc42. Cell. 2000;102(3):325–334. doi: 10.1016/s0092-8674(00)00038-6. [DOI] [PubMed] [Google Scholar]
- 17.Kool M, et al. Cutting edge: Alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181(6):3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 18.Arthur JC, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol. 2010;185(8):4515–4519. doi: 10.4049/jimmunol.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukazawa A, et al. GEF-H1 mediated control of NOD1 dependent NF-kappaB activation by Shigella effectors. PLoS Pathog. 2008;4(11):e1000228. doi: 10.1371/journal.ppat.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker KE, Parker R. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16(3):293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Scally A, Durbin R. Revising the human mutation rate: Implications for understanding human evolution. Nat Rev Genet. 2012;13(10):745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 22.Taft RA, Davisson M, Wiles MV. Know thy mouse. Trends Genet. 2006;22(12):649–653. doi: 10.1016/j.tig.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Yanaka N. Mammalian glycerophosphodiester phosphodiesterases. Biosci Biotechnol Biochem. 2007;71(8):1811–1818. doi: 10.1271/bbb.70062. [DOI] [PubMed] [Google Scholar]
- 24.Ippagunta SK, et al. The inflammasome adaptor ASC regulates the function of adaptive immune cells by controlling Dock2-mediated Rac activation and actin polymerization. Nat Immunol. 2011;12(10):1010–1016. doi: 10.1038/ni.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ippagunta SK, et al. Addendum: Defective Dock2 expression in a subset of ASC-deficient mouse lines. Nat Immunol. 2012;13(7):701–702. doi: 10.1038/ni.2389. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 27.Petkov PM, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14(9):1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engelhardt KR, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124(6):1289–1302. doi: 10.1016/j.jaci.2009.10.038. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, et al. DOCK8 regulates lymphocyte shape integrity for skin antiviral immunity. J Exp Med. 2014;211(13):2549–2566. doi: 10.1084/jem.20141307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purtha WE, Swiecki M, Colonna M, Diamond MS, Bhattacharya D. Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc Natl Acad Sci USA. 2012;109(15):E898–E904. doi: 10.1073/pnas.1118155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alberts B, et al. 2014. DNA, chromosomes, and genomes. Molecular Biology of the Cell (Garland Science, New York), 6 Ed, pp 184–185.
- 32.Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD. Deletion of nicotinamide nucleotide transhydrogenase: A new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes. 2006;55(7):2153–2156. doi: 10.2337/db06-0358. [DOI] [PubMed] [Google Scholar]
- 33.Hirota T, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44(11):1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 34.Murphy N, Grehan B, Lynch MA. Glial uptake of amyloid beta induces NLRP3 inflammasome formation via cathepsin-dependent degradation of NLRP10. Neuromolecular Med. 2014;16(1):205–215. doi: 10.1007/s12017-013-8274-6. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.