Significance
About 560 million East Asians have an impaired ability to eliminate acetaldehyde because of a point mutation in an enzyme called aldehyde dehydrogenase 2 (ALDH2). Humans with this mutation have ∼20-fold higher blood acetaldehyde levels than those with normal enzyme activity after consuming one to two units of alcoholic beverages. Because acetaldehyde is a potent carcinogen and causes behavioral impairment, its accumulation is a health risk. We identified a pharmacologic agent that recruits ALDH3A1, a closely related enzyme, to compensate for a loss of ability of ALDH2 to metabolize acetaldehyde. Pharmacologic agents that alter substrate specificity of an enzyme have not yet been described and may have wide clinical application in treating patients with impaired ability to detoxify toxic substances.
Keywords: acetaldehyde, ALDH2, ALDH3A1, Alda, ethanol metabolism
Abstract
Correcting a genetic mutation that leads to a loss of function has been a challenge. One such mutation is in aldehyde dehydrogenase 2 (ALDH2), denoted ALDH2*2. This mutation is present in ∼0.6 billion East Asians and results in accumulation of toxic acetaldehyde after consumption of ethanol. To temporarily increase metabolism of acetaldehyde in vivo, we describe an approach in which a pharmacologic agent recruited another ALDH to metabolize acetaldehyde. We focused on ALDH3A1, which is enriched in the upper aerodigestive track, and identified Alda-89 as a small molecule that enables ALDH3A1 to metabolize acetaldehyde. When given together with the ALDH2-specific activator, Alda-1, Alda-89 reduced acetaldehyde-induced behavioral impairment by causing a rapid reduction in blood ethanol and acetaldehyde levels after acute ethanol intoxication in both wild-type and ALDH2-deficient, ALDH2*1/*2, heterozygotic knock-in mice. The use of a pharmacologic agent to recruit an enzyme to metabolize a substrate that it usually does not metabolize may represent a novel means to temporarily increase elimination of toxic agents in vivo.
The aldehyde dehydrogenase (ALDH) superfamily comprises 19 enzymes that catalyze the oxidation and detoxification of a wide spectrum of short and long aliphatic and aromatic aldehydes (1, 2). Acetaldehyde is a product of ethanol metabolism, which is consumed by >80% of humans. In addition to the behavioral impairment risk, the ethanol metabolite, acetaldehyde, is a proven group 1 carcinogen (3). Above and beyond the health risk in the general population, ∼40% of East Asians [∼560 million or ∼8% of the world’s population (4, 5)] carry a point mutation in the ALDH2 gene that leads to a severe enzyme deficiency and accumulation of toxic acetaldehyde (6). After consuming two units of alcoholic beverage, blood acetaldehyde levels reach 60 μM and remain elevated for several hours in heterozygotic carriers of this mutation, whereas within 30 min, acetaldehyde levels are not detected in carriers of the wild-type enzyme (7). The inactivating glutamate 487 to lysine mutation (E487K) (8), denoted ALDH2*2 (vs. ALDH2*1 for the wild-type allele) (9), is dominant; heterozygotic ALDH2*1/*2 individuals have only 17–30% of wild-type activity (10, 11). ALDH2 deficiency is associated with severe facial flushing, longer behavioral impairment (intoxication), longer-lasting headache, nausea, and palpitations from moderate ethanol consumption compared with individuals with normal ALDH2*1/*1 (4).
Despite the unpleasant reaction to acetaldehyde accumulation, 17–27% of individuals with ALDH2*1/*2 (heterozygotes) are heavy drinkers (4, 12, 13). These heterozygotic heavy drinkers (consuming >18 alcoholic drinks/week) have greater than 80-fold increased risk for squamous cell carcinomas in the upper aerodigestive track (UADT; i.e., oral cavity and pharynx, larynx, and esophagus) compared with a ∼fourfold increase in wild-type ALDH2*1/*1 heavy drinkers (4, 13–16). Further, an elevated risk of hepatocarcinoma and its recurrence occurs among hepatitis C-infected patients with the ALDH2*2 mutation (17). Acetaldehyde levels are particularly high in the saliva after ethanol ingestion (18), leading to a significant increase in acetaldehyde-DNA adduct levels in ALDH2*1/*2 heterozygotes, even after moderate ethanol consumption (19). Because acetaldehyde is a carcinogen, and the duration and extent of exposure influences its toxicity, increasing the rate of acetaldehyde elimination, especially in ALDH2*1/*2 heterozygotes, may reduce important health risks. We therefore set out to identify a pharmacologic tool to “recruit” another member of the ALDH family to enhance the elimination of acetaldehyde. We focused on ALDH3A1 because it is highly expressed in the epithelial cell layer of the UADT, stomach, liver, and kidney (20–22). ALDH3A1 metabolizes aromatic, aliphatic medium chain aldehydes and α,β-hydroxyalkenal aldehydes, but not acetaldehyde under basal condition (21, 23). The challenge therefore was to find a pharmacologic means to enable ALDH3A1 to assist in the elimination of acetaldehyde.
Our laboratory has identified a group of small molecules, Aldas (aldehyde dehydrogenase activators), that increase the catalytic activity of ALDH2 (24). One of these molecules, Alda-1, interacts with the substrate-binding site of ALDH2 and accelerates acetaldehyde metabolism to carboxylic acid by about twofold (24, 25), probably by increasing productive interactions of the substrate with the catalytic Cys302 and reducing the Km for the NAD+ coenzyme (25). We reasoned that another small molecule may increase productive interaction of acetaldehyde with Cys243 in the catalytic site of ALDH3A1, and thus temporarily recruit this enzyme to assist the mutant ALDH2 in eliminating acetaldehyde.
Results
Activation of ALDH3A1 Metabolism of Acetaldehyde by Alda-89.
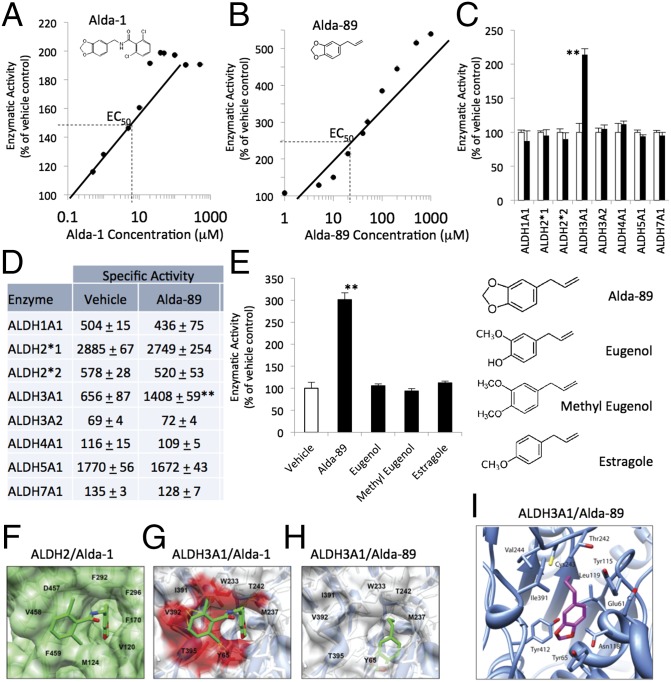
We previously showed that Alda-1 is an ALDH2-specific activator that binds to the catalytic cavity of ALDH2 (25). Alda-1 [N-(1,3-benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide, MW 324] increases acetaldehyde metabolism of ALDH2 by twofold with an EC50 of ∼6 μM (Fig. 1A and Fig. S1 A and B). Because the catalytic cavity of ALDH3A1 is bigger than that of ALDH2 (26), small aldehydes, such as acetaldehyde, are poorly metabolized by this enzyme (27). We reasoned that an Alda that binds to the ALDH3A1 catalytic cavity and reduces its size should improve the catalysis of small aldehydes, such as acetaldehyde, by ALDH3A1. Searching through a family of molecules in a similar chemical space as Alda-1, we previously identified Alda-89 as a selective activator of acetaldehyde metabolism by ALDH3A1 (28). Alda-89 [5-(2-propenyl)-1,3-benzodioxole, commonly known as safrole, MW = 162] increased acetaldehyde metabolism of ALDH3A1 by fivefold with an EC50 of ∼20 μM (Fig. 1B). Alda-89 activated ALDH3A1 with either NAD+ or NADP+ as a cofactor, but did not activate other ALDHs, including ALDH1A1, ALDH2, ALDH3A2, ALDH4A1, ALDH5A1, and ALDH7A1 (Fig. 1 C and D).
Fig. 1.
Selectivity of Alda-89 for ALDH3A1. (A) Structure and dose/response of Alda-1 activation of purified human recombinant ALDH2 and (B) Alda-89 activation of purified human recombinant ALDH3A1. Data are from three independent assays for each concentration, using NAD+ (2.5 mM) as a cofactor and 10 mM acetaldehyde as a substrate. (C) Alda-89 (20 μM) selectively activates ALDH3A1 and not seven other ALDH enzymes (5–20 μg of each enzyme, using 10 mM acetaldehyde as a substrate); data are presented as percentage of control (n = 3; **P < 0.01; bars represent the mean ± SD). (D) Same as C, but data are expressed as specific activity (μmole NADH/min/mg protein). (E) Structure activity relationship of Alda-89, eugenol, methyl eugenol, and estragole analogs (each at 50 μM) for ALDH3A1 activation. (F) Cocrystal structure of ALDH2 (green surface) with Alda-1 (green sticks) (PDB ID code 3INJ). (G) Superimposition of Alda-1 (green sticks) with clashing residues (red surface) from crystal structure of ALDH3A1 (blue ribbon/white surface; PDB ID code 3SZA). (H) A docking model of ALDH3A1 (blue ribbon/white surface) with Alda-89 (green sticks). (I) A modeled complex of ALDH3A1 with Alda-89. The position of Alda-89 was determined using DOCK 6.5. Electrostatic interactions and global constraints were calculated. Alda-89 (purple sticks, with the two oxygens labeled in red) is located within the enzymatic cavity. Cys243 (yellow stick) is the catalytic cysteine in the enzymatic cavity.
A stringent structural requirement for Alda-89 to activate ALDH3A1 was demonstrated by the lack of any activation by three closely related compounds, eugenol, methyl eugenol, and estragole (Fig. 1E). In addition, Alda-89 only activated metabolism of small aldehydes such as acetaldehyde by ALDH3A1; there was a ∼25% increased metabolism of a three-carbon (C3) aldehyde, propionaldehyde. Alda-89 did not affect ALDH3A1 metabolism of other substrates, including long-chain aliphatic aldehydes (heptaldehyde, C7; decanal, C10) and aromatic aldehydes (benzaldehyde, cinnamaldehyde) (Fig. S2 A and B). Similar to the effect of Alda-1 on ALDH2 activity (25), Alda-89 reduced the Km of ALDH3A1 for acetaldehyde by twofold, and increased its Vmax by 1.5-fold and Kcat by 50%.
Structural Comparison of ALDH2 and ALDH3A1.
To identify the molecular determinants that enable Alda-1 and Alda-89 to act as activators of ALDH2 and ALDH3A1, respectively, we compared the structure of the substrate-binding sites of these two enzymes. Superimposition of the substrate-binding site in ALDH2 (using the crystal structure of human ALDH2 bound to Alda-1; PDB ID code 3INJ) and ALDH3A1 (using human ALDH3A1 alone; PDB ID code 3SZA) demonstrated the differences in the size and shape of the catalytic cavity (Fig. 1 F and G). In ALDH2, the catalytic cavity consists mostly of aromatic amino acids (25), whereas ALDH3A1 contains more aliphatic and polar amino acids (26, 29). In ALDH2, amino acids 455–460 form a loose chain, whereas the corresponding amino acids (389–396) in ALDH3A1 form an α-helix (Fig. S3 A and B). These differences in secondary structure may contribute to why Alda-1 cannot bind to ALDH3A1. The benzodioxole group of Alda-1 makes critical contacts with the aromatic and hydrophobic amino acids (Val-120, Met-124, Phe-170, Phe-292, Phe-296, and Phe-459) in ALDH2 (Fig. 1F). There is critical π-stacking between residues Phe-170 and Phe-459 and the benzodioxole group of Alda-1. If Alda-1 bound similarly to ALDH3A1, Met-237, Thr-242, Trp-233, Ile-391, Val-392, Thr-395, and Tyr-65 of this ALDH3A1 would collide with Alda-89 (Fig. 1G; highlighted in red). These amino acids are bulkier and more polar than the corresponding residues in ALDH2. Further, π-stacking with the benzodioxole of Alda-1 cannot occur with Met-237 in ALDH3A1.
Docking Analysis of Alda-89 in ALDH3A1.
In the absence of crystal structures of Alda-89 bound to either ALDH enzymes, we analyzed Alda-89 using computational modeling. The crystal structure of the apo form of human ALDH3A1 (PDB ID code 3SZA) was the basis of the ALDH3A1/Alda-89 docking studies. The docking modeling confirmed that Alda-1 bound to ALDH2 in a very similar orientation as in the crystal structure (Fig. 1F and Fig. S3A). Docking studies of Alda-89 with ALDH3A1 showed that Alda-89 bound further into the interior of the enzymatic cavity compared with Alda-1 (Fig. 1F vs. Fig. 1H). Further inspection revealed that Alda-89 was in a cavity adjacent to the catalytic cysteine, Cys243 (Fig. 1I). The benzodioxy ring formed π-interactions with Tyr65 of ALDH3A1 (Fig. 1I). Such an interaction and requirement of the methylenedioxy ring may explain why the closely related compounds eugenol, methyl eugenol, and estragole failed to function as ALDH3A1 activators (Fig. 1E). Because the substrates of ALDH3A1 tend to be bulky aromatics (e.g., benzaldehyde), Alda-89 may fill the cavity enough to help orient the much smaller substrate, acetaldehyde, to interact productively with the ALDH3A1 catalytic amino acid, Cys243.
Metabolism of Acetaldehyde in the Presence of Alda-1 Together with Alda-89.
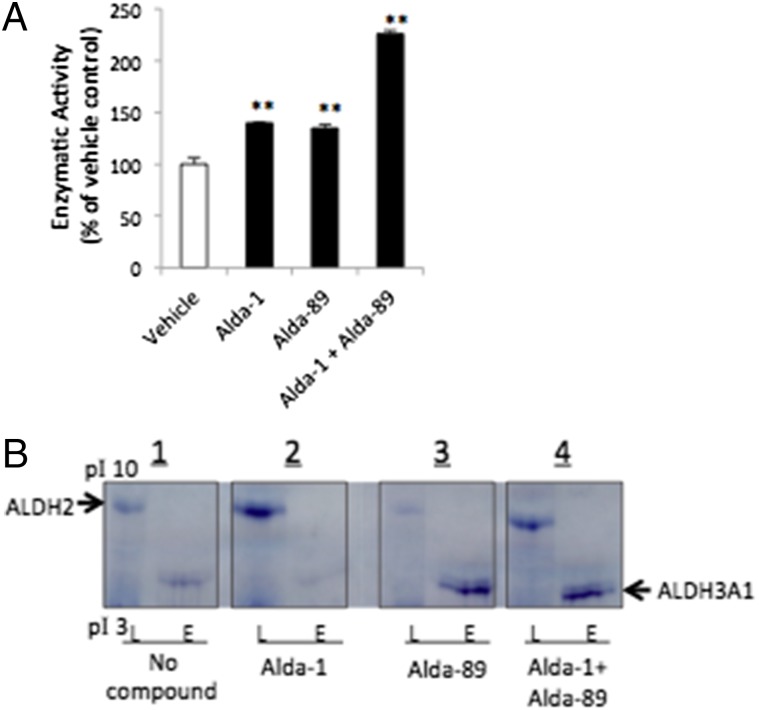
We next determined whether a faster acetaldehyde removal can be achieved by simultaneously activating ALDH2 and ALDH3A1, compared with activating each ALDH alone. When using equal amounts of recombinant human ALDH2 and ALDH3A1 in vitro, Alda-1 or Alda-89 alone each enhanced acetaldehyde metabolism to ∼140% relative to vehicle control (Fig. 2A). However, together, Alda-1 with Alda-89 increased the metabolism of acetaldehyde to 225%, relative to vehicle (Fig. 2A).
Fig. 2.
(A) The effect of a single activator or combination of Alda-1 and Alda-89 on acetaldehyde metabolism, using a mixture of ALDH2 and ALDH3A1 recombinant enzymes (15 μg each); 10 μM of Alda-1 or Alda-89 were used individually or in combination. Data for each enzyme are from three independent assays (n = 3; **P < 0.01; bars represent the mean ± SD). (B) Isoelectric focusing in-gel activity assay for ALDH2 and ALDH3A1, using tissue homogenates of mouse liver mitochondria fraction (L, 50 μg total protein/lane; rich in ALDH2) or eye (E, 100 μg total protein/lane; rich in ALDH3A1). ALDH2 and ALDH3A1, as indicated by the arrow signs, were separated by their different pI values by precast gels, using the PhastSystem isoeletric focusing (IEF) electrophoresis apparatus. ALDH activities were detected in each gel panel as blue formazan precipitation bands from reduced tetrazolium salt, which is a direct reflection of the ALDH enzyme activity. Shown is staining with (1) no ALDH activators in the staining solution (2), with Alda-1 (3), with Alda-89, or (4) with a mixture of both Alda-1 and Alda-89, 50 μM each.
We also determined the benefit of using both Aldas on acetaldehyde metabolism. Because all tissues contain several ALDH family members, we used isoelectric focusing to separate them and measured in-gel ALDH activity using NAD+-coupled phenazine methosulfate/nitroblue tetrazolium color reaction (30). The calculated pI values for mouse ALDH2 and ALDH3A1 are 7.5 and 6.5, respectively. We used mouse liver mitochondrial as an enriched ALDH2 source, and mouse eye homogenates as the source of ALDH3A1 [cornea, lens, and vitreous fluid of the eye contain very high amounts of ALDH3A1 (22)]. ALDH3A1 activity was also detected in mouse esophagus and stomach tissue (Fig. S4). ALDH2 clearly separated from ALDH3A1 by this in-gel staining technique when using acetaldehyde as the substrate (Fig. 2B1). Alda-1 (50 μM) in the staining solution selective increased ALDH2 activity in the mitochondria-enriched liver homogenate, but not in the eye homogenate (Fig. 2B2). Conversely, 50 μM of Alda-89 increased the ALDH3A1 staining (activity) only in the eye homogenate, but not in the liver homogenate (Fig. 2B3). As expected, Alda-1 together with Alda-89 increased the activity of both ALDH2 and ALDH3A1 in the two samples (Fig. 2B4), indicating that Alda-1 and Alda-89 selectively increased the activity of native (tissue-derived) ALDH2 and ALDH3A1, respectively.
Treatment with Alda-1 Together with Alda-89 Was Superior to Treatment with Alda-1 or Alda-89 Alone in Reducing Blood Ethanol and Acetaldehyde Levels After Acute Intoxication with Ethanol in Vivo.
We next determined whether enhanced acetaldehyde metabolism by both Alda-1 and Alda-89 can be achieved in vivo. In humans, liver ALDH2 is the major acetaldehyde metabolizing enzyme (7, 31). Because the first pass metabolism of ethanol occurs mainly in the stomach and liver (32), it is conceivable that simultaneous activation of ALDH3A1 in the mouth, upper gastrointestinal tract, and stomach, together with the activation of liver ALDH2, may synergistically lead to a more rapid metabolism and clearance of acetaldehyde. We therefore measured blood acetaldehyde and ethanol levels after an acute ingestion of ethanol in mice treated with the combination of Alda-1 and Alda-89 or with each alone. C57BL/6 wild-type ALDH2 mice were given a high dose of ethanol (3.3 g/kg orally), which is equivalent to binge drinking in humans (33), to produce significantly elevated levels of blood ethanol and acetaldehyde, mimicking the behavioral deficit of intoxication in humans.
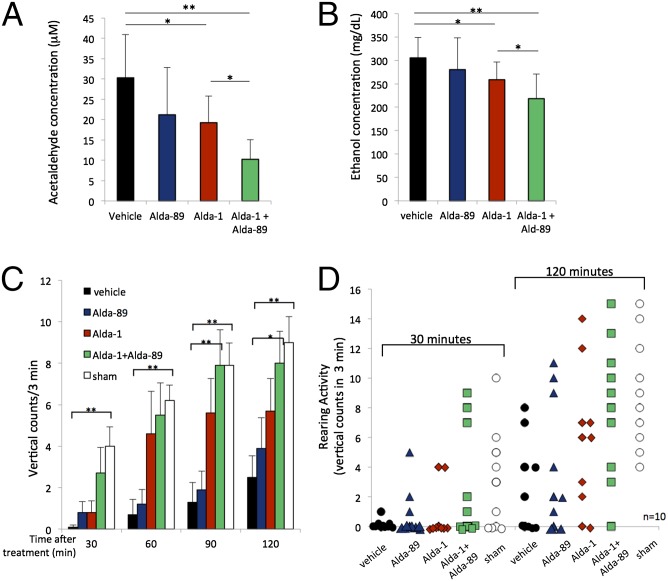
Blood acetaldehyde levels in the presence of Alda-1 alone declined by 37% (from 30 to 19 μM; Fig. 3A), relative to vehicle, when measured 45 min after ethanol ingestion (Fig. 3A). However, a more significant decline in acetaldehyde occurred when treating with Alda-1 together with Alda-89; blood acetaldehyde levels were reduced up to 66% (from 30 to 10 μM; Fig. 3A). Similarly, Alda-1 alone slightly reduced ethanol levels, mostly because of a lower extent of acetaldehyde substrate inhibition on alcohol dehydrogenase (15%; from 305 to 258 mg/dL; Fig. 3B), and this effect was doubled to 30% (from 305 to 218 mg/dL; Fig. 3B) in mice treated with Alda-1 together with Alda-89. Therefore, because acetaldehyde oxidation is the rate-limiting step in ethanol elimination (31), there was a correlation between the decline in the levels of acetaldehyde and ethanol in the blood.
Fig. 3.
Enhanced metabolism of acetaldehyde and ethanol and behavioral recovery, in vivo, in wild-type C57BL/6 mice. (A) Blood acetaldehyde levels after ethanol treatment in the presence of vehicle, Alda-89, Alda-1 alone, or by the combination of Alda-89 and Alda-1. Alda-1 reduced blood acetaldehyde levels by 37%. Alda-1/Alda-89 combination treatment reduced blood acetaldehyde levels by 66% (n = 7; *P < 0.05, **P < 0.01; bars represent the mean ± SD). (B) Blood ethanol levels after treatment with vehicle, Alda-89, Alda-1 alone, or by the combination of Alda-89 and Alda-1. Alda-1 reduced blood ethanol levels by ∼15%, whereas Alda-1/Alda-89 combination treatment reduced blood ethanol levels by ∼30% (n = 12; *P < 0.05, **P < 0.01; bars represent the mean ± SD). (C) Average rearing on back legs (vertical) counts from a total of 10 male mice per treatment group over time (0–120 min) after acute gavage of 3.3 g/kg ethanol or after sham gavage, without ethanol (sham). The average rearing events for 10 mice was recorded for a period of 3 min at the start of each indicated time (n = 10; *P < 0.05, **P < 0.01 vs. vehicle-treated group). Alda-1, Alda-89 alone or in combination (90 mg/kg each) was given via oral gavage 15 min before ethanol gavage. In the vehicle group, PEG400 in the same volume was given without either Alda. In the sham control group, saline in the same volume as ethanol was administered by gavage to the mice instead of ethanol. (D) Distribution of rearing behavior of the 10 mice in each treatment group above at 30 min and 120 min after ethanol ingestion or after sham gavage, without ethanol (sham). Note that rearing behavior also increases over time in mice that were not exposed to ethanol [empty bar (C) and symbol (D)], reflecting natural behavior suppression after handling.
Because acute ethanol and acetaldehyde toxicity results in sedation, hypoactivity, and lethargy (2, 34–36), we next measured behavioral recovery using a standard rearing activity test (37). In this behavioral test, each mouse was monitored every 30 min for its ability to rise on its hind legs. As in the previous study, vehicle, Alda-1 or Alda-89 alone, or Alda-1 together with Alda-89 were administered by oral gavage (90 mg/kg) 15 min before ethanol administration. Already at 30 min after ethanol ingestion, mice treated with the combination of Alda-1 and Alda-89 showed better behavioral recovery compared with mice treated only with Alda-1, Alda-89, or vehicle alone (Fig. 3C): they rose on their hind legs (vertical counts) an average of three times when treated with Alda-1 together with Alda-89 compared with one time after Alda-1 alone or Alda-89 alone, and not at all in the vehicle-treated group (Fig. 3C). Ninety minutes after ethanol ingestion, mice treated concomitantly with Alda-1 and Alda-89 appeared to have recovered completely, whereas the Alda-1 alone group still showed some impairment (Fig. 3C; data in Fig. 3D provide the behavior of each mouse in each treatment group). We noted a steady increase in the number of vertical counts during the 2-hour period (Fig. 3D and Table S1). These data indicate that treatment of Alda-1, together with Alda-89, resulted in a fastest recovery from ethanol/acetaldehyde toxicity; at all times, the behavior of the mice after ethanol ingestion and treatment with Alda-1 plus Alda-89 (green) was not statistically different from that of mice that did not receive any ethanol (white, Fig. 3 A and B).
Because ∼560 million East Asians carry an inactivating ALDH2*2 mutation with only residual enzyme activity, we also tested ALDH2*2 knock-in heterozygotic mice that we recently generated (38). These mice enabled us to determine whether recruiting ALDH3A1 for acetaldehyde elimination by a small-molecule activator, such as Alda-89, could be a viable strategy to compensate for the enzyme deficiency in human subjects with this mutation. The ALDH2*2 knock-in mouse was created to mimic the human ALDH2*2 mutation by homologous recombination of the mouse gene to substitute the equivalent glutamic acid by lysine at amino acid position 487 of human ALDH2 (38). These mutant mice had the expected reduced ALDH2 activity, levels and acetaldehyde detoxification capability as heterozygotic ALDH2*1/*2 humans (11, 38). In a previous study, when challenged with 4 g/kg ethanol, the heterozygotic ALDH2*1/*2 mice showed a greater than fourfold higher blood acetaldehyde concentration than that of the wild-type mice (102 vs. 24 μM, 60 min after ethanol administration) (38).
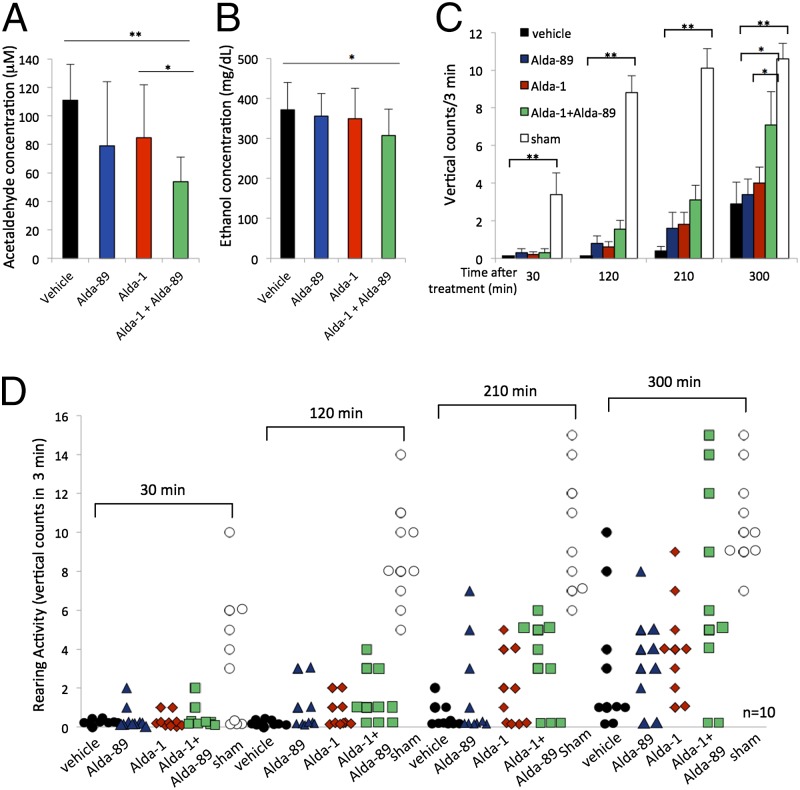
To enable monitoring behavioral difference, we administered only 2 g/kg ethanol to the ALDH2*1/*2 mice. As before, Alda-1 or Alda-89 treatments show only a trend to reduce blood acetaldehyde levels (from 111 to 79 and 85 μM, respectively; Fig. 4A). However, combined treatment of Alda-89 with Alda-1 showed a 52% reduction in blood acetaldehyde level (from 111 to 54 μM; Fig. 4A) and a 17% reduction in blood ethanol levels (from 372 to 307 mg/d; Fig. 4B). ALDH2*1/*2 mice treated with 2 g/kg were intoxicated for a much longer time than the wild-type mice treated with 3.3 g/kg ethanol, whereas the ALDH2*1/*2 group had zero total (0 average) vertical counts for 10 ALDH2*1/*2 mice vs. 25 total (2.5 average) counts for 10 wild-type mice over a period of 3 min at 120 min after ethanol ingestion (Fig. 3 C and D vs. Fig. 4 C and D and Tables S1 and S2). Compared with wild-type mice, an additional 150–180 min were required before the ALDH2*1/*2 mice recovered to a level equivalent to that of the wild-type mice (25 total counts at 120 min for wild-type and 29 total counts at 300 min for ALDH2*2; Table S1 vs. Table S2). Given the same dose of ALDH activators as the wild-type mice (90 mg/kg), activation by Alda-1 or Alda-89 alone in the ALDH2*1/*2 mice achieved only a minimal effect, whereas combined treatment of Alda-1 plus Alda-89 showed a significantly greater improved detoxification recovery beginning 120 min after ethanol exposure. At 300 min, the average count of rearing activity was seven when treated with Alda-1 together with Alda-89 compared with four for Alda-1 and three for Alda-89 and the vehicle-treated groups (Fig. 4C; P < 0.05; n = 10; Table S2). Similar results were obtained when ethanol-induced behavioral impairment was assessed using the loss of righting reflex (LORR) assay. Combined treatment of Alda-1 with Alda-89 significantly reduced LORR duration by more than 30 min compared with the vehicle-treated group (from 106 to 72 min; Fig. S5; n = 10; P < 0.01) and by more than 20 min compared with the Alda-89-treated group (from 94 to 72 min; Fig. S5; n = 10; P < 0.05). These results support our hypothesis that recruitment of ALDH3A1 by Alda-89 to accelerate acetaldehyde metabolism in vivo is possible and that Alda-89 can improve and compensate for the loss of function caused by ALDH2 deficiency.
Fig. 4.
Enhanced behavioral recovery, in vivo, in ALDH2*1/*2 C57BL/6 mice. (A) Blood acetaldehyde levels after ethanol treatment in the presence of vehicle, Alda-89, Alda-1 alone, or the combination of Alda-89 and Alda-1. Alda-1/Alda-89 combination treatment reduced blood acetaldehyde levels by 52% (n = 10; *P < 0.05, **P < 0.01; bars represent the mean ± SD). (B) Blood ethanol levels after treatment with vehicle, Alda-89, Alda-1 alone, or the combination of Alda-89 and Alda-1. Alda-1/Alda-89 combination treatment reduced blood ethanol levels by 17% (n = 10; *P < 0.05; bars represent the mean ± SD). (C) Average rearing on back legs (vertical) counts from a total of 10 male mice per treatment group over time (0–300 min) after acute gavage with 2 g/kg ethanol or sham gavage (sham). The average rearing events for 10 mice were recorded for a period of 3 min at the start of each indicated time (n = 10; *P < 0.05, **P < 0.01 vs. vehicle-treated group). Alda-1 and Alda-89 alone or in combination (90 mg/kg each) was given via oral gavage 15 min before ethanol gavage. In the vehicle group, PEG400 in the same volume was given without either Alda. In the sham control group, saline in the same volume as ethanol was administered by gavage to the mice instead of ethanol. (D) Distribution of rearing behavior of the 10 mice in each treatment group at 30, 120, 210, and 300 min after ethanol ingestion or sham gavage (sham).
Discussion
We described a means to recruit ALDH3A1, an enzyme that metabolizes bulky aromatic aldehydes, to enhance metabolism of the smaller substrate, acetaldehyde. Similar to the effect of Alda-1 on ALDH2, Alda-89 likely increased acetaldehyde metabolism by ALDH3A1 by decreasing the size of the catalytic tunnel of ALDH3A1 and increasing the chance of productive encounters between the small substrate, acetaldehyde, and the catalytic Cys in this enzyme. Enhanced acetaldehyde metabolism and behavioral recovery by a mixture of Alda-1 with Alda-89 were demonstrated in vivo in both wild-type and in the knock-in ALDH2*1/*2 mutant mice, which mimics the common mutation in East Asians. The major acetaldehyde metabolizing enzyme, ALDH2, is abundant in liver, but much less in stomach, esophagus, and saliva (20, 39). Because relatively high levels of ALDH3A1 are present in the esophagus, upper aerodigestive track, and stomach (20–22), Alda-89 may contribute to a faster elimination of acetaldehyde in organs shown to be more vulnerable to acetaldehyde-induced cancers in humans with the ALDH2*2 mutation (3, 4).
A combination of Alda-1 with Alda-89 also enhanced ethanol elimination. This is expected, as the rate of acetaldehyde oxidation to acetic acid is the rate-limiting step in ethanol elimination (31). Enhanced ethanol elimination may also be of another clinical use, as 88,000 deaths have been attributed to excessive ethanol use in the United States each year (40). Further, about 8% of all emergency room visits, or 7.6 million per year, involve ethanol use in the United States (41). Current treatment for these patients does not include accelerated metabolism of ethanol or acetaldehyde, thus requiring relatively long stays in the already overcrowded emergency rooms. Recently, using nanoparticles encapsulated with a mixture of alcohol oxidase and catalase as an oral antidote, enhanced ethanol detoxification in rodents was achieved (42). However, because both alcohol oxidase and catalase convert ethanol to acetaldehyde, a known carcinogen, adopting such a strategy to reduce blood ethanol levels may carry an increased health risk from acetaldehyde exposure, especially for ALDH2-deficient subjects. In contrast, treatment with compounds that accelerate acetaldehyde metabolism by ALDH2 (e.g., by Alda-1), together with ALDH3A1 (e.g., by Alda-89), may enhance the recovery from ethanol intoxication while also accelerating the elimination of the carcinogenic acetaldehyde, and therefore may have a better outcome for these subjects. Studies with ALDH3A1 knock-out mice (43) and ALDH3A1-overexpressing mice may further validate this strategy.
Alda-89 (safrole) used to be a common food additive, especially in soft drinks. However, since 1960, the use of safrole in food has been banned by the US Food and Drug Administration because of its genotoxicity. It should be noted that safrole is naturally found in a variety of spices, including in cinnamon and black pepper and in herbs such as basil. Nevertheless, a noncarcinogenic Alda-89–like compound should be used to enhance acetaldehyde metabolism in humans.
We provide evidence that converting the substrate specificity of ALDH3A1 by the small molecule Alda-89 recruits this enzyme to complement the inactivating mutation of ALDH2*2, a common mutation in the East Asian population. Treatment with Alda-89-like compounds may reduce the increased incidence of squamous cell carcinomas in the UADT induced by acetaldehyde genotoxicity among ALDH2*1/*2 heterozygous carriers. The ALDH2*2 mutation is not benign: In addition to UADT cancers (4), deficiency of ALDH2 has been implicated in many other human diseases including osteoporosis and cardiovascular and neurodegenerative diseases (2). Another risk factor enhanced by toxic acetaldehyde is for patients with Fanconi anemia. ALDH2 knock-out mice carrying one of the Fanconi anemia mutations have accelerated bone marrow failure and leukemia, the hallmarks of the disease (44). Similarly, accelerated progression of bone marrow failure has been reported in Japanese patients with Fanconi anemia who are also ALDH2*2 carriers (45). Recruiting ALDH3A1 to compensate for the loss of ALDH2*2 function using a small molecule such as Alda-89 may therefore represent a useful strategy to treat ALDH2 deficiency-related diseases.
Finally, our work provides an example for the use of a small molecule as a cofactor to temporarily alter the normal catalytic activity of an enzyme, and thus enhance metabolism of a toxic agent. A number of detoxifying enzymes, such as those belonging to the P450 liver enzymes, differ in their substrate preference. Indeed, a single amino acid substitution converted the specificity of one P450 isozyme from coumarin to steroid hydroxylation (46), demonstrating that changing substrate selectivity of an enzyme by mutagenesis is possible. Here we describe a pharmacologically induced change in substrate specificity that is reversible, and therefore more practical. Our work demonstrates that it is possible to identify small pharmacologic agents that recruit additional members of a family of enzymes to metabolize a toxic substance after excessive accidental exposure, or to supplement the activity in patients who have an impaired ability to detoxify the given substance.
Materials and Methods
Chemicals.
Alda-1 was synthesized as previously described (24). Alda-89, acetaldehyde, propionaldehyde, heptaldehyde, decanal, benzaldehyde, transcinnamaldehyde, and all other chemicals were purchased from Sigma Aldrich Inc. Eugenol was purchased from ACROS Organics, and methyl eugenol and estragole were purchased from Pfaltz & Bauer Inc.
ALDH Isozyme Cloning, Expression, Purification, and Enzymatic Assay.
Cloning and expression of human recombinant ALDH1A1, ALDH2*1, ALDH2*2, and ALDH5A1 isozymes have been described previously (24). Similarly, full-length ALDH cDNA clones for ALDH3A1, ALDH3A2, ALDH4A1, and ALDH7A1 isozymes were cloned and expressed as described in SI Materials and Methods. Enzymatic activity of ALDHs was determined spectrophotometrically by monitoring the reductive reaction of NAD+ to NADH at λ340 nm (24, 25). All of the assays were carried out at 25 °C in 0.1 M sodium phosphate buffer at pH 7.5, 2.5 mM NAD+, using acetaldehyde or other aldehyde substrates. ALDH-specific activities were expressed as the production of micromole NADH/min/mg protein. DMSO was used as a solvent to dissolve Alda-1, Alda-89, eugenol, methyl eugenol, and estragole.
Computational Docking Procedures for ALDH2 and ALDH3A1.
The crystal structures of ALDH2 and ALDH3A1 were obtained from the RCSB Protein Data Bank (PDB ID codes 3INL (25) and 3SZA (26). Waters and ligands were removed from the structure. Alda-1 and Alda-89 were prepared according to the ZINC (47) database protocols and docked using DOCK 6.5. The ALDH structures were kept rigid and ligands flexible. Alda-1 and Alda-89 were evaluated in up to 600 conformations and scored by van der Waals and electrostatic complementarity, using AMBER and DelPhi derived potentials, respectively, corrected for ligand desolvation. The protein was protonated to optimize the H-bond interactions and potential steric clashes.
Isoelectric Focusing and in-Gel ALDH Enzyme Activity Staining.
Separation of ALDH2 and ALDH3A1 from tissue homogenates by isoelectric focusing gel electrophoresis was carried by the use of the PhastSystem gel focusing apparatus (GE Healthcare Sciences). Mouse liver mitochondrial fraction was prepared by differential centrifugation in a buffer containing 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 5 mM MOPS at pH 7.4 according to Hoppel and coworkers (48). Enriched liver mitochondrial fraction was then solubilized by a homogenization buffer containing 0.1% Triton X-100, 10 mM DTT, 20% (vol/vol) glycerol, 100 mM Tris⋅HCl at pH 7.5, followed by centrifugation. Supernatants of eye extract were obtained from the whole eyes of killed mice, followed by homogenization in the same homogenization buffer and centrifugation. Protein concentrations of each sample were determined by a standard Bradford assay. Fifty micrograms of total liver mitochondrial protein or 100 μg total eye protein samples were loaded per lane in precast PhastGels, pI range 3–9, and separated according to manufacturer’s recommended protocol. Immediately after the completion of electrophoresis, each gel section was stained for ALDH enzyme activity in a solution containing 1 mM NAD+, 0.5 mM nitroblue tetrazolium, 1 mM phenazine methosulfate, 100 mM Tris⋅HCl at pH 7.5, and 10 mM acetaldehyde as a substrate. After 30 min staining in the dark, the staining reaction was terminated by substitution of the staining solution with H2O, as described in a previously published method (30).
In Vivo Treatment with ALDH Activators Alda-1 and Alda-89 and Behavioral Assessment.
Mice breeding and all animal procedures were approved by the Institutional Administrative Panel on Laboratory Animal Care at Stanford University. Construction of ALDH2*1/*2 knock-in mice in the C57BL/6 background has been previously described (38). Both wild-type and ALDH2*1/*2 C57BL/6 mice (male, 25–32 g) were fed standard rodent chow and water ad libitum. They were housed in a quiet room for at least 3 d with 12/12 h light-dark cycles with free access to regular diet before each experiment (conducted between 10:00 AM and 4:00 PM). Alda-1, Alda-89 (100 mM in PEG400 stock solution), and vehicle control (PEG400) were delivered by oral gavage at a dose of 90 mg/kg 15 min before ethanol administration. Wild-type and mutant mice were given 3.3 g/kg or 2 g/kg ethanol, respectively, in 19% ethanol (vol/vol; at a final volume of 340–640 μL) by oral gavage. For sham control groups, saline in the same volume of ethanol was given by oral gavage instead of ethanol. Behavioral assessment of rearing activity was conducted on the basis of a well-established and published method (37) by observing five animals as a group in a single open cage and allowing them to move freely for 3 min. The number of times each animal rose up on two hind legs against the cage was recorded during the 3-min period. The animals were rested and monitored in 30-min intervals. A total of 2 and 5 h were recorded for the ALDH2 wild-type and ALDH2*1/*2 mutant mice, respectively.
Blood Ethanol Measurement.
Blood samples (50 μL) from the retroorbital sinus were collected, 45 min after ethanol ingestion, into heparinized tubes and immediately processed by centrifugation. After centrifugation, 5 μL plasma was injected directly into the GM7 ethanol analyzer, following manufacturer’s instructions (Analox Instruments Ltd.).
Blood Acetaldehyde Measurement.
Blood acetaldehyde concentration was determined using an established HPLC method (49). Briefly, blood samples (50 μL) from the retroorbital sinus were collected, 45 min after ethanol ingestion, into heparinized tubes containing 10% perchloric acid. The samples were deproteinized and immediately processed by centrifugation. After centrifugation, the supernatant was mixed with 20% ammonium acetate, 6% thiourea, and 1.5% 1,3-cyclohexanedione for the formation of acetaldehyde fluorescent adduct decahydro-9-methylcaridine-1,8-dione, as previously described (50). Samples were analyzed using a Shimadzu HPLC system equipped with a fluorescent detector at 366 nm (excitation) and 440 nm (emission).
Statistical Analyses.
Statistical analysis for enzyme activity assays, blood acetaldehyde, and ethanol measurements conducted using standard t test. All data are expressed as means ± SD. For animal behavioral analysis, data were assessed by one-way ANOVA with Tukey’s correction and expressed as mean ± SEM. A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Thomas D. Hurley (Indiana University) for critical review of the manuscript and Dr. Natalie Zahr from the laboratories of Adolf Pfefferbaum and Edith Sullivan at SRI International and Stanford University, Department of Psychiatry and Behavioral Science, for assistance in blood ethanol measurement. This work was supported by NIH AAA11147 (to D.M.-R.) and NIH training grant NIH T32 CA09151 (to L.A.C.).
Footnotes
Conflict of interest statement: D.M.-R. and C.-H.C. are cofounders of ALDEA Pharmaceuticals. None of the research in the D.M.-R. laboratory is discussed with, supported by, or performed in collaboration with the company.
This article is a PNAS Direct Submission. D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414657112/-/DCSupplemental.
References
- 1.Vasiliou V, Pappa A. Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology. 2000;61(3):192–198. doi: 10.1159/000028400. [DOI] [PubMed] [Google Scholar]
- 2.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: New therapeutic opportunities. Physiol Rev. 2014;94(1):1–34. doi: 10.1152/physrev.00017.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baan R, et al. WHO International Agency for Research on Cancer Monograph Working Group Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007;8(4):292–293. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 4.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6(3):e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng MY, Luczak SE, Wall TL. ALDH2, ADH1B, and ADH1C genotypes in Asians: A literature review. Alcohol Res Health. 2007;30(1):22–27. [PMC free article] [PubMed] [Google Scholar]
- 6.Impraim C, Wang G, Yoshida A. Structural mutation in a major human aldehyde dehydrogenase gene results in loss of enzyme activity. Am J Hum Genet. 1982;34(6):837–841. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YC, et al. Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics. 2009;19(8):588–599. doi: 10.1097/FPC.0b013e32832ecf2e. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81(1):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida A, Ikawa M, Hsu LC, Tani K. Molecular abnormality and cDNA cloning of human aldehyde dehydrogenases. Alcohol. 1985;2(1):103–106. doi: 10.1016/0741-8329(85)90024-2. [DOI] [PubMed] [Google Scholar]
- 10.Ferencz-Biro K, Pietruszko R. Human aldehyde dehydrogenase: Catalytic activity in oriental liver. Biochem Biophys Res Commun. 1984;118(1):97–102. doi: 10.1016/0006-291x(84)91072-6. [DOI] [PubMed] [Google Scholar]
- 11.Lai CL, et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38(1):44–50. doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- 12.Chen CC, et al. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65(3):795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama A, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis. 2002;23(11):1851–1859. doi: 10.1093/carcin/23.11.1851. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama A, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19(8):1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 15.Pelucchi C, Gallus S, Garavello W, Bosetti C, La Vecchia C. Alcohol and tobacco use, and cancer risk for upper aerodigestive tract and liver. Eur J Cancer Prev. 2008;17(4):340–344. doi: 10.1097/CEJ.0b013e3282f75e91. [DOI] [PubMed] [Google Scholar]
- 16.Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res. 2007;149(6):293–297. doi: 10.1016/j.trsl.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Tomoda T, et al. Genetic risk of hepatocellular carcinoma in patients with hepatitis C virus: A case control study. J Gastroenterol Hepatol. 2012;27(4):797–804. doi: 10.1111/j.1440-1746.2011.06948.x. [DOI] [PubMed] [Google Scholar]
- 18.Väkeväinen S, Tillonen J, Agarwal DP, Srivastava N, Salaspuro M. High salivary acetaldehyde after a moderate dose of alcohol in ALDH2-deficient subjects: Strong evidence for the local carcinogenic action of acetaldehyde. Alcohol Clin Exp Res. 2000;24(6):873–877. [PubMed] [Google Scholar]
- 19.Balbo S, et al. Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol Biomarkers Prev. 2012;21(4):601–608. doi: 10.1158/1055-9965.EPI-11-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin SJ, et al. Alcohol and aldehyde dehydrogenases in human esophagus: Comparison with the stomach enzyme activities. Alcohol Clin Exp Res. 1993;17(2):376–381. doi: 10.1111/j.1530-0277.1993.tb00779.x. [DOI] [PubMed] [Google Scholar]
- 21.Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1): Biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376(Pt 3):615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: A corneal crystallin with diverse functions. Exp Eye Res. 2007;84(1):3–12. doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Wymore T, et al. Molecular recognition of aldehydes by aldehyde dehydrogenase and mechanism of nucleophile activation. Proteins. 2004;57(4):758–771. doi: 10.1002/prot.20256. [DOI] [PubMed] [Google Scholar]
- 24.Chen CH, et al. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Miller S, et al. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17(2):159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khanna M, et al. Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J Biol Chem. 2011;286(50):43486–43494. doi: 10.1074/jbc.M111.293597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King G, Holmes R. Human ocular aldehyde dehydrogenase isozymes: Distribution and properties as major soluble proteins in cornea and lens. J Exp Zool. 1998;282(1-2):12–17. [PubMed] [Google Scholar]
- 28.Banh A, et al. A novel aldehyde dehydrogenase-3 activator leads to adult salivary stem cell enrichment in vivo. Clin Cancer Res. 2011;17(23):7265–7272. doi: 10.1158/1078-0432.CCR-11-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimble-Hill AC, Parajuli B, Chen CH, Mochly-Rosen D, Hurley TD. Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones. J Med Chem. 2014;57(3):714–722. doi: 10.1021/jm401377v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang CP, et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human small intestine. Alcohol Clin Exp Res. 2012;36(12):2047–2058. doi: 10.1111/j.1530-0277.2012.01836.x. [DOI] [PubMed] [Google Scholar]
- 31.Zakhari S. Overview: How is alcohol metabolized by the body? Alcohol Res Health. 2006;29(4):245–254. [PMC free article] [PubMed] [Google Scholar]
- 32.Baraona E. Site and quantitative importance of alcohol first-pass metabolism. Alcohol Clin Exp Res. 2000;24(4):405–406. [PubMed] [Google Scholar]
- 33.Carson EJ, Pruett SB. Development and characterization of a binge drinking model in mice for evaluation of the immunological effects of ethanol. Alcohol Clin Exp Res. 1996;20(1):132–138. doi: 10.1111/j.1530-0277.1996.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 34.Isse T, Matsuno K, Oyama T, Kitagawa K, Kawamoto T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol Clin Exp Res. 2005;29(11):1959–1964. doi: 10.1097/01.alc.0000187161.07820.21. [DOI] [PubMed] [Google Scholar]
- 35.Isse T, et al. Paired acute inhalation test reveals that acetaldehyde toxicity is higher in aldehyde dehydrogenase 2 knockout mice than in wild-type mice. J Toxicol Sci. 2005;30(4):329–337. doi: 10.2131/jts.30.329. [DOI] [PubMed] [Google Scholar]
- 36.Quertemont E, Tambour S, Tirelli E. The role of acetaldehyde in the neurobehavioral effects of ethanol: A comprehensive review of animal studies. Prog Neurobiol. 2005;75(4):247–274. doi: 10.1016/j.pneurobio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Dere E, et al. Connexin30-deficient mice show increased emotionality and decreased rearing activity in the open-field along with neurochemical changes. Eur J Neurosci. 2003;18(3):629–638. doi: 10.1046/j.1460-9568.2003.02784.x. [DOI] [PubMed] [Google Scholar]
- 38.Zambelli VO, et al. Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med. 2014;6(251):ra118. doi: 10.1126/scitranslmed.3009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: From fish to mammals. Chem Biol Interact. 2001;130-132(1-3):181–191. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 40. Centers for Disease Control and Prevention. Fact sheets—Alcohol use and health. Available at www.cdc.gov/alcohol/fact-sheets/alcohol-use.htm. Accessed February 8, 2015.
- 41.McDonald AJ, 3rd, Wang N, Camargo CA., Jr US emergency department visits for alcohol-related diseases and injuries between 1992 and 2000. Arch Intern Med. 2004;164(5):531–537. doi: 10.1001/archinte.164.5.531. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, et al. Biomimetic enzyme nanocomplexes and their use as antidotes and preventive measures for alcohol intoxication. Nat Nanotechnol. 2013;8(3):187–192. doi: 10.1038/nnano.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassen N, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: Cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J Biol Chem. 2007;282(35):25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475(7354):53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 45.Hira A, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122(18):3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindberg RL, Negishi M. Alteration of mouse cytochrome P450coh substrate specificity by mutation of a single amino-acid residue. Nature. 1989;339(6226):632–634. doi: 10.1038/339632a0. [DOI] [PubMed] [Google Scholar]
- 47.Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoppel C, DiMarco JP, Tandler B. Riboflavin and rat hepatic cell structure and function. Mitochondrial oxidative metabolism in deficiency states. J Biol Chem. 1979;254(10):4164–4170. [PubMed] [Google Scholar]
- 49.Ung-Chhun NS, Collins MA. Estimation of blood acetaldehyde during ethanol metabolism: A sensitive HPLC/fluorescence microassay with negligible artifactual interference. Alcohol. 1987;4(6):473–476. doi: 10.1016/0741-8329(87)90088-7. [DOI] [PubMed] [Google Scholar]
- 50.Peng GS, et al. Involvement of acetaldehyde for full protection against alcoholism by homozygosity of the variant allele of mitochondrial aldehyde dehydrogenase gene in Asians. Pharmacogenetics. 1999;9(4):463–476. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.