Significance
Readthrough-promoting drugs cause amino acid insertion at premature termination codons (PTCs), and thus have broad potential as a therapeutic approach to inherited disorders attributable to nonsense mutations. Because the mechanism involved in the insertion of near-cognate tRNAs at nonsense codons is unknown, we have identified the yeast translation errors ensuing from nonsense suppression occurring either inherently or enhanced by drugs or mutations that compromise termination fidelity. Our analyses of the products of nonsense suppression provide insights into the rules that govern readthrough at PTCs and delineate specific nonstandard Watson–Crick codon/anticodon base pairings critical to this process. These results should enable predictions of the likelihood of obtaining functional full-length readthrough products, and thus better clinical outcomes, with therapeutic nonsense suppression.
Keywords: translation termination, nonsense codon readthrough, therapeutic nonsense suppression
Abstract
Premature termination codons (PTCs) in an mRNA ORF inactivate gene function by causing production of a truncated protein and destabilization of the mRNA. Readthrough of a PTC allows ribosomal A-site insertion of a near-cognate tRNA, leading to synthesis of a full-length protein from otherwise defective mRNA. To understand the mechanism of such nonsense suppression, we developed a yeast system that allows purification and sequence analysis of full-length readthrough products arising as a consequence of endogenous readthrough or the compromised termination fidelity attributable to the loss of Upf (up-frameshift) factors, defective release factors, or the presence of the aminoglycoside gentamicin. Unlike classical “wobble” models, our analyses showed that three of four possible near-cognate tRNAs could mispair at position 1 or 3 of nonsense codons and that, irrespective of whether readthrough is endogenous or induced, the same sets of amino acids are inserted. We identified the insertion of Gln, Tyr, and Lys at UAA and UAG, whereas Trp, Arg, and Cys were inserted at UGA, and the frequency of insertion of individual amino acids was distinct for specific nonsense codons and readthrough-inducing agents. Our analysis suggests that the use of genetic or chemical means to increase readthrough does not promote novel or alternative mispairing events; rather, readthrough effectors cause quantitative enhancement of endogenous mistranslation events. Knowledge of the amino acids incorporated during readthrough not only elucidates the decoding process but also may allow predictions of the functionality of readthrough protein products.
The information in an mRNA ORF can be repurposed during translation by several different types of recoding events, including leaky scanning, initiation at noncanonical start codons, ribosome frame-shifting, or stop codon readthrough (1). Stop codon readthrough, or nonsense suppression, occurs when a ribosome positioned with a nonsense codon in its A-site incorporates an amino acid into the nascent polypeptide chain instead of terminating translation by promoting the hydrolysis and release of that polypeptide from the P-site peptidyl-tRNA (2, 3). Two release factors mediate termination in eukaryotes: eRF1 (eukaryotic release factor 1), a protein that directly recognizes the three nonsense codons (UAA, UAG, or UGA), and eRF3, a GTPase that binds eRF1 and stimulates its activity (2, 4). Although translation termination is the kinetically favored event when a nonsense codon occupies the ribosomal A-site, recognition of that nonsense codon by eRF1 is subject to competition from near-cognate aminoacyl-tRNAs (2). Productive binding of a near-cognate aminoacyl-tRNA to an A-site harboring a nonsense codon bypasses termination and results in stop codon readthrough. Readthrough of a normal termination codon (NTC) leads to translation elongation into the mRNA 3′-UTR, generating a C-terminally extended polypeptide, whereas readthrough of a premature termination codon (PTC) allows continued translation elongation in the proper reading frame and synthesis of a full-length polypeptide. Nonsense suppression at PTCs occurs at a much higher frequency than at NTCs (2, 5, 6), and this difference may well be a consequence of the association of termination-stimulating factors with the 3′-UTR mRNP (messenger ribonucleoprotein) complex that abuts the NTC, but not a PTC (7). Conditions that alter the fidelity of translation termination or decoding by an aminoacyl-tRNA, including the binding of small-molecule drugs to the ribosome or mutations in the genes encoding the release factors or the nonsense-mediated decay (NMD)-specific Upf (up-frameshift) proteins, have all been shown to enhance nonsense suppression (2, 3, 5, 8–10).
The misreading of a nonsense codon during readthrough could, in principle, be mediated by an incorrect tRNA with either single (near-cognate tRNA) or multiple (noncognate tRNA) nonstandard Watson–Crick codon/anticodon base pairs. The mechanism that minimizes the incorporation of incorrect aminoacyl-tRNAs during translation elongation has two components: selection of a tRNA complexed to an elongation factor and “kinetic proofreading” after GTP hydrolysis by the elongation factor (11). For a given codon in the A-site of the ribosome, the fate of an incoming aminoacyl-tRNA is based on its extent of base pairing and the detection of that base pairing by the surrounding rRNA (2, 12). A cognate tRNA is able to elicit GTP hydrolysis, as can a near-cognate tRNA, even though doing so introduces a translation error. However, a noncognate tRNA is unable to elicit GTP hydrolysis, and thus cannot be “accommodated” by the A-site (12). Recent crystal structures of ribosomes have revealed that nonstandard Watson–Crick base pairs can be tolerated in the A-site, and that it is the shape of the base pairs that is pivotal and not the number, or types, of the hydrogen bonds that they form (12). Furthermore, “geometrical mimicry” by nonstandard Watson–Crick pairing has been proposed as a mechanism that favors incorporation of near-cognate tRNAs (12, 13).
To understand how translational fidelity is maintained by the ribosome during termination, it is important to know which mismatches are allowed, even if they occur at a low frequency. Hence, the decoding events occurring during readthrough of PTCs are important phenomena that need to be elucidated. Further, from a therapeutic perspective, the identity of the amino acids that get incorporated at PTCs may be critical in predicting the potential functionality of the readthrough product when readthrough is induced pharmacologically. Several previous studies have sought to identify the amino acids incorporated at a PTC (14–23). In vitro studies in rabbit reticulocyte lysates identified Trp, Arg, and Cys incorporation at a UGA codon (20). Studies addressing the insertion of amino acids at UAA and UAG codons in viral mRNAs revealed only the presence of Gln (20, 24). However, given the nature of these studies, it has been difficult to judge the relative frequencies of insertion or to conclude that the same tRNAs would be involved during readthrough in vivo. Furthermore, the low amount of readthrough product, together with less sensitive detection techniques, could have obscured the identification of low-abundance amino acid insertions, as in the case of the study addressing insertion in place of UAA. A recent study in Saccharomyces cerevisiae tried to address these issues in vivo (25). However, to ensure maximal capture of the readthrough products, this study combined two different readthrough-inducing conditions (upf1∆ and [PSI+]). Although this approach did help to identify the insertion of three different amino acids at all three PTCs, the interpretation of the results was not clear-cut because the frequencies of insertions reflected the combined effects of three independent readthrough events, namely, endogenous readthrough, UPF1 deletion, and a defect in eRF3.
Here, we have investigated the mechanisms of near-cognate tRNA insertion during PTC readthrough in yeast by identifying the amino acids that are inserted upon readthrough of each of the three nonsense codons. First, we describe an in vivo reporter system that allows for detection, purification, and mass spectrometric analyses of the endogenous unprogrammed readthrough product from WT cells without any treatment. Having identified the amino acids that get inserted at a PTC in WT cells, we compared and characterized the readthrough products that are synthesized when termination fidelity is compromised (in the absence of Upf factors, loss of functional translation termination factors, or conformational change in the decoding center by gentamicin binding). Our study provides a comprehensive analysis of termination readthrough from a PTC under different readthrough-inducing conditions and elucidates the nature of the nonstandard base pairings that are favored under physiological conditions in the cell.
Results
Reporter System for Studying Readthrough.
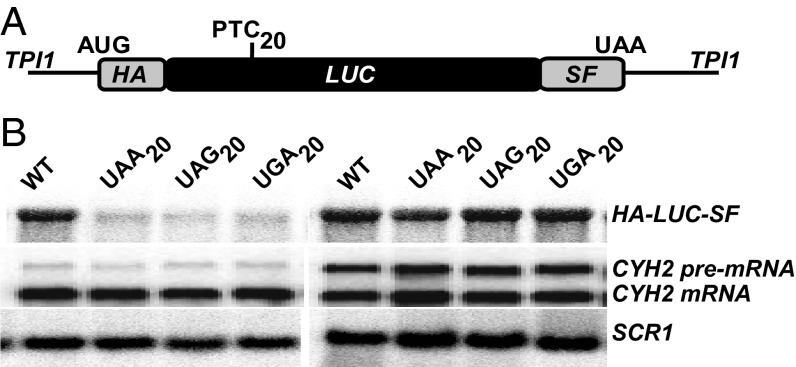
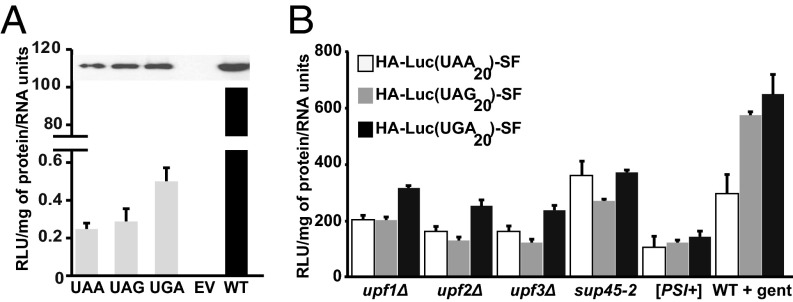
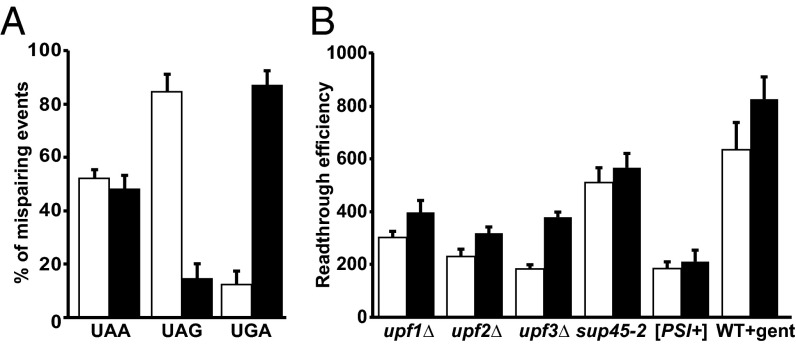
To identify the specific amino acids incorporated when PTCs are subject to readthrough, we constructed a luciferase gene (LUC) reporter system with several key attributes, including the ability to yield highly purified readthrough products in sufficient quantities for MS even when those products are expressed at low levels and the ability to derive unambiguous sequencing information from well-resolved peptides. Starting with a codon optimized firefly LUC gene, we inserted an in-frame 3× HA tag at the N terminus and adjacent StrepII and FLAG (SF) tags at the C terminus of the luciferase ORF and directed transcription of the construct with the potent TPI1 promoter (Fig. 1A). Three separate alleles were constructed with each of the possible PTCs inserted at codon 20 of the LUC ORF (Table S1), rendering each respective mRNA an NMD substrate (Fig. 1B). Readthrough of each of the LUC PTCs in WT [PSI−] cells resulted in the translation of full-length luciferase that also retains activity (Fig. 2A), indicating that these mRNAs are susceptible to basal levels of readthrough under normal growth conditions. The efficiency of readthrough from each PTC under different conditions known to compromise termination fidelity (the absence of Upf factors, the presence of defective translation termination factors, or the presence of the aminoglycoside gentamicin) was then assessed as a measure of luciferase activity (normalized to total protein content of the sample as well as to the level of the LUC mRNA) (Fig. 2B). As expected (26), readthrough was observed from all three nonsense codons, with UGA being the most permissive (Fig. 2B). Maximal readthrough was observed when strains were either defective in translation termination factor eRF1 (sup45-2; for UAA) or treated with gentamicin (for UAG and UGA) (Fig. 2B). Taken together, these results demonstrate that the HA-LUC(PTC20)-SF constructs act as bona fide NMD reporters and validate their use as a quantitative readthrough reporter system.
Fig. 1.
Reporters for studying termination readthrough at PTCs. (A) Schematic representation of the HA-LUC(PTC20)-SF reporter with an in-frame N-terminal HA tag, followed by the luciferase ORF and C-terminal SF tags. Nucleotide substitutions encoding PTCs (UAA, UAG, or UGA) were inserted at the position of amino acid 20 of luciferase. (B) Northern blot analysis of the HA-LUC-SF mRNA expressed in either WT [PSI−] (Left) or upf1∆ [PSI−] (Right) cells. Blots were reprobed for CYH2 transcripts as an internal NMD control. SCR1 RNA was used as a loading control.
Fig. 2.
Readthrough efficiency from HA-LUC(PTC20)-SF reporters. (A, Top) Full-length luciferase readthrough products from WT [PSI−] cells expressing HA-LUC(PTC20)-SF reporters. Samples were immunoprecipitated using the N terminus HA tag, followed by Western blot analysis with anti-FLAG antibody. (A, Bottom) Readthrough products are active as observed by luciferase assay. Luciferase activity is expressed as relative luciferase units per microgram of protein/RNA units, and the activity of WT protein is set to 100%. (B) Readthrough efficiency measured as luciferase activity from the HA-LUC(PTC20)-SF reporters in response to various readthrough effectors. Activity is represented as the percentage of increase of WT [PSI−] for each codon (n = 3; error bars represent SD from the mean).
Characterization of the Endogenous Readthrough Product from PTCs.
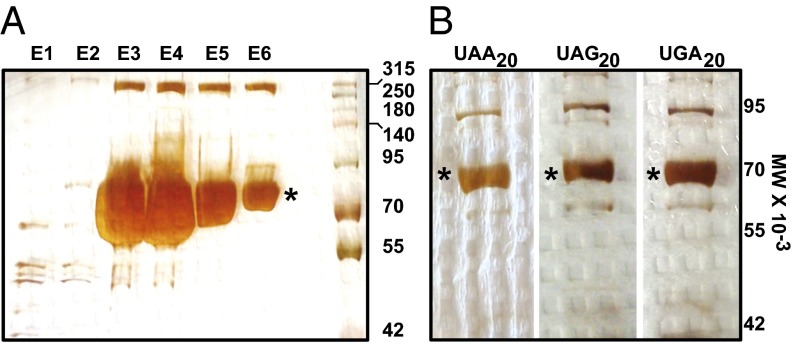
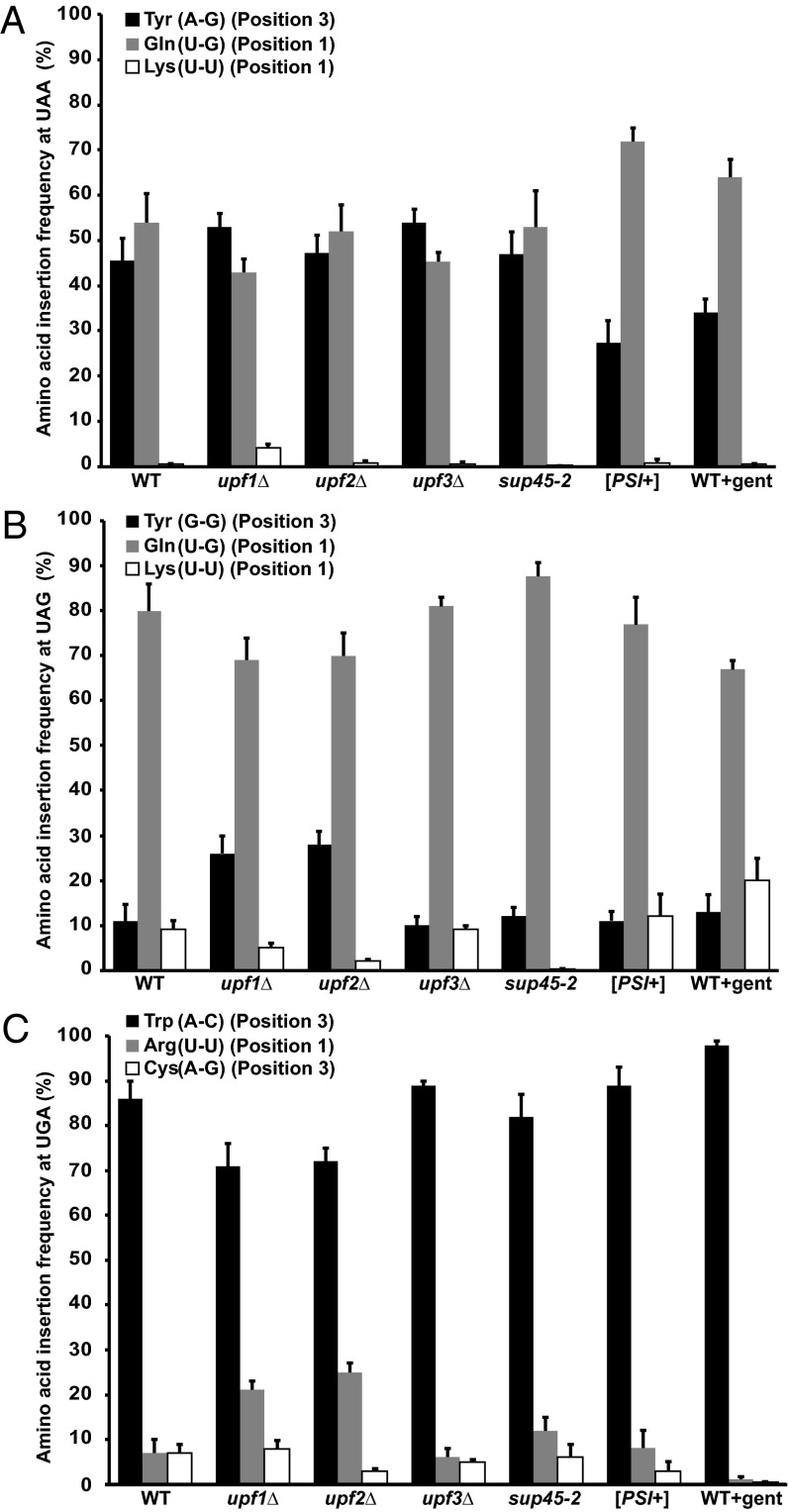
To gain insight into the misreading events that occur when a termination codon is present at the A-site of the ribosome, it is important, first, to characterize these events when the ribosome is not predisposed to any other readthrough stimulus. Accordingly, to ensure that the readthrough observed was solely a readout of basal readthrough events, we expressed the HA-LUC(PTC20)-SF reporters in WT cells with a [PSI−] background [i.e., cells devoid of the prion form of Sup35 (eRF3)]. Hence, any detectable readthrough in these cells is unlikely to be attributable to Sup35 aggregation (27). Strep-Tactin (IBA) purification of WT luciferase (Fig. 3A) and the readthrough products expressed from all three PTC alleles was followed by gel electrophoretic analysis to resolve the full-length products. Following silver staining of the gel, the band corresponding to full-length luciferase was excised and subjected to endo-LysC digestion and liquid chromatography-tandem MS analysis. Analyses of the readthrough products purified from WT [PSI−] cells showed that when UAA was the PTC, three amino acids were incorporated at that position, namely, Gln, Lys, and Tyr (Fig. 4A). Three independent experiments revealed that the frequencies of insertion of Tyr and Gln were similar (45.5 ± 5% and 54 ± 7%, respectively) but that Lys was inserted at a much lower frequency (0.5 ± 0.1%) (Fig. 4A). The same sets of amino acids were inserted when readthrough from a UAG occurred in WT [PSI−] cells, but with different frequencies. Gln was identified as the predominant amino acid, with an insertion frequency of 80 ± 6% (Fig. 4B). Tyr (11 ± 4%) and Lys (9 ± 2%), on the other hand, had similar insertion frequencies (Fig. 4B). With UGA as the PTC, we identified the incorporation of Trp, Arg, and Cys (Fig. 4C and Fig. S1). Trp was inserted at a higher frequency (86 ± 4%) than both Arg (7 ± 3%) and Cys (7 ± 2%) (Fig. 4C). The identification of Trp, Arg, and Cys as a consequence of UGA readthrough confirms observations from earlier studies (20). For UAG readthrough products, identification of Gln as the most abundant amino acid suggests that an earlier study with viral mRNAs that identified only Gln, and not Lys or Tyr, might reflect the lower frequencies of insertion for the latter two amino acids. This observation suggests that our approach for identifying the amino acids inserted during nonsense suppression is suitable for the detection of low-frequency events.
Fig. 3.
Strep-Tactin purification of HA-LUC-SF and HA-LUC(PTC20)-SF products. (A) Silver-stained SDS/PAGE gel showing Strep-Tactin purification of the luciferase reporter protein from WT [PSI−] cells. (B) Silver-stained gel showing the readthrough products purified from upf1∆ [PSI−] cells expressing HA-LUC(PTC20)-SF reporters. The asterisks denote full-length WT (A) and readthrough (B) luciferase products.
Fig. 4.
Amino acids inserted at PTCs during termination readthrough. HA-LUC(PTC20)-SF protein products purified from WT [PSI−] cells, upfΔ [PSI−] mutants, termination factor mutants, or WT [PSI−] cells treated with gentamicin (WT + gent) were subjected to MS analyses. The numbers denote the frequency (±SD of the mean) of insertion of the amino acids at UAA (A), UAG (B), or UGA (C) (n = 3). The type and position of mispairing in the codon are represented for each amino acid inserted.
Readthrough in the Absence of Upf Factors.
NMD requires the regulatory factors, Upf1, Upf2, and Upf3 (7). In yeast, the loss of any of these factors results in the stabilization and increased accumulation of PTC-containing mRNAs, with little or no effect on the abundance and stability of most WT transcripts (7). In addition to their role in NMD, the yeast Upf factors control the fidelity of translation termination, as manifested by increased nonsense suppression when these factors are mutated or deleted (10, 28). This effect on nonsense suppression is largely attributable to NMD regulation of the uORF (upstream open reading frame)-containing ALR1 mRNA and the altered intracellular Mg+2 concentration that occurs when this mRNA has been stabilized (29). Recent work in yeast suggests that the nonsense suppression phenotypes of Upf-deficient cells may also include direct effects of the Upf factors on the regulation of translation termination (30). Thus, as expected, our analyses showed that the loss of any of the three Upf factors led to increased readthrough from the HA-LUC(PTC20)-SF constructs (Fig. 2B). Because we observed a distinct pattern of amino acid insertion for endogenous readthrough in WT [PSI−] cells, we assayed the readthrough products arising in the absence of the Upf factors for their amino acid incorporation patterns at PTCs. Readthrough products were purified from all three PTC-containing reporters expressed in upf1Δ (Fig. 3B and Fig. S2), upf2Δ, or upf3Δ cells that were also [PSI−] to ensure that the insertions observed were due solely to loss of the Upf factors. Readthrough from UAA in the absence of each of the Upf factors showed the insertion of Tyr, Gln, and Lys in the position of the PTC (Fig. 4A). The frequencies of insertion of the amino acids were similar in all three deletion strains, with equal distribution of Tyr [upf1Δ (53 ± 3%), upf2Δ (47.2 ± 4%), and upf3Δ (54 ± 3%)] and Gln [upf1Δ (43 ± 3%), upf2Δ (52 ± 6%), and upf3Δ (45.4 ± 2%)] and a very low frequency of insertion for Lys [upf1Δ (4 ± 1%), upf2Δ (0.8 ± 0.5%), and upf3Δ (0.6 ± 0.4%)] (Fig. 4A). With UAG as a PTC, the same set of amino acids was inserted (i.e., Tyr, Gln, Lys) (Fig. 4B). Insertion of Gln was found to be predominant in all three strains [upf1Δ (69 ± 5%), upf2Δ (70 ± 5%), and upf3Δ (81 ± 2%)], with Lys being the least favored insertion [upf1Δ (5 ± 1%), upf2Δ (2 ± 0.5%), and upf3Δ (9 ± 1%)]. However, the frequency of insertion of Tyr was lower in the upf3Δ strain (9 ± 1%) than observed for the upf1Δ and upf2Δ strains (26 ± 4% and 28 ± 3%, respectively). Analysis of the readthrough products from UGA as the PTC showed the insertion of Arg, Trp, and Cys, similar to amino acid insertions observed from WT cells (Fig. 4C). Trp insertion was favored [upf1Δ (71 ± 5%), upf2Δ (72 ± 3%), and upf3Δ (89 ± 1%)] over insertion of either Arg [upf1Δ (21.1 ± 2%), upf2Δ (25 ± 2%), and upf3Δ (6 ± 2%)] or Cys [upf1Δ (7.9 ± 2%), upf2Δ (3 ± 0.5%), and upf3Δ (5 ± 0.5%)].
The results of Fig. 4 indicate that upf1Δ and upf2Δ cells exhibited similar amino acid insertion patterns for UAG and UGA, which were distinct from those insertion patterns manifested by upf3Δ cells (the frequencies of insertion of Tyr and Gln at UAG and Trp and Arg at UGA differed significantly, with P < 0.001, as determined by one-way ANOVA followed by post hoc Tukey analyses). It is not clear why deletion of Upf3 would affect readthrough differently than the other Upf factors, but the observation supports the notion that enhanced readthrough in the absence of Upf factors is not likely to be exclusively due to increased magnesium levels in the cell. Rather, the Upf factors might be affecting readthrough in more than one way, independently (Upf3) or in concert (Upf1 and Upf2).
Readthrough in the Presence of Defective Translation Termination Factors.
Translation termination is mediated by eRF1 and eRF3 (Sup45 and Sup35 in yeast), and defects in these factors are known to increase readthrough at PTCs (8, 31). The sup45-2 strain is a temperature-sensitive eRF1 mutant that fails to bind the ribosome upon a temperature shift to 37 °C, leading to increased readthrough activity (8). Readthrough from the HA-LUC(PTC20)-SF constructs in sup45-2 [PSI−] cells increased approximately fivefold after a 30-min shift to 37 °C (Fig. S3). Readthrough in sup45-2 [PSI−] cells that were not temperature-shifted was similar to readthrough of WT cells (Fig. S3). Characterization of the readthrough products purified from sup45-2 [PSI−] cells after 30 min at 37 °C showed that when UAA was the premature stop codon, Tyr, Gln, and Lys were inserted with frequencies of 46.9 ± 5%, 53 ± 8%, and 0.03 ± 0.05%, respectively (Fig. 4A). Gln (87.7 ± 3%) was the predominant insertion at a UAG codon, whereas Tyr was inserted with a frequency of 12 ± 2% and Lys was the least favored insertion (0.3 ± 0.1%) (Fig. 4B). Readthrough at a UGA resulted in the insertion of Trp (82 ± 5%), Arg (12 ± 3%), and Cys (6 ± 3%) (Fig. 4C).
Because the [PSI+] epigenetic state in yeast decreases translation termination efficiency because the prion form of Sup35 (eRF3) aggregates, we also characterized the readthrough activity and readthrough products from [PSI+] cells. These cells showed increased readthrough compared with [PSI−] cells (Fig. 2B), and mass spectrometric analysis of the readthrough products purified from [PSI+] cells showed the incorporation of the same set of amino acids at PTCs as in [PSI−] cells. UAA readthrough products in [PSI+] cells showed over twofold more insertion of Gln (72 ± 3%) than Tyr (27.3 ± 5%) (Fig. 4A). Similar to sup45-2 [PSI−] cells, Lys was the least favored insertion (0.7 ± 1%). At a UAG, [PSI+] cells showed Gln insertion at a frequency of 77 ± 6% and an equal distribution of Tyr (11 ± 2%) and Lys (12 ± 5%) (Fig. 4B). Trp was inserted at a frequency of 89 ± 4% in UGA readthrough products, whereas Cys insertion was observed at 3 ± 0.5%. Interestingly, Arg insertion (8 ± 3%) was an order of magnitude lower than Arg insertion found in sup45-2 [PSI−] cells (Fig. 4C).
In light of the joint roles played by eRF1 and eRF3 in termination (2) it is of interest to determine whether defects in either factor yield comparable readthrough results. For readthrough from UAA, defects in eRF1/Sup45 resembled those defects seen in WT [PSI−] cells, whereas [PSI+] strains exhibited prevalent Gln insertion rather than the approximately equal ratio of Tyr/Gln insertions observed under the other readthrough-inducing conditions. Amino acid insertion frequencies at UAG and UGA in sup45-2 strains were also similar to WT [PSI−], whereas [PSI+] resembled the Upf factor deletion category for UAG insertions and WT [PSI−] for UGA insertions. The resemblance in amino acid insertion frequencies between sup45-2, [PSI+], and WT [PSI−] suggests that the basal level of readthrough that occurs in the WT [PSI−] cells may have its basis in the reduced eRF binding at PTCs thought to occur because of their distance from the normal 3′-UTR of the mRNA (7). The [PSI+] strains resemble the WT [PSI−] readthrough profile for all but UAA. It is not clear at this point what leads to this difference, but it should be noted that our understanding of the [PSI+] prion form of Sup35 is just emerging (32).
Readthrough in the Presence of an Aminoglycoside.
Having characterized the distribution of amino acids incorporated at PTCs in different genetic conditions that predispose the ribosome to read through a PTC, we then compared the corresponding events when a small-molecule drug is used to induce readthrough. Gentamicin, a commonly used aminoglycoside, can suppress PTCs and restore protein function in vivo by promoting readthrough (2, 3). Binding of gentamicin to the ribosome decoding center can promote termination readthrough by induction of A-site structural changes (33). As in previous experiments, we used WT [PSI−] cells to ensure that the readthrough observed is solely due to gentamicin treatment. WT [PSI−] cells treated with varying concentrations of gentamicin showed an increase in readthrough from the HA-LUC(PTC20)-SF reporters, as measured by luciferase activity (34) (Fig. S4). Readthrough products were purified from WT [PSI−] cells treated with gentamicin at a final concentration of 100 μg/mL and the frequencies of amino acid insertion determined as above. Amino acids inserted at the UAA codon included Gln (64 ± 4%), Tyr (34 ± 3%), and Lys (0.5 ± 0.1%) (Fig. 4A). Gln was inserted with a frequency of 67 ± 2%, Tyr with a frequency of 13 ± 4%, and Lys with a frequency of 20 ± 5% when UAG was the PTC (Fig. 4B). Readthrough products from UGA showed predominant insertion of Trp (98 ± 1%), followed by Arg (1.2 ± 0.5%) and then Cys (0.4 ± 0.1%) (Fig. 4C). Even though gentamicin treatment resulted in the insertion of the same amino acids observed in other conditions, the relative frequencies of the respective insertions were significantly different from any other condition (P < 0.001, as determined by one-way ANOVA with post hoc Tukey analysis). The most dramatic of these differences were observed for Tyr and Gln insertion at UAA, Lys and Gln insertion at UAG, and Trp insertion at UGA. These observations suggest that the conformational changes at the A-site due to gentamicin binding induce readthrough in a way that prefers the selection of some near-cognate tRNAs over others, even though cellular tRNA abundance should be unaffected.
Discussion
Protein synthesis is a highly efficient and accurate process that allows an organism to translate genomic information into functional protein products. Despite its apparent accuracy, translation has a misincorporation rate of 10−3–10−4 (11). Like other steps in gene expression where the maintenance of fidelity is crucial (e.g., DNA replication, DNA transcription), translation relies on the complementarity of nucleotides to choose the right substrate. The selection of cognate aminoacyl-tRNAs is based on codon/anticodon base pairing and is central to maintaining translation fidelity. Although failures in translational fidelity generally lead to nonfunctional proteins, not all mistranslation events have deleterious effects (35). For example, the [PSI+] state in yeast appears to increase adaptability to environmental cues in parallel with increased readthrough of NTCs (36).
Stop codon readthrough can also be beneficial to a cell when the readthrough event takes place at a PTC. Ordinarily, the occurrence of a PTC in an ORF will lead to the formation of a truncated, nonfunctional polypeptide and a vast reduction in the affected mRNA as a consequence of NMD, and will essentially yield a null allele (37, 38). These consequences are at least partially overridden when the PTC is translated (i.e., read through by the ribosome), leading to in-frame elongation and production of the full-length protein product. Stop codon readthrough at both PTCs and NTCs takes place when recognition of the termination codon by a class I release factor is superseded by recognition of a near-cognate aminoacyl-tRNA. Readthrough is known to be enhanced by genetic and pharmacological effectors, and the phenomenon has been exploited for therapeutic purposes (2, 3), but the mechanism of near-cognate tRNA selection and its enhancement by readthrough-inducing agents is unknown. Here, we have used readthrough of PTCs in yeast reporter genes to determine (i) which amino acids are inserted in place of a termination codon when the ribosome reads through a premature stop codon, (ii) whether readthrough induced under different conditions has varying effects on amino acid insertions, and (iii) whether the pattern of near-cognate tRNA insertion in response to novel readthrough effectors can be predicted.
Several important general conclusions can be drawn from our analysis of the readthrough events in response to readthrough effectors. First, our comparisons of the readthrough products from WT [PSI−] cells with cells subjected to various modes of readthrough enhancement indicate that although readthrough efficiency is enhanced by genetic and pharmacological modulations, the nature of the readthrough product is similar. The same set of amino acids gets inserted for each of the PTCs (Tyr, Gln, and Lys for UAA and UAG; Arg, Trp, and Cys for UGA) regardless of the mechanism causing readthrough. Second, the deciding factor for insertion of a near-cognate tRNA is the codon sequence itself, and not the readthrough effector. Increased readthrough efficiencies do not generate any novel misreading events. Rather, they lead to quantitative enhancement of the basal translation remodeling events that are permitted on the ribosome, leading us to speculate that the known effects of termination codon sequence context on readthrough efficiency will also not affect the nature of the amino acid inserted. Third, the frequency of insertion of the amino acids at a particular codon is affected by the nature of the readthrough effector, independent of tRNA abundance in the cell. Hence, changing the sequence context and other parameters known to affect readthrough (e.g., the identity of the nucleotide 3′ to the PTC) will most likely determine the frequency with which an amino acid gets inserted at the PTC.
Our analyses of readthrough products also provided insight into the codon positions that would tolerate mispairing during near-cognate tRNA insertion. These studies demonstrated that near-cognate tRNA insertion at a PTC occurs by mispairing at either position 3 of the codon (the “wobble” position) or at position 1, and that utilization of these two positions is comparable for endogenous readthrough (Fig. 5A) or when readthrough was enhanced by specific effectors (Fig. S5). More specifically, under multiple conditions, readthrough of UAA led to an equal distribution of position 1 and position 3 mispairing, whereas readthrough of UAG favored mispairing at position 1 and readthrough of UGA favored mispairing at position 3. We did not observe any amino acid insertions that could be a result of a codon position 2 mismatch (Fig. 4 and Table S2). However, we cannot rule out the possibility that any amino acids that may have been inserted by position 2 mispairing (e.g., Leu or Ser for UAA and UAG or Leu, Ser, and Trp for UAG) could not be detected because their abundance was below our detection level of 0.1%.
Fig. 5.
Readthrough from position 1 and position 3 mispairing. (A) Comparison of position 1 (white bar) or position 3 (black bar) mispairing for each termination codon leading to amino acid insertion at the PTC for WT [PSI−] cells. (B) Comparison of readthrough efficiency (measured as luciferase activity per milligram of protein/RNA units) from either position 1 (white bar) or position 3 (black bar) under multiple conditions. Readthrough efficiency is represented as the percentage of increase over WT.
For codon position 3 mispairing, the incorporation of Tyr at UAA and UAG codons (with A-G and G-G mispairing) and the insertion of Cys and Trp (with A-G and A-C mispairing) at UGA suggest that nonstandard Watson–Crick base pairings are allowed in the third position of the codon, with A-C favored over A-G and G-G (Fig. 4). In all instances with codon position 1 mispairing, the nonstandard Watson–Crick U-G mispairing was observed to be predominant (Fig. 4). Our observation of a prevalent U-G mispairing at codon position 1 is supported by crystallographic studies of conformational rearrangements in the 70S ribosome decoding center (12, 13). These studies showed that when a U-G mispairing occurs at either position 1 or position 2 of the codon, the decoding center adopts a standard Watson–Crick geometry resembling the Watson–Crick geometry of C-G base pairing. High-frequency position 1 insertions observed during PTC readthrough from UAA and UAG can therefore be attributed to U-G mispairing that becomes favored by virtue of its geometrical mimicry of a standard base-pairing event. U-G mispairing has also been reported as a major contributor to amino acid misincorporation during translation elongation (39). Interestingly, U-C mispairing was never observed for any of the PTCs in our analysis, because we never observed the presence of Gly (for UGA) or Glu (from UAA or UAG). Given that the structure of the base pairs is a key determinant of mispairing, it is possible that U-C mispairing at position 1 of a nonsense codon is not geometrically favored. The absence of Glu or Gly insertion, however, is not because of low abundance of the corresponding tRNAs (40). Consistent with other experiments showing a lack of correlation between absolute tRNA abundance and translation elongation rates (41), the insertion of a near-cognate tRNA at a PTC is determined not by tRNA abundance, but by how well the A-site can accommodate the base pairing. Taken together, position 1 or position 3 mispairings are favored when UAA and UAG are present at the A-site of the ribosome. Position 1 mispairings are favored when a U-G nonstandard Watson–Crick mispairing is involved, and U-C mispairing does not seem to be allowed. Position 3 mispairings were observed with multiple nonstandard Watson–Crick pairs (A-C > G-G > A-G). Furthermore, comparison of readthrough efficiencies from either codon position 1 or position 3 mispairing (Fig. 5B) shows higher readthrough from position 3 mispairing events. Higher readthrough from position 3 mispairing might explain why readthrough from UGA, with predominant position 3 mispairing, occurs more readily than it does with the other two terminators. Taken together, our study shows that the decoding center of the ribosome accommodates alternative noncanonical base-pairing events during termination, allowing expansion of the genetic code.
An understanding of the decoding events taking place during premature termination may provide useful insights into the utilization of this process for therapeutic purposes. For example, knowledge of the amino acids inserted at a specific PTC should help to predict the likelihood of success with a readthrough-inducing small molecule, particularly when the targeted PTC would lead to insertion of a novel amino acid in an essential functional site of a protein. Further, because premature termination is less efficient than normal termination (2, 3, 7) it is plausible that some miscoding events that are allowed in the context of a PTC may not happen during normal termination (when there appears to be more efficient eRF sampling and the ribosome does not pause as long) (3). It will thus be of interest to compare the amino acid insertions at NTCs to determine if the rules of near-cognate tRNA insertion are different.
Materials and Methods
Plasmid Construction.
The wild-type (HA-LUC-SF) and the PTC-containing luciferase (HA-LUC(PTC20)-SF) constructs comprise, from 5′ to 3′: the TPI1 promoter, followed by an N-terminal 3× Hemagglutinin tag fused in-frame with the firefly LUC gene (without/with an in-frame stop codon at LUC position 20), in-frame C-terminal StrepII and FLAG tags, and the TPI1 3′-UTR. The ORF spans from the 3×HA tag to the StrepII/FLAG tags. Plasmids harboring either HA-LUC-SF or HA-LUC(PTC20)-SF were generated by PCR and standard molecular cloning techniques. In each case, oligonucleotides (listed in Table S1) containing restriction sites were used for PCR amplification, and the resulting fragments were inserted into yEplac181 after digestion of the respective restriction sites. Premature termination codons (UAA, UAG, or UGA) were inserted at codon 20 of the LUC ORF using site-directed mutagenesis and oligonucleotides described in Table S1.
RNA and Protein Analyses.
Procedures for RNA analysis, protein analysis, and luciferase assays were as described previously (29) or in SI Materials and Methods. Readthrough products were purified according to instructions provided by the manufacturer of Strep-Tactin resin (IBA). Details are provided in SI Materials and Methods. Procedures used for mass spectrometric analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Andrei A. Korostelev and Christopher R. Trotta for helpful discussions. This work was supported by NIH Grant R37 GM27757-35 (to A.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424127112/-/DCSupplemental.
References
- 1.Gesteland RF, Atkins JF. Recoding: Dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 2.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–425. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: Mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 5.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6(7):1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci USA. 1995;92(12):5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kervestin S, Jacobson A. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stansfield I, Kushnirov VV, Jones KM, Tuite MF. A conditional-lethal translation termination defect in a sup45 mutant of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1997;245(3):557–563. doi: 10.1111/j.1432-1033.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- 9.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2(1):e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16(10):5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaher HS, Green R. Fidelity at the molecular level: Lessons from protein synthesis. Cell. 2009;136(4):746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westhof E, Yusupov M, Yusupova G. Recognition of Watson-Crick base pairs: Constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014;6:19. doi: 10.12703/P6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. New structural insights into the decoding mechanism: Translation infidelity via a G·U pair with Watson-Crick geometry. FEBS Lett. 2013;587(13):1848–1857. doi: 10.1016/j.febslet.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Fearon K, McClendon V, Bonetti B, Bedwell DM. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem. 1994;269(27):17802–17808. [PubMed] [Google Scholar]
- 15.Weiss WA, Friedberg EC. Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J Mol Biol. 1986;192(4):725–735. doi: 10.1016/0022-2836(86)90024-0. [DOI] [PubMed] [Google Scholar]
- 16.Pure GA, Robinson GW, Naumovski L, Friedberg EC. Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. J Mol Biol. 1985;183(1):31–42. doi: 10.1016/0022-2836(85)90278-5. [DOI] [PubMed] [Google Scholar]
- 17.Lin JP, Aker M, Sitney KC, Mortimer RK. First position wobble in codon-anticodon pairing: Amber suppression by a yeast glutamine tRNA. Gene. 1986;49(3):383–388. doi: 10.1016/0378-1119(86)90375-6. [DOI] [PubMed] [Google Scholar]
- 18.Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J Mol Biol. 1971;58(2):439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- 19.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29(23):4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1990;87(22):8860–8863. doi: 10.1073/pnas.87.22.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urban C, Beier H. Cysteine tRNAs of plant origin as novel UGA suppressors. Nucleic Acids Res. 1995;23(22):4591–4597. doi: 10.1093/nar/23.22.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerfass K, Beier H. The leaky UGA termination codon of tobacco rattle virus RNA is suppressed by tobacco chloroplast and cytoplasmic tRNAs(Trp) with CmCA anticodon. EMBO J. 1992;11(11):4167–4173. doi: 10.1002/j.1460-2075.1992.tb05510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Translational readthrough of an amber termination codon during synthesis of feline leukemia virus protease. J Virol. 1985;55(3):870–873. doi: 10.1128/jvi.55.3.870-873.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA. 1985;82(6):1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchet S, Cornu D, Argentini M, Namy O. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42(15):10061–10072. doi: 10.1093/nar/gku663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol. 1995;251(3):334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 27.Serio TR, Lindquist SL. [PSI+]: An epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 28.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20(13):4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson MJ, Jacobson A. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 2010;24(14):1491–1495. doi: 10.1101/gad.1930710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He F, Ganesan R, Jacobson A. Intra- and intermolecular regulatory interactions in Upf1, the RNA helicase central to nonsense-mediated mRNA decay in yeast. Mol Cell Biol. 2013;33(23):4672–4684. doi: 10.1128/MCB.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stansfield I, Eurwilaichitr L, Akhmaloka, Tuite MF. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol Microbiol. 1996;20(6):1135–1143. doi: 10.1111/j.1365-2958.1996.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 32.Baudin-Baillieu A, et al. Genome-wide translational changes induced by the prion [PSI+] Cell Reports. 2014;8(2):439–448. doi: 10.1016/j.celrep.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 33.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 34.Altamura N, et al. Tobramycin is a suppressor of premature termination codons. J Cyst Fibros. 2013;12(6):806–811. doi: 10.1016/j.jcf.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: Friend or foe? Trends Biochem Sci. 2014;39(8):355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29(12):691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19(6):594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Shah B, Bondarenko PV. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry. 2013;52(45):8165–8176. doi: 10.1021/bi401002c. [DOI] [PubMed] [Google Scholar]
- 40.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141(2):344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20(2):237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.