Significance
Embryos depend on maternally deposited RNA until zygotic transcription activates. In many species, genome activation coincides with cell cycle lengthening and cellular motility, which collectively comprise the midblastula transition (MBT). A long-standing model is that MBT onset is controlled by titration of a maternally loaded inhibitor against exponentially increasing DNA. To identify MBT inhibitors, we developed an assay using Xenopus egg extract that recapitulates transcriptional activation only above a DNA-to-cytoplasm ratio similar to MBT embryos and identified histones H3 and H4 as inhibitors. Changing histone levels quantitatively shifts the DNA concentration required for transcription in vitro and alters the onset of both zygotic transcription and cell cycle lengthening in vivo. Our work strongly supports histones as a titrated MBT inhibitor.
Keywords: early vertebrate development, systems biology, cell size control, maternal zygotic transition, transcription activation
Abstract
During early development, animal embryos depend on maternally deposited RNA until zygotic genes become transcriptionally active. Before this maternal-to-zygotic transition, many species execute rapid and synchronous cell divisions without growth phases or cell cycle checkpoints. The coordinated onset of transcription, cell cycle lengthening, and cell cycle checkpoints comprise the midblastula transition (MBT). A long-standing model in the frog, Xenopus laevis, posits that MBT timing is controlled by a maternally loaded inhibitory factor that is titrated against the exponentially increasing amount of DNA. To identify MBT regulators, we developed an assay using Xenopus egg extract that recapitulates the activation of transcription only above the DNA-to-cytoplasm ratio found in embryos at the MBT. We used this system to biochemically purify factors responsible for inhibiting transcription below the threshold DNA-to-cytoplasm ratio. This unbiased approach identified histones H3 and H4 as concentration-dependent inhibitory factors. Addition or depletion of H3/H4 from the extract quantitatively shifted the amount of DNA required for transcriptional activation in vitro. Moreover, reduction of H3 protein in embryos induced premature transcriptional activation and cell cycle lengthening, and the addition of H3/H4 shortened post-MBT cell cycles. Our observations support a model for MBT regulation by DNA-based titration and suggest that depletion of free histones regulates the MBT. More broadly, our work shows how a constant concentration DNA binding molecule can effectively measure the amount of cytoplasm per genome to coordinate division, growth, and development.
The midblastula transition (MBT) is the first major developmental transition in many fast-developing organisms. Before the MBT, proteins encoded by maternally supplied RNAs drive the early, synchronous cell divisions of the embryo. At the MBT, the cell cycles lengthen to incorporate growth phases, and the zygotic genome becomes transcriptionally active (1, 2). In addition, the MBT is often associated with the emergence of cell cycle checkpoints, cell motility, and in Drosophila, cellularization of the syncytium.
The onset of the MBT during early development has been attributed to two general classes of mechanisms. The first class posits that embryos monitor the amount of time since fertilization or count the number of divisions. The second class posits that embryos monitor changes in the DNA-to-cytoplasm ratio in individual cells. Before the MBT, synchronously dividing cells of the embryo do not grow, and thus, the DNA-to-cytoplasm ratio doubles as the cell volume is halved with each division. In D. melanogaster, some genes respond to the timer mechanism, and other genes, including those that control cell cycle lengthening and syncytium cellularization, respond to the DNA-to-cytoplasm ratio (3–7). In Xenopus, the degradation of the cell cycle regulator cyclin E and the emergence of the mitotic checkpoint have been attributed to timer mechanisms (8, 9). However, the DNA-to-cytoplasm ratio controls critical MBT events, including zygotic genome activation, cell cycle lengthening, and onset of cellular motility (1, 2, 8).
One prominent model for how frog embryos sense the DNA-to-cytoplasm ratio is that a maternally contributed inhibitor is deposited at a set concentration in the cytoplasm and then titrated against the exponentially increasing amount of DNA. The MBT is triggered when the DNA concentration crosses a threshold, at which the inhibitor no longer saturates the DNA (1). Since this model was proposed, several potential factors have been suggested to regulate the MBT, including chromatin components and modifiers, signaling proteins, replication factors, metabolic components, and transcription factors.
Factors regulating chromatin assembly were the first proposed to control MBT timing. Consistent with this hypothesis, chromatin assembly can compete with transcriptional machinery before the MBT (1, 10–12). In vitro, histones can inhibit transcription from ssDNA (13), and coinjection of histones with exogenous DNA into Xenopus embryos reduces the amount of transcription from that exogenous DNA template (10). Taken together, these experiments led to the hypothesis that histone levels regulate the timing of transcriptional activation in the Xenopus embryo (10–13). However, the effects of histone levels on the transcriptional activation of endogenous genes and other MBT-related events, including cell cycle lengthening, were not assessed. It, therefore, remains unclear whether histone or some other chromatin component acts as a titratable inhibitor controlling MBT timing. Indeed, other chromatin binding proteins have been proposed to be the titratable factor at the MBT. The chromatin modifier DNA (cytosine-5-)-methyltransferase 1 (xDnmt1) inhibits transcription in pre-MBT embryos. However, because xDnmt1 mRNA has been observed only in one-half of the embryo, xDnmt1 is unlikely to be responsible for sensing the DNA-to-cytoplasm ratio in the entire embryo (14–16).
In addition, two sets of factors have recently been implicated in S-phase lengthening at the Xenopus MBT. One set is the protein phosphatase 2A (PP2A) with its regulatory subunit B55α. PP2A opposes the cell cycle arrest in response to replication stress and is limiting for rapid replication at high DNA concentrations in vitro (17). However, it is unclear if PP2A is limiting in vivo. The other set of proposed S-phase regulators is the replication factors Cut5, RecQ4, Treslin, and Drf1. Overexpression of these factors together induces additional rapid cell cycles in vivo (18). However, these factors become unstable at the MBT, and therefore, it is unclear whether they become limiting by titration or instead, degradation in response to another MBT-initiating signal. Moreover, although both sets of factors may contribute to cell cycle lengthening at the MBT, it is unclear how they would control transcriptional activation. Interestingly, other essential replication components, including dNTPs, also become limiting near the MBT (19), suggesting that more than one pathway may impinge on S-phase lengthening at this stage.
Many genes near the MBT are activated by specific early transcription factors, which include Zelda in Drosophila and Nanog, Pou5f1 (Oct4), and SoxB1 in zebrafish (20–26). In Drosophila, a proposed site-specific repressor, Grainyhead, might be the titrated factor, because it competes with the transcriptional activator Zelda for the same binding sites (21). Thus, increasing the number of binding sites through replication might serve to titrate Grainyhead or similar inhibitors to allow activator binding. However, titration based on specific binding sites cannot explain the observation that injection of exogenous DNA lacking specific transcription factor binding sites is sufficient to activate genome-wide zygotic transcription in Xenopus (1). Thus, although necessary and clearly important, specific transcription factors do not explain the dependence of many transcripts on the ratio of DNA to cytoplasm.
Therefore, although several MBT regulators have been identified, it is unclear which, if any, of these titrate against the genome to control MBT timing. Here, we developed a cell-free system that recapitulates transcriptional activation dependent on the DNA-to-cytoplasm ratio. Using this system, we biochemically purified an inhibitory activity and identified histones H3 and H4 as responsible for transcriptional inhibition in our assay. We show that manipulation of H3/H4 levels quantitatively shifts the amount of DNA required for transcriptional activation in vitro. In vivo, an ∼50% reduction of H3 shifts the MBT earlier by one cell cycle as assayed by global transcription and cell cycle lengthening, and increasing the amount of H3/H4 in the embryo shortens post-MBT cell cycles. Taken together, our results support a critical role for histones H3 and H4 in sensing the DNA-to-cytoplasm ratio in vivo to trigger the MBT.
Results
Transcription in Xenopus Egg Extract Is Sensitive to the DNA-to-Cytoplasm Ratio.
To study the mechanism of zygotic genome activation in early embryos, we developed a cell-free system using Xenopus laevis egg extracts that allowed us to directly manipulate the DNA-to-cytoplasm ratio and measure the transcriptional response. We added varying concentrations of purified sperm chromatin into a constant volume of cytosolic extracts from unfertilized Xenopus eggs supplemented with 32P-α-UTP to measure new RNA synthesis (Fig. 1A). The sperm chromatin was rapidly decondensed into membrane-bound nuclei by the extract (27) and induced transcription dependent on DNA concentration. Moreover, the threshold DNA concentration required for transcriptional activation in the extract was strikingly similar to the DNA concentration present in embryos at the MBT (∼50 ng/µL) (Fig. 1 B and C).
Fig. 1.
In vitro transcription is sensitive to the DNA-to-cytoplasm ratio. (A) Experimental design for B and C. Variable quantities of sperm chromatin are added to a constant volume of extract. (B) A representative autoradiograph showing transcription above a threshold DNA concentration (50 ng DNA per 1 µL extract). Transcription was quantified using the indicated boxed band corresponding to the Pol III transcript OAX (in the text and SI Appendix, Figs. S2 and S3). (C) Semilog plot of mean transcriptional response to increasing amounts of sperm chromatin shows transcription at or above 50 ng sperm DNA per 1 µL extract. (D) Semilog plot of titration of λ-phage DNA into egg extract that activates transcription from reporter sperm chromatin (present at ∼15 ng/µL) at a similar DNA concentration as in C.
Although most genes are not transcribed before the MBT, some essential developmental genes, including critical signaling components, escape this global repression in Xenopus (28–31). To determine if our transcripts were sensitive to the DNA-to-cytoplasm ratio rather than simply increasing in proportion to the amount of template, we measured transcription in extracts of different volumes containing the same number of sperm transcriptional templates (SI Appendix, Fig. S1). We measured a similar DNA-to-cytoplasm ratio threshold, indicating that the observed transcripts are, indeed, responsive to this ratio and that the observed DNA concentration dependence was not an artifact of the detection limit of our assay.
The predominant transcript from sperm chromatin was used to quantify the transcriptional response (Fig. 1B). This transcript is a 180- to 220-nt single-stranded Pol III transcript based on its sensitivity to polymerase inhibitors and specific RNases (SI Appendix, Fig. S2). These observations are consistent with our major transcript being from the satellite repeat OAX, which has previously been observed to be highly expressed from sperm chromatin in Xenopus egg extract (32). To confirm the identity of this transcript, we performed quantitative RT-PCR on samples containing above-threshold amounts of sperm DNA with or without the RNA polymerase inhibitors α-amanitin and actinomycin D. We observed a >70-fold increase in OAX transcription, whereas another previously reported sperm transcript of small size, 5S RNA, remained uninduced (SI Appendix, Fig. S3).
In Xenopus embryos, the addition of nonspecific DNA is sufficient to induce transcription (1). To test whether our cell-free system depends on specific DNA sequences, we added λ-phage DNA to extracts and assayed transcription from a subthreshold number of reporter sperm nuclei. We found that λ-DNA was sufficient to induce transcription from the sperm nuclei in a dose-dependent manner with a threshold similar to the sperm chromatin alone (Fig. 1D). The λ-DNA was not transcribed, likely because of a lack of suitable Xenopus promoters. Our experiments are consistent with a previous report showing that transcription from a single-stranded reporter was induced by the addition of λ-DNA (12). These experiments indicate that titration of the transcriptional inhibitory activity is unlikely to be DNA sequence-specific and does not require the titrating DNA to be a template for transcription.
A Titratable Transcriptional Inhibitor Is Present in Xenopus Egg Extract.
Because DNA binding proteins are likely candidates for a DNA titratable inhibitor of transcription, we tested whether we could deplete the inhibitory activity from extracts using DNA as an affinity reagent. To do this, we coupled DNA from the pBluescript plasmid to magnetic beads and added these DNA beads to an extract to achieve a DNA concentration five times higher than the transcription threshold. We then removed the DNA beads and any associated factors and measured the transcriptional response of sperm chromatin titrated into this extract. We found that extracts depleted using DNA beads activated transcription at lower concentrations of sperm DNA than mock depleted controls (Fig. 2 A and B). These results are consistent with the removal of an inhibitory factor by the DNA beads.
Fig. 2.
A titratable factor inhibits transcription in Xenopus egg extract. (A and C) Schematics of experiments corresponding to B and D, respectively. DNA beads are used to bind, deplete, and transfer the inhibitory activity in egg extracts. (B) Extracts depleted of a transcriptional inhibitor with DNA-coated beads (purple curve) activate transcription at a lower sperm concentration than extracts exposed to beads not coupled to DNA (green curve). (D) DNA-coated beads preincubated in an extract are unable to induce transcription in a naïve extract (orange curve) compared with DNA beads preincubated with buffer (green curve). Transcription is measured from reporter sperm chromatin (present at 15 ng/µL). All error bars represent SEM.
We next tested whether the inhibitory activity removed from the extract with DNA beads could alter the transcriptional response when added to a fresh extract (Fig. 2C). DNA beads sufficiently saturated with inhibitory activity from a first extract should be unable to titrate out additional inhibitor in a second extract. In contrast, control DNA beads treated with buffer should be competent to titrate out the inhibitor and thereby, induce transcription. Because pBluescript is not transcribed in extracts, we added subthreshold amounts of reporter sperm chromatin to measure transcription. The DNA beads that were pretreated with extract were unable to induce transcription, consistent with a transcriptional inhibitor binding to the DNA beads (Fig. 2D). This loss of titration ability was not caused by degradation or permanent modification of the DNA on the beads, because boiling DNA beads after incubation in the first extract restored their ability to titrate inhibitor from a second extract (SI Appendix, Fig. S4).
Identification of Histones H3 and H4 as the Inhibitory Factor in Vitro.
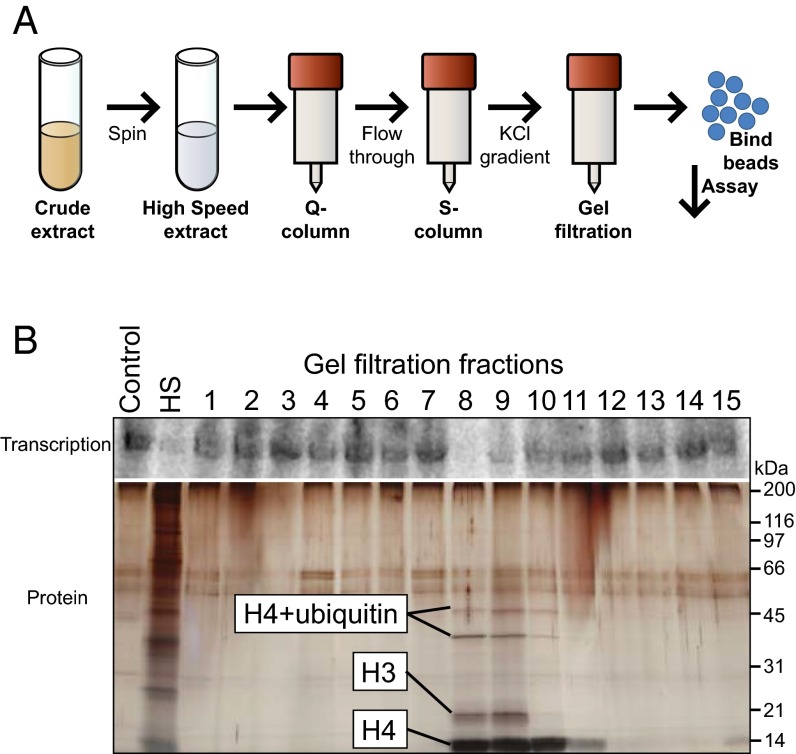
To identify the transcriptional inhibitor, we sought to biochemically fractionate the egg extract and follow the inhibitory activity using our DNA bead assay. Throughout the fractionation, we added and removed DNA beads from each fraction and then assayed the DNA beads’ ability to induce transcription in naïve extract (Fig. 2C). We found that the inhibitory factor was present in high-speed, clarified extract and stable through multiple chromatographic steps (Materials and Methods, Fig. 3A, and SI Appendix, Fig. S5). Two fractions from the purification’s final size exclusion step exhibited the bulk of the inhibitory activity and contained four protein bands as observed by silver staining. These bands were identified by MS as histones H3 and H4 with the slower mobility bands containing histone H4 and ubiquitin, consistent with posttranslational modification of the histone H4 (Fig. 3B and SI Appendix, Table S1). These results indicate that H3 and H4 are likely responsible for the inhibitory activity in our assay.
Fig. 3.
Isolation of histones H3 and H4 as the inhibitory factor. (A) Schematic of the final purification protocol resulting in the gels shown in B. (B, Upper) Autoradiograph of the transcriptional inhibition activity of control (buffer), high-speed (HS) extract, and individual fractions from the final-size exclusion step of the purification. Fractions 8 and 9 show inhibitory activity. (B, Lower) Silver stain gel of the proteins bound to the beads in each fraction. A more detailed description of the purification can be found in Materials and Methods and SI Appendix, Fig. S5.
Normalized H3 and H4 Concentrations Are Constant Through the MBT.
The titration hypothesis predicts that the titratable factor should be present in the embryo at a relatively constant concentration but should be depleted from the cytoplasm over the course of the cleavage divisions. To test this, we examined the concentrations of H3 and H4 in whole embryos by Western blot at time points starting immediately after fertilization until ∼4 h post-MBT. We find that total embryo concentrations for both H3 and H4 normalized to tubulin loading controls do not increase over this time period (Fig. 4). Our result is consistent with a quantitative whole-proteome developmental time course, which also indicates that H3 and H4 levels do not change through early development (33). Previous measurements have shown that histone protein is produced during the early cleavage division (34), which suggests the possibility that histone turnover is regulated to achieve the constant total H3 and H4 levels observed in this and other studies. However, during the early cleavage divisions, which lack growth, the DNA concentration increases exponentially so that cytoplasmic histones would be rapidly depleted approaching the MBT. Consistent with this model, a recent study has shown that H4 is depleted from the cytoplasm and accumulates in the nuclear fraction over the course of development (35).
Fig. 4.
H3 and H4 concentrations are constant relative to tubulin before the MBT. Normalized histones (A) H3 and (B) H4 protein levels are constant through the MBT. Both proteins are normalized to tubulin and an extract control. For each time point, n = 3; error bars represent SEM.
Histones H3 and H4 Inhibit Transcription in Xenopus Egg Extract in a Dosage-Dependent Manner.
If histones H3 and H4 are the inhibitory factors titrated during division, then increasing the histone concentration in the extract should increase the threshold amount of DNA required for transcription. To test this, we added bacterially produced H3/H4 at varying concentrations to extracts containing a range of concentrations of sperm chromatin or sperm chromatin and λ-DNA. We found that exogenous H3/H4 increased the amount of DNA required to activate transcription in a dosage-dependent manner (Fig. 5A). These results are consistent with previous findings that addition of exogenous histones is sufficient to inhibit transcription from reporters in Xenopus extracts (12, 13). Transcriptional inhibition by H3/H4 is not specific to sperm chromatin, because histone addition also inhibited transcription from Xenopus U6 plasmid DNA (SI Appendix, Fig. S6A). Moreover, we observed a linear trend over a fourfold concentration range in the relationship between the amount of histone H3 in the extract and the amount of DNA required to activate transcription (Fig. 5B). This apparently linear relationship between H3/H4 concentration and the DNA threshold strongly supports H3 and H4 as the titrated factors, because in the simplest titration model, doubling the amount of inhibitor should precisely double the amount of DNA required to overcome that inhibition.
Fig. 5.
H3/H4 tetramers set the threshold for transcriptional activation in extract. (A) Increasing the levels of histones H3 and H4 in egg extracts increases the concentration of sperm chromatin required to induce transcription. H3/H4 addition resulted in total concentrations equal to 1×, 1.6×, 2.1×, and 2.8× the untreated extract as measured using a Western blot for H3. (B) The concentration of DNA required for transcriptional activation (defined as the first point above 1.5× background) scales roughly linearly with histone concentration for both sperm chromatin (blue circles) and λ-phage DNA (red diamonds). Each point represents a titration as depicted in A. Points above the dashed line had thresholds beyond the detection limit. (C) Depletion of histones H3/H4 with DNA beads reduces the amount of DNA required to activate transcription (purple curve) compared with the control (green curve), and adding back H3 and H4 restores the transcriptional inhibition below the control threshold (blue curve). Dep, depleted. (D) Relationship between histone concentration and the amount of DNA required for transcriptional activation as described in B for the add-back experiments depicted in C. Histone H3 concentrations were quantified by Western blot.
Histones H3 and H4 assemble into nucleosomes with equimolar stoichiometry to histones H2A and H2B. We tested whether the inhibitory capacity of histones in our assay was limited to H3/H4 by adding exogenous H2A/H2B dimers to extracts and assaying for transcription. We found that H2A/H2B dimers were able to inhibit transcription, which is consistent with a previous report showing that H2A/H2B addition inhibited transcription in Xenopus extract (13). However, H3/H4 tetramers are approximately two times as efficient at inhibiting transcription as H2A/H2B dimers (as determined by how much is needed to reduce transcription from sperm chromatin by 50%) (SI Appendix, Fig. S6 B and C). The greater inhibitory activity of H3/H4 likely reflects the higher affinity of the H3/H4 tetramer for DNA compared with the H2A/H2B dimer (36, 37) and the more rapid exchange of H2A/H2B in chromatin compared with H3/H4 (38). We note that, whereas both histone pairs can inhibit transcription in vitro on their own, in the embryo, the nucleosome may be the ultimate inhibitory complex.
Histone ubiquitination is important for transcriptional regulation (39), and a fraction of the histones that we isolated from extracts seems to be ubiquitinated. However, the bacterially purified histones that we use for the histone addition experiments are not posttranslationally modified before addition to the extract. Thus, although histone modifications, including ubiquitination, may be necessary for the transcriptional inhibition activity, their acquisition in our system is likely rapid and not rate-limiting.
To test whether depleting histones from extract would reduce the amount of DNA required for transcriptional activation, we removed histones using DNA beads, because histone immunodepletion was inefficient (Fig. 2A). After histones are removed in this manner, transcription was initiated at lower DNA concentrations (Fig. 5C). Although a large number of proteins is depleted by this manipulation, returning H3/H4 concentration to mock-depleted levels by addition of purified H3/H4 tetramers restored the DNA threshold to mock-depleted levels (Fig. 5 C and D). Taken together, our data support histones as the titrated transcriptional inhibitor.
Histone Levels Set the Timing of Specific MBT Events in Vivo.
To test our histone titration model in vivo, we reduced the histone H3 concentration in embryos using previously characterized H3 morpholinos (40). Morpholino injection at the one-cell stage reduced H3 levels by 40–56% as measured at 7.5 h postfertilization, the time at which we begin to observe large cell cycle lengthening in our assay (SI Appendix, Fig. S7). New histone synthesis is known to occur in the early embryo (34, 41), but total H3 levels are constant during the pre-MBT period (Fig. 4A) (33). This result suggests that approximately one-half of the relevant pool of histone H3 is turned over during embryogenesis and replaced by new translation before the MBT to maintain constant concentration.
To assay the effects of H3 levels on genome-wide transcriptional activation, we coinjected 32P-α-UTP along with the H3 morpholino at the one-cell stage and measured total embryonic transcription from pools of five embryos starting at 6 h after fertilization. Both H3 morphant and control embryos display a similar set of transcripts exhibiting a greater diversity of molecular weights than we observed in our in vitro system (SI Appendix, Fig. S8). Critically, H3 morphant embryos advanced zygotic transcription compared with control embryos from the same clutch, consistent with our in vitro results (Fig. 6A and SI Appendix, Fig. S8). This result shows that histone levels modulate the timing of zygotic genome activation in otherwise unperturbed Xenopus embryos. More specifically, the titration model predicts that a 50% reduction in H3 should shift the timing of transcriptional activation by a single-cell cycle, because the amount of DNA doubles with each division. Consistent with this quantitative model, we observe a shift in transcriptional activation by ∼30 min, the duration of a pre-MBT cell cycle (Fig. 6A).
Fig. 6.
Reduction of histone H3 causes premature initiation of the MBT in vivo. (A) H3 morphant embryos (red) have increased transcription at earlier time points compared with controls (blue) as measured by the total 32P-UTP incorporation in microinjected embryos (SI Appendix, Fig. S8). (B) The cumulative effect of longer cell cycles results in larger cells and therefore, lower cell density in the H3 morphant embryos (H3 MOs) compared with control-injected embryos (Control). Data are shown for 215 min after the ninth division (n = 3 embryos). (C) H3 morphant embryos appear normal until near the MBT, when they display increased cell cycle lengthening and larger cells compared with controls. The location of the zoomed panels is marked by the black boxes. (D) H3 morphant embryos die at gastrulation. (E) H3 morphant embryos display early cell cycle lengthening compared with control embryos. Cell cycle duration lengthens one cycle early in H3 morphant embryos compared with control morpholino. Circles represent embryos that did not reach a 15th division before the movie limit. All data are shown for >15 cells per embryo; there are three control embryos and four H3 morphant embryos from the same clutch. Division times were manually annotated from movies with a 1-min frame rate (Materials and Methods). The box height represents the 25th to 75th percentiles, and the centerline represents the median. The whiskers extend to the farthest data point that is not an outlier. Outliers are plotted as diamonds. Results from two similar experiments are shown in SI Appendix, Fig. S9. Div, division; ns, P value > 0.01 between H3 morphant and control embryos in consecutive cell cycles. *P values < 0.01 between H3 morphant and control embryos in the same cell cycle.
Although our results indicate that histone levels modulate the onset of zygotic genome activation, it is unclear whether histone levels would also affect other aspects of the MBT, such as cell cycle lengthening and onset of cellular motility. To examine the effect of histone levels on cell cycle control, we measured the duration of cell division cycles in H3 morphant and control embryos from the same clutch at single-cell resolution. We observed gradual but significant increases of cell cycle durations starting at the 10th cycle followed by a sharper increase near the onset of the MBT at cycles 12 and 13. For all comparisons, H3 morphant embryos had longer cell cycles than control embryos at the same division cycle. Slower cell cycles in the H3 morphants also resulted in larger cells (Fig. 6 B and C). As in the case of transcriptional activation, an ∼50% reduction of H3 shifted cell cycle lengthening and asynchrony one cycle earlier (Fig. 6 C and E, SI Appendix, Fig. S9, and Movie S1).
Our single-cell analysis of cell cycle durations indicates that, whereas H3 morphants are advanced one cycle compared with control embryos (P < 0.01, all comparisons), the kinetics of cell cycle lengthening are highly similar. For example, a comparison of the distributions of cell cycle durations shown in Fig. 6E indicates that 12th cycle H3 morphant embryos have similar cell cycles to 13th cycle controls. Moreover, 13th cycle morphants are similar to 14th cycle controls. Indeed, for all morphant cycles before the 14th cycle, cell cycle durations are similar to the following cycle in controls (P > 0.01, all comparisons) (Fig. 6E). These results indicate that, although H3 morphants go on to die at gastrulation (40) (89 of 92 morphant vs. 3 of 31 control embryos) (Fig. 6D), they undergo a quantitatively normal MBT that is shifted by a single-division cycle. Therefore, H3 morphant embryos behave precisely as predicted by the histone titration model, implying that cell cycle lengthening is not caused by some nonspecific developmental defect.
To further examine how H3 depletion affected MBT onset, we examined cellular motility in dissociated blastomeres. Embryo dissociation conditions can influence the timing of motility onset (2, 42, 43); thus, we examined motility using two distinct methods. The embryos were dissociated either at the 16- to 32-cell stage or immediately before examination (Materials and Methods). The onset of cellular motility was similar in dissociated blastomeres from H3 morphant and control embryos (SI Appendix, Fig. S10 and Movies S2 and S3). The ability of H3 morphant cells to initiate motility demonstrates that H3 morphant embryos were competent to initiate multiple aspects of the MBT before undergoing apoptosis. Our data also suggest that the timing of motility onset is controlled by a process that is separate from transcriptional activation and cell cycle lengthening. This finding is consistent with previous observations that motility does not depend on zygotic protein synthesis and can precede other aspects of the MBT (44).
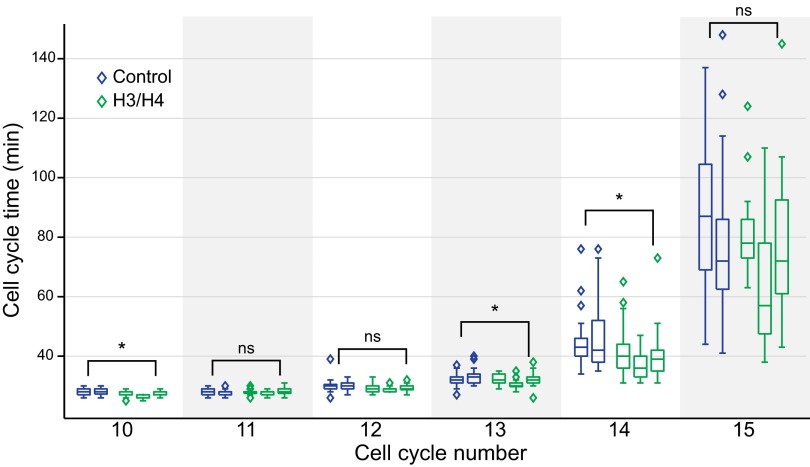
The histone titration model predicts that doubling histone concentration should delay the MBT by a single-cell cycle. To test this, we microinjected exogenous H3/H4 into single-cell embryos. However, all attempts to double H3/H4 concentrations in vivo resulted in cleavage defects and early embryonic death. We, therefore, injected H3/H4 concentrations that increased H3 by 10–25% as measured 7.5 h postfertilization. This smaller manipulation resulted in a significant shortening of the immediate post-MBT cell cycles, which is consistent with predictions of the histone titration model (Fig. 7 and SI Appendix, Fig. S11). After one or two shortened division cycles post-MBT, cell cycle durations returned to normal. This manipulation was not toxic, because embryos survived to the end of the assay at the neurula stage. Taken together, our in vivo results support histones as the titrated factor controlling cell cycle lengthening and activation of zygotic transcription at the MBT (Fig. 8).
Fig. 7.
Increasing H3 and H4 shortens early post-MBT cell cycles. Injection increased H3 concentration by ∼25% as measured 7.5 h postfertilization. Time-lapse movies were analyzed and are presented as in Fig. 6E. Results from two additional H3/H4 injection experiments are shown in SI Appendix, Fig. S11. ns, P value > 0.01, and *P values < 0.01 between H3 morphant and control embryos in the same cell cycle.
Fig. 8.
Model of histone titration measuring the DNA-to-cytoplasm ratio up to the MBT. Schematic showing titration of histones, which are at constant concentration in the early embryo, against the exponentially increasing amount of DNA.
Discussion
Here, we developed an in vitro assay that recapitulated DNA-to-cytoplasm ratio-dependent transcription and used this assay to identify histones H3 and H4 as the major titrated transcriptional inhibitors in extract. Consistent with the histone titration model, alteration of H3 levels quantitatively shifted the MBT in vivo. In addition to our results presented here, several other observations support histones as a DNA-titrated MBT inhibitor. In pre-MBT Xenopus embryos, histones are in excess and directly bind DNA, and their constant levels are not tied to DNA synthesis (34, 41). Moreover, histones are repressive to exogenous transcription in Xenopus embryos and extract (10, 12). Because nucleosomal histones are present across the genome, histone titration could directly regulate the diverse zygotic gene set associated with the MBT (30, 33). Finally, histones bind DNA nonspecifically, which is consistent with the observation that many different DNA sources can shift the timing of the MBT (1).
Coordination of Multiple MBT Events.
The degree of interdependence between zygotic genome activation and cell cycle lengthening at the MBT is unclear. Substantial inhibition of transcription in Xenopus embryos had no effect on cell cycle elongation at the MBT (2), suggesting a lack of strong coupling between these events. Furthermore, the acquisition of S-phase checkpoints, another aspect of the MBT, likely does not depend on transcription (45). However, early transcription at the MBT has been shown to control G1 acquisition in zebrafish embryos (46) and the number of syncytial divisions in Drosophila (3–5, 7).
Our results show that histone levels affect both cell cycle lengthening and transcriptional activation, suggesting a common regulatory mechanism. A threshold for S-phase lengthening might result from a lack of available histones to rapidly package newly synthesized DNA. Titration of the histone pool could lead to a cell cycle delay, which in turn, might allow sufficient time for transcript elongation. Alternatively, histones might affect transcription directly, so that cell cycle lengthening occurs because of the expression of cell cycle inhibitor(s). For example, in Drosophila, the cell cycle activator Cdc25 is destabilized by the zygotic transcription of tribbles (4). A similar mechanism may regulate cell cycle lengthening in Xenopus, where some zygotic transcript(s) could destabilize the replication factors Cut5, RecQ4, Treslin, and Drf1, which become limiting for rapid S-phase progression (18). Alternatively, histones, replication factors, and other molecules might act semiredundantly to ensure precise and coordinated timing of the many disparate MBT events.
Chromatin-Based Mechanism of Transcriptional Activation.
One explanation for how depletion of available histones might affect zygotic transcription would be through changes in local chromatin structure. In this model, pre-MBT chromatin would be densely occupied by histones to inhibit transcriptional machinery from assembling on and/or moving through the DNA. As histones are titrated by the increasing concentration of DNA, nucleosome occupancy would drop to provide greater access for transcription factors and RNA polymerases. Although histones bind throughout the genome, only specific transcripts become activated at the MBT, implying that additional regulators refine the global histone signal. Transcription factors that bind before widespread genome activation, such as Zelda in Drosophila, are critical regulators of early gene expression. Such early transcription factors might interact with histone titration signals to specify the locations of early nucleosome loss that primes sites for transcription (21, 25, 26).
More general changes in chromatin architecture might also play a role in zygotic genome activation. Although chromatin remodeling and histone-modifying enzymes do not seem to be rate-limiting in our in vitro assay, they may influence MBT chromatin structure by repositioning, evicting, or modifying nucleosomes to promote transcription. xDmnt1 is known to modify transcriptional output at the MBT through DNA modification (14, 16), and changes in histone methylation and RNA polymerase II occupancy are detected at that time (47, 48). In many organisms, including Xenopus, there is exchange of maternal histones with zygotic variants at the MBT (40), suggesting that specific variants or subpools of maternal histones may play different roles during the MBT. The Drosophila maternal variant of linker histone H1 (dBigH1) has been suggested to inhibit premature zygotic genome activation (49), and the zebrafish maternal H2A variant, H2af1o, has been linked to cell cycle synchrony (50). Thus, multiple proteins and pathways may be required to convert the histone titration signal into a regulated and coherent transcriptional response regulating multiple cellular events and pathways.
DNA Titration Sensors Coordinating Growth and Division.
Our work shows how a titration sensor can measure the DNA-to-cytoplasm ratio to set the timing of a developmental decision (Fig. 8). More broadly, any cellular process that alters either the numerator or denominator of the DNA-to-cytoplasm ratio can be monitored by a DNA-based titration mechanism. Titration of a regulator supplied at constant concentration allows the genome to serve as a yardstick for measuring the number of regulatory molecules per cell, which is proportional to the cytoplasmic volume. Before the MBT, cells become smaller with each division, so that the number of histones per cell decreases until reaching a threshold. In growing cells, a titration mechanism could measure the amount of a constant concentration regulator accumulating in proportion to cytoplasmic volume. Thus, DNA titration mechanisms could be used to coordinate growth-dependent cell cycle events with cell size, which has been suggested for DNA replication in bacteria and yeast (51–54).
The initial observation leading to the MBT titration hypothesis was the relationship between ploidy and the number of rapid pre-MBT division cycles. In a wide variety of species, haploids undergo one more and tetraploids undergo one fewer rapid division cycle, so that the MBT is traversed at a constant DNA-to-cytoplasm ratio (55). The DNA-to-cytoplasm ratio at the Xenopus MBT is part of a broad linear relationship between cell size and ploidy across eukaryotes in both endoreduplicated and experimentally manipulated contexts (53, 56), which may reflect widespread use of titration mechanisms measuring the DNA-to-cytoplasm ratio. However, the molecular identity of the titrated regulator is unlikely to be conserved, because outside early embryonic development and histone and DNA synthesis are tightly coupled. Cells have many candidate titrated factors; most proteins are found at a nearly constant concentration through the cell division cycle, and many of these interact with DNA (57–60). Here, we identify histone titration as a specific mechanism in what we expect to be an important class of DNA titration-based sensors coordinating transcription, growth, and division.
Materials and Methods
Xenopus Egg Extract.
Preparation of extracts was performed as previously described (61); 750 µM CaCl2 was added to release the extracts into interphase. To monitor new transcription, 50 nL/µL 10 mCi/mL or 12.5 nL/µL 40 mCi/mL 32P-α-UTP (BLU507H or BLU507Z; Perkin-Elmer) was added to each extract. Clarified extracts were prepared as in the work by Desai et al. (62). Briefly, crude extracts not containing 32P-α-UTP or CaCl2 were spun for a second time at 50,000 rpm in a Beckman SW55Ti rotor for 2 h at 4 °C. The clear middle layer was extracted using a 22-gauge needle.
Sperm Chromatin Preparation.
Sperm chromatin was prepared as previously described (61). Briefly, X. laevis testes were dissected and macerated, and the sperm was pelleted. The sperm was demembranated with lysolecithin and resuspended in storage buffer [250 mM sucrose, 15 mM Hepes, pH 7.4, 1 mM EDTA, 0.5 mM spermidine trihydrochloride, 0.2 mM spermine tetrahydrochloride, 1 mM DTT, 0.3% BSA, 30% (wt/vol) glycerol]. The stock concentration of sperm was measured using a hemocytometer. Stocks were flash-frozen in liquid nitrogen and stored at −80 °C until use.
DNA Immobilization.
pBluescript DNA was cut using EcoRI and BamHI (New England Biolabs) and end-filled using klenow (New England Biolabs) and biotin-dATP (Invitrogen). DNA was coupled to streptavadin dynabeads (65305; Invitrogen) following the protocol outlined in ref. 63. For factor and histone depletion assays, ∼250 ng/µL DNA on beads was incubated in interphase extract at 4 °C for 1–2 h and then removed. In assays that transfer the factor(s) from one extract or fraction to another, DNA beads were first incubated in an excess of extract or fraction for 1–2 h and then added to a fresh extract at ∼50 ng/µL.
In Vitro Transcription Assay.
Interphase extracts containing 32P-α-UTP were prepared as described above and distributed into 20-µL aliquots with or without DNA and bacterially purified histones as indicated. Histones were purified as previously described (64, 65). In cases where sperm chromatin was used as a reporter, it was used at a concentration of 15 ng/µL, with the remainder of the DNA coming from either λ-phage (New England Biolabs) or pBluescript plasmid attached to dynabeads. Extracts were incubated at 16 °C for 1.5 h with frequent mild agitation to prevent extract settling. RNAs were extracted using either Qiagen RNAeasy Kits or Tripure (Roche) following the manufacturers’ instructions. Both RNA purification methods produced similar results. The entire RNA sample was run on a 5% (wt/vol) acrylamide and 8 M urea gel in 89 mM Tris-borate and 2 mM EDTA. Gels were dried, exposed to a phosphor screen overnight, and imaged on a Typhoon 9400 Variable Mode Imager (GE Healthcare). Pixel intensity within the band denoted in Fig. 1C was quantified after background subtraction using ImageJ (National Institutes of Health). Background subtraction was performed using an identical region below the end of the same lane. All titration series were normalized to a reaction lacking DNA. The threshold for transcriptional activation was defined as the first point 1.5-fold above the no-DNA sample.
Inhibitor Purification.
High-speed supernatant was diluted fivefold in dilution buffer (2 mM MgCl2, 0.1 mM CaCl2, 10 mM K-Hepes, pH 7.7, 5 mM K-EGTA, pH 7.7) containing 20 mM KCl. Filtered extract was passed through a Q-Sepharose Fast Flow Column (GE Healthcare) and bound to an S-Sepharose Fast Flow Column (GE Healthcare). The S column was then washed with 5 column volumes 400 mM KCl and eluted with a linear gradient from 400 mM to 1 M KCl in dilution buffer. Activity-containing fractions were pooled and concentrated (10,000 molecular weight; Amicon), and then, they were separated over a Superdex 200 Column in dilution buffer containing 1 M KCl. The inhibitory activity of each fraction was assayed by first dialyzing each sample into cytostatic factor extract buffer (CSF-XB; 100 mM KCl, 50 mM sucrose, 2 mM MgCl2, 0.1 mM CaCl2, 10 mM K-Hepes, pH 7.7, 5 mM K-EGTA, pH 7.7) supplemented with 1 mM PMSF and 10 mM β-mercaptoethanol overnight and then incubating DNA-coupled dynabeads in each fraction for 1–2 h at 4 °C. After recovering the beads with a magnet, we used one-half of the beads to assay the protein content by SDS/PAGE and silver staining, and the other one-half was used to assay the inhibitory activity associated with the beads as described above.
Measurement of Histone Levels.
Fertilized embryos were allowed to develop at 20.0 °C ± 0.5 °C and collected as indicated. For each sample, 10 embryos were pooled and flash-frozen in liquid nitrogen. Embryos were rapidly thawed and lysed in 65 mM Tris, pH 7.5, 10 mM EDTA, 1 mM PMSF, 10 µg/mL aprotinin, 1 µg/mL pepstatin, and 5 mM DTT. Embryos were centrifuged at 20,800 × g for 20 min at 4 °C to remove cellular debris and yolk. Equal protein concentrations were separated by SDS/PAGE and transferred to PVDF membrane (Bio-Rad Laboratories) in transfer buffer of 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.3, 0.1% SDS, and 20% methanol. Membranes were incubated overnight in rabbit anti-H3 (1:1,000; 04–928; Millipore), 2 μg/mL rabbit anti-H4 (ab7311; Abcam), or 0.25 μg/mL mouse antitubulin (T9026; Sigma-Aldrich), washed, and then incubated for a minimum of 1 h in Alexa Fluor 488- or Alexa Fluor 647-conjugated goat anti-rabbit or anti-mouse antibodies (1:2,500; Invitrogen). Fluorescence was detected using a gel imager (Typhoon 9400 Variable Mode Imager; GE Healthcare) and quantified in ImageJ. For extract experiments, 1 µL extract was loaded per lane. For developmental time points, either 40 or 60 ng total protein was loaded. For morpholino experiments, a volume equivalent to 0.125–2 embryos was loaded as noted. Histone concentrations were normalized to tubulin loading controls. Extract experiments were normalized to untreated or mock-depleted extracts as appropriate. Developmental time points were normalized to an extract standard.
Microinjection.
H3 morpholinos (which target both H3.2 and H3.3) and control morpholinos were previously described by Szenker et al. (40). We microinjected embryos using a Harvard Apparatus PLI-100. Embryos were injected four times distributed equally around the animal cap with ∼5 nL relevant solution per injection. Control and H3 morpholinos were used at a concentration of 1 mM in water. In experiments using radiolabeled UTP, morpholinos were diluted with an equal volume of 40 mCi/mL 32P-α-UTP (BLU007Z; Perkin-Elmer). H3/H4 tetramers were injected at 1.1–1.4 ng/nL 50% CSF XB. Embryos were allowed to recover in 3–5% Ficoll and visually inspected before use in all experiments.
In Vivo Transcription Assay.
Embryos were injected as described above with 32P-α-UTP and morpholinos and collected at time points as labeled. 32P-α-UTP injection quantity was measured by scintillation counts on a Beckman-Coulter LS6500 and did not vary by greater than 15% between control and morphant embryos. RNAs were purified, visualized, and quantified as described for the in vitro transcription assay. Individual radiolabeled bands were measured and then summed to quantify total transcription. Each time series was normalized to its own 6.5-h value.
Cell Density Quantification.
To control for the effects of variation in embryo size, the number of cells was counted inside a square with size that was normalized by total embryo size. Cells inside were included if more than one-half of the visible cell was within the box. P values were calculated using a two-tailed t test with unequal variance between the indicated distributions.
Cell Cycle Duration Measurement.
To generate the data presented in Figs. 6 and 7 and SI Appendix, Figs. S9 and S11, we made and analyzed movies as follows. Movies were started at ∼2 h postfertilization (after the first or second cleavage). Embryos were allowed to develop in 1/3× Marc's modified Ringer's solution (MMR) containing 3–5% Ficoll until ∼4 h postfertilization, when they were changed to 1/2× modified Barth's saline for the duration of their development. Images were captured every 1 min on a Leica MZ16FA Stereomicroscope at 24× magnification using Image-Pro software. Movies were begun at the two- or four-cell stage, and divisions were counted to determine the frame number of the eighth cleavage. Then, ∼30 individual cells were selected from the visible portion of each embryo after the eighth cleavage. The intercleavage period was determined by manually tracking individual cells and noting the frame number at which the cleavage furrow visibly transected the entire cell. When daughter cells did not divide concurrently, the division time of the earliest dividing daughter was always used, and that cell was followed for the remainder of the movie. When the cleavage could not be observed, as in cases where the cleavage plane did not intersect with the embryo surface, the cell was omitted from analysis.
Box plots were generated using the first and third quartiles (q1 and q3, respectively; 25th and 75th percentiles) as the box and the median as the centerline. Whisker lengths correspond to the farthest data point that was not an outlier. Outliers were defined as points above or below q3 + 1.5(q3 − q1) or q1 − 1.5(q3 − q1), respectively. P values were calculated using a two-tailed t test with unequal variance between the indicated distributions. At least 15 cells were counted for each embryo at each cell cycle.
Cell Motility Assay (Method 1).
We followed the method in ref. 2 with slight modifications. Embryos were fertilized and allowed to develop at 19 °C in 5% Ficoll and 1/2× MMR until the 16- to 32-cell stage, at which point they were transferred to a 0.5% agarose-containing Petri dish, where vitelline membranes were manually removed. After 30 min, embryos were placed in fresh dissociation media (100 mM sodium isethionate, 20 mM sodium pyrophosphate, 20 mM d-glucose, pH 9.0) in agar-bottomed Petri dishes, where dissociated blastomeres continued to divide and spread out laterally. At indicated time points, dissociated blastomeres from one or two embryos were gently transferred to chambered slides (0.7 × 0.7 cm; Nunc Lab-Tek II Chambered Coverglass) for time-lapse analysis.
Cell Motility Assay (Method 2).
Fertilized embryos developed in 5% Ficoll and 1/2× MMR until 15 min before the indicated time points, when the vitelline membranes were manually removed. Demebranated embryos were gently transferred to fresh dissociation media for 5 min and then, chambered slides. Embryos were gently pipetted three to four times to fully dissociate blastomeres.
Cell Motility Measurement.
Dissociated blastomeres were imaged in phase contrast on an inverted Leica DMI6000 Microscope using a 10× objective. One image was taken every 5 s for 60–90 s alternating between H3 morphant and control embryos. Dissociated blastomeres were scored as motile or nonmotile using the parameters derived from ref. 66. We scored two motile subtypes: finger-like protrusions and bleb and/or circus movements. Cells exhibiting both phenotypes were scored as bleb and/or circus movement. Large vegetal blastomeres were excluded.
Quantitative PCR.
Transcriptionally active samples contained >50 ng/μL sperm DNA. In other samples, sperm plus 45 μM H3 and H4 or sperm plus 20 ng/μL α-amanitin and 5 ng/μL actinomycin-D were added to inhibit transcription. We used polymerase-inhibited controls with identical quantitates of sperm DNA to control for the effects of any potential genomic DNA contamination, because OAX and other Pol III transcripts are derived from high copy-repetitive elements. RNA was extracted as described, and treated with DNase (TURBO DNase; Ambion) followed by isolation with a minElute RNA Cleanup Kit (Qiagen); 5 μg DNase-treated RNA was reverse-transcribed with random hexamer primers using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocols. The resulting cDNA was amplified using gene-specific primers tested for similar efficiency, and levels of target transcripts were measured with quantitative RT-PCR using SYBR Green Master Mix (11760; Invitrogen) on an ABI Prism 7900HT and cycle thresholds (CTs) determined automatically. Gene expression values were normalized to the reference gene ODC, which has stable expression in all extract conditions tested. Relative expression levels were calculated with the ΔΔCT method (67). Fold changes are reported relative to transcription-blocked controls. Each quantitative PCR experiment graphed in SI Appendix, Fig. S3 consists of two technical replicates from two experimental replicates (different experiments from the same extract) in two to three biological replicates (from different extracts).
The following X. laevis primers were used: OAX_forward, CTCCAGTCTGCTGTGCCTAA; OAX_reverse, GAAGGCGGATGATGGACTAC; 5S_rRNA_forward, CCCTGAAAGTGCCTGATCTC; 5S_rRNA_reverse, AAGCCTACGACACCTGGTATTC; U6_forward, TGCTTCGGCAGCACATATAC; U6_reverse, ATGGAACGCTTCACGAATTT; ODC_forward, GGAAAGCAACCTTGGCAAGA; and ODC_reverse, AGGCCACACTGGCAACTCA.
RNA Characterization.
For the polymerase inhibition experiments, α-amanitin or actinomycin-D at labeled concentrations was added to extracts before incubation at 16 °C. To test nuclease sensitivity, RNAs were extracted and treated for 1 h at room temperature with 100 ng/µL inactive RNase H-D10A, RNase H, RNase III, or RNase A in 1× PBS with 2 mM MgCl2 or 0.1 units/µL DNase I in 10 mM Tris, pH 7.4, 2.5 mM MgCl2, and 0.1 mM CaCl2. Transcript size was measured by SYBR Gold (Invitrogen) staining of the unlabeled low-range ssRNA ladder (New England Biolabs) before autoradiography and aligning images in Adobe Illustrator.
Heat Inactivation of Inhibitor.
After incubation with crude extract, DNA beads were recovered, washed, and placed into 10 µL CSF XB. Beads were either placed at 95 °C or left on ice for 10 min and added to extracts at the indicated concentrations without additional washing.
XlU6 plasmid was a gift from Philippe Carbon (Institut de Biologie Moleculaire du CNRS, Strasbourg Cedex, France) (68).
Supplementary Material
Acknowledgments
We thank T. Stearns, M. Simon, J. Ferrell, I. R. Lehman, J. Baker, and members of the laboratories of A.F.S. and J.M.S. for discussion. We also thank G. Anderson, A. Wills, and C. Reid for technical advice; M. Krasnow for sharing equipment; P. Carbon (Université de Strasbourg) for reagents; and B. French for aid in figure design. This work was supported by National Institutes of Health Grants T32GM007276 (to A.A.A.), F32GM108295 (to D.J.), and R21HD073772 (to A.F.S. and J.M.S.), and the Burroughs Wellcome Fund (Career Awards at the Scientific Interface; to J.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413990112/-/DCSupplemental.
References
- 1.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 2.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 3.Di Talia S, et al. Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Curr Biol. 2013;23(2):127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell JA, O’Farrell PH. Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr Biol. 2013;23(2):118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell JA, Shermoen AW, Yuan K, O’Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev. 2012;26(7):714–725. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136(12):2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung HW, Spangenberg S, Vogt N, Großhans J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr Biol. 2013;23(2):133–138. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Clute P, Masui Y. Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev Biol. 1995;171(2):273–285. doi: 10.1006/dbio.1995.1280. [DOI] [PubMed] [Google Scholar]
- 9.Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc Natl Acad Sci USA. 1996;93(5):2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995;14(8):1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prioleau MN, Huet J, Sentenac A, Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77(3):439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 12.Almouzni G, Méchali M, Wolffe AP. Competition between transcription complex assembly and chromatin assembly on replicating DNA. EMBO J. 1990;9(2):573–582. doi: 10.1002/j.1460-2075.1990.tb08145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almouzni G, Méchali M, Wolffe AP. Transcription complex disruption caused by a transition in chromatin structure. Mol Cell Biol. 1991;11(2):655–665. doi: 10.1128/mcb.11.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135(7):1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- 15.Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Dev Biol. 2002;243(1):155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- 16.Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14(3):313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Murphy CM, Michael WM. Control of DNA replication by the nucleus/cytoplasm ratio in Xenopus. J Biol Chem. 2013;288(41):29382–29393. doi: 10.1074/jbc.M113.499012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341(6148):893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vastag L, et al. Remodeling of the metabolome during early frog development. PLoS ONE. 2011;6(2):e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5(5):e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345(2):248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanodia JS, et al. Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys J. 2012;102(3):427–433. doi: 10.1016/j.bpj.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson JC, Watson JD, Crews ST. Drosophila melanogaster Zelda and Single-minded collaborate to regulate an evolutionarily dynamic CNS midline cell enhancer. Dev Biol. 2012;366(2):420–432. doi: 10.1016/j.ydbio.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503(7476):360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341(6149):1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- 27.Philpott A, Leno GH, Laskey RA. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65(4):569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- 28.Collart C, et al. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141(9):1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol. 2011;357(2):478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan MH, et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23(1):201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129(24):5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- 32.Wolffe AP. Transcriptional activation of Xenopus class III genes in chromatin isolated from sperm and somatic nuclei. Nucleic Acids Res. 1989;17(2):767–780. doi: 10.1093/nar/17.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, et al. Quantitative proteomics of Xenopus laevis embryos: Expression kinetics of nearly 4000 proteins during early development. Sci Rep. 2014;4:4365. doi: 10.1038/srep04365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adamson ED, Woodland HR. Histone synthesis in early amphibian development: Histone and DNA syntheses are not co-ordinated. J Mol Biol. 1974;88(2):263–285. doi: 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang WL, et al. Phosphorylation and arginine methylation mark histone H2A prior to deposition during Xenopus laevis development. Epigenetics Chromatin. 2014;7:22. doi: 10.1186/1756-8935-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton DR, et al. The interaction of core histones with DNA: Equilibrium binding studies. Nucleic Acids Res. 1978;5(10):3643–3663. doi: 10.1093/nar/5.10.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oohara I, Wada A. Spectroscopic studies on histone-DNA interactions. II. Three transitions in nucleosomes resolved by salt-titration. J Mol Biol. 1987;196(2):399–411. doi: 10.1016/0022-2836(87)90700-5. [DOI] [PubMed] [Google Scholar]
- 38.Kimura H, Cook PR. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153(7):1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17(22):2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 40.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Reports. 2012;1(6):730–740. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Adamson ED, Woodland HR. Changes in the rate of histone synthesis during oocyte maturation and very early development of Xenopus laevis. Dev Biol. 1977;57(1):136–149. doi: 10.1016/0012-1606(77)90360-8. [DOI] [PubMed] [Google Scholar]
- 42.Johnson KE. Circus movements and blebbing locomotion in dissociated embryonic cells of an amphibian, Xenopus laevis. J Cell Sci. 1976;22(3):575–583. doi: 10.1242/jcs.22.3.575. [DOI] [PubMed] [Google Scholar]
- 43.Minoura I, Nakamura H, Tashiro K, Shiokawa K. Stimulation of circus movement by activin, bFGF and TGF-beta 2 in isolated animal cap cells of Xenopus laevis. Mech Dev. 1995;49(1-2):65–69. doi: 10.1016/0925-4773(94)00303-5. [DOI] [PubMed] [Google Scholar]
- 44.Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48(3):399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- 45.Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: Studies in Xenopus. Cell. 1990;61(5):811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- 46.Zamir E, Kam Z, Yarden A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol Cell Biol. 1997;17(2):529–536. doi: 10.1128/mcb.17.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akkers RC, et al. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17(3):425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vastenhouw NL, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez-Montero S, Carbonell A, Morán T, Vaquero A, Azorín F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013;26(6):578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Yue HM, et al. Oocyte-specific H2A variant H2af1o is required for cell synchrony before midblastula transition in early zebrafish embryos. Biol Reprod. 2013;89(4):82. doi: 10.1095/biolreprod.113.108043. [DOI] [PubMed] [Google Scholar]
- 51.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219(5158):1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 52.Hill NS, Kadoya R, Chattoraj DK, Levin PA. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 2012;8(3):e1002549. doi: 10.1371/journal.pgen.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Curr Biol. 2012;22(9):R350–R359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 2009;7(9):e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masui Y, Wang P. Cell cycle transition in early embryonic development of Xenopus laevis. Biol Cell. 1998;90(8):537–548. [PubMed] [Google Scholar]
- 56.Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev Camb Philos Soc. 2001;76(1):65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- 57.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 58.Oliva A, et al. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3(7):e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 62.Desai A, Deacon HW, Walczak CE, Mitchison TJ. A method that allows the assembly of kinetochore components onto chromosomes condensed in clarified Xenopus egg extracts. Proc Natl Acad Sci USA. 1997;94(23):12378–12383. doi: 10.1073/pnas.94.23.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1(5):2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 64.Guse A, Fuller CJ, Straight AF. A cell-free system for functional centromere and kinetochore assembly. Nat Protoc. 2012;7(10):1847–1869. doi: 10.1038/nprot.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272(3):301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 66.Satoh N, Kageyama T, Sirakami K. Motility of dissociated embryonic cells in xenopus laevis: Its significance to morphogenetic movements. Dev Growth Differ. 1976;18(1):55–67. doi: 10.1111/j.1440-169X.1976.00055.x. [DOI] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68.Krol A, Carbon P, Ebel JP, Appel B. Xenopus tropicalis U6 snRNA genes transcribed by Pol III contain the upstream promoter elements used by Pol II dependent U snRNA genes. Nucleic Acids Res. 1987;15(6):2463–2478. doi: 10.1093/nar/15.6.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.