Significance
By RNA profiling of 10 stages of maize anthers plus mature pollen, we found two distinct classes of phased small-interfering RNAs (phasiRNAs): 21-nt premeiotic phasiRNAs, after germinal and somatic cell specification, and 24-nt meiotic phasiRNAs coordinately accumulated during meiosis and persist into pollen. Sequencing of RNA from five male-sterile, anther developmental mutants—ocl4, mac1, ms23, msca1, and ameiotic1—demonstrated the involvement of specific somatic layers. Premeiotic phasiRNAs require a functional epidermis, whereas meiotic phasiRNAs require a normal tapetum. Mammalian germ cells express “prepachytene” or “pachytene” PIWI-interacting RNAs (piRNAs). Whereas differences in biogenesis indicate independent origins, grass phasiRNAs and mammalian piRNAs share developmental timing, a lack of obvious targets, and an impact on male fertility, suggesting a possible evolutionary convergence.
Keywords: phasiRNAs, anther development, piRNAs, tapetum, tasiRNAs
Abstract
Maize anthers, the male reproductive floral organs, express two classes of phased small-interfering RNAs (phasiRNAs). PhasiRNA precursors are transcribed by RNA polymerase II and map to low-copy, intergenic regions similar to PIWI-interacting RNAs (piRNAs) in mammalian testis. From 10 sequential cohorts of staged maize anthers plus mature pollen we find that 21-nt phased siRNAs from 463 loci appear abruptly after germinal and initial somatic cell fate specification and then diminish, whereas 24-nt phasiRNAs from 176 loci coordinately accumulate during meiosis and persist as anther somatic cells mature and haploid gametophytes differentiate into pollen. Male-sterile ocl4 anthers defective in epidermal signaling lack 21-nt phasiRNAs. Male-sterile mutants with subepidermal defects—mac1 (excess meiocytes), ms23 (defective pretapetal cells), and msca1 (no normal soma or meiocytes)—lack 24-nt phasiRNAs. ameiotic1 mutants (normal soma, no meiosis) accumulate both 21-nt and 24-nt phasiRNAs, ruling out meiotic cells as a source or regulator of phasiRNA biogenesis. By in situ hybridization, miR2118 triggers of 21-nt phasiRNA biogenesis localize to epidermis; however, 21-PHAS precursors and 21-nt phasiRNAs are abundant subepidermally. The miR2275 trigger, 24-PHAS precursors, and 24-nt phasiRNAs all accumulate preferentially in tapetum and meiocytes. Therefore, each phasiRNA type exhibits independent spatiotemporal regulation with 21-nt premeiotic phasiRNAs dependent on epidermal and 24-nt meiotic phasiRNAs dependent on tapetal cell differentiation. Maize phasiRNAs and mammalian piRNAs illustrate putative convergent evolution of small RNAs in male reproduction.
Diverse small RNAs exist in male reproductive cells of animals and plants. In animals, PIWI proteins, a subfamily of the ARGONAUTEs, and their interacting piRNAs are required for spermatogenesis; mutants defective for the PIWI-encoding genes fail to produce mature sperm (1–3). Whereas most Drosophila piRNAs are repeat-derived and function to silence transposable elements (TEs) (4, 5), mammalian piRNAs predominantly map to unique intergenic regions and have unclear but essential roles during gonad development. Based on their expression timing, different sizes, and distinctive PIWI partners, mammalian piRNAs are further classified as prepachytene or pachytene (6). Given the continuum of developmental stages in the testes, the prepachytene class is characteristic of gonads in which no cells have reached pachytene, whereas the pachytene-associated small RNAs are characteristic of gonads in which the most advanced germ line cells have reached this meiotic stage and all prior stages are also present in the more immature zone of the gonad.
In flowering plants, the anther is equivalent to the mammalian testes in that it consists of multiple somatic cell types required to support the premeiotic, meiotic, and postmeiotic haploid cells. In contrast to the continuum of mammalian gonads, however, an entire anther progresses through sequential developmental landmarks, and in maize, meiosis is synchronous within the organ. A second major difference between plants and animals is that the haploid meiotic products of plants are microspores, which undergo mitotic divisions to produce the three-celled gametophyte. Two of the gametophytic cells are sperm—later involved in double fertilization—and the third cell is a metabolically active, haploid vegetative cell. Like their mammalian counterparts, the plant germ line also contains repeat- and nonrepeat-derived small RNAs. In Arabidopsis pollen, TE-derived small-interfering RNAs (siRNAs) expressed in the vegetative nuclei reinforce silencing after transfer to sperm nuclei (7). Additionally, rice inflorescences produce 21- and 24-nt phased, secondary siRNAs (phasiRNAs) from nonrepeat regions (8, 9).
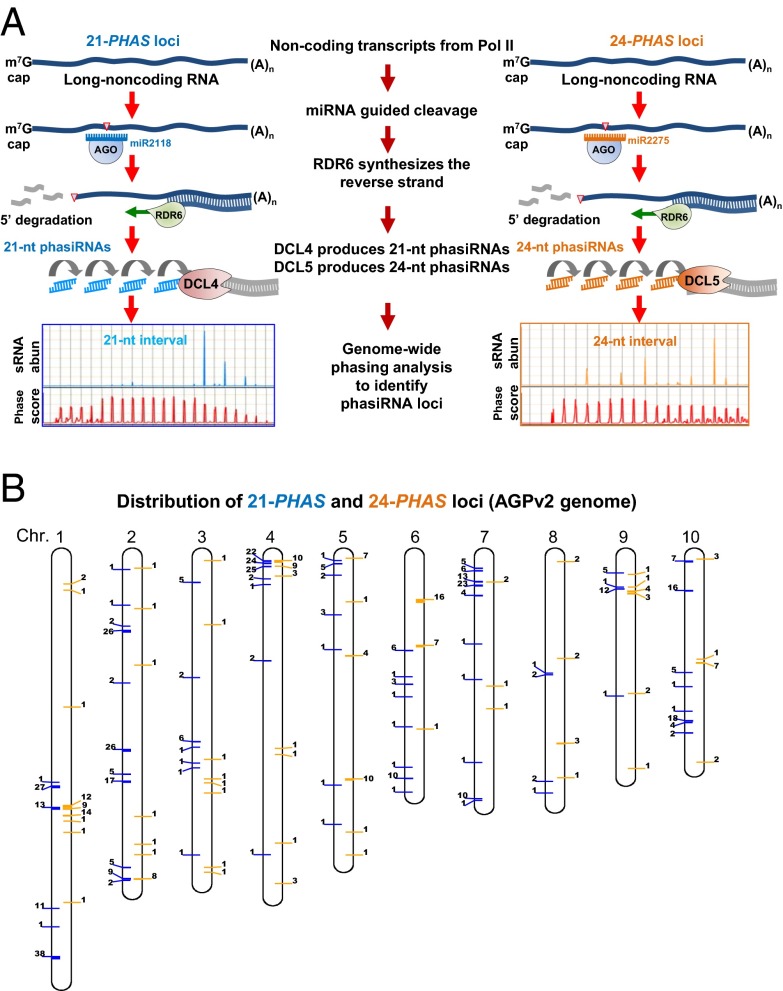
A key step in the production of many plant secondary siRNAs is cleavage of their precursors directed by a 22-nt microRNA (miRNA). In the case of grass phasiRNAs, their mRNA precursors—“PHAS” transcripts—are transcribed by RNA polymerase II, capped, and polyadenylated. These long noncoding precursor transcripts are internally cleaved, guided by 22-nt miR2118 to generate the 21-nt phasiRNAs or by miR2275 for the 24-nt phasiRNA (Fig. 1A) (10). In rice and putatively in maize, RDR6 recognizes the cleaved, uncapped 3′ fragments of these transcripts and synthesizes a second strand, forming double-stranded RNA (11). Subsequent processing by DCL4 and DCL5 generates 21- and 24-nt phasiRNAs, respectively (9). Both dicers exhibit sequential slicing activity, starting precisely at the 11th nucleotide of the miRNA binding site. This activity generates populations of regularly spaced, phased siRNAs from each PHAS precursor (12).
Fig. 1.
Genome-wide identification of 21-nt and 24-nt phasiRNAs loci in maize. (A) PhasiRNA biogenesis pathways, 21-nt phasiRNAs (Left) and 24-nt phasiRNAs (Right), result in loci with characteristic phased small RNA patterns. (B) Distribution of 21-PHAS (left side of the chromosome, blue bar) and 24-PHAS (right side of the chromosome, orange bar) loci on 10 maize chromosomes. Loci within 500,000 bp are clustered together; the number adjacent to each bar represents the number of loci in that particular cluster.
Although plants lack PIWI-clade ARGONAUTEs that bind piRNAs, the plant AGO family has diversified extensively, and there are plant-specific AGO proteins. MEL1, a rice homolog of Arabidopsis AGO5, mainly localizes to the cytoplasm of premeiotic cells (13). Recently MEL1 was shown to selectively bind 21-nt phasiRNAs (14). mel1 loss of function mutants have abnormal tapetum and aberrant pollen mother cells (PMCs, the final differentiated state before the start of meiosis) that arrest in early meiosis (13), suggesting that 21-nt phasiRNAs are crucial for male fertility.
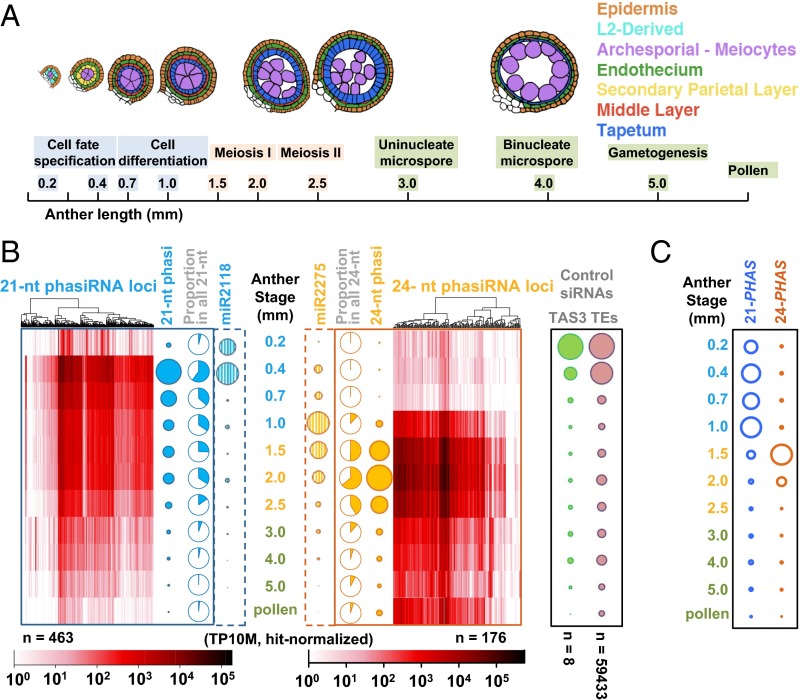
As a monoecious plant with large cohorts of synchronously developing flowers, maize (Zea mays) is particularly useful for studying male reproduction; anthers are readily dissected and staged using length as a proxy for developmental events (Fig. 2A) (15). We used this developmental regularity to characterize the spatiotemporal patterns of phasiRNA accumulation. Small RNA sequencing (sRNA-seq) and RNA-seq were applied to 11 sequential wild-type (fertile) stages, ranging from the initial step of cell fate specification in anther primordia through pollen production. We demonstrated that both phasiRNAs and their precursor transcripts show striking spatiotemporal regulation.
Fig. 2.
The 21-nt premeiotic and 24-nt meiotic phasiRNAs are developmentally regulated. (A) Anthers at 10 different lengths (developmental stages) were analyzed plus pollen. Above, a schematic of cell patterns in a single lobe of the anther; cell types in colors shown in key (Right). (B) Heat maps depicting the abundances of 21-nt premeiotic phasiRNAs from 463 loci (Left) and 24-nt meiotic phasiRNAs from 176 loci (Right) at each stage. Hierarchical clustering was based on similarity of expression pattern. (Left) Solid bubbles represent the total abundance of phasiRNAs; blue pie charts represent the proportion of 21-nt phasiRNAs from all 21-nt sRNAs at each stage; striped blue bubbles represent miR2118 abundances (trigger of premeiotic phasiRNA). (Right) The solid and striped orange bubbles represent 24-nt phasiRNA and miR2275 abundances, respectively; orange pie charts represent 24-nt phasiRNAs from all 24-nt sRNAs. Controls (Far Right): green represents TAS3-derived ta-siRNAs, and pink represents all TE-associated siRNAs mapped to the first 100 Mb of maize chromosome 1 (as a proxy for the whole genome). (C) Quantification of 21-PHAS and 24-PHAS precursor transcripts by RNA-seq.
Results
Temporal Regulation of Premeiotic and Meiotic phasiRNAs.
To explore the dynamics of small RNA populations in male reproductive organs of maize, 32 sRNA libraries from 11 sequential stages of W23 fertile anthers (Dataset S1) were sequenced deeply to allow accurate and sensitive identification of phasiRNAs. The phasiRNAs were then mapped to the genome by computational, genome-wide scans (Dataset S2) (16), identifying 463 21-PHAS and 176 24-PHAS loci; both classes of loci are distributed on all 10 maize chromosomes (Fig. 1B and Fig. S1). These loci correspond to unique or low-copy genomic regions (Fig. S2). This distinguishes the 24-nt phasiRNAs from plant DCL3-dependent, 24-nt heterochromatic siRNAs (hc-siRNAs) (17), which are largely derived from repetitive elements, primarily TEs.
Both 21-nt and 24-nt phasiRNAs exhibit striking temporal regulation (Fig. 2B) distinct from the timing of either TAS3-derived 21-nt trans-acting siRNAs (ta-siRNAs) or 24-nt hc-siRNAs derived from TEs (Fig. 2B and Dataset S2) (10). Few phasiRNAs were observed at 0.2 mm when germinal and initial somatic fate setting starts from pluripotent stem cells (18, 19). After 24 h at the 0.4-mm stage when premeiotic archesporial (AR) and three somatic cell types—epidermis, endothecium, and bipotent secondary parietal layer (SPL)—exist, 21-nt phasiRNAs peak in quantity and diversity, comprising 60% of all 21-nt RNAs (Fig. 2B). Most 21-nt phasiRNAs are present for approximately 1 wk (0.4-mm to 2.0-mm stages), but decline steadily from 0.7 mm when all SPL cells have divided, producing middle layer and tapetal daughter cells. In contrast, most 24-nt phasiRNAs are undetectable until 1.0 mm, when all cell types are specified and the postmitotic ARs start meiotic preparation as PMCs (Fig. 2B). The 24-nt phasiRNAs peak from 1.5 to 2.0 mm, coincident with meiotic progression through prophase I to metaphase I, and when somatic cells continue differentiating for postmeiotic supporting roles; at this peak, 24-nt phasiRNAs reached 64% of all 24-nt RNAs (Fig. 2B). Most 24-nt phasiRNAs are present when meiosis finishes (2.5 mm), then decline in abundance but remain detectable in mature pollen, 2 wk later. PhasiRNA dynamics were validated with three biological replicates and by RNA hybridization (Fig. S3). Based on their expression timing, we named the two size classes premeiotic (21-nt) and meiotic (24-nt) phasiRNAs to highlight the parallels with mammalian gonad piRNAs.
PhasiRNA synthesis requires both miRNAs and PHAS precursors (Fig. 1A). miR2118 family members were abundant at 0.2 mm, peaked at 0.4 mm, then vanished by 0.7 mm. In contrast, miR2275 family members peaked at 1.0 mm (Fig. 2B). Both miRNA families accumulate to their peak before that of the corresponding phasiRNA burst. RNA-seq from all 11 anther stages demonstrated that 21-PHAS precursor transcripts are highly expressed from 0.4 to 1.0 mm, whereas 24-PHAS transcripts peak in 1.5-mm anthers (Fig. 2C and Fig. S4). Collectively, all three components exhibit tight timing in developing anthers: miRNA triggers precede coordinate deployment of PHAS precursors and their phasiRNA products.
Epidermis Is Necessary and Sufficient for Premeiotic phasiRNA Biogenesis.
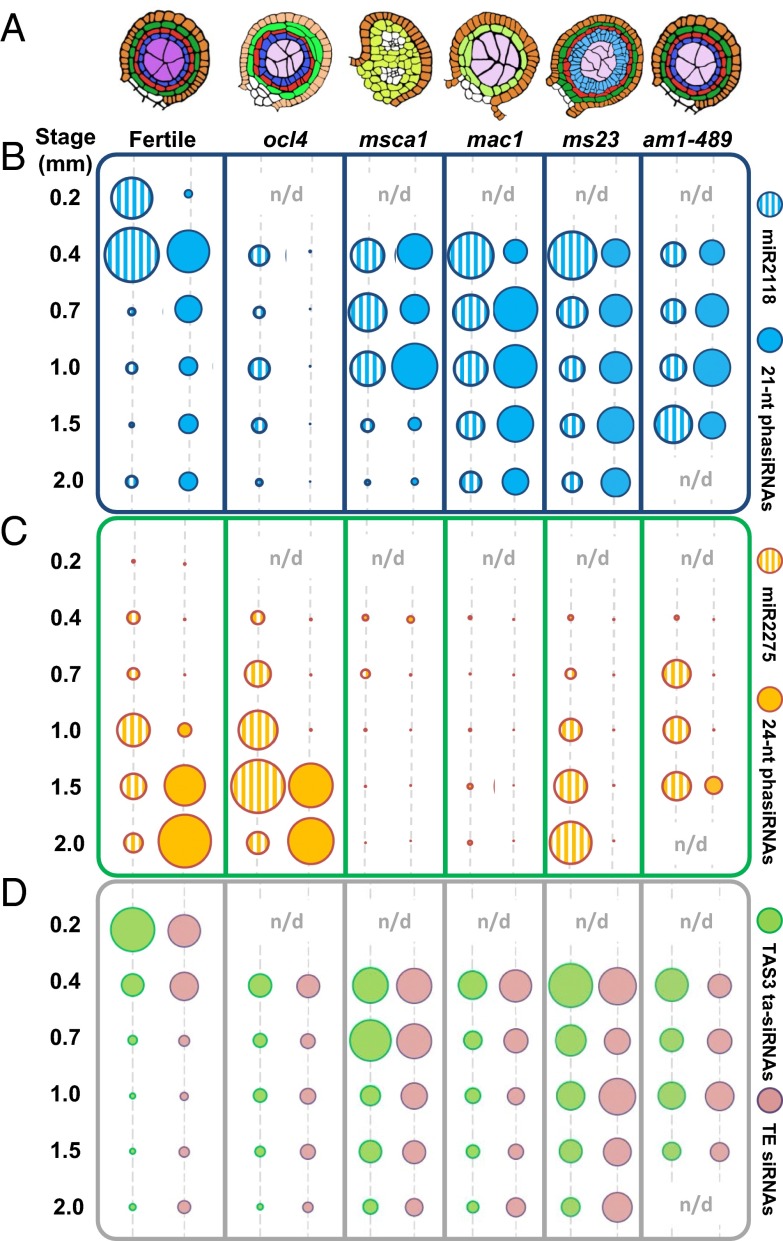
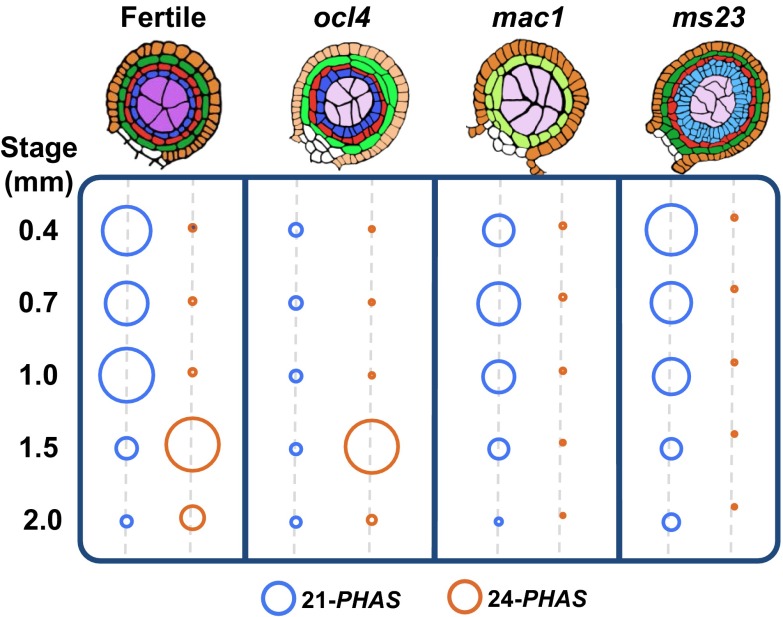
To gain insight into cell type contributions to phasiRNA production, RNAs were analyzed from developmental mutants defective in specific anther cell types (Fig. 3A and Dataset S3) (20). OCL4 (OUTER CELL LAYER 4) is an epidermal-specific transcription factor repressing periclinal divisions in the adjacent subepidermal endothecial cells (21), presumably through a mobile signal (22). ocl4 anthers lack all 21-nt premeiotic phasiRNAs despite containing reduced but robust levels of the miR2118 trigger (Fig. 3B). Because ocl4 accumulates other RDR6/DCL4 products such as TAS3 ta-siRNAs (Fig. 3D), the defect could be in the production of 21-PHAS precursors. Indeed, RNA-seq of 0.4- to 2.0-mm anthers showed that ocl4 lacks 21-PHAS transcripts (Fig. 4 and Fig. S4). ocl4 has nearly normal timing and abundances of miR2275, 24-PHAS precursors, and 24-nt meiotic phasiRNAs (Figs. 3C and 4), indicating their independence from both epidermal regulation and the premeiotic phasiRNA pathway. Furthermore, in msca1 (male sterile converted anther 1), in which mutant organs retain anther shape but no anther lobe cell types exist except the epidermis (23), there were near-normal levels of premeiotic phasiRNAs, with prolonged, elevated levels of miR2118 (Fig. 3B and Fig. S3B). We conclude, based on ocl4 and msca1 analysis, that a differentiated anther epidermis is necessary and sufficient for premeiotic phasiRNA biogenesis.
Fig. 3.
Impact of maize male-sterile mutants on the accumulation of miRNA triggers, PHAS precursors, and phasiRNAs. (A) Illustration of cell layer organization in fertile, ocl4, msca1, mac1, ms23, and ameiotic1 anther lobes at 0.7 mm. Color key as in Fig. 2A. (B) Quantification of 21-nt phasiRNAs and miR2118 in fertile, ocl4, msca1, mac1, ms23, and am1-489 mutants; colors as in Fig. 2B. All dots with the same color were normalized together (and across the genotypes) to permit comparison across all time points and genotypes. (n/a, not available) (C) Quantification of 24-nt phasiRNAs and miR2275 triggers in fertile and mutant anthers. (D) TAS3-derived ta-siRNAs and TE-derived siRNAs are shown as control sRNA.
Fig. 4.
Impact of maize male-sterile mutants on the accumulation of PHAS precursors profiled by RNA-seq. Quantification of 21-PHAS and 24-PHAS in fertile, ocl4, mac1, and ms23; colors as in Fig. 2C.
A Tapetal Layer, but Not Meiocytes, Is Required for Producing Meiotic phasiRNAs.
To further explore cell requirements, additional male-sterile mutants were analyzed. mac1 (multiple archesporial cells 1) mutants have excessive AR cells that mature and start meiosis, but typically the mutant anthers have only a single, undifferentiated subepidermal cell population (22); ms23 (male sterile 23) mutants have a normal endothecium and middle layer but pretapetal cells divide periclinally, forming an abnormal, undifferentiated bilayer (Fig. 3A) (24). The mac1 and ms23 anthers as well as msca1 lack meiotic phasiRNAs (Fig. 3C and Fig. S3B), suggesting that a specified tapetal layer is required for meiotic phasiRNAs. Interestingly, whereas mac1 lacks both miR2275 and 24-PHAS transcripts, ms23 retains nearly normal miR2275 levels but is missing 24-PHAS precursors (Fig. 4 and Fig. S4). Together with the conclusions from analysis of ocl4, these data suggest that in both premeiotic and meiotic phasiRNA pathways, the production of the miRNA triggers and the PHAS precursors are separately regulated.
To test whether normal meiocytes are required for phasiRNAs production, we sequenced anther sRNAs from ameiotic1 (am1), in which somatic lobe cells are normal but PMCs or meiocytes are defective (25). Two am1 alleles, am1-489 (PMCs conduct mitosis instead of meiosis) and am1-praI (meiocytes arrest in prophase I), have premeiotic and meiotic phasiRNAs (Fig. 3 B and C and Fig. S3C). Additionally, RDR6 transcripts, a shared biogenesis factor for both phasiRNA types, are not detected in fertile germinal cells by in situ hybridization (19). Therefore, phasiRNA production does not require normally differentiated germinal cells.
Spatial Distribution of phasiRNA Pathway Components.
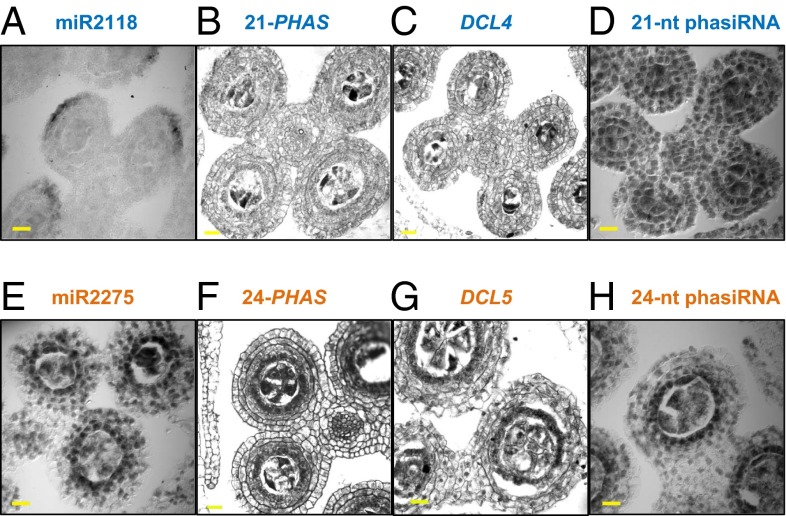
To further examine the spatial distribution of phasiRNAs, in situ localizations were performed on fertile anthers. miR2118 accumulated in a distal epidermal arc at 0.4 mm (Fig. 5A and Fig. S5A), precisely where the OCL4 transcription factor is expressed (21). The 21-PHAS precursors and DCL4 transcripts are slightly enriched in AR cells, whereas 21-nt premeiotic phasiRNAs localized in all anther cell layers (Fig. 5 B–D and Fig. S5A). Key biogenesis components for the 24-nt meiotic phasiRNAs were enriched in the tapetum as well as meiocytes (Fig. 5 E–H and Fig. S5 B and C). Therefore, the in situ results further support the distinct niches of the epidermis and tapetum in phasiRNA biogenesis. In addition, the separation of components required for biogenesis of premeiotic phasiRNAs suggests movement of one or more factors. The later-appearing meiotic phasiRNAs require tapetal differentiation, where biogenesis components colocalize. Tapetal cells are crucial for anther function: They secrete nutrients to support meiosis and later build the outer pollen coat. Because ARs and PMCs contain meiotic phasiRNAs, we speculate that these RNAs may be an additional type of “cargo” that tapetal cells supply to developing meiocytes.
Fig. 5.
Localization of phasiRNA biogenesis components in developing anthers. Small RNA in situ hybridization with a probe for (A) miR2118, (D) premeiotic phasiRNA from 21-PHAS_NO142, (E) miR2275, and (H) meiotic phasiRNA from 24-PHAS_NO132. Regular mRNA in situ hybridization with probes for (B) 21-PHAS_NO142, (C) DCL4, (F) 24-PHAS_NO132, and (G) DCL5. (Scale bar, 20 μm, for all images.)
phasiRNAs Lack Sequence Complementarity to TEs.
Plant miRNAs and ta-siRNAs trigger target mRNA cleavage; such cleaved sites can be validated in bulk using parallel analysis of RNA ends (PARE) (26). To investigate possible targets of phasiRNAs, we constructed PARE libraries from several anther stages and mature pollen (Dataset S1). Sequencing confirmed cleavage of 21- and 24-PHAS precursors by miR2118 and miR2275, respectively (Dataset S4). From the ∼9 million predicted phasiRNA-target pairs of the 1,000 most abundant premeiotic phasiRNAs, fewer than 1% showed a PARE signal. For the gene targets that showed PARE signals, GO analysis did not yield significant enrichment in any particular pathway or process (Dataset S4). These results are consistent with an earlier conclusion from rice that 21-nt premeiotic phasiRNAs lack obvious targets (9). Furthermore, an absence of premeiotic or meiotic phasiRNAs in ocl4, mac1, and ms23 does not result in TE transcript accumulation (Dataset S5). The massive complexity of phasiRNAs and the lack of obvious target or association with transposons suggest that phasiRNAs function distinctively from miRNAs, ta-siRNAs, or hc-siRNAs.
Discussion
Binding Partners of phasiRNAs.
Although plants lack PIWI-clade ARGONAUTEs that bind piRNAs, the plant AGO family has diversified extensively with 10 AGO members in Arabidopsis (27), 17 in maize (28, 29), and 19 in rice (Fig. S6) (30). Some AGO members are specifically expressed in flowers (29, 30) and are further enriched in either somatic or germinal cells of anthers (19, 31, 32). Presumably, this AGO expansion reflects a functional diversification of plant small RNAs for roles specific to anther developmental stages and cell types.
Recently it has been shown that the rice Argonaute MEL1 (13) binds 21-nt phasiRNAs (14). The closest homolog of MEL1 in maize is AGO5c (Fig. S6A). Maize AGO5c is highly expressed in 0.7-mm anthers, after premeiotic phasiRNAs peak (Dataset S6). Therefore, AGO5c is likely the binding partner of premeiotic phasiRNAs in maize. The binding partner of meiotic phasiRNAs has not yet been reported. Based on transcriptional analysis of laser-microdissected cell types (19, 32), plus the RNA-seq and microarray profiling of different anther stages (Dataset S6), we found that the expression profile of maize AGO18b matches the expression timing of meiotic phasiRNAs. Both AGO18b transcripts and proteins are enriched in the tapetal and meiotic cells (29). Because it mirrors the distribution and timing of meiotic phasiRNAs, and like them is a recently evolved gene absent in dicots, AGO18b is strongly implicated as the partner of the meiotic phasiRNAs.
Proposed Functions of phasiRNAs.
Although phasiRNAs lack sequence complementarity to TEs, they may have the capacity for genome surveillance of reproductive somatic and/or germinal cell transcripts, similar to what has been reported for Caenorhabditis elegans piRNAs (also known as 21U-RNAs) (33). In flowering plants, TE silencing pathways are heavily redundant to ensure genome integrity. For example, even in the maize rdr2/mop1 mutant in which 24-nt hc-siRNAs are missing, there are only modest changes in TE expression (34). Given their large genomes containing many repetitive elements, the grasses may have evolved additional pathways operating through the phasiRNAs to regulate the TEs. It is also plausible that the phasiRNAs guard the anther somatic and germinal cell genomes against attack by pathogens such as viruses, fungi, or oomycetes, or even protect against horizontal transfer or retropositioning of their nucleic acids such as TEs.
Alternatively, phasiRNAs may serve as mobile signals coordinating anther development. Anthers lack an organizing center, in contrast to the meristem regions of shoots and roots. Meristems organize a continuum of developmental stages displaced from the stem cell population, whereas anthers “self-organize” tissue layers and the entire organ progresses through development as one unit with high fidelity and temporal regularity (15, 18, 32). The potential movement of phasiRNAs from the site of biogenesis to neighboring cell layers (Fig. S7) is reminiscent of the TE-derived siRNAs in Arabidopsis pollen, produced in vegetative nuclei, and transported into sperm nuclei (7). Rather than participating in TE silencing, however, phasiRNAs may coordinate cell-type–specific expression by an as yet unknown pathway. RDR6 is responsible for the production of both 21-nt and 24-nt phasiRNAs in rice (11). Interestingly, the RDR6-dependent trans-acting siRNAs in Arabidopsis demonstrated relatively high mobility (35), further support for the concept that phasiRNAs could act as mobile signals within developing anthers.
Although both miR2275 and meiotic phasiRNAs have only been reported in grass species (8–10), miR2118 is present in dicots. The primary miR2118 targets in dicots are NB-LRR pathogen-defense genes; the 21-nt phasiRNAs produced from the NB-LRR mRNAs function in trans and in cis, and they are expressed constitutively (16). Therefore, miR2118 and the 21-nt phasiRNAs it triggers have evolved distinct functions in dicot and grass lineages, representing to our knowledge the first case of neofunctionalization among plant miRNAs. One of the two major subgroups with the NB-LRR gene family, the TIR-NB-LRRs, is not found in grass genomes, perhaps hinting at an origin for the miR2118-targeted 21-PHAS precursors. The origin of miR2275 is unknown, but DCL5 is most similar to DCL3, and was earlier named DCL3b (9, 36). Both miR2275 and DCL5 are absent from dicot genomes, suggesting their recent derivation within the grasses or within related monocots.
Putative Convergent Evolution of Grass phasiRNAs and Mammalian piRNAs.
Male reproduction in mammals is also characterized by a high abundance of two classes of small RNAs with accumulation patterns tightly restricted to specific cell types and developmental stages. These small RNAs are known as PIWI-interacting RNAs, or piRNAs. Maize phasiRNAs that we have described and more generally those of grasses share notable similarities with mammalian piRNAs (Table 1), an intriguing case of putative convergent evolution to produce novel classes of small RNAs in male germinal cells and somatic tissues. PhasiRNAs and mammalian piRNAs both exist in two size classes: The shorter size class occurs premeiotically and the longer size accumulates during meiosis. Thus far, neither the grass phasiRNAs nor the majority of mammalian piRNAs have a defined role. This parallelism is an evolutionary puzzle, as is the origin of miR2275, DCL5, and the meiotic phasiRNAs in grasses.
Table 1.
Parallels between grass phasiRNAs and mammalian piRNAs
| Characteristic | Grass (maize) phasiRNA* | Mammalian piRNA† | ||
| Organ | Anther | Testis | ||
| Developmental stage | Premeiotic | Meiotic | Prepachytene | Pachytene |
| Peak timing | 0.4-mm anther | 2.0-mm anther | 12.5 dpp | 17.5 dpp |
| Size | 21 nt | 24 nt | 26–27 nt | 29–30 nt |
| AGO partner | AGO5c‡ | AGO18b‡ | MILI, MIWI2 | MIWI |
| Master regulator | miR2118 | miR2275 | Unknown | A-myb |
| Number of loci | 463 | 176 | ∼900 | ∼100 |
| Abundance | Very high | |||
| Distribution | Present on all chromosomes; most loci are clustered | |||
| Precursor | Noncoding, Pol II transcripts | |||
| Single copy? | Nearly all | |||
| Repeat associated? | Few | |||
| Impacts on fertility | Misregulated in sterile mutants; biogenesis defects can cause sterility | |||
| Targeting | The majority lack complementarity to TEs or other loci | |||
| Function(s) | Unknown | Mainly unknown; a subset silence TEs | ||
What mammals and flowering plants share is a high investment in their progeny. Fertilized embryos are retained within the maternal body and supported by nutritive accessory organs (placenta or endosperm) that do not exist in predecessor taxa. We consider it likely that the piRNAs of mammals and the phasiRNAs of the grasses are contributors to the quality of the male contribution in reproduction, healthy sperm. Despite the fundamental differences between mammalian testes and grass anthers, the parallels in evolving two classes of piRNAs and phasiRNAs, in developmental timing before and during meiosis, the very high abundance, the numerous loci, and lack of obvious mRNA targets suggest that there are considerable evolutionary advantages in each kingdom for these systems for producing small RNAs during male reproduction.
Experimental Procedures
Plant Materials.
Fertile anthers of the W23 inbred line, mac1 and msca1, were introgressed five times into W23, ocl4 in the A188 inbred background, ms23 in the ND101 background, ameiotic1-489 (50% B73 + 25% A619 + 25% mixed other or unknown), and am1-praI allele (75% A619 + 25% mixed other or unknown) were grown in Stanford, CA under greenhouse conditions. Anthers were dissected and measured using a micrometer as previously described (15).
RNA Isolation, Library Construction, and Illumina Sequencing.
Total RNA was isolated using Tri reagent (Molecular Research Center) or Plant RNA reagent (Invitrogen). Small RNA and RNA-seq libraries were constructed using TruSeq Small RNA Sample Prep kits and TruSeq RNA Sample Prep kits (Illumina). PARE libraries were constructed as previously described (37). All libraries were sequenced on an Illumina HiSeq 2000 instrument at the Delaware Biotechnology Institute.
Data Handling and Bioinformatics.
Ninety-six small RNA libraries were constructed and sequenced, using input materials and generating read counts as described in Dataset S1. Approximately 2 billion small RNA sequences were obtained after removing adapters and low-quality reads, with lengths between 18 and 34 nt. After excluding those matching to structural RNAs (tRNA or rRNA loci), ∼1.5 billion small RNA reads were mapped perfectly (no mismatches) back to the reference genome of maize, version AGPv2 (38). Mapping was performed using Bowtie (39). Any read with more than 50 perfect matches (“hits”) to the genome was excluded from further analysis. Abundances of small RNAs in each library were normalized to “TP10M” (transcripts per 10 million) based on the total count of genome-matched reads in that library.
Genome-wide phasing analysis was performed as previously described (16). To achieve maximum sensitivity, all small RNA libraries were combined to create a union set for detection of the phased distribution of small RNAs. Analysis of phasing was performed in fixed intervals from 19 to 25 nt. Only the 21- and 24-nt intervals generated a result that was significantly higher than background. As a final check of loci with phasing scores higher than or equal to 25, the scores and abundances of small RNAs from each high-scoring locus were graphed and checked visually to remove false positives such as miRNA or unfiltered t/rRNA loci. This yielded 463 loci generating 21-nt premeiotic phasiRNAs and 176 loci generating 24-nt meiotic phasiRNAs.
Forty-four RNA-seq libraries were made from 0.4- and 0.7-mm anthers of W23 (wild type), ocl4, and mac1. After trimming, RNA-seq reads were mapped to the reference genome using TopHat (40). Abundances of RNA-seq reads in each library were normalized to TP10M based on the total genome-matched reads of that library.
Five PARE libraries were made from materials described in Dataset S1. Data analysis and target validation were performed as previously described (4). In brief, we defined two windows flanking each predicted target site (i): a small window “WS” of 5 nt (cleavage site, ±2 nt), and (ii) a large window “WL” of 31 nt (cleavage site, ±15 nt). Cleavage sites were filtered to retain only those for which WS/WL ≥ 0.5 in the PARE library to remove noisy signals. Target prediction and scoring was done using CleaveLand2 (41).
In Situ Hybridization.
Small RNAs were detected using locked-nucleic acid (LNA) probes synthesized by Exiqon. Samples were vacuum fixed using 4% (vol/vol) paraformaldehyde and submitted to the histology laboratory at the A. I. DuPont Hospital for Children for paraffin embedding. We followed published protocols for the prehybridization, hybridization, posthybridization, and detection steps (42). Probe sequences for hybridization are listed in Dataset S7.
For PHAS locus and gene transcripts, in situ hybridizations were performed as previously described (19). Probes were synthesized from PCR fragments amplified from genomic DNA followed by transcription using the DIG RNA Labeling kit (T7/SP6) (Roche). Probe sequences for hybridization are listed in Dataset S7.
Confocal Microscopy.
Confocal images were taken with a Zeiss LSM780 using a C-Apochromat 40× (N.A. = 1.3) oil immersion objective lens at the Delaware Biotechnology Institute. Sections were excited at 458 nm and autofluorescence was detected using a 578– to 674-nm band pass detector. We also used the same laser for in situ hybridizations, using differential interference contrast.
Small RNA Detection with Splinted Ligation-Mediated miRNA Detection.
miRNAs and phasiRNAs were detected using the USB miRNAtect-It miRNA labeling and detection kit (Affymetrix) as previously described (43, 44). Each experiment used 10 µg of total RNA. Analyses were performed using oligonucleotides listed in Dataset S6.
Phylogenetic Analysis.
Protein sequences of 17 AGOs in maize, 19 in rice, and 10 in Arabidopsis were downloaded from the National Center for Biotechnology Information and aligned using MEGA6 (45). The evolutionary history was inferred using the neighbor-joining method by MEGA6 (45) and configured by Figtree (tree.bio.ed.ac.uk/software/figtree/).
Supplementary Material
Acknowledgments
We thank Tim Kelliher and Rachel Egger for dissecting 0.2-mm anthers, Sandra Mathioni for library construction, and Dong-Hoon Jeong for the small RNA blot. This project was supported by the US National Science Foundation Plant Genome Research Program. J.Z. is a Life Science Research Foundation postdoctoral fellow, sponsored by the Gordon and Betty Moore Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE52290 for RNA-seq data, GSE52293 for small RNA data, and GSE52297 for PARE data).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418918112/-/DCSupplemental.
References
- 1.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 2.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 4.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 5.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 6.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19(8):1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song X, et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69(3):462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- 10.Arikit S, Zhai J, Meyers BC. Biogenesis and function of rice small RNAs from non-coding RNA precursors. Curr Opin Plant Biol. 2013;16(2):170–179. doi: 10.1016/j.pbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Song X, et al. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 2012;71(3):378–389. doi: 10.1111/j.1365-313X.2012.05001.x. [DOI] [PubMed] [Google Scholar]
- 12.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Nonomura K, et al. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19(8):2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komiya R, et al. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78(3):385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 15.Kelliher T, Walbot V. Emergence and patterning of the five cell types of the Zea mays anther locule. Dev Biol. 2011;350(1):32–49. doi: 10.1016/j.ydbio.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 18.Kelliher T, Walbot V. Hypoxia triggers meiotic fate acquisition in maize. Science. 2012;337(6092):345–348. doi: 10.1126/science.1220080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelliher T, Walbot V. Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 2014;77(4):639–652. doi: 10.1111/tpj.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timofejeva L, et al. Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3 (Bethesda) 2013;3(2):231–249. doi: 10.1534/g3.112.004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernoud V, et al. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009;59(6):883–894. doi: 10.1111/j.1365-313X.2009.03916.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang CJ, et al. Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development. 2012;139(14):2594–2603. doi: 10.1242/dev.077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaubal R, et al. The transformation of anthers in the msca1 mutant of maize. Planta. 2003;216(5):778–788. doi: 10.1007/s00425-002-0929-8. [DOI] [PubMed] [Google Scholar]
- 24.Chaubal R, et al. Two male-sterile mutants of Zea Mays (Poaceae) with an extra cell division in the anther wall. Am J Bot. 2000;87(8):1193–1201. [PubMed] [Google Scholar]
- 25.Nan GL, et al. Global transcriptome analysis of two ameiotic1 alleles in maize anthers: Defining steps in meiotic entry and progression through prophase I. BMC Plant Biol. 2011;11:120. doi: 10.1186/1471-2229-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26(8):941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 27.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian Y, et al. Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 2011;30(7):1347–1363. doi: 10.1007/s00299-011-1046-6. [DOI] [PubMed] [Google Scholar]
- 29.Zhai L, et al. Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J Integr Plant Biol. 2014;56(11):1042–1052. doi: 10.1111/jipb.12205. [DOI] [PubMed] [Google Scholar]
- 30.Kapoor M, et al. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics. 2008;9:451. doi: 10.1186/1471-2164-9-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh M, et al. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23(2):443–458. doi: 10.1105/tpc.110.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, et al. Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda) 2014;4(6):993–1010. doi: 10.1534/g3.113.009738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HC, et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150(1):78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia Y, et al. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5(11):e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Felippes FF, Ott F, Weigel D. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 2011;39(7):2880–2889. doi: 10.1093/nar/gkq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margis R, et al. The evolution and diversification of Dicers in plants. FEBS Lett. 2006;580(10):2442–2450. doi: 10.1016/j.febslet.2006.03.072. [DOI] [PubMed] [Google Scholar]
- 37.Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods. 2014;67(1):84–90. doi: 10.1016/j.ymeth.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 38.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 39.Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10(11):R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25(1):130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javelle M, Timmermans MC. In situ localization of small RNAs in plants by using LNA probes. Nat Protoc. 2012;7(3):533–541. doi: 10.1038/nprot.2012.006. [DOI] [PubMed] [Google Scholar]
- 43.Jeong DH, Green PJ. Methods for validation of miRNA sequence variants and the cleavage of their targets. Methods. 2012;58(2):135–143. doi: 10.1016/j.ymeth.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Jeong DH, et al. Massive analysis of rice small RNAs: Mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell. 2011;23(12):4185–4207. doi: 10.1105/tpc.111.089045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.