Significance
The detection of specific genes in fixed cells was first accomplished in 1969 by Gall and Pardue. The development of analogous methods applicable to living cells is now at hand. At the forefront of this advance (2013–2014), we and other investigators have used transcription activator-like effectors (TALEs) conjugated with fluorescent proteins to tag genomic loci in live cells. More recently, the CRISPR/Cas9 system has provided a more flexible approach to targeting specific loci. In this paper, we describe the labeling of human genomic loci in live cells with three orthogonal CRISPR/Cas9 components, allowing multicolor detection of genomic loci with high spatial resolution, which provides an avenue for barcoding elements of the human genome in the living state.
Keywords: 4D nucleome, telomeres, pericentromeric DNA, chromosomes
Abstract
The intranuclear location of genomic loci and the dynamics of these loci are important parameters for understanding the spatial and temporal regulation of gene expression. Recently it has proven possible to visualize endogenous genomic loci in live cells by the use of transcription activator-like effectors (TALEs), as well as modified versions of the bacterial immunity clustered regularly interspersed short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system. Here we report the design of multicolor versions of CRISPR using catalytically inactive Cas9 endonuclease (dCas9) from three bacterial orthologs. Each pair of dCas9-fluorescent proteins and cognate single-guide RNAs (sgRNAs) efficiently labeled several target loci in live human cells. Using pairs of differently colored dCas9-sgRNAs, it was possible to determine the intranuclear distance between loci on different chromosomes. In addition, the fluorescence spatial resolution between two loci on the same chromosome could be determined and related to the linear distance between them on the chromosome’s physical map, thereby permitting assessment of the DNA compaction of such regions in a live cell.
A major advance in molecular cell biology occurred with the introduction of in situ nucleic acid hybridization in cytological preparations (1–3). The importance of this method, which has evolved into ever-more sensitive versions over the years, cannot be overstated. Subsequently, methods were introduced for fluorescently labeling specific chromosomal sites in live cells (4, 5) and, more recently, yet other approaches have been developed. The first of these newer methods used transcription activator-like effectors (TALEs) conjugated with fluorescent proteins to label specific chromosomal loci in living cells (6–8; reviewed in ref. 9). The second method arose from a repurposing of the bacterial immunity clustered regularly interspersed short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system for gene editing in eukaryotic cells (10–14), in which programmable DNA recognition and cleavage of targeted genes has been achieved by using the Cas9 nuclease in collaboration with target site-customized single-guide RNAs (sgRNAs).
In parallel with its deployment for gene editing, the CRISPR/Cas9 system also has been used for sequence-specific gene regulation using nuclease-inactive Cas9 (dCas9) (15), with this version of Cas9 subsequently applied to the visualization of genomic loci in live cells through fusion with a fluorescent protein (16, 17). Resolving different interchromosomal or intrachromosomal loci within the nucleus with CRISPR technology has remained challenging, however, because of the need for dual labels. Here we describe a multicolor CRISPR system to specifically and differentially label various pairs of chromosomal loci simultaneously, allowing estimation of the interlocus distances in living human cells.
Results
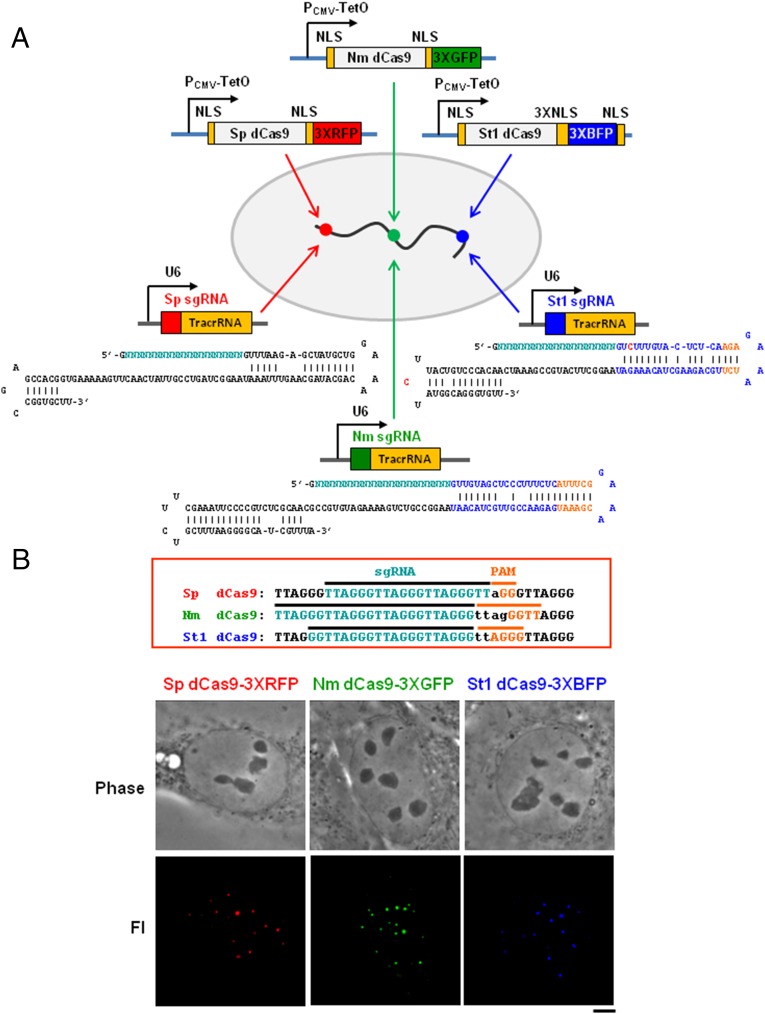
The fluorescent labeling of endogenous genomic DNA by CRISPR using Streptococcus pyogenes (Sp) dCas9-GFP has greatly simplified study of the spatial organization of the genome in live cells, owing to the simplicity with which recognition complexes can be programmed to target a wide array of different genomic sequences. Although genomic labeling with a single color is possible with the Sp dCas9 system (16, 17), multiple orthogonal labels, which have not been described previously, are needed to determine the relative position and movement of pairs of loci during cellular processes of interest. To address these needs, we chose to optimize for genomic labeling orthogonal Cas9 variants from three bacterial species, S. pyogenes, Neisseria meningitidis (Nm), and Streptococcus thermophilus (St1), which have been used for editing and gene regulation in human cells without cross-talk in cognate sgRNA binding (18). A schematic of the key components and overall multicolor CRISPR labeling strategy is shown in Fig. 1A. The three sites depicted on the chromosome are simply hypothetical loci, colored to convey the particular fluorescent protein. Catalytically inactive forms of the Cas9 endonuclease (notated in all figures as “dCas9”; Materials and Methods) from three bacterial species fused to green fluorescent protein (GFP), red fluorescent protein (RFP), or blue fluorescent protein (BFP) were expressed from tetracycline-regulated plasmids, and sgRNAs cognate with each Cas9 ortholog were designed for the human telomere DNA repeat and expressed with U6 promoters.
Fig. 1.
The multicolor CRISPR labeling system exploiting Cas9 orthologs. (A) Designs of dCas9 (“d” for “nuclease-dead”) ortholog-fluorescent proteins: S. pyogenes Sp dCas9, N. meningitidis Nm dCas9 and S. thermophilus St1 dCas9 fused to NLSs and under the control of the CMV-TetO promoter. The cognate sgRNAs Sp sgRNA, Nm sgRNA, and St1 sgRNA are under the control of the U6 promoter, and their sequences are shown below, with the mutations in red and hairpin extensions in orange. Directed by the appropriate sgRNA, different chromosomal loci are expected to become fluorescently labeled with any one of the three spectral versions of dCas9. In the diagram, such hypothetical targeted sequences are indicated by the color matching a particular dCas9/sgRNA ortholog. (B) sgRNAs and PAM sequences designed for targeting telomeres by each of the dCas9 orthologs Sp, Nm, and St1. The panels below are phase-contrast (Upper) and fluorescent signals for telomeres (Lower) images for human U2OS cells expressing the indicated dCas9-FPs and cognate sgRNAs. (Scale bar: 5 μm.)
Also shown in Fig. 1A are the guide RNA sequences of each sgRNA, as well as the vicinal protospacer adjacent motif (PAM) elements essential for Cas9 recognition of one strand of the telomeric repeat, the DNA sequence that we chose for the initial development and optimization of the method. Fig. 1B shows the labeling of telomeres in the highly aneuploid human U2OS cell line using the three Cas9 orthologs Sp, Nm, and St1, fused to RFP, GFP, and BFP, respectively, along with their cognate sgRNAs. Numerous fluorescent foci were observed with each pair of dCas9-FPs and sgRNAs. These results were obtained following a comprehensive optimization of the system, which was critical to obtain robust labeling of a genomic locus from each dCas9-FP. Optimized parameters included the choice of the promoter driving the expression of dCas9 (Fig. S1A), number of fluorescent proteins fused in tandem (Fig. S1B), length of guide RNAs (Fig. S2A), PAM sequence choice (Figs. S2B and S3C), choice of the sequence composition of the sgRNAs (Figs. S3 A and B and S4 B and C), and number of nuclear localization signals (NLSs) (Fig. S4A).
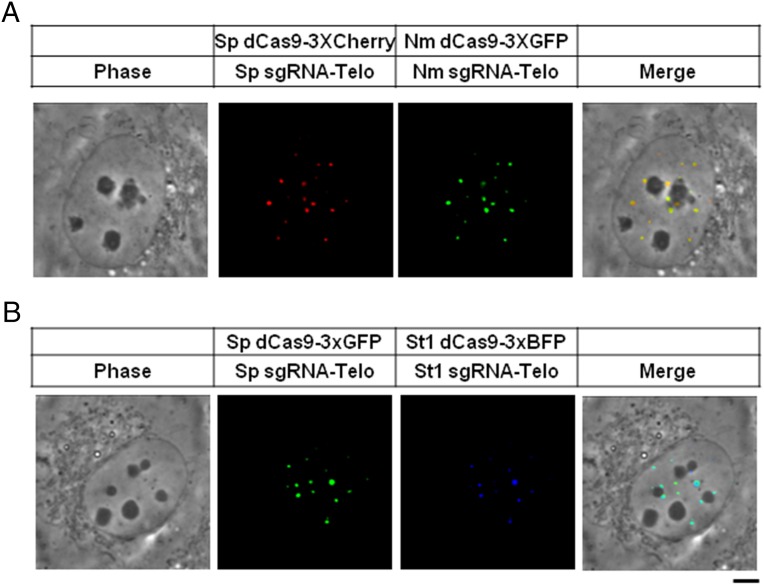
To determine whether two orthogonal CRISPR systems can be applied simultaneously to label a given chromosomal locus in a live cell, we coexpressed Sp dCas9-3xCherry and Nm dCas9-3xGFP along with their cognate sgRNAs targeting one strand of the human telomeric repeat. As shown in Fig. 2A, this resulted in identical patterns of discrete nuclear foci with the two colors displaying complete spatial coincidence, indicating that both Sp dCas9 and Nm dCas9 can find and co-occupy a given telomere. Similar results were obtained by coexpression of Sp dCas9-3xGFP and St1 dCas9-3xBFP in conjunction with their cognate sgRNAs (Fig. 2B). These results also indicate that the expression levels of the two orthogonal systems are sufficiently similar such that neither is at such vast excess that it saturates a given telomeric repeat.
Fig. 2.
Application of multicolor CRISPR to telomere labeling in live cells. (A) Sp dCas9-3xCherry (Middle Left) and Nm dCas9-3xGFP (Middle Right) were coexpressed in U2OS cells together with their cognate sgRNAs (Sp sgRNA-Telo and Nm sgRNA-Telo) targeting to telomeres. Shown are phase-contrast images (Left) and the merged fluorescence image (Right). (B) Sp dCas9-3xGFP (Middle Left) and St1 dCas9-3XBFP (Middle Right) were coexpressed in U2OS cells together with their cognate sgRNAs (Sp sgRNA-Telo and St1 sgRNA-Telo) targeting to telomeres. Shown are phase-contrast images (Left) and the merged fluorescence image (Right). (Scale bar: 5 μm.)
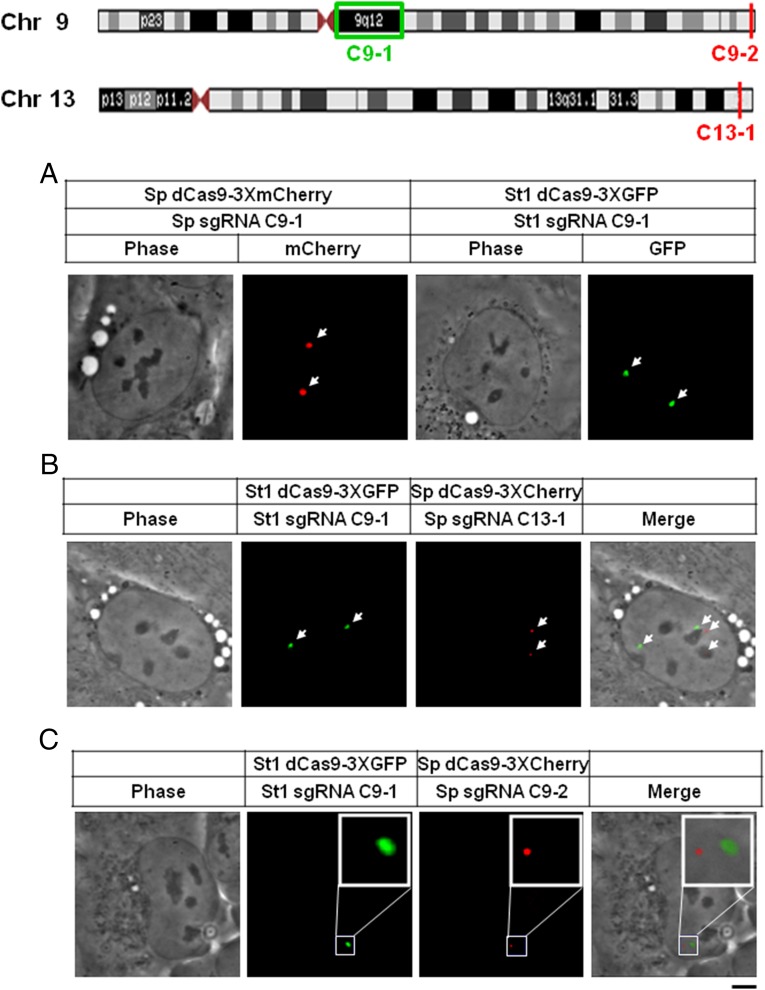
We next explored the application of this dual-color system to labeling of different repeated, chromosome-specific sequences, which would allow assessment of the interchromosomal or intrachromosomal spatial relationships between such loci. On the basis of bioinformatics mining of repeats throughout the human genome (Materials and Methods), we first chose those diagrammed in Fig. 3. C9-1 and C9-2 are pericentromeric and subtelomeric repeats, respectively, located specifically on chromosome 9. C13-1 is a subtelomeric repeat on the long arm of chromosome 13 that is present solely on that chromosome. We designed sgRNAs for each of these repeats and expressed them in U2OS cells or diploid RPE-1 cells, together with dual color pairs of dCas9/sgRNA. Fig. 3A shows the labeling of C9-1 with Sp or St1 dCas9 orthologs with cognate sgRNAs in RPE-1 cells, a diploid human cell line (Fig. S5). Two specific foci were observed with each pair of dCas9-FPs and sgRNAs, indicating that these cells are in G1, which was confirmed by a 3D view of the chromosome 9 pericentromeric locus C9-1 (Fig. S6). The proximity of two interchromosomal loci, C9-1 and C13-1, is shown in Fig. 3B. The two copies of each chromosomal locus were detected as a pair of foci, and each pair was clearly quite far apart from the other in the 3D nuclear volume as regards this interchromosomal situation. Fig. 3C addresses the intrachromosomal propinquity of C9-1 and C9-2, revealing them to be ∼2 μm apart. This cytological distance corresponds to the known distance of 75 megabase pairs (Mbp) between these two loci on the physical map of chromosome 9. As far as we know, this is the first time such an interrogation of two endogenous intrachromosomal loci has been made in a live cell.
Fig. 3.
Interchromosomal and intrachromosomal loci labeling by multicolor CRISPR. Pericentromeric satellite DNA in human chromosomes 9 (C9-1) and repeat sequences unique to chromosome 9 (C9-2) or chromosome 13 (C13-1) are shown at the top. (A) Plasmids encoding Sp dCas9-3XCherry or St1 dCas9-3XGFP were cotransfected into RPE-1 cells with cognate sgRNAs (Sp sgRNA C9-1 or St1 sgRNA C9-1) targeting to pericentromeric satellite DNA in human chromosome 9. Phase-contrast images (Left and Middle Right) and fluorescent images (Middle Left and Right) are shown. (B and C) St1 dCas9-3xGFP and Sp dCas9-3XCherry were coexpressed in RPE-1 cells with their cognate sgRNAs (St1 sgRNA C9-1 and Sp sgRNA C13-1) (B) or in U2OS cells with cognate sgRNAs (St1 sgRNA C9-1 and Sp sgRNA C9-2) (C). Shown are phase-contrast (Left in B and C), GFP (Middle Left in B and C), Cherry (Middle Right in B and C), and the merged fluorescence (Right in B and C) images. (Scale bar: 5 μm.)
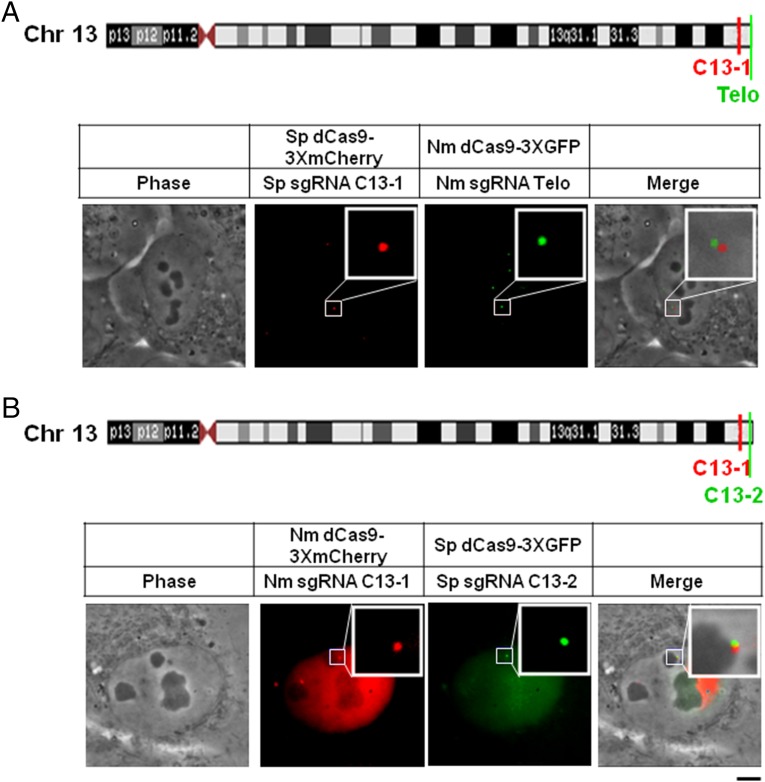
We next wanted to test whether this method might have an even greater degree of interlocus resolution on a chromosome. For this, we turned to two sites that are even more closely situated than the pair interrogated in Fig. 3C. Fig. 4A shows the location of the aforementioned repeat C13-1 in relation to the telomere of the long arm of this chromosome, constituting a distance of 2 Mbp on the physical map, in contrast to the 75- Mbp distance between the loci examined in Fig. 3C. When we expressed a dual-color pair of dCas9 and cognate sgRNAs targeting this latter pair of loci, we found two very proximal foci (Fig. 4A), indicating that this labeling method reports intrachromosomal distances in live cells that are compatible with the chromosome’s physical map.
Fig. 4.
Spatial resolution of subtelomeric loci in chromosome 13 and the adjacent telomere. (A) Diagram of the locations of C13-1 and the telomere on the long arm of chromosome 13. Sp dCas9-3xCherry (Middle Left) and Nm dCas9-3xGFP (Middle Right) were coexpressed in U2OS cells together with their cognate sgRNAs (Sp sgRNA-C13-1 and Nm sgRNA-Telo). Shown are a phase-contrast image (Left), the Cherry and GFP fluorescence images (Middle), and the merged image (Right). (B) Diagram of the locations of C13-1 and C13-2. Nm dCas9-3xCherry (Middle Left) and Sp dCas9-3xGFP (Middle Right) were coexpressed in U2OS cells together with their cognate sgRNAs (Nm sgRNA-C13-1 and Sp sgRNA-C13-2). Shown are a phase-contrast image (Left), the Cherry and GFP fluorescence images (Middle), and the merged fluorescence image (Right). In the right panel, most of the uniformly dispersed fluorescence background (Middle Left) was removed by increasing the threshold to facilitate observation of the merged signal. (Scale bar: 5 μm.)
We also targeted a second pair of loci in the subtelomeric region of the long arm of chromosome 13, the aforementioned C13-1 and a vicinal repeat, C13-2, which lies 1.9 Mbp away (Fig. 4B). Here an extremely close proximity of the two signals was observed, with a striking overlapping zone at the interface (see the yellow zone in “Merge”). The fact that these two pairs of loci with very similar distances between them (1.9 Mbp vs. 2.0 Mbp) are nonetheless observed as distinctly more distal (C13-1 and telomere) or proximal (C13-1 and C13-2) indicates that the dual-color CRISPR method that we have developed can discriminate different degrees of chromatin compaction over distances as short as 1.9–2.0 Mbp on the physical map.
Discussion
In this study, we have developed a CRISPR-based multicolor labeling system that enables the imaging of multiple endogenous genomic DNAs simultaneously and allows measurement of the proximity of different genomic loci in live cells. By optimizing three orthogonal Cas9 systems with different PAM specificities, our system also provides users with expanded targeting flexibility overall for the labeling of genomic loci.
This system has a number of potential applications. The multicolor CRISPR labeling method described here promises to be a useful tool for probing dynamic interactions of intrachromosomal and interchromosomal domains during cell cycle progression, during epigenetic regulation, or in response to cellular stimuli. We are presently applying this approach to map the intrachromosomal locations of repeated sequences that are unique to each human chromosome. In principal, the method also can be used to interrogate nuclear lamina-associated domains and chromosome capture-based topologically associating domains, and to permit the visualization of such events as translocations and cancer-associated chromosome shattering and rearrangements (i.e., chromothripsis) in live cells.
As with any method, there are important technical points. One such point is the method’s sensitivity of detection, because CCD cameras operate near or at the quantum efficiency limit, and the brightness of each fluorescent protein is also limited. Based on our previous TALEColor study (7) and other considerations, we estimate that 150–200 FP molecules need to be stationed on a given chromosomal site to create a detectable signal (i.e., ∼50–70 dCas9-FP/sgRNA complexes when using 3XGFP). At the time that this manuscript was submitted for publication, a protein-tagging system (SunTag) was reported in which a repeating peptide array can recruit up to 24 copies of GFP (19). If successfully deployed, this would significantly enhance the CRISPR/dCas9 chromosome labeling signals and extend the detection limit.
Along with sensitivity, another consideration of course is the method’s spatial resolution. We detected two pairs of chromosomal loci that lie 1.9 or 2.0 Mbp in the DNA physical map with a standard fluorescence microscopy system in which the diffraction-limited spatial resolution was at the classical Abbé limit of ∼0.2 μm (Fig. 4). One can only imagine the extent to which this multicolor CRISPR/Cas9 method for detecting the propinquity of intrachromosomal or interchromosomal loci might be enhanced by superresolution microscopy, although formidable background issues might need to be addressed, as has been the case when combining fixed-cell FISH with superresolution microscopy (20). With respect to mapping intrachromosomal loci, the compaction of DNA in the interphase chromosome has been a subject of research on the nucleus for decades. As mentioned earlier, in two cases of intrachromosomal loci that we studied (Fig. 4), where the distance between loci on the DNA physical map differed by only 0.1 Mbp, the microscopically resolved distances differed to a much greater degree, suggesting a significant difference in DNA compaction in the two regions. Thus, the dual-color CRISPR method described here may provide a new tool in the study of interphase DNA compaction in live cells, particularly for examining genomic regions that may have unusual chromatin structure.
There is resurgent interest in the repeated sequence elements of the human genome as their “post-Human Genome Project” characterization continues (21). For many applications, we envision the need to identify sets of repeated DNA sequences that are unique to a single locus on a chromosome or are present at multiple sites on only one chromosome in the complement (i.e., as a bar code). We have already identified such sequences in the present study (C9-1, C9-2, and C13-1), and are currently compiling a much larger set of these and testing them in our multicolor CRISPR/Cas9 system. With the method described here, it also should be feasible to carry out dual-color labeling of two single-copy genomic loci using tiled arrays of sgRNAs across suitable regions of each locus and their different-colored dCas9 orthologs or, using the same strategy, between a single-copy locus and a repeated sequence lying nearby or more distant. In addition, Sp Cas9 was recently adapted for programmable RNA recognition and cleavage (22). The simultaneous use of Sp Cas9 for RNA recognition and other Cas9 orthologs for DNA recognition could provide a new tandem toolkit for studying the 4D nucleome and the regulation of eukaryotic gene expression across a broad landscape of cell types and stages of development, differentiation, and human disease.
Materials and Methods
Construction of Cas9 Expression Plasmids.
Human codon-optimized dCas9 (nuclease-dead) from S. pyogenes (23), N. meningitidis, and S. thermophilus (18) were fused to 1XGFP, 2XGFP, 3XGFP, 3Xcherry, or 3XBFP and then subcloned into pHAGE-DEST lentiviral vectors. To optimize the promoters for U2OS and RPE-1 cells, the EF1α promoter in the pHAGE- EF1α-DEST vector was replaced by EFS, SFFV, and CMV-TetO promoters, respectively, resulting in pHAGE-EFS-DEST, pHAGE-SFFV-DEST, and pHAGE-TO-DEST. To optimize nuclear localization, 2X SV40 NLSs were fused to S. pyogenes dCas9 and N. meningitidis dCas9, and up to 6X SV40 NLSs were fused to S. thermophilus dCas9. A list of the Cas9 fusion proteins constructed is presented in Table S1. All of the plasmids reported here will be deposited at Addgene.
Construction of sgRNA Expression Vectors.
The sgRNA expression vector is based on the pLKO.1 lentiviral expression plasmid containing the CcdB gene between two BbsI sites for inserting guide sequences into the sgRNAs. An optimized sgRNA (16) for S. pyogenes Cas9 was subcloned into pLKO.1-Hygro, resulting in pLH-SpsgRNA2. Nm sgRNA mutants for N. meningitidis Cas9 were subcloned into pLKO.1-Hygro, resulting in pLH-NmsgRNAm1 and pLH-NmsgRNA1.1. St1 sgRNA mutants for S. thermophilus Cas9 were subcloned into pLKO.1-Hygro, resulting in pLH-St1sgRNAm1, pLH-St1sgRNAm7, pLH-St1sgRNA1.1, pLH-St1sgRNA2.1, and pLH-St1sgRNA3.1.
A rapid-guide RNA expression plasmid construction protocol was optimized as follows. A pair of oligodeoxynucleotides (2 μM) was denatured at 95 °C for 3 min and then cooled to room temperature. Then a 10-μL reaction mixture of oligos (4 pM), sgRNA vectors (100 ng), BbsI (4 units), T7 ligase (300 units), and ATP (1 mM) in CutSmart Buffer (New England Biolabs) was incubated at 37 °C for 10 min in the single tube and then directly subjected to transformation using CcdB as a counterselection. The sgRNA vectors are listed in Table S2; guide RNA sequences, in Table S3.
Cell Culture and Transfection.
U2OS cells were cultured at 37 °C in DMEM (Life Technologies) supplemented with 10% (vol/vol) FBS. RPE-1 cells were kindly provided by Dr. Yumi Uetake (Department of Molecular, Cell, and Cancer Biology, University of Massachusetts Medical School) and cultured at 37 °C in DMEM:F12 medium supplemented with 10% (vol/vol) FBS. The RPE-1 cell line was karyotyped by Quest Diagnostics. For imaging, cells were grown on 35-mm glass-bottom dishes (MatTek). In experiments with U2OS cells, 150 ng of dCas9 plasmid and 750 ng of sgRNA plasmid were cotransfected using Lipofectamine 2000 (Life Technologies), after which the cells were incubated for another 48 h. For RPE-1 cells, 50 ng of dCas9 plasmid and 250 ng of sgRNA plasmids were cotransfected using Lipofectamine LTX (Life Technologies).
Fluorescence Microscopy.
The microscope stage incubation chamber was maintained at 37 °C (24), and phase-contrast and fluorescence microscopy was performed as described previously (7). RFP was excited at 556/20 nm (wavelength/bandwidth), and its emission was collected in a 630/91-nm channel. GFP was excited at 470/28 nm, and its emission was collected in a 512/23-nm channel. BFP was excited at 387/11 nm, and its emission was collected using a 464/23-nm filter. Imaging data were acquired and analyzed with MetaMorph acquisition software (Molecular Devices). Thresholds were set based on the ratios of nuclear focal signals to background nucleoplasmic fluorescence.
Mining for Chromosome-Specific Repeats.
The human reference genome hg19 was downloaded from the UCSC genome browser (genome.ucsc.edu). The gaps (regions labeled with “Ns”) in chromosomes 9 and 13 were replaced with randomly generated nucleotides. The Tandem Repeat Finder bioinformatics tool (25) was used to identify tandem repeats in chromosomes 9 and 13. Highly conserved repeats with copy number >100 were selected as candidates for CRISPR labeling. 23-mers in the tandem repeats ending with GG were used for designing Sp sgRNAs for C9-1, C9-2, C13-1, and C13-2; 28-mers ending with GCTT were used for designing Nm sgRNAs for C13-1; and 26-mers ending with GGAA were used for designing St1 sgRNA for C9-1.
The detailed parameters for each targeted repeats are as follows: C9-2 is located in subtelomeric region q34.3 of chromosome 9 with the location chr9: 140459676–140463065, and contains 115 copies of sgRNA target sites. C13-1 consists of 177 copies of sgRNA target sites, located in subtelomeric region q34 of chromosome 13 with the location chr13: 112930173–112968847. C13-2 consists of three neighboring tandem repeats in q34 of chromosome 13 chosen to achieve a combined 102 copies of sgRNA target sites with the following locations: chr13: 114793685–114795158, with 22 copies of target sites; chr13: 114848979–114852850, with 57 copies of target sites; and chr13: 114903631–114905572, with 23 copies of target sites. The BLAT alignment tool (26) was used to verify the chromosome specificity of these sgRNA target sites in the human genome. Finally, C9-1 was a tandem array of GGAAT repeats, which are highly concentrated in the pericentromeric region of chromosome 9 (27). Details of all guide RNA sequences are provided in Table S3.
Supplementary Material
Acknowledgments
We thank Yumi Uetake (University of Massachusetts Medical School) for the RPE-1 cells, Anne Higgins (Quest Diagnostics) for RPE-1 cell karyotyping, Claire Vourc’h (Institut Albert Bonniot) for the C9-1 sequences, and Jeffrey Nickerson (University of Massachusetts Medical School) for help with confocal microscopy. This research was supported in part by US National Science Foundation Grant MCB 0445841 (to T.P.).
Footnotes
Conflict of interest statement: The authors are named inventors on a patent application related to this work filed by the University of Massachusetts.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420024112/-/DCSupplemental.
References
- 1.Gall JG, Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci USA. 1969;63(2):378–383. doi: 10.1073/pnas.63.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John HA, Birnstiel ML, Jones KW. RNA-DNA hybrids at the cytological level. Nature. 1969;223(5206):582–587. doi: 10.1038/223582a0. [DOI] [PubMed] [Google Scholar]
- 3.Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci USA. 1969;64(2):600–604. doi: 10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11(6):250–257. doi: 10.1016/s0962-8924(01)02000-1. [DOI] [PubMed] [Google Scholar]
- 5.Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12(6):439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 6.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol. 2013;20(11):1321–1324. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 7.Ma H, Reyes-Gutierrez P, Pederson T. Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors. Proc Natl Acad Sci USA. 2013;110(52):21048–21053. doi: 10.1073/pnas.1319097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanisch K, et al. Targeting and tracing of specific DNA sequences with dTALEs in living cells. Nucleic Acids Res. 2014;42(6):e38. doi: 10.1093/nar/gkt1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pederson T. Repeated TALEs: Visualizing DNA sequence localization and chromosome dynamics in live cells. Nucleus. 2014;5(1):28–31. doi: 10.4161/nucl.28143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5(2):163–172. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markaki Y, et al. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture: 3D structured illumination microscopy of defined chromosomal structures visualized by 3D (immuno)-FISH opens new perspectives for studies of nuclear architecture. BioEssays. 2012;34(5):412–426. doi: 10.1002/bies.201100176. [DOI] [PubMed] [Google Scholar]
- 21.Altemose N, Miga KH, Maggioni M, Willard HF. Genomic characterization of large heterochromatic gaps in the human genome assembly. PLOS Comput Biol. 2014;10(5):e1003628. doi: 10.1371/journal.pcbi.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141(1):219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson MR, Pederson T. RNA traffic and localization reported by fluorescence cytochemistry. In: Richter JD, editor. Analysis of mRNA Formation and Function. Academic; New York: 1997. pp. 341–359. [Google Scholar]
- 25.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eymery A, Souchier C, Vourc’h C, Jolly C. Heat shock factor 1 binds to and transcribes satellite II and III sequences at several pericentromeric regions in heat-shocked cells. Exp Cell Res. 2010;316(11):1845–1855. doi: 10.1016/j.yexcr.2010.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.