Significance
Perennial plants display seasonal cycles of growth. For example, in the trees of boreal temperate forests, growth must cease prior to the advent of winter and cold hardiness must be acquired to survive extreme low temperature. Growth cessation and activation of transcriptional programs underlying adaptive responses associated with cold hardiness are photoperiodically controlled. We show that the evolutionarily conserved protein FD implicated in the control of flowering mediates photoperiodic control of seasonal growth in trees by forming a complex with FLOWERING LOCUS T (FT) protein. FD genes of hybrid aspen display neofunctionalization and, in contrast to Arabidopsis, have evolved functions that are independent of their interaction with FT, such as transcriptional control of the adaptive response and bud maturation pathways in trees.
Keywords: growth cessation, bud set, seasonal growth, adaptive response, hybrid aspen
Abstract
A complex consisting of evolutionarily conserved FD, FLOWERING LOCUS T (FT) proteins is a regulator of floral transition. Intriguingly, FT orthologs are also implicated in developmental transitions distinct from flowering, such as photoperiodic control of bulbing in onions, potato tuberization, and growth cessation in trees. However, whether an FT–FD complex participates in these transitions and, if so, its mode of action, are unknown. We identified two closely related FD homologs, FD-like 1 (FDL1) and FD-like 2 (FDL2), in the model tree hybrid aspen. Using gain of function and RNAi-suppressed FDL1 and FDL2 transgenic plants, we show that FDL1 and FDL2 have distinct functions and a complex consisting of FT and FDL1 mediates in photoperiodic control of seasonal growth. The downstream target of the FT–FD complex in photoperiodic control of growth is Like AP1 (LAP1), a tree ortholog of the floral meristem identity gene APETALA1. Intriguingly, FDL1 also participates in the transcriptional control of adaptive response and bud maturation pathways, independent of its interaction with FT, presumably via interaction with ABSCISIC ACID INSENSITIVE 3 (ABI3) transcription factor, a component of abscisic acid (ABA) signaling. Our data reveal that in contrast to its primary role in flowering, FD has dual roles in the photoperiodic control of seasonal growth and stress tolerance in trees. Thus, the functions of FT and FD have diversified during evolution, and FD homologs have acquired roles that are independent of their interaction with FT.
The evolutionarily conserved protein FLOWERING LOCUS T (FT) plays a key role in the control of flowering in plants (1). Because FT lacks DNA binding activity, its interaction with transcription factors, such as FD (2–4) and more recently identified BRANCHED1 (BRC1) (5), is critical for the formation of protein complexes to control flowering via transcriptional control of downstream targets [e.g., floral meristem identity genes, transcription factors APETALA1 (AP1) and OsMADS1]. Whereas the FT–FD complex promotes flowering at the shoot apical meristem (4), BRC1 appears to delay floral transition at the axillary meristem (5). These findings indicate that depending upon its interaction partner, FT-containing complexes can have distinct roles. The structure of FT–FD complex elucidated in rice has shown that a 14-3-3 protein mediates the interaction between rice FT homolog HEADING DATE 3a (Hd3a) and FD homolog OsFD1 via the C-terminally located SAP (serine alanine proline) motif in OsFD1 to generate the active nuclear localized florigen activation complex (3).
Interestingly, FT homologs are also involved in the control of diverse developmental transitions distinct from flowering, such as tuberization in potatoes (6), bulb formation in onions (7), stomatal opening (8), and photoperiodic control of seasonal growth in trees (9–11). These observations suggest that complexes of FT are not only important to the control of flowering but have a broader functionality. However, the mechanisms underlying the functional diversity of the FT complexes and how they can participate in the control of developmental pathways distinct from flowering are not well understood because, in contrast to FT, FD or BRC1 (or other interactors of FT), which provide DNA binding ability to the complexes of FT and thus are crucial for the function of these complexes, have not been well characterized in pathways distinct from flowering. Thus, the role of complexes of FT and their mode of action in these pathways remain poorly understood.
Cessation of growth before the onset of winter is essential for the survival of trees growing in temperate and boreal forests during subsequent periods of low temperatures (12, 13). Growth cessation is a photoperiodically controlled process (14, 15). In “short days” (SDs), when day length falls below the critical threshold allowing growth, elongation growth ceases (14–16). Moreover, the lamina of the last leaf primordia formed before the perception of SDs is aborted, and its stipules develop into bud scales that enclose the embryonic leaves inside a bud at the apex (17). Thus, bud set results in termination of the emergence of new leaves. SDs also concomitantly induce transcriptional changes resulting in the so-called “adaptive response,” a metabolic shift toward the accumulation of vegetative proteins and acquisition of cold hardiness that protect the shoot apical meristem and embryonic leaves (18).
Several studies have shown that the tree ortholog of Arabidopsis FT plays an important role in photoperiodic control of growth cessation (9–11). For example, down-regulation of FT2 expression by SDs is a key early event in the induction of growth cessation (9). Importantly, functional studies have shown that overexpression of FT2 or its paralog FT1 in hybrid aspen (P. tremula × tremuloides) attenuates SD responses and abolishes growth cessation, whereas suppression of FT expression leads to earlier growth cessation than in WT plants (9–11). The MADS box transcription factor LAP1 (Like-APETALA1), a tree homolog of AP1, has been recently identified as a target of SDs downstream of FT (19). Like FT, LAP1 overexpression delays SD-mediated growth cessation, whereas its down-regulation causes early growth cessation in SDs (19). These findings clearly demonstrate that as in flowering, FT plays a central role in photoperiodic control of seasonal growth in trees.
In addition to FT, FD is a key component of the FT–FD complex, given its role in selection of downstream targets by the FT–FD complex. However, in contrast to FT, there are few studies addressing the role of FD other than in flowering (8, 20). Therefore, to improve understanding of the photoperiodic control of seasonal growth, we initiated analysis of FD homologs in the model tree hybrid aspen. Functional analyses in hybrid aspen of the two closely related FD homologs FD-like 1 (FDL1) and FD-like 1 (FDL2) show that they have distinct roles. We show that FDL1 interacts with FT2 to form a complex that mediates in photoperiodic control of seasonal growth. Most previously published data suggest that FD functions primarily in a complex with FT (2–4, 20). However, we show that FDL1 has additional roles in trees, independent of its interaction with FT. Thus, the function of the FT–FD complex has diversified during evolution, and FD homologs have acquired novel roles that are independent of its interaction with FT.
Results
FDL1 and FDL2 Have Distinct Functions.
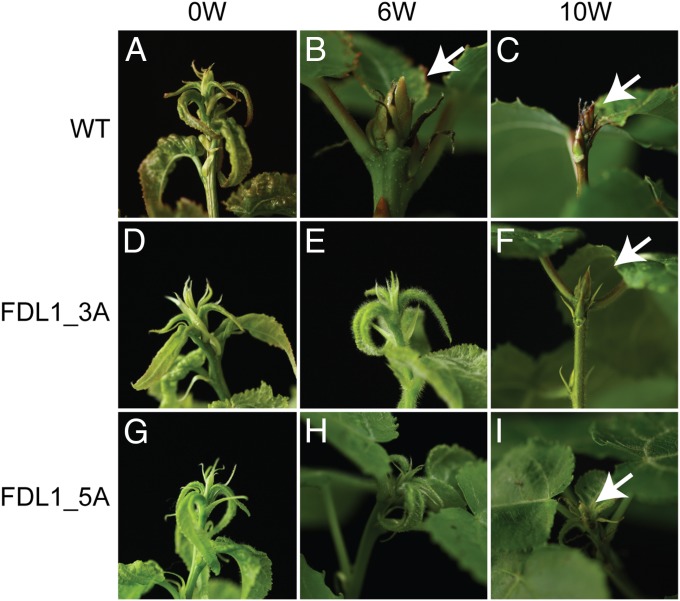
We cloned full-length cDNAs for two highly similar FD-like genes, FDL1 and FDL2, from hybrid aspen, which encode 168-aa and 302-aa proteins, respectively. The larger size of FDL2 is due to an insertion at the C terminus (Fig. S1). Both FDL1 and FDL2 are highly similar to FD proteins from other plants and contain the functionally important conserved threonine (T)/SAP motif at the C terminus (20) (Fig. S1). To investigate the functions of FDL1 and FDL2, we generated hybrid aspen plants overexpressing FDL1 (FDL1oe) or FDL2 (FDL2oe), as well as plants in which FDL1 (FDL1RNAi) or FDL2 (FDL2RNAi) expression was reduced (Fig. S2). In contrast to FDL1oe, FDL2oe plants were severe dwarfs when grown in long days (Fig. S3). We then investigated the function of FDL1 and FDL2 in photoperiodic control of growth cessation. Although WT plants clearly set buds, we saw no evidence of bud set in FDL1oe plants after 6 wk of SDs (Fig. 1). The FDL1oe plants eventually set buds after 10 wk of SDs, indicating that they can respond to SDs, but more slowly than WT plants (Fig. 1). The perception of SDs results in cessation of new leaf formation; thus, the number of leaves formed between initiation of SD treatment and bud set provides a sensitive measure of SD response (16, 19). Using this assay, we observed that FDL1oe plants formed more, whereas FDL1RNAi plants formed fewer, leaves than WT controls between initiation of the SD treatment and growth cessation (Fig. S4A). Thus, FDL1 overexpression leads to a delay, whereas FDL1 down-regulation leads to a faster SD response compared with the WT. In contrast to FDL1 transgenic plants, neither FDL2oe nor FDL2RNAi plants displayed any significant difference from WT in SD-mediated growth cessation (Fig. S4 B and C). Thus, despite high similarity, FDL1 and FDL2 have distinct functions, and FDL1, but not FDL2, mediates in photoperiodic control of growth.
Fig. 1.
Bud formation in WT and FDL1oe (lines 3A and 5A) plants. A, D, and G represent plants growing in long days (LD). WT plants had ceased growth and developed buds (B and C), but the FDL1oe plants had not (E and H). (F and I) FDL1oe plants set buds after 10 wk (W) of SDs. Arrows indicate apical buds.
FDL1 and FDL2 Can Interact with FT.
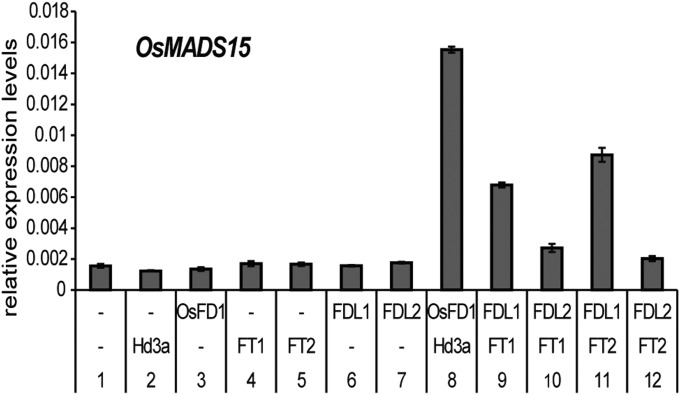
Despite being highly similar, FDL1, but not FDL2, is involved in photoperiodic control of growth. This observation promoted us to investigate the molecular basis of the functional difference between FDL1 and FDL2. We investigated whether the difference between FDL1 and FDL2 stems from differences in their ability to interact with FT or, alternatively, whether both FDLs can interact with FT but the FDL1–FT complex differs in function from the FDL2–FT complex. To differentiate between these two possibilities, we used two approaches. First, we used bimolecular fluorescence complementation (BiFC) (21) to investigate interaction between hybrid aspen FT and FDL proteins (Fig. S5). BiFC assays indicate that YFP fluorescence is observed only when FT1 or FT2 fused to C-terminal YFP is coexpressed with FDL1 or FDL2 fused to N-terminal YFP (Fig. S5, top four rows), but not when FT1 or FT2 fused to C-terminal YFP is coexpressed with N-terminal YFP or when FDL1 or FDL2 fused to N-terminal YFP is coexpressed with C-terminal YFP controls. Thus, both hybrid aspen FT1 and FT2 could interact with FDL1 and FDL2. Second, we used a rice protoplast system developed previously, in which the activation of OsMADs15 (a rice AP1 homolog) expression, a downstream target of the FT–FD complex, is used as a transcriptional read-out for the ability of FT and FD to act together (3). We expressed FT1 and FT2 and FDL1 and FDL2 alone or together. As controls, we expressed Hd3a or OsFD1 alone (as a negative control) or coexpressed Hd3a and OsFD1 cDNAs (as a positive control) in rice protoplasts. Hd3a and OsFD1 coexpression activated OsMADS15 transcription, unlike expression of Hd3a or OsFD1 alone, indicating that activation of OsMADs15 depends on the expression of both proteins [Fig. 2, compare lane 8 with lanes 2 and 3 in agreement with previous data (3)]. When coexpressed with hybrid aspen FT1 and FDL1, but not FDL2, activated OsMADS15 transcription (Fig. 2, compare lane 9 with lane 10). Similarly, when cotransformed with hybrid aspen FT2, only FDL1 activated OsMADS15 (Fig. 2, compare lane 11 with lane 12). In contrast, the expression of FT1, FT2, FDL1, or FDL2 alone did not lead to activation of OsMADs15 expression. Moreover, FDL1 (but not FDL2) was also able to activate OsMADS15 transcription when coexpressed with rice Hd3a (Fig. S6A). Expression of hybrid aspen FT1, FT2, FDL1, and FDL2 cDNAs in the protoplasts was confirmed by RT-PCR (Fig. S6B). Taken together, these results show that although FDL1 and FDL2 can interact with FT1 and FT2, as demonstrated by BiFC assays, the FDL1–FT and FDL2–FT complexes differ from each other, because only FT–FDL1 can activate OsMADS15 expression in the transcriptional read-out assays. Importantly, the differences between the FDL1–FT and FDL2–FT complexes suggested by the transcriptional read-out assays support the observed phenotypic differences between FDL1 and FDL2 transgenic plants, showing that FDL1, but not FDL2, mediates photoperiodic control of growth. To confirm the interaction of FDL1 with FT, we expressed hybrid aspen FDL1 cDNA in the Arabidopsis thaliana fd-2 mutant (Fig. S7). FDL1 expression (Fig. S7A) could partially suppress the late flowering phenotype of the fd-2 mutant (Fig. S7B), indicating that, indeed, FDL1 can function like Arabidopsis FD and also supporting the hypothesis that FDL1 interaction with FT proteins is important for its function in photoperiodic control of growth.
Fig. 2.
Analysis of interaction between hybrid aspen FD and FT homologs. FDL1 interacts with hybrid aspen FT1 and FT2 to activate OsMADS15 expression when coexpressed in rice protoplasts. Plasmid DNA for expressing FT and FD homologs from rice or poplar was transformed into rice protoplasts, and OsMADS15 induction was assayed by quantitative RT-PCR analysis. cDNAs expressed and the OsMADS15/ubiquitin expression ratio 24 h after the transformation are shown on the x and y axes, respectively. Error bars indicate SDs of triplicate measurements.
FDL1 Mediates in the Photoperiodic Regulation of LAP1.
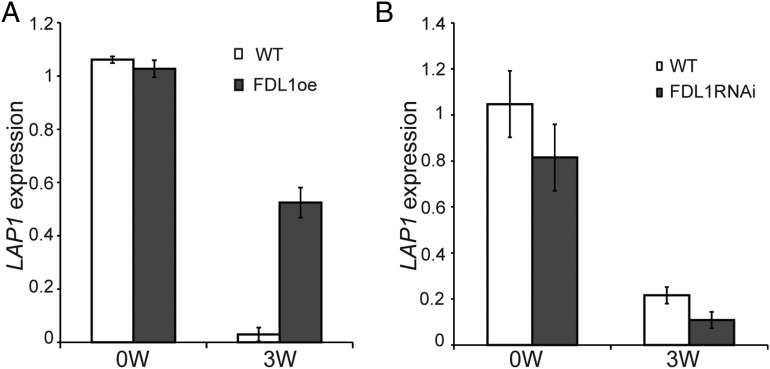
LAP1, a tree AP1 homolog, is targeted by SDs and is proposed to act downstream of FT in the photoperiodic control of growth in hybrid aspen (19). Moreover, inability of SDs to down-regulate LAP1 expression underlies the attenuation of growth cessation responses in FT overexpressors (19). These findings prompted us to investigate the regulation of LAP1 by SDs in FDL1oe and FDL1RNAi plants (Fig. 3). LAP1 expression was reduced after 3 wk of SD treatment in WT apices, whereas SD-induced down-regulation of LAP1 was severely attenuated in the FDL1oe plants (Fig. 3A). Conversely, there was a greater reduction in LAP1 expression after SD treatment in the FDL1RNAi plants than in WT controls (Fig. 3B). Thus, like FT, FDL1 clearly mediates in the photoperiodic control of LAP1 expression.
Fig. 3.
FDL1 mediates in the photoperiodic control of LAP1 expression. Expression of LAP1 in the WT and FDL1oe plants (A) and expression of LAP1 in the WT and FDL1RNAi plants after SDs (B). The duration of SDs is shown (in weeks) on the x axis, and LAP1 expression (relative to the reference gene TIP41-like, average for three biological replicates ± SE) is shown on the y axis.
SD Treatment Up-Regulates FDL1 Expression.
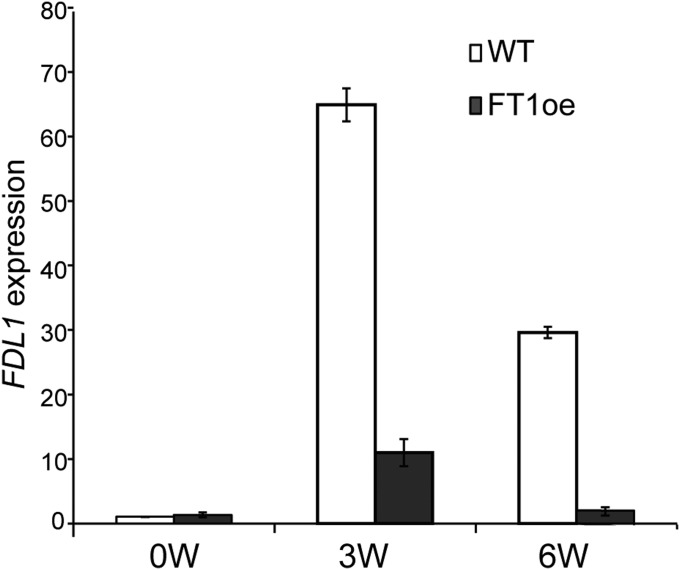
Down-regulation of FT2 expression by SDs leads to growth cessation (9, 11), but the possibility that SDs may also down-regulate FDL1 expression has not been tested. The observed attenuation of the growth cessation response in FDL1oe plants prompted us to investigate whether SD-induced growth cessation also involves down-regulation of FDL1 expression in the apex. Although FDL1 is clearly expressed in long days in the apex (Fig. 4 and Fig. S8), unexpectedly, we discovered that FDL1 expression is not down-regulated but up-regulated after SD treatment (Fig. 4). Importantly, this up-regulation of FDL1 expression after SD treatment was attenuated in FT1oe plants in which SD perception is defective (Fig. 4). These results indicate that SDs modulate FDL1 expression.
Fig. 4.
Photoperiodic control of FDL1 expression. SD induction of FDL1 expression is attenuated in FT1oe plants. The duration of SDs is shown (in weeks) on the x axis. Induction of FDL1 expression in SDs (relative to the reference gene TIP41-like, average for three biological replicates ± SE) is plotted relative to expression in long days (0 wk SDs) on the y axis.
FDL1 Mediates in Transcriptional Regulation of Adaptive Response and Bud Maturation Pathways.
The up-regulation of FDL1 expression after SD treatment suggested that it has functions in addition to its role in photoperiodic control of growth. Induction of FDL1 after SDs coincides with bud maturation and activation of the adaptive response (18). Therefore, we tested the hypothesis that FDL1 mediates SD-induced changes in the adaptive response and bud maturation pathways by monitoring expression of marker genes for the two pathways (18, 22, 23) in the WT and FDL1 transgenic plants. In WT plants, the expression of adaptive response markers, such as OSMOTIN (OSM) and LATE EMBRYOGENESIS ABUNDANT (LEA), was up-regulated after SD treatment, in accordance with earlier results (18) (Fig. 5A). Overexpression or down-regulation of FDL1 clearly affected regulation of these markers after SDs. OSM and LEA were more strongly expressed in FDL1oe plants than in WT plants after SDs (Fig. 5A). In contrast, the FDL1RNAi plants displayed the opposite pattern (i.e., OSM and LEA were expressed at a lower level than in WT plants after SDs) (Fig. 5B).
Fig. 5.
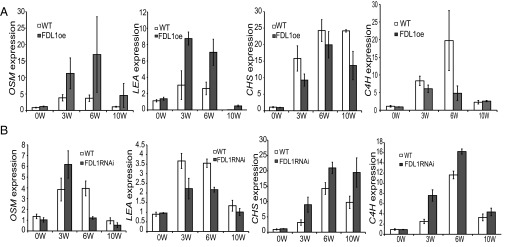
FDL1 mediates in SD-controlled adaptive response and bud maturation pathways. Expression pattern of markers for adaptive response (LEA and OSM) and bud maturation (CHS and C4H) in FDL1oe (A; line) and FDL1RNAi (B; line) apices are compared with WT plants. Expression of the cited genes is shown relative to the reference gene TIP41-like on the y axis (average for three biological replicates ± SE), and duration of SDs (in weeks) is shown on the x axis.
As mentioned, SDs induce bud maturation concomitantly with the adaptive response, and accumulation of phenylpropanoids in the bud scales is a good marker for this process (18). Accordingly, expression of bud maturation markers, such as CHALCONE SYNTHASE (CHS) and CINNAMATE ACID 4-HYDROXLASE (C4H), enzymes of the phenylpropanoid pathway, was up-regulated after SDs in WT plants (Fig. 5A), and the buds were darker in color (Fig. 1). Further, up-regulation of these markers was reduced in the FDL1oe plants at one or more time points after SDs (Fig. 5A), and their buds were greener (Fig. 1), whereas the FDL1RNAi plants again exhibited opposite changes in their expression (Fig. 5B). Thus, FDL1 mediates in transcriptional control of the adaptive response and bud maturation pathways.
ABI3 and FDL1 Interact and Share Overlapping Targets in Adaptive Response and Bud Maturation Pathways.
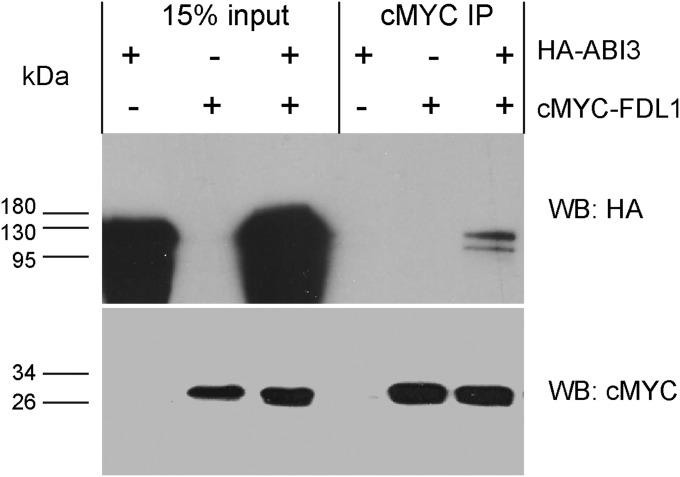
FDL1 belongs to the A-group of basic leucine zipper domain (bZIP) transcription factors, which are known to interact with other transcription factors, including ABI3 (24). ABI3 has been implicated in the transcriptional control of the adaptive response and bud maturation pathways (18). Interestingly, the phenotype of FDL1oe plants (e.g., green buds) (Fig. 1) is reminiscent of ABI3 overexpressors (18). Therefore, we tested the hypothesis that FDL1 and ABI3 could interact and function in the same pathway. To test the interaction between FDL1 and ABI3, we coexpressed HA-tagged ABI3 and c-Myc epitope-tagged FDL1 (c-Myc–FDL1) in Arabidopsis protoplasts. Our data indicate that c-Myc–FDL1 can coimmunoprecipitate HA-ABI3 (Fig. 6). We then confirmed the FDL1–ABI3 interaction using BiFC assays. BiFC assays demonstrate that YFP fluorescence is observed only when ABI3 fused to C-terminal YFP is coexpressed with FDL1 fused to N-terminal YFP (Fig. S9, top row). In contrast no YFP fluorescence is observed when ABI3 fused to C-terminal YFP or FDL1 fused to C-terminal YFP is coexpressed with N-terminal or C-terminal YFP controls. Thus, the BiFC assays also demonstrate that FDL1 and ABI3 can interact. The interaction between FDL1 and ABI3, and the phenotypic similarity of FDL1 and ABI3 plants, led us to investigate whether FDL1 and ABI3 could mediate in regulation of the same set of downstream targets. After SD treatment, changes in the expression of adaptive response and bud maturation marker genes in ABI3-overexpressing (ABI3oe) plants, relative to WT responses, were similar to those changes in expression observed in the FDL1oe plants (Fig. S10) (i.e., OSM and LEA were more strongly expressed, whereas C4H and CHS were more weakly expressed). These findings suggest that ABI3 and FDL1 impinge on the same set of downstream target genes of the adaptive response and bud maturation pathways.
Fig. 6.
Interaction of FDL1 and ABI3 proteins. c-Myc–FDL1 and HA-tagged ABI3 (HA-ABI3) were coexpressed in Arabidopsis protoplasts derived from cell suspension cultures. c-Myc–FDL1 was immunoprecipitated (IP) from the protein extracts using anti–c-Myc antibody, and HA-ABI3 was then assayed with anti-HA antibody by Western blot (WB). (Lower) c-Myc–FDL1 bound to the beads was revealed using anti–c-Myc antibody.
Discussion
FDL1 Mediates in Photoperiodic Control of Growth.
Previously demonstrated functions of FD homologs are largely confined to the control of flowering (2–4). Delayed growth cessation in FDL1oe plants and early growth cessation in FDL1RNAi plants in SDs demonstrate that the hybrid aspen FD homolog FDL1 mediates photoperiodic control of growth, a process distinct from flowering. Intriguingly, overexpression of FDL1 delays the SD response even when FT2 expression is highly reduced. This observation raises the possibility that FDL1 could act independent of FT in photoperiodic control of growth in contrast to flowering, in which FD interaction with FT is essential (2, 4). However, several lines of evidences suggest the contrary. First, FDL1 interacts with FTs and mediates in the regulation of the same downstream target, LAP1, as FT (19). Second, FDL1RNAi plants display early growth cessation in SDs like FTRNAi plants (9). Third, FDL1 overexpression delays growth cessation in SDs (when FT expression is highly reduced) but cannot abolish the response to SDs, suggesting the dependence of FDL1 on FT2 in photoperiodic control of growth. Also, SDs induce FDL1 expression in the WT, and if FDL1 acted independent of FT, SDs would be unable to induce growth cessation. Additionally, FDL1 suppresses the Arabidopsis fd-2 mutant phenotype, which would indicate that FDL1 must interact with FT like Arabidopsis FD. Finally, FT lacks DNA binding ability and must interact with a transcription factor to control gene expression, and all of the evidence would suggest that this transcription factor is FDL1 in photoperiodic control of growth. Therefore, we conclude that FDL1 interaction with FT mediates in photoperiodic control of growth and the delayed growth cessation in FDL1oe plants results from supraoptimal levels of FDL1 overcoming the lack of FT in SDs. Thus, the function of FD homologs is not limited to flowering control.
Functional Divergence of FDL1 and FDL2.
Most plants contain multiple FD homologs (20), but their functions, apart from flowering, are not well characterized. FDL1 and FDL2 are highly similar to FD homologs from other plants and contain the conserved S/TAP motif necessary for 14-3-3 protein-mediated interaction with FT (3, 4). In agreement with the role of this motif in mediating interaction between FT and FD, both FDL1 and FDL2 can interact with FT1 and FT2. However, intriguingly, transcriptional read-out assays suggest that FT–FDL1 and FT–FDL2 differ from each other. These results lead us to hypothesize that the difference in FDL1 and FDL2 functions may not be due to differences between them to interact with FT. Rather, it is more likely that FDL1–FT and FDL2–FT complexes have distinct roles. Indeed, FT complexes can have distinct roles depending upon their interaction partner, as shown for FT–FD and FT–BRC1 complexes (5). Importantly, this hypothesis is supported by the differential photoperiodic responses of FDL1 and FDL2 transgenics, as well as the growth habits of these transgenics under long days. Currently, what underlies the differences between FDL1–FT and FDL2–FT complexes is not entirely clear. However, it is worth noting that the 3D structure of FDL2 could be distinct from FDL1 due to the presence of a sequence at the C terminus in FDL2 that FDL1 lacks (Fig. S1). As a result of this terminal extension, the bZIP domain of FDL2 is much longer than FDL1, which can contribute to the transcriptional differences between these two FDLs. It is not unusual for similar proteins to have distinct functions, as is already evident in the FT/TFL family (25). Nevertheless, these observations raise the possibility that despite high overall similarity and ability to interact with FT, not all members of the FD family or FT–FD complexes have the same functions.
Neofunctionalization of FDL Genes in Hybrid Aspen.
Like hybrid aspen, Arabidopsis also has two closely related FD paralogs, both of which are involved in FT-mediated control of flowering (26). In contrast, neofunctionalization has occurred in the FD family in hybrid aspen. For example, FDL1 interacts with FT and mediates in photoperiodically controlled processes retaining the ancestral features of FD, whereas FDL2 can interact with FT but has acquired a role distinct from FDL1. Interestingly, FD partner FT has also undergone gene duplication in Populus and the two FT paralogs, like FD, are proposed to act in distinct processes (11). Whereas FT1 is proposed to be involved in flowering, FT2 mediates photoperiodic control of seasonal growth (11). However, differential function of FT paralogs is related to differential tissue and temporal expression patterns, with FT1 being expressed during flowering in the apex and FT2 during active growth in summer (11). In contrast, FDL1 and FDL2 have largely overlapping patterns of expression (Fig. S8); thus, neofunctionalization of FDL2 is presumably due to structural differences resulting from insertion at the C terminus in FDL2.
Control of LAP1 by the FT–FDL1 Module Is Involved in Photoperiodic Regulation of Growth.
The targets of the FT–FD module in pathways distinct from flowering are not known as yet. Therefore, the mechanisms whereby this regulatory module mediates in bulbing or seasonal growth in trees are not well understood. AP1 homolog LAP1 is a target of SDs, and its down-regulation is essential for SD-mediated growth cessation (19). Our data indicate that FDL1, like FT, mediates in the photoperiodic regulation of LAP1 (Fig. 3), suggesting that LAP1 is a downstream target of the FT–FDL1 module in photoperiodic control of growth in hybrid aspen trees. Interestingly, the FT–FD module also acts on AP1 in control of stomatal opening (8). Thus, the FT–FD module and its downstream target, AP1/LAP1, are conserved in the regulation of developmental pathways distinct from flowering, such as photoperiodic control of growth.
FT-Independent Role of FDL1.
The function of FD has been explored primarily in flowering (2–4) and in photoperiodic control of seasonal growth (this study), in which FD functions together with FT. Moreover, FD homologs can also function in pathways other than flowering (e.g., stomatal opening in Arabidopsis and overexpression of the rice FD homolog OsFD2 modulate leaf development) (8, 20). However, in contrast to these roles of FD, FDL1 functions in adaptive response and bud maturation is independent of its interaction with FT, because FT expression is negligible after SDs. Therefore, the independence of FDL1 from interaction with FT in these SD responses contrasts with OsFD2’s modulation of leaf development in rice, because control of the latter appears to involve OsFD2 interaction with the FT homolog Hd3a (20). Thus, not only can FD homologs function in processes other than flowering but, more importantly, some of these functions may be independent of their interaction with FT even in FD homologs, such as FDL1, that retain certain ancestral features of FD.
FDL1 Is Involved in SD-Mediated Transcriptional Control of the Adaptive Response and Bud Maturation.
As yet, little is known about the functions of FD homologs that are independent of their interaction with FT. Our data demonstrate that FDL1 participates in transcriptional control of the adaptive response pathway independent of FT. Interestingly, FDL1 transgenics share several phenotypic similarities with transgenic plants in which the expression of ABI3, a signaling intermediate in abscisic acid (ABA) responses, is perturbed (ref. 18 and this study). The phenotypic similarity between FDL1oe and ABI3oe plants, together with their overlapping downstream targets and coimmunoprecipitation and BiFC assays, suggests that FDL1 and ABI3 are components of a regulatory network underlying the SD-mediated transcriptional control of the adaptive response and bud maturation pathway. Such an interaction could also allow FDL1-mediated integration of photoperiodic and hormonal (ABA) signaling in control of the pathway.
FDL1 Mediates Coordination of Growth Cessation and Other SD-Controlled Responses.
Intriguingly, FT has antagonistic effects on FDL1 expression [e.g., up-regulation of FDL1 after SDs mirrors down-regulation of FT2, increased expression of FT (as in FT-overexpressing plants) suppresses SD-mediated increase in FDL1 expression]. To our knowledge, such an antagonistic effect of FT on FD has not been previously recorded, but it provides a possible mechanism for temporal coordination of the induction of growth cessation with concomitant transcriptional activation of the bud maturation and adaptive response pathways by the same environmental cues (SDs). In the long days, FDL1 and FT act together to promote growth. Subsequently, when down-regulating FT upon the shift to SDs, SDs may up-regulate FDL1 expression, and in the resulting absence (or low levels) of FT, FDL1 can potentially be freed to interact with other factors (e.g., ABI3) involved in these pathways.
Conclusion
Based on our data, we present a model for the coordination of seasonal growth and adaptive response pathways by the photoperiodic signal in trees. In long days, FT interaction with FDL1 prevents growth cessation by maintaining the expression of LAP1. Perception of SDs promotes growth cessation by down-regulating the expression of FT and LAP1, the downstream target of the FT–FDL1 complex. Simultaneously, SDs induce FDL1 expression, which, together with ABI3, activates the adaptive response pathway, thereby temporally coordinating these two processes. Hence, hybrid aspen FDL1 has dual roles in photoperiodic control of pathways distinct from flowering, and, moreover, at least of some these functions of FDL1 are independent of its interaction with FT. Thus, FD homologs in hybrid aspen contrast to Arabidopsis, in which FD (and its paralog FDP) are both involved in the control of the same process, namely, flowering via interaction with FT (26). Although we have elucidated the role of FD, it remains to be seen whether tree homologs of BRC1 also have a role in SD-induced growth cessation or other related processes in the future. In summary, our results extend the functional repertoire of FD homologs and open new avenues to explore the function of FD in the future.
Experimental Procedures
Plant Material, Growth Conditions, and Tissue Sampling.
WT hybrid aspen (Populus tremula × tremuloides, clone T89) and transgenic plants grown in soil were subjected to SDs (8-h day/16-h night, 20 °C during day/15 °C at night), and SD responses were investigated (19), as described in SI Experimental Procedures. Tissue samples for gene expression analyses were collected from shoot apices after 0, 3, 6, and 10 wk of SDs, frozen in liquid nitrogen, and stored at −80 °C, and RNA was isolated as described in SI Experimental Procedures.
Construction of Transgenic Plants and Plasmid Constructs.
The cloning of FDL1 and FDL2 cDNAs, construction of FDL1oe and FDL2oe and FDL1RNAi and FDL2RNAi, and expression of FDL1 cDNA in the Arabidopsis fd-2 mutant are described in SI Experimental Procedures.
Transient Expression Assays in Rice Protoplasts.
Transient expression in rice protoplasts was performed as described previously (3) and is described in detail in SI Experimental Procedures.
Transient Expression and Coimmunoprecipitation Assay in Arabidopsis Protoplasts.
An Arabidopsis cell suspension culture derived from Col-0 roots was used in all experiments. Protoplast isolation and transient transfection were carried out as described (27), followed by coimmunoprecipitation performed using protein extracts from transfected protoplasts, as described in detail in SI Experimental Procedures.
RNA Isolation and Quantitative RT-PCR Analysis.
Total RNA was extracted using an Aurum Total RNA Kit (Bio-Rad). RNA (5 μg) was treated with RNase-free TURBO DNase (Life Technologies, Ambion), and 1 μg was then utilized for cDNA synthesis using an iScript cDNA Synthesis Kit (BioRad). In all experiments, TIP41-like was selected by GeNorm software (Biogazelle) (28) as a reference gene. Quantitative RT-PCR experiments were conducted using LightCycler 480 SYBR Green I Master mix and a Light Cycler 480 II instrument (both supplied by Roche). The ∆-cq method was used to calculate relative expression values of genes of interest.
BiFC Assay.
For the BiFC assay, full-length cDNA of FDL1, ABI3, and hybrid aspen FTs was cloned in pUC-SPYNE or pUC-SPYNE vector (21). Arabidopsis protoplasts were transfected as described (27), and YFP fluorescence was visualized 24 h after transfection on a Carl Zeiss LSM780 confocal microscope.
Supplementary Material
Acknowledgments
We thank Ingela Sandström for technical assistance, Dr. Gergely Molnar for advice on transient expression and coimmunoprecipitation experiments, and Dr. Urs Fischer for helpful comments. This work was funded by grants from Vetenskapsrådet, Berzelii, Trees and Crops for the Future, and the Knut and Alice Wallenberg Foundation (to R.P.B.). S.T. was funded by BIOIMPROVE.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423440112/-/DCSupplemental.
References
- 1.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 2.Abe M, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 3.Taoka K, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476(7360):332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 4.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 5.Niwa M, et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell. 2013;25(4):1228–1242. doi: 10.1105/tpc.112.109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro C, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- 7.Lee R, Baldwin S, Kenel F, McCallum J, Macknight R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat Commun. 2013;4:2884. doi: 10.1038/ncomms3884. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita T, et al. FLOWERING LOCUS T regulates stomatal opening. Curr Biol. 2011;21(14):1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 9.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312(5776):1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 10.Hsu CY, Liu Y, Luthe DS, Yuceer C. Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell. 2006;18(8):1846–1861. doi: 10.1105/tpc.106.041038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu CY, et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc Natl Acad Sci USA. 2011;108(26):10756–10761. doi: 10.1073/pnas.1104713108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiser CJ. Cold Resistance and Injury in Woody Plants: Knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science. 1970;169(3952):1269–1278. doi: 10.1126/science.169.3952.1269. [DOI] [PubMed] [Google Scholar]
- 13.Petterle A, Karlberg A, Bhalerao RP. Daylength mediated control of seasonal growth patterns in perennial trees. Curr Opin Plant Biol. 2013;16(3):301–306. doi: 10.1016/j.pbi.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Nitsch JP. Photoperiodism in woody plants. Proc Am Soc Hortic Sci. 1957;70:526–544. [Google Scholar]
- 15.Olsen JE, et al. Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J. 1997;12(6):1339–1350. [Google Scholar]
- 16.Howe GT, Gardner G, Hackett WP, Furnier GR. Phytochrome control of short-day-induced bud set in black cottonwood. Physiol Plant. 1996;97(1):95–103. [Google Scholar]
- 17.Rohde A, et al. ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell. 2000;12(1):35–52. doi: 10.1105/tpc.12.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruttink T, et al. A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell. 2007;19(8):2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azeez A, Miskolczi P, Tylewicz S, Bhalerao RP. A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr Biol. 2014;24(7):717–724. doi: 10.1016/j.cub.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 20.Tsuji H, Nakamura H, Taoka K, Shimamoto K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 2013;54(3):385–397. doi: 10.1093/pcp/pct005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40(3):428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 22.Karlberg A, et al. Analysis of global changes in gene expression during activity-dormancy cycle in hybrid aspen apex. Plant Biotechnol. 2010;27(1):1–16. [Google Scholar]
- 23.Resman L, et al. Components acting downstream of short day perception regulate differential cessation of cambial activity and associated responses in early and late clones of hybrid poplar. Plant Physiol. 2010;154(3):1294–1303. doi: 10.1104/pp.110.163907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakoby M, et al. bZIP Research Group bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 25.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA. 2005;102(21):7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25(3):820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dóczi R, Hatzimasoura E, Bögre L. Mitogen-activated protein kinase activity and reporter gene assays in plants. Methods Mol Biol. 2011;779:79–92. doi: 10.1007/978-1-61779-264-9_5. [DOI] [PubMed] [Google Scholar]
- 28.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):H0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.