Significance
Here we demonstrate that Hes1, an important target of Notch signaling, modulated pathogenesis of osteoarthritis by using Col2a1-CreERT;Hes1fl/fl mice. Adamts5 and Mmp13, catabolic enzymes that break down cartilage matrix, were induced by Hes1. Additionally, microarray analysis and ChIP-seq revealed novel Hes1 target genes, including Il6 and Il1rl1, coding a receptor for IL-33. CaMK2δ was activated during osteoarthritis development. CaMK2δ formed a protein complex with Hes1, and switched it from a transcriptional repressor to a transcriptional activator to induce cartilage catabolic factors.
Keywords: osteoarthritis, Hes1, CaMK2
Abstract
Notch signaling modulates skeletal formation and pathogenesis of osteoarthritis (OA) through induction of catabolic factors. Here we examined roles of Hes1, a transcription factor and important target of Notch signaling, in these processes. SRY-box containing gene 9 (Sox9)-Cre mice were mated with Hes1fl/fl mice to generate tissue-specific deletion of Hes1 from chondroprogenitor cells; this deletion caused no obvious abnormality in the perinatal period. Notably, OA development was suppressed when Hes1 was deleted from articular cartilage after skeletal growth in type II collagen (Col2a1)-CreERT;Hes1fl/fl mice. In cultured chondrocytes, Hes1 induced metallopeptidase with thrombospondin type 1 motif, 5 (Adamts5) and matrix metalloproteinase-13 (Mmp13), which are catabolic enzymes that break down cartilage matrix. ChIP-seq and luciferase assays identified Hes1-responsive regions in intronic sites of both genes; the region in the ADAMTS5 gene contained a typical consensus sequence for Hes1 binding, whereas that in the MMP13 gene did not. Additionally, microarray analysis, together with the ChIP-seq, revealed novel Hes1 target genes, including Il6 and Il1rl1, coding a receptor for IL-33. We further identified calcium/calmodulin-dependent protein kinase 2δ (CaMK2δ) as a cofactor of Hes1; CaMK2δ was activated during OA development, formed a protein complex with Hes1, and switched it from a transcriptional repressor to a transcriptional activator to induce cartilage catabolic factors. Therefore, Hes1 cooperated with CaMK2δ to modulate OA pathogenesis through induction of catabolic factors, including Adamts5, Mmp13, Il6, and Il1rl1. Our findings have contributed to further understanding of the molecular pathophysiology of OA, and may provide the basis for development of novel treatments for joint disorders.
Endochondral ossification is an essential process not only for skeletal formation, but also for development of osteoarthritis (OA), which is the most prevalent form of joint disease (1–3). Recent studies involving experimental mouse models with surgically induced instability in the knee joints have shown that OA is initiated by production of proteinases like matrix metalloproteinase-13 (MMP13) and a disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 5 (ADAMTS5), which are induced by various molecules and signals including Runx2, C/EBP-β, HIF-2α, and NF-κB (4–10).
Notch is a single-pass transmembrane cell-surface receptor that plays a crucial role in cell-fate determination by regulating differentiation and apoptosis during embryogenesis and postnatal development (11, 12). In mammals, the Notch signaling pathway is regulated by several molecules including Notch ligands (Delta-like1, -3, -4, or Jagged1 and -2), Notch receptors (Notch1 to -4), a transcriptional effector Rbpj (recombination signal binding protein for Ig kappa J), and target transcription factors of the Hes (hairy and enhancer of split)/Hey (hairy/enhancer-of-split related with YRPW motif) family (13). Notch signaling is initiated when Notch ligands on a cell surface bind to Notch receptors on adjacent cells. Upon ligand binding, the Notch receptor is cleaved by proteinases, such as a disintegrin and metalloproteinase (Adam), and subsequently by a γ-secretase complex; these successive cleavages release the Notch intracellular domain (ICD) into the cytoplasm. The ICD then translocates to the nucleus and binds to the transcriptional effector Rbpj to form a transcriptional activator that induces target genes such as basic helix–loop–helix transcription factors in the Hes/Hey family.
Previously we reported that Notch signaling regulates skeletal formation and OA development (14). In mouse limb growth plates, ICDs of Notch1 and Notch2 are translocated into the nucleus as the chondrocytes differentiate and this translocation also occurs as OA progresses in articular cartilage (14). Tissue-specific knockout of Rbpj in chondrocyte progenitors disrupts the terminal stage of endochondral ossification in limb cartilage, and knockout of Rbpj in adult articular cartilage after skeletal growth causes resistance to OA development in the knee joint (14). Moreover, Notch ICDs facilitate endochondral ossification by inducing Hes1 in cultured chondrocytes (14).
Herein we describe the roles of Hes1 during skeletal formation and OA development using conditional knockout mice. We also examined the underlying mechanism of Hes1-mediated OA development.
Results
Hes1 Is Dispensable During Skeletal Formation.
To examine the physiological role of Hes1 in skeletal formation, we generated tissue-specific knockout mice to conditionally inactivate Hes1 in chondrocyte progenitors; specifically, SRY-box containing gene 9 (Sox9)-Cre mice, which have an internal ribosome entry site and a Cre recombinase gene inserted into the 3′ untranslated region of the Sox9 (15), were mated with mice homozygous for a floxed Hes1 allele (Hes1fl/fl) (16). Although the conditional knockout (Sox9-Cre;Hes1fl/fl) mice died in the perinatal period, the embryos [embryonic day (E) 18.5] showed no obvious abnormality compared with Hes1fl/fl littermates (Fig. S1 A and B). The two genotypes were similar with regard to the length of each limb and vertebra (Fig. S1 C and D). No obvious differences were evident between genotypes with regard to Safranin-O staining specimens or immunofluorescence of three marker proteins [type X collagen (Col10a1), Adamts5, or Mmp13]; nevertheless, Hes1 was efficiently knocked down (Fig. S1E). We further investigated expression of mRNAs encoding six Hes/Hey family members that are known targets of Notch signaling. Notably, only Hes1 mRNA was markedly lower in primary costal chondrocytes of Hes1-knockdown mice than in those of control mice; moreover, compensatory up-regulation of other Hes/Hey family members was not evident (Fig. S1F). All these data at E18.5 indicate that Hes1 is dispensable during skeletal formation in the embryonic stage.
Hes1 Modulates OA Development.
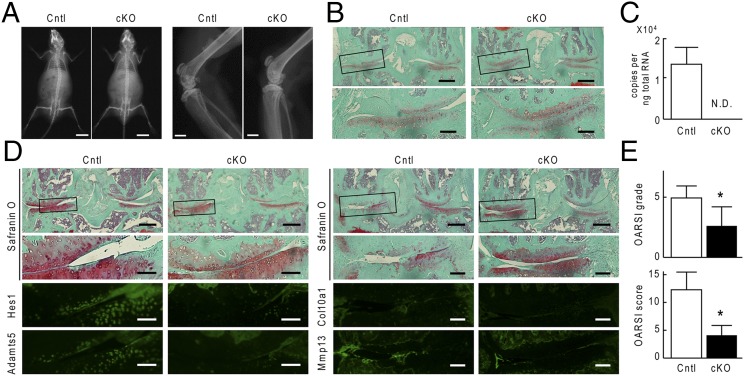
We next examined the contribution of Hes1 to OA development. The Sox9-Cre;Hes1fl/fl mice died in the perinatal period and could not be used for this study. Therefore, we generated inducible conditional knockout mice by mating Col2a1-CreERT mice with Hes1fl/fl mice (Col2a1-CreERT;Hes1fl/fl), in which Hes1 deletion was induced by tamoxifen injection; specifically, the Col2a1-CreERT construct encoded a Cre recombinase fused to a mutated ligand binding domain of the human estrogen receptor and was driven by the type II collagen (Col2a1) promoter, the Cre fusion protein therefore translocated into nuclei and caused Hes1 deletion only when the estrogen antagonist tamoxifen was administered (17). We injected tamoxifen into 7-wk-old Col2a1-CreERT;Hes1fl/fl mice and Hes1fl/fl littermates daily for 5 d. At 8 wk, 2 d after the last tamoxifen injection, we created an OA model by surgically destabilizing knee joints (18). The Col2a1-CreERT;Hes1fl/fl mice developed and grew normally without abnormality in the skeleton, articular cartilage, or joints (Fig. 1 A and B). Real-time RT-PCR confirmed efficient knockout of Hes1 in articular chondrocytes (Fig. 1C). At 4 wk and at 8 wk after surgical induction of OA, Col2a1-CreERT;Hes1fl/fl knee joints differed from Hes1fl/fl knee joints with regard to cartilage degradation; specifically, cartilage degradation was suppressed in the Col2a1-CreERT;Hes1fl/fl joints (Fig. 1D). This suppression was associated with decreases in expression of Adamts5 and of Mmp13, but not of Col10a1 (Fig. 1D). Quantification by grading systems (19) confirmed that knockdown of Hes1 caused significant resistance to OA development (Fig. 1E). Histomorphometric analyses of subchondral bones in the knee joints revealed no difference between the two genotypes, indicating that the resistance to OA caused by Hes1 knockdown was not a result of alteration in bone morphology or joint biomechanics (Table S1).
Fig. 1.
OA development in Col2a1-CreERT;Hes1fl/fl (cKO) and Hes1fl/fl (Cntl) mice. We performed daily tamoxifen injections to both genotypes for 5 d at 7-wk-old. (A) Plain radiographs of the entire bodies (Left) and knee (Right) of Cntl and cKO littermates (8-wk-old). [Scale bars, 10 mm (Left), 1 mm (Right).] (B) Safranin-O staining of mouse knee joints in 8-wk-old Cntl and cKO littermates above. Boxed regions in the upper panels indicate the regions enlarged in the lower panels. (Scale bars, 400 μm and 100 μm for low and high magnification images, respectively.) (C) mRNA levels of Hes1 in articular chondrocytes from Cntl and cKO littermates (16-wk-old). Data are expressed as means ± SD of three mice per group. N.D., not detected. (D) Cartilage degradation assessed by Safranin-O staining or immunofluorescence with antibodies to Hes1, Adamts5, Col10a1, and Mmp13 in mouse knee joints 4 wk (Left) and 8 wk (Right) after surgery to create a model of OA in 8-wk-old Cntl and cKO littermates. Boxed regions in the upper panels indicate the regions enlarged in the lower panels. (Scale bars, 400 μm and 100 μm for low and high magnification images, respectively.) (E) OARSI grading of OA development. Data are expressed as means ± SD of six mice per group. *P < 0.05 vs. Cntl.
Transcriptional Regulation of Adamts5 and Mmp13 by Hes1.
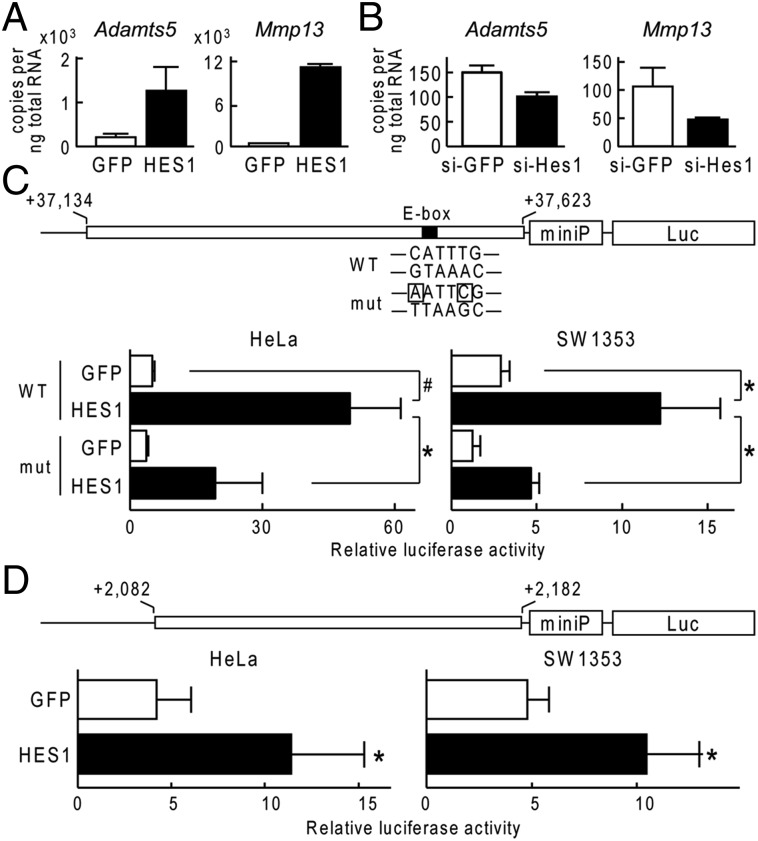
To investigate the mechanisms underlying Hes1-mediated regulation of OA, we initially determined whether Hes1 directly regulated transcription of Adamts5, Mmp13, or both; these proteins were down-regulated in articular cartilage of the Col2a1-CreERT;Hes1fl/fl mice during OA development. In cultures of ATDC5 cells, expression of each Adamts5 and Mmp13 was increased when HES1 was overexpressed (Fig. 2A); conversely, expression of both decreased following siRNA-mediated knockdown of Hes1 (Fig. 2B). To search for Hes1-binding sites around Adamts5 and Mmp13, we performed ChIP and deep sequencing (ChIP-seq) with human chondrosarcoma cell line, SW1353, which expressed 3xFLAG-tagged HES1 and anti-FLAG antibody. Around the ADAMTS5 gene, a significant peak (fold-enrichment ≧ 10, and false discovery rate ≦ 0.01) was identified in intron7 (Fig. S2). Luciferase assays revealed that HES1 transfection markedly increased the transactivation of a luciferase reporter gene containing this ADAMTS5 intron7 region fused to a minimal promoter (miniP) (Fig. 2C). The region included an E-box and a consensus sequence for Hes1 binding; moreover, mutation of the E-box suppressed Hes1-mediated transactivation of luciferase reporter genes (Fig. 2C). Around the MMP13 gene, a high but not significant peak was identified in intron4 (Fig. S3). HES1 transfection markedly increased the transactivation of a luciferase reporter gene containing this MMP13 intron4 region fused to miniP (Fig. 2D), but this region did not contain an identifiable consensus sequence for Hes1 binding.
Fig. 2.
Mechanism of Adamts5 and Mmp13 induction by Hes1. (A) mRNA levels of Mmp13 and Adamts5 in ATDC5 cells that were transfected with GFP or HES1. Cells were cultured for 2 d after lipofection and then cultured in a pellet for additional 10 d. (B) mRNA levels of Mmp13 and Adamts5 in ATDC5 cells that were transfected with siRNA for GFP (si-GFP) or Hes1 (si-Hes1). Cells were cultured as above. (C) Luciferase activities following the transfection of GFP or HES1 into HeLa or SW1353 cells with a reporter construct containing a wild-type (WT) fragment from ADAMTS5 or a fragment with a mutation in the E-box (mut), as indicated (Upper). Data are shown as means ± SD of three wells per group. #P < 0.01, *P < 0.05. (D) Luciferase activities following the transfection of GFP or HES1 into HeLa or SW1353 cells with a reporter construct containing a fragment from MMP13 as indicated in the Top panel. All data are shown as means ± SD of three wells per group. *P < 0.05 vs. GFP.
Hes1 Alters Gene Expression in Chondrocytes.
To identify molecules besides Adamts5 and Mmp13 that function downstream of Hes1 during OA development, we used mRNA samples from ATDC5 cells that overexpressed HES1 or a GFP control to perform microarray analysis; we identified 1,956 genes that were up-regulated (fold-increase > 4) in HES1-overexpressing cells and 390 that were down-regulated (fold-increase < 4). The top 30 up-regulated and top 30 down-regulated genes are shown in Tables S2 and S3.
To identify other candidates among the Hes1-regulated genes that may mediate the catabolic effect of Hes1, we searched for genes associated with mechanical stress or inflammation, which are phenomena important to OA development. Based on a gene ontology analysis, five of the up-regulated genes (fold-increase > 4) were associated with mechanical stress (Table S4), and 40 up-regulated genes (fold-increase > 4) were associated with inflammation (Table S5). Based on ChIP-seq analysis, none of the loci encoding mechanical stress-related candidates was associated with a significant peak that indicated a Hes1 binding site. However, two inflammation-related candidate genes were associated with significant peaks; a region near IL6 and another in intron 1 of IL1RL1 were each associated with a significant peak (Figs. S4 and S5).
IL6, a cytokine known to function in inflammation, is widely implicated in inflammation-associated diseases, including rheumatoid arthritis (RA) (20–22) and OA (23–25). IL-1 receptor-like 1, encoded by IL1RL1, is a receptor for IL-33. IL1RL1 is induced by proinflammatory stimuli, and IL1RL1, like IL-6, is implicated in inflammation-associated disease states (26–28).
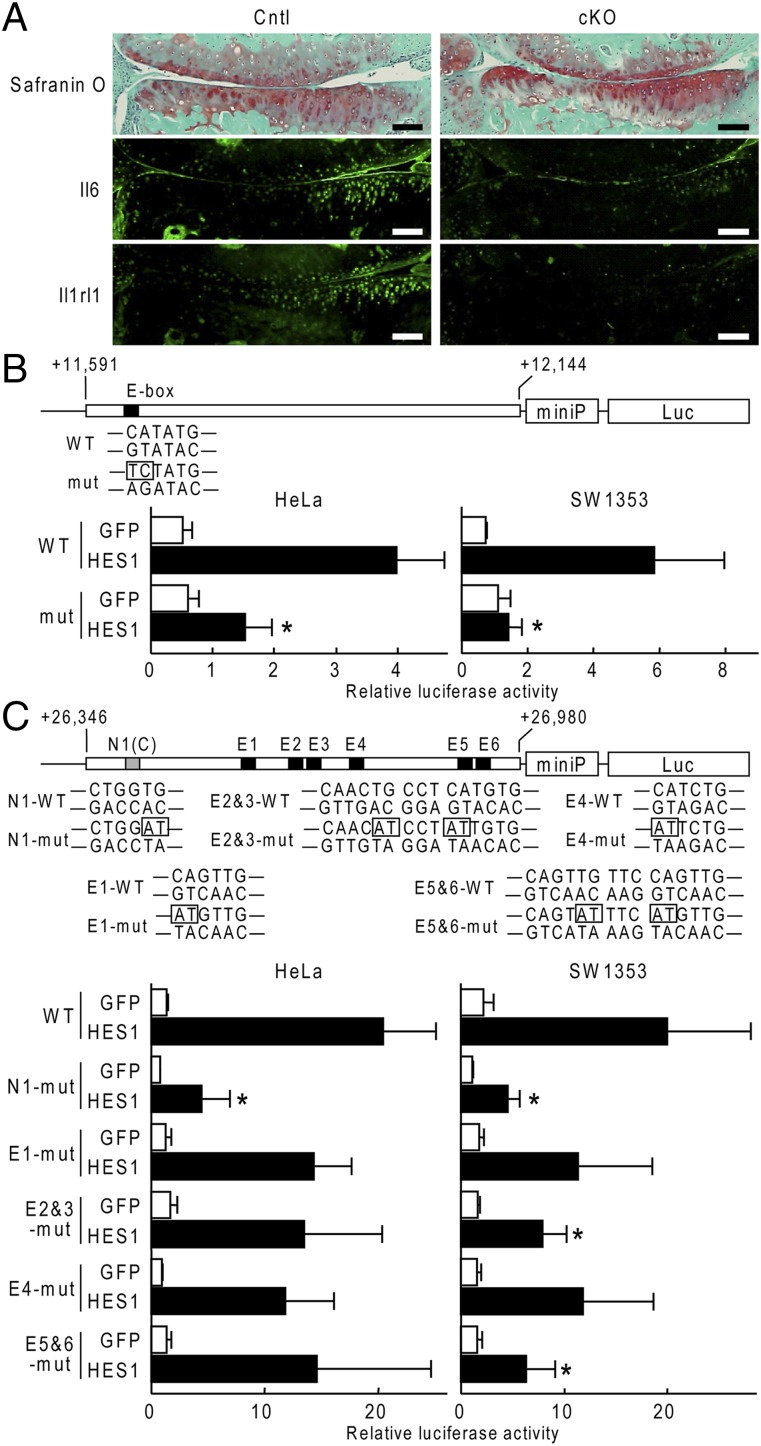
Expression of each Il6 and Il1rl1 was lower in Col2a1-CreERT;Hes1fl/fl joints than in the Hes1fl/fl joints (Fig. 3A). To examine the Hes1-dependent transcriptional regulation of these genes, we cloned each putative Hes1-responsive region identified by the ChIP-seq analysis into a luciferase reporter construct with miniP. In luciferase assays, HES1 overexpression led to marked transactivation of reporters containing the region from IL6 (Fig. 3B). This region included an E-box, and mutation of this E-box resulted in significant suppression of Hes1-mediated transactivation in both HeLa and SW1353 cells (Fig. 3B). The putative Hes1-responsive region from IL1RL1 included an N-box (N1) and six E-boxes (E1 to -6) (Fig. 3C). E-boxes E2 and E3 were adjacent to each other, as were E5 and E6 (Fig. 3C). In luciferase assays, HES1 overexpression led to marked transactivation of reporters containing the region from IL1RL1 (Fig. 3C).When we mutated each of these boxes individually, only mutations of N1 significantly decreased transactivation in both HeLa and SW1353 cells (Fig. 3C).
Fig. 3.
Transcriptional induction of Il6 and Il1rl1 by Hes1. (A) Immunofluorescence from antibodies to Il6 or Il1rl1 in mouse knee joints 4 wk after creating a surgical OA model in 8-wk-old Hes1fl/fl (Cntl) or Col2a1-CreERT;Hes1fl/fl (cKO) littermates. (Scale bars, 100 μm.) (B) Luciferase activities following transfection of GFP or HES1 into HeLa or SW1353 cells with a reporter construct containing a wild-type (WT) fragment from IL6 or a fragment with a mutation in the E-box (mut), as indicated (Upper). Data are shown as means ± SD of three wells per group. *P < 0.05 versus HES1 in WT. (C) Luciferase activities following the transfections of GFP or HES1 into HeLa or SW1353 cells with a reporter construct containing a wild-type (WT) fragment from IL1RL1 or a fragment with a mutation in N1, E1, E2&3, E4, or E5&6, as indicated in (Upper). Data are shown as means ± SD of three wells per group. *P < 0.05 vs. HES1 in WT.
Hes1 Cooperates with CaMK2δ to Regulate Downstream Molecules.
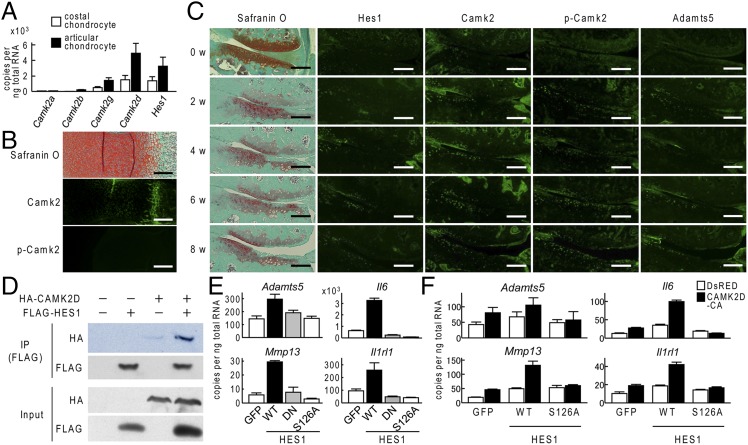
Like other Hes/Hey family proteins, Hes1 can act as a transcriptional repressor (29). However, our data indicated that Hes1 functioned as a required transcriptional activator of Adamts5, Mmp13, Il6, and Il1rl1 during OA development. To understand this putative Hes1-dependent transcriptional activation, we investigated calcium/calmodulin-dependent protein kinase 2 (CaMK2), which is the only factor known to change Hes1 from a transcriptional repressor to transcriptional activator (30). We initially looked at expression of four CaMK2 subtypes (CaMK2α, -β, -γ, and -δ) along with expression of Hes1 in mouse primary costal and articular chondrocytes. Among the four subtypes, CaMK2δ was most strongly expressed in these chondrocytes, and CaMK2δ expression, like Hes1 expression, was higher in articular chondrocytes than in costal chondrocytes (Fig. 4A). CaMK2 proteins were weakly expressed in limb cartilage of mouse embryos, and active CaMK2—which is phosphorylated at Thr-287—was barely evident (Fig. 4B). Notably, in articular cartilage from an experimental model mouse of OA, expression of CaMK2 and phosphorylation of CaMK2 increased along with Hes1 expression during OA progression (Fig. 4C). Coimmunoprecipitation assay confirmed the binding of Hes1 and CaMK2δ proteins (Fig. 4D). Expression of each Adamts5, Mmp13, Il6, and Il1rl1 was enhanced by doxycycline-induced overexpression of Hes1 in ATDC5 cells (Fig. 4E). However, similar enhancement was not evident with overexpression of either two Hes1 mutants, dnHES1 or HES1 S126A; dnHES1 is a dominant-negative mutant that cannot bind to DNA, but can dimerize with endogenous wild-type Hes1 to form a non-DNA–binding heterodimer complex (31); HES1 S126A was mutated in the serine residue of the CaMK2 phosphorylation site to inhibit Hes1-CaMK2 cooperation (30) (Fig. 4E). Hes1 and a constitutively active form of CaMK2δ were either individually overexpressed or co-overexpressed in ATDC5 cells via lentiviral vectors; induction of Adamts5, Mmp13, Il6, or Il1rl1 expression was greater with co-overexpression than with overexpression of either individual gene; however, the S126A mutant abrogated any Hes1-dependent or CaMK2δ-dependent induction (Fig. 4F).
Fig. 4.
Regulation of Hes1 transcriptional activity by CaMK2. (A) mRNA levels of Camk2α, Camk2β, Camk2γ, Camk2δ, and Hes1 in mouse primary costal chondrocytes or articular chondrocytes that were cultured for 5 d. (B) Safranin-O staining and immunofluorescence with antibodies to Camk2 and phosphorylated Camk2 (p-Camk2) in proximal tibias of mouse embryos (E18.5). (Scale bars, 200 μm.) (C) Time course of degradation in mouse knee joint cartilage in 8-wk-old mice following surgery to induce osteoarthritis as shown by Safranin-O staining or immunofluorescence from Hes1, Camk2, p-Camk2, or Adamts5. (Scale bar, 100 μm.) (D) Whole-cell lysates from SW1353 cells with the indicated plasmids were immunoprecipitated with anti-FLAG antibody and the resulting immunoprecipitate was immunobloted with the anti-HA antibody (Top panel), and with the anti-FLAG antibody (second panel). The same whole-cell lysates (Input) were immunoblotted with the anti-HA antibody (third panel), with the anti-FLAG antibody (fourth panel). (E) mRNA levels of Adamts5, Mmp13, Il6, and Il1rl1 in ATDC5 cells transfected via lentivirus with wild-type (WT) HES1, a dominant-negative form of HES1 (DN), kinase domain mutated (S126A) form of HES1, or GFP. All constructs were doxycycline inducible. Cells were cultured for 2 d after the doxycycline induction and then cultured in a pellet for additional 10 d. (F) mRNA levels of Adamts5, Mmp13, Il6, or Il1rl1 in ATDC5 cells that were transfected via lentivirus with doxycycline inducible WT HES1 or S126A mutant, and further cotransfected with DsRED-Express2 or continuous active form(CA) of CaMK2δ. Data are shown as means ± SD of three wells per group.
Discussion
The present study demonstrates that Hes1 modulated OA development in cooperation with CaMK2 via induction of catabolic factors including Adamts5, Mmp13, Il6, and Il1rl1. Considering that, in chondrocytes, Hes1 is the most abundantly expressed Hes/Hey family member among those downstream of Notch signaling, these findings are consistent with our previous finding that Rbpj-dependent Notch signaling facilitates OA development (14). However, the mice with a conditional knockout of Hes1 in chondrocyte progenitors showed no obvious disorders in skeletal growth, but those with a similar Rbpj conditional knockout showed impaired skeletal growth that was mainly caused by Mmp13 insufficiency (14). Other Hes/Hey family members were barely expressed in chondrocytes, and none was up-regulated in Hes1-deficient cells (Fig. S1F); therefore, the discrepancy between the conditional Hes1 and Rbpj phenotypes may have been to the result of other molecules induced by Rbpj-dependent Notch signaling during skeletal formation.
Hes1 is an essential mediator of Notch signaling, and generally Hes1 mediates Notch signaling by repressing expression of Hes1 target genes (29). However, data from in vivo and in vitro analyses showed that Hes1 overexpression led to up-regulation of downstream molecules, including Adamts5, Mmp13, Il6, and Il1rl1, and conversely that Hes1 suppression led to down-regulation of the same molecules. To understand this Hes1-mediated transcriptional activation, we focused on CaMK2, which is known to switch Hes1 function from transcriptional repression to transcriptional activation (30). In the present study, we observed concurrent increases in Hes1 and CaMK2 expression during OA development in vivo, and the coimmunoprecipitation of both proteins in chondrocytes. Furthermore, Hes1 and CaMK2 cooperated to induce at least four downstream molecules; moreover, a serine-to-alanine mutation of the CaMK2 phosphorylation site in Hes1 abrogated Hes1-dependent and CaMK2-dependent induction of each downstream gene. Taken together, these data indicated that CaMK2 caused Hes1 to switch from a transcriptional repressor to a transcriptional activator and thereby enhanced expression of catabolic factors in articular cartilage. CaMK2-mediated switching of Hes1function was originally discovered in an in vitro assay (30), but the data presented here constitutes, to our knowledge, the first in vivo evidence that CaMK2 causes Hes1 to function as a transcriptional activator. We did not attempt to identify any proteins other than Hes1 and CaMK2 in the activating complex. The Hes1-responsive regions that contained consensus sequences for Hes1 binding were identified in ADAMTS5, IL6, and IL1RL1 via ChIP-seq, and each was functionally confirmed via luciferase assays. In contrast, the putative Hes1-responsive region identified near MMP13 did not contain a consensus sequence for Hes1 binding. These results indicate that the protein complex that induced Mmp13 transcription may have differed from the complexes that induced the other three genes, and that Hes1 binding to the MMP13 gene may have been indirect and mediated by other transcription factors.
CaMK2 is a broadly distributed Ser/Thr protein kinase that is activated by Ca2+/Calmodulin (CaM) binding (32). It has essential roles in synaptic plasticity and memory (32); nevertheless, recent findings indicate that CaMK2 is activated by cyclic mechanical stress loading and that CaMK2 activation induces aggrecan expression in articular chondrocytes (33, 34). These recent findings indicate that CaMK2 may be activated by excessive mechanical stress loading in articular cartilage. Indeed, our data indicate that CaMK2 expression and CaMK2 phosphorylation increased as OA progressed. Moreover, expression of both unphosphorylated and phosphorylated CaMK2 was higher in articular cartilage than in embryonic limb cartilage, and notably mechanical stress loads are also higher in articular cartilage (Fig. 4 A–C). Taken together, previous and present data may indicate that therapeutic modulation of CaMK2 activity may be a potent treatment for OA.
In the present study, the ChIP-seq analysis and the microarray analyses identified IL6 and Il1rl1 as previously unrecognized targets of Hes1. IL-6 is primarily produced at sites of inflammation, and is widely implicated in inflammation-associated pathophysiological states. In an experimental OA model, IL6 plays an essential role as a downstream target of HIF-2α, a key transcription factor that induces cartilage degradation (25). IL1RL1 and its ligand IL33 are associated with inflammation-related arthritis and various immune and inflammatory diseases, including RA, systemic lupus erythematosus, ankylosing spondylitis, and Sjogren syndrome (28, 35–37). Down-regulation of these two genes by Hes1 knockout may be essential in suppression of OA development in Col2a1-CreERT;Hes1fl/fl mice, because intra-articular manipulation can induce inflammatory response in knee joints in our OA model. Furthermore, two recent studies show that inhibition of Notch signaling ameliorates experimentally induced inflammatory arthritis (38, 39). Together, these data indicate that the Notch-Hes1 signaling pathway may play essential roles in inflammatory arthritis and in OA.
In conclusion, we have found that, in cooperation with CaMK2, Hes1 modulated OA development through induction of catabolic factors, including Adamts5, Mmp13, Il6, and Il1rl1. Our findings contributed to further understanding of the molecular pathophysiology of OA, and may provide the basis for development of novel treatments for OA and RA.
Materials and Methods
Cell Cultures.
We cultured HeLa (Riken BRC) and SW1353 (American Type Culture Collection) cells in DMEM with 10% (vol/vol) FBS; ATDC5 cells (Riken BRC) were cultured in DMEM and F-12 (1:1) with 5% (vol/vol) FBS. We performed experiments involving these cell lines as previously described (40). Lentiviruses were packaged in Lenti-X 293T cells (Clontech). To establish stable transgenic cell lines, ATDC5 cells were cultured with 250 μg/mL geneticin and 1 μg/mL puromycine after lentiviral transduction. We used 2 μg/mL doxycycline to activate the Tet-On inducible expression system. Primary costal chondrocytes were isolated from ribs of C57BL/6 neonates, and primary articular chondrocytes were isolated from 6-d-old C57BL/6 mice, as described previously (41).
Construction of Expression Vectors.
We prepared expression constructs for the luciferase assay and for coimmunoprecipitation assays in pCMV-HA (Clontech) and pCMV-3Tag-1B (Agilent Technologies), respectively; we created the dnHES1 and HES1 S126A mutants, as described previously (30, 31). We used piGENEmU6 (iGENE Therapeutics) to construct an siRNA vector targeting nucleotides 618–638 of mouse Hes1. The Tet-On 3G system (Clontech) was used to generate Hes1 and CaMK2δ inducible expression constructs, which were introduced into ATDC5 cells via lentiviruses. DNA sequencing was used to verify each construct.
Luciferase Assays.
We used PCR and human genomic DNA as template to amplify the putative Hes1-responsive elements from ADAMTS5 (in intron 7, from +37,134 to +37,623 bp relative to the transcription start site), MMP13 (in intron4, +2,082 to +2,182 bp), IL6 (in 3′ fragment, +11,591 to +12,144 bp), and IL1RIL1 (in intron1, +26,346 to +26,980 bp); we cloned each PCR product individually into a pGL4.23[luc2/minP] vector (Promega). We used PCR to create mutant constructs, and used the Dual-Luciferase Reporter Assay System (Promega) to perform luciferase assays; all data are shown as a ratio of firefly luciferase activity to Renilla luciferase activity.
Coimmunoprecipitation Assay.
We used the EZ view Red Protein A and anti-FLAG M2 Affinity Gels (Sigma) according to the manufacturer’s protocol for the coimmunoprecipitation assays.
ChIP-Seq Assay.
We used SW1353 cells that expressed 3xFLAG-tagged Hes1 for the ChIP-seq assays. For immunoprecipitation, we used a monoclonal antibody that recognizes FLAG (Clone M2; Sigma-Aldrich). All protocols for Illumina/Solexa sequence preparation, sequencing, and quality control were provided by Takara Biogenomics. We used Model-based Analysis of ChIP-Seq (42) to analyze sequence data, and the University of California Santa Cruz genome browser (43) for visualization. The ChIP-seq data have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE60006.
Microarray Analysis.
Total RNA was extracted from ATDC5 cells that overexpressed GFP or HES1. SurePrint G3 Mouse GE 8 × 60K Microarrays (Agilent) and total RNA samples were used according to the manufacturer’s instructions for all microarray analyses. The microarray data have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE60737.
Mice.
We performed all experiments according to a protocol approved by the Animal Care and Use Committee of the University of Tokyo. Col2a1-CreERT mice were provided to us by Fanxin Long (Washington University in St. Louis, St. Louis, MO) (17). To generate Sox9-Cre;Hes1fl/fl mice or Col2a1-CreERT;Hes1fl/fl mice, Hes1fl/fl mice were mated with Sox9-Cre mice or Col2a1-CreERT mice, respectively; the resulting Sox9-Cre;Hes1fl/+ mice or Col2a1-CreERT;Hes1fl/+ mice were then mated with Hes1fl/fl mice. To confirm the Hes1 knockout, we isolated articular chondrocytes from 16-wk-old Hes1fl/fl and Col2a1-CreERT;Hes1fl/fl animals.
Osteoarthritis Experiments.
Tamoxifen (Sigma; 100 μg per gram of body weight) was intraperitoneally injected to 7-wk-old Col2a1-CreERT;Hes1fl/fl mice and the Hes1fl/fl littermates daily for 5 d. We then performed a surgical procedure on 8-wk-old male mice, as described previously, to create an experimental model of OA (14, 18); the mice were analyzed 8 wk after surgery. We used the Osteoarthritis Research Society International (OARSI) system (19) to quantify OA severity; all OARSI assessments were made by a single observer who was blinded to the treatment groups. We made histomorphometric measurements of the subchondral bones in eight optical fields as described in the American Society for Bone and Mineral Research nomenclature report (44).
Other Analyses.
We performed real-time RT-PCR, Western blotting, and histological analyses as described previously (9, 14, 40, 45). Primer sequences and antibody information are available upon request.
Statistical Analyses.
We used the unpaired two-tailed Student's t test to assess the statistical significance of experimental data. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Fanxin Long for generously providing Col2a1-CreERT mice, and J. Sugita, R. Yamaguchi, and H. Kawahara for technical assistance. This study was supported by a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Grants 23390358 and 24592222) and by the Nakatomi Foundation (T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.F.W. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE60006 (ChIP-seq) and GSE60737 (microarray)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419699112/-/DCSupplemental.
References
- 1.Kawaguchi H. Endochondral ossification signals in cartilage degradation during osteoarthritis progression in experimental mouse models. Mol Cells. 2008;25(1):1–6. [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Moskowitz RW. The burden of osteoarthritis: Clinical and quality-of-life issues. Am J Manag Care. 2009;15(8) Suppl:S223–S229. [PubMed] [Google Scholar]
- 4.Glasson SS, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434(7033):644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 5.Hirata M, et al. C/EBPβ and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF-2α as the inducer in chondrocytes. Hum Mol Genet. 2012;21(5):1111–1123. doi: 10.1093/hmg/ddr540. [DOI] [PubMed] [Google Scholar]
- 6.Kamekura S, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi H, et al. Transcriptional induction of ADAMTS5 protein by nuclear factor-κB (NF-κB) family member RelA/p65 in chondrocytes during osteoarthritis development. J Biol Chem. 2013;288(40):28620–28629. doi: 10.1074/jbc.M113.452169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Little CB, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60(12):3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito T, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med. 2010;16(6):678–686. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 10.Stanton H, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434(7033):648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 11.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 12.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat Neurosci. 2005;8(6):709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 13.D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosaka Y, et al. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc Natl Acad Sci USA. 2013;110(5):1875–1880. doi: 10.1073/pnas.1207458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102(41):14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135(15):2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- 17.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol. 2007;308(1):93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamekura S, et al. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthritis Cartilage. 2005;13(7):632–641. doi: 10.1016/j.joca.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Pritzker KP, et al. Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthritis Cartilage. 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Dayer JM, Choy E. Therapeutic targets in rheumatoid arthritis: The interleukin-6 receptor. Rheumatology (Oxford) 2010;49(1):15–24. doi: 10.1093/rheumatology/kep329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca JE, Santos MJ, Canhão H, Choy E. Interleukin-6 as a key player in systemic inflammation and joint destruction. Autoimmun Rev. 2009;8(7):538–542. doi: 10.1016/j.autrev.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Nishimoto N, Kishimoto T. Interleukin 6: From bench to bedside. Nat Clin Pract Rheumatol. 2006;2(11):619–626. doi: 10.1038/ncprheum0338. [DOI] [PubMed] [Google Scholar]
- 23.Doss F, et al. Elevated IL-6 levels in the synovial fluid of osteoarthritis patients stem from plasma cells. Scand J Rheumatol. 2007;36(2):136–139. doi: 10.1080/03009740701250785. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko S, et al. Interleukin-6 and interleukin-8 levels in serum and synovial fluid of patients with osteoarthritis. Cytokines Cell Mol Ther. 2000;6(2):71–79. doi: 10.1080/13684730050515796. [DOI] [PubMed] [Google Scholar]
- 25.Ryu JH, et al. Interleukin-6 plays an essential role in hypoxia-inducible factor 2α-induced experimental osteoarthritic cartilage destruction in mice. Arthritis Rheum. 2011;63(9):2732–2743. doi: 10.1002/art.30451. [DOI] [PubMed] [Google Scholar]
- 26.Kashiwakura J, et al. Interleukin-33 synergistically enhances immune complex-induced tumor necrosis factor alpha and interleukin-8 production in cultured human synovium-derived mast cells. Int Arch Allergy Immunol. 2013;161(Suppl 2):32–36. doi: 10.1159/000350424. [DOI] [PubMed] [Google Scholar]
- 27.Milovanovic M, et al. IL-33/ST2 axis in inflammation and immunopathology. Immunol Res. 2012;52(1-2):89–99. doi: 10.1007/s12026-012-8283-9. [DOI] [PubMed] [Google Scholar]
- 28.Xu WD, Zhang M, Zhang YJ, Ye DQ. IL-33 in rheumatoid arthritis: Potential role in pathogenesis and therapy. Hum Immunol. 2013;74(9):1057–1060. doi: 10.1016/j.humimm.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 29.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 30.Ju BG, et al. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119(6):815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Ström A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11(23):3168–3181. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stratton MM, Chao LH, Schulman H, Kuriyan J. Structural studies on the regulation of Ca2+/calmodulin dependent protein kinase II. Curr Opin Struct Biol. 2013;23(2):292–301. doi: 10.1016/j.sbi.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimazaki A, Wright MO, Elliot K, Salter DM, Millward-Sadler SJ. Calcium/calmodulin-dependent protein kinase II in human articular chondrocytes. Biorheology. 2006;43(3-4):223–233. [PubMed] [Google Scholar]
- 34.Valhmu WB, Raia FJ. myo-Inositol 1,4,5-trisphosphate and Ca(2+)/calmodulin-dependent factors mediate transduction of compression-induced signals in bovine articular chondrocytes. Biochem J. 2002;361(Pt 3):689–696. doi: 10.1042/0264-6021:3610689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XL, et al. Elevated serum level of IL-33 and sST2 in patients with ankylosing spondylitis: Associated with disease activity and vascular endothelial growth factor. J Investig Med. 2013;61(5):848–851. doi: 10.2310/JIM.0b013e31828deed2. [DOI] [PubMed] [Google Scholar]
- 36.Mok MY, et al. Serum levels of IL-33 and soluble ST2 and their association with disease activity in systemic lupus erythematosus. Rheumatology (Oxford) 2010;49(3):520–527. doi: 10.1093/rheumatology/kep402. [DOI] [PubMed] [Google Scholar]
- 37.Zhao L, et al. Potential contribution of interleukin-33 to the development of interstitial lung disease in patients with primary Sjogren’s Syndrome. Cytokine. 2013;64(1):22–24. doi: 10.1016/j.cyto.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Jiao Z, et al. Blockade of Notch signaling ameliorates murine collagen-induced arthritis via suppressing Th1 and Th17 cell responses. Am J Pathol. 2014;184(4):1085–1093. doi: 10.1016/j.ajpath.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Park JS, et al. Inhibition of Notch signalling ameliorates experimental inflammatory arthritis. Ann Rheum Dis. 2015;74(1):267–274. doi: 10.1136/annrheumdis-2013-203467. [DOI] [PubMed] [Google Scholar]
- 40.Saito T, Ikeda T, Nakamura K, Chung UI, Kawaguchi H. S100A1 and S100B, transcriptional targets of SOX trio, inhibit terminal differentiation of chondrocytes. EMBO Rep. 2007;8(5):504–509. doi: 10.1038/sj.embor.7400934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3(8):1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dempster DW, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirata M, et al. C/EBPbeta promotes transition from proliferation to hypertrophic differentiation of chondrocytes through transactivation of p57. PLoS ONE. 2009;4(2):e4543. doi: 10.1371/journal.pone.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.