Abstract
Pathogen reduction (PR) systems for platelets, based on chemically induced cross-linking and inactivation of nucleic acids, potentially prevent transfusion transmission of infectious agents, but can increase clinically significant bleeding in some clinical studies. Here, we documented the effects of PR systems on microRNA and mRNA levels of platelets stored in the blood bank, and assessed their impact on platelet activation and function. Unlike platelets subjected to gamma irradiation or stored in additive solution, platelets treated with Intercept (amotosalen + ultraviolet-A [UVA] light) exhibited significantly reduced levels of 6 of the 11 microRNAs, and 2 of the 3 anti-apoptotic mRNAs (Bcl-xl and Clusterin) that we monitored, compared with platelets stored in plasma. Mirasol (riboflavin + UVB light) treatment of platelets did not produce these effects. PR neither affected platelet microRNA synthesis or function nor induced cross-linking of microRNA-sized endogenous platelet RNA species. However, the reduction in the platelet microRNA levels induced by Intercept correlated with the platelet activation (p < 0.05) and an impaired platelet aggregation response to ADP (p < 0.05). These results suggest that Intercept treatment may induce platelet activation, resulting in the release of microRNAs and mRNAs from platelets. The clinical implications of this reduction in platelet nucleic acids secondary to Intercept remain to be established.
Keywords: MicroRNA, pathogen reduction, platelet, platelet function, transfusion medicine
Introduction
Platelets contain a diverse array of biologically important ribonucleic acid (RNA) species, including messenger RNAs (mRNAs) and mRNA-regulatory microRNAs. Inherited from their megakaryocytic precursors [1], up to a third of all human genes are present in platelets at the mRNA level [2]. Platelets possess the essential components of the translational machinery to translate their mRNAs into proteins [3–7] both in the circulation and during storage in the blood bank [8].
Platelets also contain an abundant and diverse array of mRNA-regulatory microRNAs [9–13] – small, 19- to 24-nucleotide (nt)-long non-coding RNA species – as well as a functioning microRNA pathway with the ability to synthesize microRNAs and mediate their function [11]. Bray and colleagues (i) described a link between the level of a microRNA (i.e. miR-96) and the platelet reactivity [14], (ii) reported a correlation between microRNA–mRNA coexpression profiles and platelet reactivity [13], and (iii) showed that platelet microRNAs can repress platelet protein expression [8, 13]. This recent evidence supports a role for microRNAs in regulating platelet function.
Current pathogen reduction (PR) systems for platelets rely on chemically induced cross-linking and inactivation of nucleic acids [15]. Two of these systems, Intercept and Mirasol, have been proven to be effective at reducing the levels of disease-causing agents. However, the SPRINT randomized controlled trial (RCT) found that PR causes loss of a proportion of the PR-treated platelets, resulting in reduced platelet dose per component, but not increased bleeding, because a 10 000/µl platelet count in patients with hypoproliferative thrombocytopenia is maintained through increased platelet transfusions [16]. Accordingly, PR of platelets was recommended to prevent the transmission of known and emerging pathogens containing nucleic acids, as well as the vast majority of cases of transfusion-associated sepsis (TAS) – a febrile to fatal reaction to transfusion of platelets contaminated with bacteria [17].
Subsequently, the RCT of Kerkoffs et al. [18] found a significant increase in bleeding in recipients of pathogen-reduced (versus non-pathogen-reduced) platelets. Other complications were also reported in recipients of pathogen-reduced platelets, including (i) six patients suffering of eye disorders (three patients with retinal hemorrhage), compared with none in the reference group, in the euroSPRITE trial aimed to evaluate the therapeutic efficacy and safety of Intercept [19], (ii) two patients suffering from intracranial bleedings (one patient died), compared with none in the reference group, in the RCT assessment of Mirasol [20], and (iii) one patient who died of intracranial bleeding after going off-protocol in the Intercept-treated group [18]. Although the prevalence of these complications did not attain statistical significance, when compared with their respective reference group, they tend to support a negative effect of PR on platelets. A meta-analysis of all completed RCTs reported a significant increase in all and in clinically significant bleeding when the analysis [21, 22] integrated the results of the safety report of the SPRINT trial [23], albeit not when it [22] used the findings from the initial report of that RCT [16]. A recent Cochrane review by Butler et al., in which 10 RCTs were analyzed, also concluded that there was no difference in clinically significant or severe bleeding between pathogen-reduced and standard platelets, but that there were significantly reduced responses following the transfusion of pathogen-reduced platelets, with a requirement for more platelet transfusions [24].
PR systems were developed before platelets were known to contain significant amounts of nucleic acids, such as mRNAs and microRNAs. To explain the observed increase in bleeding secondary to PR [18, 21, 22], we posited that PR could reduce the microRNA content of platelets through cross-linking and inactivation of their double-stranded duplexes of microRNA strands of 16–27 nt in length (median: 22 nt) [25], microRNA precursors (pre-microRNAs; 41–180 nt, with a median of 83 nt) [25], and/or primary microRNAs (pri-microRNAs; a significant fraction have lengths between 1 and 10 kb) [26], leading to reduced synthesis of microRNAs [27], provided that PR completely inhibited PCR detection of amplicons, amplified from platelet-derived mitochondrial DNA, between 868 and 1248 bp in length [28]. Alternatively, platelet activation secondary to PR [29–33] could alter the platelet content in microRNAs, as suggested by the up- and down-regulation of microRNAs observed in thrombin-activated platelets [9]. Either mechanism could result in altered microRNA content and/or function in PR-treated platelets, impairment of protein synthesis during their storage in the blood bank [8], and impairment of their function [29, 31, 34], which could increase bleeding [18, 21] despite the compensatory platelet transfusions given to restore the platelet losses [29, 31] caused by PR. To test our hypothesis, we tested whether platelet microRNAs are reduced by PR, the two postulated mechanisms for the reduction, and the effect of the reduction on platelet function.
Methods
The methods are described in more detail in the Supplementary information methods section.
Study design
The main objective of this study was to document the effects of three PR strategies on platelet microRNAs, mRNAs, activation, and function. Therefore, we designed our study accordingly, so that each of the Irradiation, mirasol and intercept groups could be compared with their most relevant control groups. For that purpose, 50 single-donor (apheresis) PCs (n = 10 per group) were collected, processed, and stored according to applicable standards [35]. The PCs were subjected to the following five treatments: control (platelets stored in donor plasma); additive solution (platelets stored in 65% storage solution for platelets [SSP+; MacoPharma] and 35% donor plasma); Irradiation (platelets treated with gamma irradiation [30 Gy] and stored in donor plasma); mirasol (platelets treated with riboflavin and ultraviolet-B (UVB) light and stored in donor plasma); or intercept (platelets stored in SSP+ – the same medium as in the additive solution group, which was found to be a suitable additive solution for intercept platelets [31] – and treated with amotosalen and UVA light). Although both SSP+ and InterSol are recommended for use with intercept platelets by Cerus Corporation, we used SSP+ because it causes less platelet activation during storage than InterSol [36]. All treatments were applied according to the standard procedures or the manufacturer's instructions without modification. Sterility tests performed on PCs stored for more than 24 hours were negative.
Platelet isolation, RNA extraction and analysis by quantitative polymerase chain reaction qPCR)
Platelets were isolated from platelet-rich plasma (PRP) samples collected from the PCs, according to a published protocol [11]. The number of contaminating leukocytes in the platelet preparations after negative CD45+ selection routinely yielded a single contaminating leukocyte per 3.2 × 106 platelets [11]. Total RNA, including small RNAs, was extracted using mirVana miRNA isolation reagent, according to the supplier's instructions (Life Technologies, Carlsbad, CA).
Megaplex RT primers (Human Pool Set v3.0; Life Technologies, Carlsbad, CA) were used to reverse transcribe 11 selected microRNAs [9], which were quantified on days 1, 4, and 7 after treatment (n = 10 for each group) by qPCR using TaqMan miRNA Expression Assays (Life Technologies) and U6 small nuclear RNA [snRNA]) as a reference gene [37]. Nine of the 11 investigated microRNAs were selected among the 20 most abundant human platelet microRNAs, as assessed by qPCR [9], whereas the two remaining microRNAs belong to the Let-7 family that was reported to be predominantly expressed in platelets in a recent RNA-Seq study [10]. We focused on the most abundant microRNAs in order to obtain the most accurate measurements and because they are also more likely to fulfill an important role in modulating platelet activation and function.
The absolute number of copies of the pro-survival Bcl-2-like protein 1 (BCL2L1; also called Bcl-xl) and B-cell CLL/lymphoma 2 (BCL2) mRNAs, and of the anti-apoptotic Clusterin (CLU) platelet mRNAs were determined at 1.5 and 20 hour after treatment (n = 6 for controls and n = 4 for each treatment group) by qPCR using an eight-point standard curve in duplicates on a StepOnePlus qPCR apparatus (Life Technologies).
Analyses of microRNA synthesis, function, and cross-linking
Platelet microRNA biogenesis and function were assessed in Dicer and RISC activity assays, as described previously [11, 38].
To detect the presence of cross-linked RNA adducts in PR-treated platelets, 50 ng of total platelet RNA were dephosphorylated using antarctic phosphatase at 37 °C for 15 min. The enzyme was inactivated by heating at 65 °C for 5 min, and 10 ng of RNA were subjected to phosphorylation using [32-P]-ATP for 1 hour at 37 °C. Phosphorylated RNAs were directly resolved by denaturing polyacrylamide gel electrophoresis (PAGE) 12% and autoradiography.
Platelet activation and function tests
Samples collected from the PCs on days 1, 4, and 7 after treatment were also analyzed for platelet CD62P (P-selectin) surface expression, a marker of platelet activation [39], by flow cytometry.
PRP samples obtained from the PCs on days 1, 4, and 7 underwent ADP, TRAP (thrombin-receptor activating peptide), and collagen-induced turbidimetric platelet aggregation tests.
Statistical analysis
Fold changes in microRNA levels were analyzed with DataAssist software (Life Technologies) for relative qPCR quantification, and the data presented as recommended by Van Peer et al. [40]. A difference was considered significant when the two-sided p value remained <0.05 after correction for 132 comparisons (4 treatments × 11 microRNAs × 3 time points).
Data from the three anti-apoptotic mRNAs, Dicer and RISC activity assays, platelet activation and aggregation tests, platelet volume and platelet count in PCs, were compared across five groups at each time point (day 1, 4, or 7 of storage) by non-parametric analysis of variance (ANOVA), followed by a Kruskal–Wallis test with correction for multiple comparisons.
The existence of a linear correlation between the relative change in the percentage of activated platelets and the microRNA fold change was tested by a general linear model using SAS 9.3 software (Software Intelligence Corporation, Spring Valley, CA).
Results
Altered microRNA profiles in stored platelets
Treatments of stored platelets with additive solution and Irradiation significantly (p < 0.05) reduced the level of only one microRNA each (miR-223 and let-7e, respectively), and they did so only on day 7 of storage (i.e. beyond the clinically relevant storage period of 5 days for untreated platelets, but corresponding to the shelf life of PR-treated platelets) (Figure 1A and K, respectively). Mirasol did not significantly reduce the level of any of the 11 tested microRNAs (Figure 1), although the level of let-7e was 56% lower in mirasol-treated platelets than in controls on day 7 (Figure 1K). Platelet microRNA levels remained stable in the control samples over 7 days of storage (Supplementary Figure S2).
Figure 1.
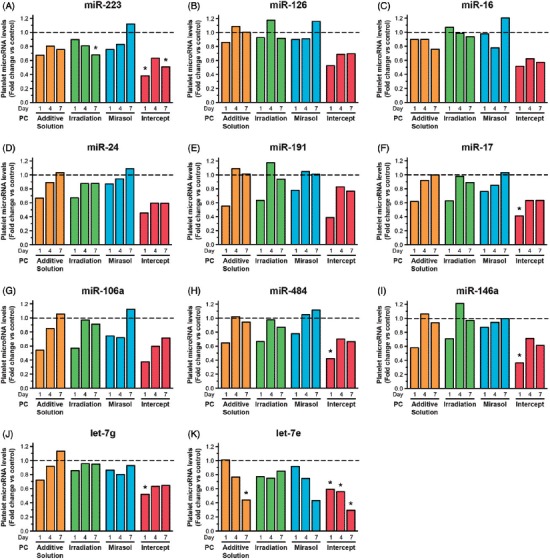
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on platelet microRNA levels. The figure shows the median fold change for each individual platelet microRNA level, in each of the four treatment groups (additive solution, Irradiation, mirasol, or intercept) and at each time point (i.e. day 1, 4, or 7 of storage after treatment), compared with its level in the control group at the same time point, which is set at 1.0 (—) (n = 10 donors per group). All comparisons were two-sided. ∗p < 0.05, namely a statistically significant reduction in the level of the specific microRNA for the corresponding treatment group versus the control group at the particular time point and following correction for multiple comparisons.
In contrast, Intercept significantly reduced the level of 6 (54.5%) of the 11 tested microRNAs on day 1 (Figure 1A, F, and H–K), 1 microRNA on day 4 (Figure 1K), and 2 microRNAs on day 7 (Figure 1A and K). Let-7e was reduced by as much as 70% by day 7 (Figure 1K). The microRNA profile of platelets stored in additive solution resembled that of the platelets treated with intercept (Figure 1), perhaps because platelets were stored in the same additive solution (SSP+).
Intercept causes deregulation of the pro-survival gene Bcl-xl and the anti-apoptotic gene CLU
The platelet count of PCs differed across the five groups on days 1, 4, and 7 of storage (p < 0.001 in all cases). Consistent with previous reports [29, 31], mirasol and intercept platelet counts (103/µl) were significantly lower than the control (mean ± standard deviation; SD): on day 1 (1024 ± 103, 1052 ± 81 and 1339 ± 181), on day 4 (983 ± 122, 997 ± 85 and 1296 ± 200), and on day 7 (985 ± 118, 979 ± 78 and 1304 ± 205), respectively.
Mice devoid of Bcl-xl exhibit a reduction in platelet life span from 5 days to 24 hours [41]. Because PR causes early platelet losses [29, 31] and that inhibition of Bcl-xl induces platelet apoptosis [42], we investigated whether PR reduces the level of pro-survival Bcl-xl and BCL2 mRNAs, as well as that of the anti-apoptotic mRNA CLU during the initial 24 hours after treatment. The number of copies of the relatively abundant Bcl-xl and CLU mRNAs differed across the five groups at 1.5 hour post-treatment (p = 0.022 and p = 0.032, respectively); Bcl-xl also differed at 20 hours (p = 0.025) post-treatment (Figure 2A and B). At both 1.5 and 20 hours, only the number of Bcl-xl copies in intercept platelets differed from that in controls (p < 0.05). Similarly, at 1.5 hours, only the number of CLU copies in the intercept platelets differed from that in the controls (p < 0.05). The level of the less abundant BCL2 mRNA was not significantly reduced in any of the five groups at 1.5 and 20 hours after treatment compared with the control group (Figure 2C). These results suggest that the lower platelet count subsequent to intercept treatment may be related, at least in part, to changes in platelet anti-apoptotic gene expression.
Figure 2.
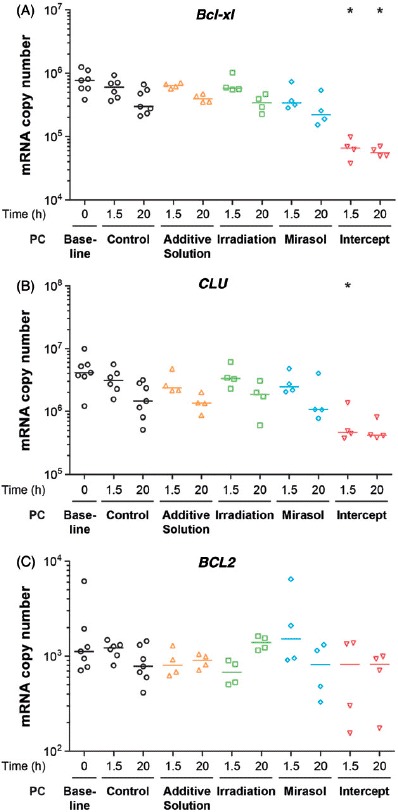
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on platelet apoptosis-related mRNAs. The figure shows the absolute number of copies of Bcl-xl (A), CLU (B), and BCL2 (C) mRNAs in platelets isolated from samples collected from PCs at 1.5 and 20 hours after treatment with additive solution, Irradiation, mirasol, intercept, or no treatment (control). For each group and time point, the figure shows the individual data (n = 4–6 donors per group), along with their median. All comparisons were two-sided. ∗p < 0.05, namely a statistically significant decrease in the number of mRNA copies at 1.5 or 20 hours for the corresponding group, compared with the control group at the same time point).
PR does not affect platelet microRNA synthesis
Supplementary Figure S3 shows the standardization and validation of our Dicer activity assays. Platelet Dicer activity did not differ across the five groups on day 1, 4, or 7 of storage (Figure 3), and immunoblot analyses showed no difference in Dicer protein levels among the five groups (data not shown). These results suggest that the reduction in platelet microRNA levels induced by PR treatment is not related to a compromised microRNA synthesis.
Figure 3.
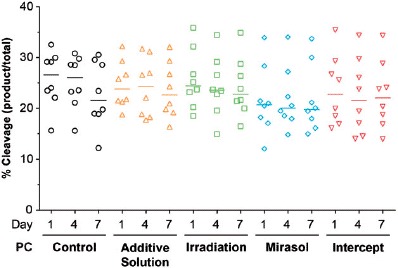
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on platelet microRNA synthesis. The figure shows the percentage of pre-microRNA substrate cleaved into mature microRNA by platelet Dicer, which is known to mediate platelet microRNA biogenesis [11]. Dicer activity assays were performed by coincubating a 32P-labeled pre-microRNA substrate (32P-labeled pre-let-7a-3) with protein extracts from platelets collected from PCs at 1, 4, and 7 days of storage after treatment with additive solution, Irradiation, mirasol, intercept, or no treatment (control). For each group and time point, the figure shows the individual data (n = 8 donors per group), along with their median.
PR does not induce formation of cross-linked RNA adducts
To determine if the PR-induced reduction in platelet microRNA levels may be related to cross-linking of endogenous RNA, we visualized platelet RNA by 32P-labeling and denaturing PAGE. Separated according to their molecular weight, platelet RNA affected by intra or intermolecular cross-linking events would be expected to migrate differently and exhibit a modified band pattern and/or intensity. These analyses did not reveal any modified band pattern and/or intensity indicative of the formation of a new and/or cross-linked adduct involving RNA species shorter than 300 nt that could be attributed to PR treatment (Figure 4). Similar results were obtained when platelet RNA was not dephosphorylated prior to labeling (Supplementary Figure S5).
Figure 4.
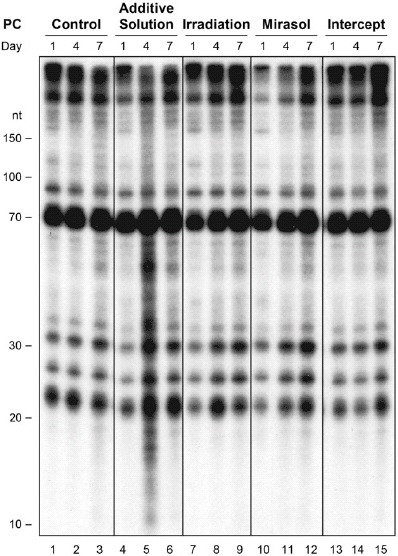
Storage in additive solution or treatment with Irradiation, mirasol, or intercept does not induce formation of cross-linked adducts of endogenous platelet RNAs. The figure shows the lack of cross-linked adducts of endogenous platelet RNAs that can be attributed to treatment with additive solution, Irradiation, mirasol or intercept, as compared with no treatment (control), at any time point (day 1, 4, or 7 of storage) by total RNA extraction, dephosphorylation, 32P labeling, and analysis by denaturing PAGE. Compare with Supplementary Figure S4, which shows the results without dephosphorylation.
PR does not affect platelet microRNA function
Supplementary Figure S5 shows the standardization and validation of our RISC activity assays. Platelet RISC activity did not differ across the five groups on day 1, 4, or 7 of storage (Figure 5), although it was slightly increased in the additive solution and intercept samples. Immunoblot analyses showed no differences in the level of Ago2 proteins, the core effector of platelet microRNA function [11], among the five groups (data not shown). These results suggest that platelet Ago2•microRNA function may not be altered upon PR treatment.
Figure 5.
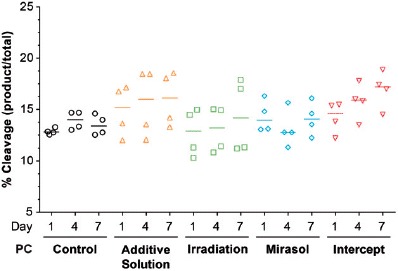
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on the ability of platelets to mediate microRNA function. The figure shows the percentage of miR-223 sensor RNA substrate cleaved by platelet Ago2•miR-223 complexes, which is known to mediate platelet microRNA function [11]. RISC activity assays were performed by coincubating a 32P-labeled RNA sensor perfectly complementary to miR-223 with protein extracts from platelets collected from PCs at 1, 4, and 7 days of storage after treatment with additive solution, Irradiation, mirasol, intercept, or no treatment (control). For each group and time point, the figure shows the individual data (n = 4 donors per group), along with their median.
PR causes platelet activation which correlates with the observed reduction in platelet microRNAs
The percentage of platelets expressing CD62P, a marker of platelet activation [39], on their surface differed across all five groups on day 1 (p < 0.001) and day 4 (p < 0.001) of storage (Figure 6). On day 1, the additive solution and intercept groups, and less so the Irradiation group, demonstrated greater CD62P surface expression compared with the control group (p < 0.05). The activation level of the mirasol platelets was similar to that of the controls. All four treatment groups, including mirasol, showed more activation than the control group on day 4 (p < 0.05), while on day 7 no treatment group's activation level differed significantly from that of the control group.
Figure 6.
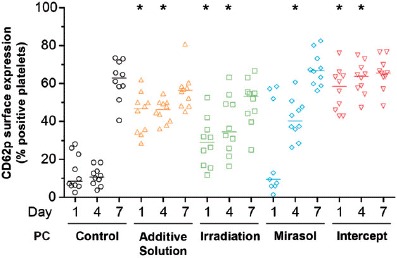
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on the activation level of platelets. The figure shows the percentage of platelets expressing CD62P on their surface by flow cytometry analysis of samples collected from the PCs on day 1, 4, or 7 for each group (storage in additive solution, treatment with Irradiation, mirasol or intercept, or control). For each group, the figure shows the individual data (n = 10 donors per group) obtained at each time point, along with the median. All comparisons were two-sided. ∗p < 0.05, namely a statistically significant decrease in the corresponding treatment (compared with the control) group on the same day of storage.
For the intercept-treated platelets (which showed both platelet activation (Figure 6) and reduced platelet microRNA levels (Figure 1), general linear models demonstrated significant linear correlations between the relative change in platelet activation and the corresponding microRNA fold change for 8 of the 11 microRNAs tested (p < 0.05): miR-126, miR-16, miR-17, miR-191, miR-484, miR-106a, miR-146a, and let-7g. No linear correlation was detected for the other three treatments that did not produce significant reductions in platelet microRNA levels (Figure 1).
PR impairs some aspects of platelet function
The aggregation response of platelets to ADP differed across the five groups on day 1 (p < 0.001), day 4 (p < 0.001), and day 7 (p < 0.001) of storage (Figure 7A). Whereas the aggregation response of mirasol platelets did not differ from that of controls on day 1, it was virtually absent on day 4 (p < 0.001 compared with controls), and it remained so on day 7 (p < 0.001). The aggregation response of both the intercept and additive solution platelets was almost absent already on day 1 (p < 0.001 compared with controls), and it remained so on days 4 (p < 0.05) and 7 (p < 0.001).
Figure 7.
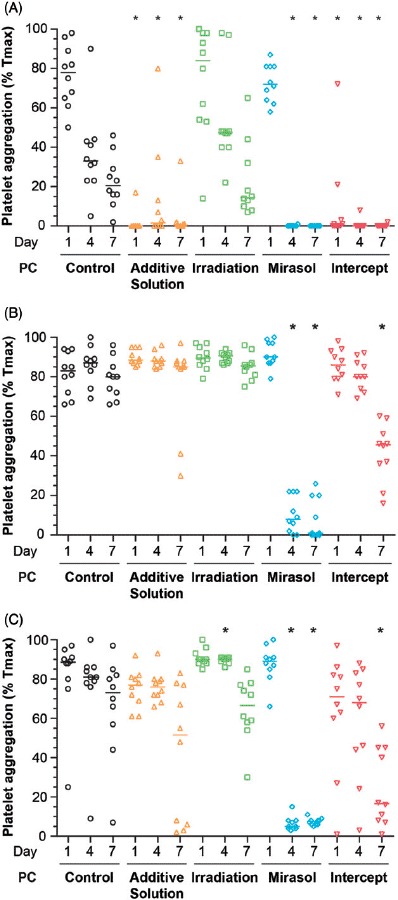
Effect of storage in additive solution or treatment with Irradiation, mirasol, or intercept on platelet aggregation. The figure shows the aggregation response to 20 µM ADP (A), 40 µM TRAP (B) and 4 µg/ml collagen (C) of samples collected from the PCs on day 1, 4, or 7 for each group (control, additive solution, Irradiation, mirasol, or intercept). Individual results from each PC, expressed as a percentage of maximal transmission (Tmax), are shown along with the median (n = 10 donors per group). All comparisons were two-sided. ∗p < 0.05, namely a statistically significant difference in aggregation in the corresponding treatment (compared with the control) group on the same day of storage.
The aggregation response to TRAP differed across the five groups on day 4 (p < 0.001) and 7 (p < 0.001; Figure 7B). On day 4, only the mirasol group differed from the control group (p < 0.001), while on day 7 both mirasol and intercept platelets differed from the controls (p < 0.001). The aggregation response to collagen also differed across the five groups on days 4 (p < 0.001), and 7 (p < 0.001; Figure 7C). These data indicate that PR impairs the aggregation response of platelets.
Additive solution and intercept reduce platelet volume on day 1
Platelet activation is associated, in part, with changes in mean platelet volume [43]. The mean platelet volume did differ across all five groups on days 1 (p < 0.001) and 7 (p = 0.005) of storage (Supplementary Figure S6). On day 1, both additive solution and intercept platelets had a smaller volume (p < 0.05) compared with the controls, but on day 7 no group differed significantly from the control group following adjustment for multiple comparisons. The observed decrease in mean platelet volume may potentially be explained by the platelet activation and release of microparticles induced by additive solution and intercept treatments.
Additive Solution and Intercept enhance the total amount of platelet microparticle-derived miR-223
Knowing that platelet activation is associated with the release of microparticles [44], and having recently demonstrated that activated platelets may release microRNAs through microparticles [45], we reasoned that activated stored platelets may also release microRNAs through such microparticles in the supernatant. To verify that possibility, we collected the supernatant fraction from a small number of PCs in each of the five groups and analyzed their total content in microparticular miR-223 [45], one of the most abundant platelet microRNAs [10, 11]. These analyses unveiled a 30–86% increase in the total amount of miR-223 in the microparticles released from additive solution and intercept-treated platelets, compared with the control platelets (Supplementary Figure S7). This increase, associated to additive solution and intercept-treated platelets, reached up to 3.8-fold compared with the baseline values versus 2.2-fold in the control group. The total amount of microparticular miR-223 released by platelets subjected to Irradiation or mirasol treatment was not higher to that observed in control platelets. These observations, which need to be corroborated by analyzing a larger number of samples, suggest that the release of microRNA-containing microparticles by PR-treated platelets may be a mechanism by which the microRNA content of PR-treated platelets may be modulated.
Discussion
PR systems have been developed and proposed to reduce the risk of transmission of infectious agents by transfusion of blood components, based on their PR efficacy in vitro. However, deleterious effects of PR on platelet counts and function have also been reported [16, 18, 21, 22]. In this study, we report a reduction in the microRNA content, and in the anti-apoptotic mRNA content, of platelets treated with intercept. This reduction in microRNA content coincided with platelet activation and impairment of some aspects of platelet function. Our findings may provide at least a partial explanation for the platelet losses [16, 24, 29, 31] known to occur with intercept, and for the increased bleeding secondary to PR with intercept in the RCT of Kerkhoffs et al. [18] and the meta-analysis of all intercept RCTs by Vamvakas [22]. Recently, a Cochrane systematic review reported benefit from non-PR-treated (compared with PR-treated) platelet transfusions for a range of laboratory outcomes, and it concluded that results from ongoing or new RCTs are required to determine if there are clinically important differences in bleeding risk between PR-treated and non-PR-treated platelet transfusions [24].
Our study is the first experimental demonstration of an altered (i.e., reduced) microRNA content in platelets treated for PR, such as with intercept. Although 6 of the 11 studied microRNAs (54%) were reduced by intercept already on day 1, no such effect was observed with gamma-irradiation (which is similar to UV light and has a long safety record). Because the reduction in platelet microRNA content was absent from both the control and Irradiation groups, and was observed only with intercept (Figure 1), it may well be clinically relevant. We found no published reports attributing a regulatory role vis-à-vis platelet function to the six microRNAs (and perhaps other) reduced by intercept. Although the regulatory properties of these six microRNAs have been inferred from studies performed in nucleated cells, recent studies tend to support a regulatory role for platelet microRNAs [14] and Ago2•microRNA complexes [11] in platelet biology [13, 14]; the definitive proof remains to be presented.
Platelets treated with either intercept or mirasol showed a similar reduction in platelet count on day 1 of storage. However, only intercept platelets exhibited a decrease in anti-apoptotic mRNAs during the initial 24 hours, suggesting that platelet losses secondary to intercept can be ascribed, in part, to increased platelet apoptosis due to deregulation of the pro-survival gene Bcl-xl and the anti-apoptotic gene CLU by intercept. Deregulation of these platelet mRNAs, which might also be consecutive to alterations in the level of specific microRNAs [46], may result in impaired protein synthesis during storage in the blood bank [8], and a correspondingly impaired function of the PR-treated platelets. This is a plausible mechanism to account for both the in vitro results reported here and the clinical bleeding reported previously [18, 21, 22], although (i) the occurrence and/or physiologic importance of protein synthesis during platelet storage in the blood bank [8], (ii) the role of microRNAs in regulating mRNA translation, function, and reactivity of anucleate platelets [47], and (iii) the clinical relevance of our platelet activation and aggregation findings, remain to be established by further research.
Our study is also the first experimental confirmation of the prediction [28] that amotosalen does not decrease the microRNA content of PR-treated platelets, either through reduced microRNA synthesis or formation of cross-linked adducts. Statistically, amotosalen forms adducts in double-stranded regions of nucleic acids every 100 base-pairs on average [28], with larger nucleic-acid molecules being more likely to be modified by amotosalen than are smaller molecules. One to 10-kb long pri-microRNAs may be modified by amotosalen, but the possibility that they contribute to the pool of mature platelet microRNAs is unlikely. Whereas platelets harbor Argonaute 2 and Dicer complexes that are active in mediating mature microRNA function and formation from pre-microRNA substrates, respectively [11, 38], they lack the nuclear pri-microRNA processing enzyme Drosha [11]. Although we cannot exclude a possible role for unprocessed pri-microRNAs in platelet function, these observations suggest that mature microRNAs and pre-microRNAs are the most likely to play a role in modulating platelet function. Our cross-linking analyses suggest that RNA species shorter than 300 nt, such as mature microRNAs and pre-microRNAs, may not be cross-linked to a significant extent, and are thus unlikely direct targets for amotosalen.
We propose that the reduction of the microRNA content observed on day 1 in intercept-treated platelets may be due to the release of microRNAs from the activated PR-treated platelets via microparticles. This possibility is supported by the observed decrease in platelet microRNA levels [9] and release of microparticles containing microRNAs [45] upon platelet activation with thrombin, and by the increased amount of microparticular miR-223 in the supernatant of intercept-treated platelets. The proposed mechanism is also supported by (i) the observed reduction in mean platelet volume caused by intercept on day 1 of storage (Supplementary Figure S6), which coincided with the fact that intercept-treated platelets were the most activated on day 1 (Figure 6), and (ii) the observed correlation between the platelet activation and the reduction in platelet microRNA levels induced by intercept. Although this latter correlation does not prove the existence of a causal link, it does not argue against it. Consistent with these reasoning, mirasol-treated platelets were the least activated on day 1, with an activation level similar to that of controls, and they demonstrated the least affected microRNA profile compared with the controls (Figure 1).
As with microRNAs, the reduction in apoptotic mRNA levels in PR-activated platelets could be explained by their possible release through microparticles [44], which have been shown to contain mRNAs [48]. PR treatment-induced deregulation of pro-survival gene expression in stored platelets may explain the known platelet loss and increased bleeding secondary to administration of intercept-treated platelets, as reported clinically. This possibility is supported by a recent study reporting that inhibition of Bcl-xl and BCL2 induces platelet apoptosis, depletes intracellular calcium, and prevents platelet stimulation by physiological agonists [42].
Although both intercept and mirasol have been reported to cause increased platelet activation and reduced platelet aggregation to varying extents [29–33], with negative results reported as well [49, 50], intercept inactivates mitochondrial DNA [51] while mirasol may not affect mitochondrial function [52]. Because of the contribution of mitochondria to platelet function [53, 54], and the reduced platelet microRNA content secondary to intercept (Figure 1), we might have expected more of a functional impairment with intercept than with mirasol, as we have seen when using ADP as an agonist (Figure 7A).
Several aspects pertaining to the storage solution have to be taken into account when analyzing and interpreting the results from our platelet aggregation tests. The differential plasma content (between 35% and 100%) and fibrinogen concentration of the PCs is unlikely to have exerted confounding effects in our ADP-induced aggregation tests, since, when citrated platelet-rich plasma isolated from PCs is diluted with additive solution at different plasma/additive solution ratios, the platelet response to 20 µmol/l of ADP is not affected by plasma concentration as low as 30% (data not shown). A recent study by Nogawa et al. (2013) [36] has confirmed the findings previously reported by Keuren et al. (2006) [55] and demonstrated that additive solution containing magnesium and potassium causes less platelet activation during storage than additive solution without magnesium and potassium, such as intersol used for intercept. Hence, the importance of using the same additive solution (i.e. SSP+) for both additive solution and intercept PC preparations. We cannot exclude the possibility that the additive solution may have desensitized intercept platelets to ADP, as reported for platelets stored in PAS-II or composol [55]. It is also important to consider that weak platelet aggregation responses to ADP are in accordance with high percentages of platelet activation and associated ADP secretion, which results in refractoriness to subsequent stimulation by ADP [55]. The clinical relevance of the impaired aggregation response to ADP that intercept produced in our study remains to be determined.
The finding of reduced platelet microRNA and mRNA content in intercept, but not mirasol, platelets suggest that mirasol may not be as “harsh” on platelets as intercept. Unlike mirasol, intercept uses a compound absorbing device to remove residual amotosalen and its photometabolites. This renders the process rather complex, during which platelets come into contact with several different surfaces that may induce platelet surface changes and loss of granule contents.
Intercept treatment also involves storage in additive solution. Although smaller and, for the most part, non-significant, the changes in platelet microRNA levels, platelet activation, and ADP-induced platelet aggregation induced by additive solution paralleled those induced by intercept, suggesting an important contribution of the additive solution to the effects observed with intercept. This reasoning may partly explain the milder effects observed in mirasol-treated platelets, which were stored in plasma in the present study in order to facilitate comparisons with control platelets also stored in plasma. This a priori study design, however, has the disadvantage to limit comparisons between treatment groups using different storage solutions. Although mirasol platelets have been approved for storage in either plasma or additive solution, the incentive to blood operators to reserve plasma for other uses may mean that mirasol platelets will likely be prepared in additive solution. In that case, additive solution would be expected to exacerbate the effects associated with mirasol treatment.
A technical limitation of our study is that when platelets were isolated from the PCs, the platelet population that we actually recovered and studied could have been influenced by the degree of platelet activation and rate of cell death, which may have varied between treatments. Platelets most severely affected (i.e. activated) or killed by PR may not have been fully recovered, whereas less severely affected platelets or platelets undergoing apoptosis may have been sampled. Such treatment-dependent variations in the platelet population isolated from the PCs (e.g. differential changes in cell density, due to the formation of platelet aggregates, and compromised structural integrity) may thus limit the interpretation of our data. Additionally, we used relative quantification, the most frequently used real-time qPCR method [56], to determine platelet microRNA levels [37]. Our findings may thus need to be confirmed by absolute quantification, i.e. the exact copy number of each platelet microRNA [56].
Although the exact role of microRNAs in platelets remains to be established [8], their recognized importance in regulating mRNA translation [57] supports the possibility that PR-induced reduction in platelet microRNA levels may affect platelet function. In that context, it would be tempting to speculate that the impaired function of the intercept-treated platelets, which may contribute to the increased bleeding observed in some recipients of intercept-treated (versus non-pathogen-reduced) platelets [18, 21, 22], may be related to the reduction in platelet microRNA levels secondary to intercept treatment. Therefore, it may not be prudent to pursue the implementation of PR of platelets until PR is demonstrated not to cause functional impairment of the treated platelets owing to a reduction in the platelet microRNA content.
Acknowledgements
We thank Anna Pfister and Anette Schleifer for technical assistance, Achim Jung for the preparation of the samples and Susanne Seifert-Hitzler for logistic support.
Declaration of interest
B. L. was supported by a PhD studentship from the Canadian Blood Services. This work was supported by Grant nos. LIO-202231 and LIO-203061 from the County Council of Östergötland, Sweden (to A. O.) and Grant no. 286777 from the Canadian Blood Services/Canadian Institutes of Health Research (CIHR) Blood Utilization and Conservation Initiative via Health Canada (to P. P.). The views expressed herein do not necessarily represent the view of the federal government. There was no involvement of a pharmaceutical/other company in funding this study. The authors declare that they have no conflicts of interest.
Authorship contributions
A. O., W. E. H., E. C. V., and P. P. conceived and coordinated the study; A. O., W. E. H. and P. P. designed and planned the experiments; W. E. H. coordinated the sampling and treatment of the platelet concentrates; A. O., C. U. M., P. L., A. C., B. L., and P. H. performed the experiments; E. B. provided new analytical tools and expertise; A. O. and E. C. V. performed the statistical analyses; all authors analyzed the data and edited/commented on the manuscript; A. O., W. E. H., E. V. C., and P. P. wrote the manuscript.
References
- 1.Roth GJ, Hickey MJ, Chung DW, Hickstein DD. Circulating human blood platelets retain appreciable amounts of poly (A)+ RNA. Biochem Biophys Res Commun. 1989;160(2):705–710. doi: 10.1016/0006-291x(89)92490-x. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118(14):e101–e111. doi: 10.1182/blood-2011-03-339705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ts'ao CH. Rough endoplasmic reticulum and ribosomes in blood platelets. Scand J Haematol. 1971;8(2):134–140. doi: 10.1111/j.1600-0609.1971.tb01964.x. [DOI] [PubMed] [Google Scholar]
- 4.Kieffer N, Guichard J, Farcet JP, Vainchenker W, Breton-Gorius J. Biosynthesis of major platelet proteins in human blood platelets. Eur J Biochem. 1987;164(1):189–195. doi: 10.1111/j.1432-1033.1987.tb11010.x. [DOI] [PubMed] [Google Scholar]
- 5.Booyse FM, Rafelson ME., Jr Studies on human platelets. I. synthesis of platelet protein in a cell-free system. Biochim Biophys Acta. 1968;166(3):689–697. doi: 10.1016/0005-2787(68)90376-6. [DOI] [PubMed] [Google Scholar]
- 6.Weyrich AS, Dixon DA, Pabla R, Elstad MR, McIntyre TM, Prescott SM, Zimmerman GA. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc Natl Acad Sci U S A. 1998;95(10):5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evangelista V, Manarini S, Di Santo A, Capone ML, Ricciotti E, Di Francesco L, Tacconelli S, Sacchetti A, D'Angelo S, Scilimati A, et al. De novo synthesis of cyclooxygenase-1 counteracts the suppression of platelet thromboxane biosynthesis by aspirin. Circ Res. 2006;98(5):593–595. doi: 10.1161/01.RES.0000214553.37930.3e. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein LC, Bray PF. MicroRNAs in platelet production and activation. Blood. 2011;117(20):5289–5296. doi: 10.1182/blood-2011-01-292011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osman A, Falker K. Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes. Platelets. 2011;22(6):433–441. doi: 10.3109/09537104.2011.560305. [DOI] [PubMed] [Google Scholar]
- 10.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16(9):961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruchova H, Merkerova M, Prchal JT. Aberrant expression of microRNA in polycythemia vera. Haematologica. 2008;93(7):1009–1016. doi: 10.3324/haematol.12706. [DOI] [PubMed] [Google Scholar]
- 13.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, Chen J, McKnight GS, Lopez JA, Yang L, et al. Platelet microRNA–mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondkar AA, Bray MS, Leal SM, Nagalla S, Liu DJ, Jin Y, Dong JF, Ren Q, Whiteheart SW, Shaw C, et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: Suggested role for platelet microRNA. J Thromb Haemost. 2010;8(2):369–378. doi: 10.1111/j.1538-7836.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wollowitz S. Fundamentals of the psoralen-based Helinx technology for inactivation of infectious pathogens and leukocytes in platelets and plasma. Semin Hematol. 2001;38(4 Suppl 11):4–11. doi: 10.1016/s0037-1963(01)90118-0. [DOI] [PubMed] [Google Scholar]
- 16.McCullough J, Vesole DH, Benjamin RJ, Slichter SJ, Pineda A, Snyder E, Stadtmauer EA, Lopez-Plaza I, Coutre S, Strauss RG, et al. Therapeutic efficacy and safety of platelets treated with a photochemical process for pathogen inactivation: The SPRINT trial. Blood. 2004;104(5):1534–1541. doi: 10.1182/blood-2003-12-4443. [DOI] [PubMed] [Google Scholar]
- 17.Klein HG, Anderson D, Bernardi MJ, Cable R, Carey W, Hoch JS, Robitaille N, Sivilotti ML, Smaill F. Pathogen inactivation: Making decisions about new technologies. Report of a consensus conference. Transfusion. 2007;47(12):2338–2347. doi: 10.1111/j.1537-2995.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- 18.Kerkhoffs JL, van Putten WL, Novotny VM, Te Boekhorst PA, Schipperus MR, Zwaginga JJ, van Pampus LC, de Greef GE, Luten M, Huijgens PC, et al. Clinical effectiveness of leucoreduced, pooled donor platelet concentrates, stored in plasma or additive solution with and without pathogen reduction. Br J Haematol. 2010;150(2):209–217. doi: 10.1111/j.1365-2141.2010.08227.x. [DOI] [PubMed] [Google Scholar]
- 19.van Rhenen D, Gulliksson H, Cazenave JP, Pamphilon D, Ljungman P, Kluter H, Vermeij H, Kappers-Klunne M, de Greef G, Laforet M, et al. Transfusion of pooled buffy coat platelet components prepared with photochemical pathogen inactivation treatment: The euroSPRITE trial. Blood. 2003;101(6):2426–2433. doi: 10.1182/blood-2002-03-0932. [DOI] [PubMed] [Google Scholar]
- 20.Mirasol Clinical Evaluation Study Group A randomized controlled clinical trial evaluating the performance and safety of platelets treated with MIRASOL pathogen reduction technology. Transfusion. 2010;50(11):2362–2375. doi: 10.1111/j.1537-2995.2010.02694.x. [DOI] [PubMed] [Google Scholar]
- 21.Vamvakas EC. Meta-analysis of the randomized controlled trials of the hemostatic efficacy and capacity of pathogen-reduced platelets. Transfusion. 2011;51(5):1058–1071. doi: 10.1111/j.1537-2995.2010.02925.x. [DOI] [PubMed] [Google Scholar]
- 22.Vamvakas EC. Meta-analysis of the studies of bleeding complications of platelets pathogen-reduced with the Intercept system. Vox Sanguinis. 2012;102(4):302–316. doi: 10.1111/j.1423-0410.2011.01555.x. [DOI] [PubMed] [Google Scholar]
- 23.Snyder E, McCullough J, Slichter SJ, Strauss RG, Lopez-Plaza I, Lin JS, Corash L, Conlan MG. Clinical safety of platelets photochemically treated with amotosalen HCl and ultraviolet A light for pathogen inactivation: The SPRINT trial. Transfusion. 2005;45(12):1864–1875. doi: 10.1111/j.1537-2995.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 24.Butler C, Doree C, Estcourt LJ, Trivella M, Hopewell S, Brunskill SJ, Stanworth S, Murphy MF. Pathogen-reduced platelets for the prevention of bleeding. Cochrane Database Syst Rev. 2013;3:CD009072. doi: 10.1002/14651858.CD009072.pub2. [DOI] [PubMed] [Google Scholar]
- 25.Fang Z, Du R, Edwards A, Flemington EK, Zhang K. The sequence structures of human microRNA molecules and their implications. PLoS One. 2013;8(1):e54215. doi: 10.1371/journal.pone.0054215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hitzler W, Vamvakas EC. Platelet microRNA profiles and the effect of pathogen reduction on platelet function. Clin Lab. 2011;57(7–8):451–454. [PubMed] [Google Scholar]
- 28.Bruchmuller I, Janetzko K, Bugert P, Mayaudon V, Corash L, Lin L, Kluter H. Polymerase chain reaction inhibition assay documenting the amotosalen-based photochemical pathogen inactivation process of platelet concentrates. Transfusion. 2005;45(9):1464–1472. doi: 10.1111/j.1537-2995.2005.00553.x. [DOI] [PubMed] [Google Scholar]
- 29.Apelseth TO, Bruserud O, Wentzel-Larsen T, Bakken AM, Bjorsvik S, Hervig T. In vitro evaluation of metabolic changes and residual platelet responsiveness in photochemical treated and gamma-irradiated single-donor platelet concentrates during long-term storage. Transfusion. 2007;47(4):653–665. doi: 10.1111/j.1537-2995.2007.01167.x. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Pujol S, Tonda R, Lozano M, Fuste B, Lopez-Vilchez I, Galan AM, Li J, Goodrich R, Escolar G. Effects of a new pathogen-reduction technology (Mirasol PRT) on functional aspects of platelet concentrates. Transfusion. 2005;45(6):911–919. doi: 10.1111/j.1537-2995.2005.04350.x. [DOI] [PubMed] [Google Scholar]
- 31.Johnson L, Loh YS, Kwok M, Marks DC. In vitro assessment of buffy-coat derived platelet components suspended in SSP+ treated with the Intercept blood system. Transfusion Med. 2013;23(2):121–129. doi: 10.1111/tme.12020. [DOI] [PubMed] [Google Scholar]
- 32.Carvalho H, Alguero C, Santos M, de Sousa G, Trindade H, Seghatchian J. The combined effect of platelet storage media and intercept pathogen reduction technology on platelet activation/activability and cellular apoptosis/necrosis: Lisbon-RBS experience. Transfusion and apheresis science: Official journal of the World Apheresis Association: Official journal of the European Society for Haemapheresis. 2006;34(2):187–192. doi: 10.1016/j.transci.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 33.Tauszig ME, Picker SM, Gathof BS. Platelet derived cytokine accumulation in platelet concentrates treated for pathogen reduction. Transfus Apher Sci. 2012;46(1):33–37. doi: 10.1016/j.transci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Picker SM, Speer R, Gathof BS. Functional characteristics of buffy-coat PLTs photochemically treated with amotosalen-HCl for pathogen inactivation. Transfusion. 2004;44(3):320–329. doi: 10.1111/j.1537-2995.2003.00590.x. [DOI] [PubMed] [Google Scholar]
- 35.Bekanntmachung der Richtlinien zur Gewinnung von Blut und Blutbestandteilen und zur Anwendung von Blutprodukten (Hämotherapie) gemäß §§ 12 und 18 des Transfusionsgesetzes (TFG) (Änderungen und Ergänzungen 2010) vom 4. Mai 2010, Herausgegeben vom Bundesministerium der Justiz, Bundesanzeiger Nr. 101a. Jahrgang 62, 9. Juli 2010 (ISSN 0720-6100)
- 36.Nogawa M, Naito Y, Chatani M, Onodera H, Shiba M, Okazaki H, Matsuzaki K, Satake M, Nakajima K, Tadokoro K. Parallel comparison of apheresis-collected platelet concentrates stored in four different additive solutions. Vox Sang. 2013;105(4):305–312. doi: 10.1111/vox.12064. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Perron MP, Landry P, Plante I, Provost P. Detection of human dicer and argonaute 2 catalytic activity. Methods Mol Biol. 2011;725:121–141. doi: 10.1007/978-1-61779-046-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyant TL, Smith PC, Brown B, Kantor AB. Whole blood microvolume laser scanning cytometry for monitoring resting and activated platelets. Platelets. 2001;12(5):309–318. doi: 10.1080/09537100120068206. [DOI] [PubMed] [Google Scholar]
- 40.Van Peer G, Mestdagh P, Vandesompele J. Accurate RT-qPCR gene expression analysis on cell culture lysates. Sci Rep. 2012;2:222. doi: 10.1038/srep00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, Hilton AA, Ellis S, Kelly PN, Ekert PG, Metcalf D, Roberts AW, et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128(6):1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 42.Vogler M, Hamali HA, Sun XM, Bampton ET, Dinsdale D, Snowden RT, Dyer MJ, Goodall AH, Cohen GM. BCL2/BCL-X(L) inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood. 2011;117(26):7145–7154. doi: 10.1182/blood-2011-03-344812. [DOI] [PubMed] [Google Scholar]
- 43.Bath PM, Butterworth RJ. Platelet size: Measurement, physiology and vascular disease. Blood Coagul Fibrinolysis. 1996;7(2):157–161. [PubMed] [Google Scholar]
- 44.Boilard E, Nigrovic PA, Larabee K, Watts GF, Coblyn JS, Weinblatt ME, Massarotti EM, Remold-O'Donnell E, Farndale RW, Ware J, et al. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science. 2010;327(5965):580–583. doi: 10.1126/science.1181928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, Provost P. Activated platelets can deliver mRNA regulatory Ago2•microRNA complexes to endothelial cells via microparticles. Blood. 2013;122(2):253–261. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 46.Ple H, Maltais M, Corduan A, Rousseau G, Madore F, Provost P. Alteration of the platelet transcriptome in chronic kidney disease. Thromb Haemost. 2012;108(4):605–615. doi: 10.1160/TH12-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stakos DA, Gatsiou A, Stamatelopoulos K, Tselepis AD, Stellos K. Platelet microRNAs: From platelet biology to possible disease biomarkers and therapeutic targets. Platelets. 2013;24(8):579–589. doi: 10.3109/09537104.2012.724483. [DOI] [PubMed] [Google Scholar]
- 48.Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood. 2012;119(26):6288–6295. doi: 10.1182/blood-2011-12-396440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Rhenen DJ, Vermeij J, Mayaudon V, Hind C, Lin L, Corash L. Functional characteristics of S-59 photochemically treated platelet concentrates derived from buffy coats. Vox Sang. 2000;79(4):206–214. doi: 10.1159/000056732. [DOI] [PubMed] [Google Scholar]
- 50.Janetzko K, Lin L, Eichler H, Mayaudon V, Flament J, Kluter H. Implementation of the Intercept blood system for platelets into routine blood bank manufacturing procedures: Evaluation of apheresis platelets. Vox Sang. 2004;86(4):239–245. doi: 10.1111/j.0042-9007.2004.00419.x. [DOI] [PubMed] [Google Scholar]
- 51.Bruchmuller I, Losel R, Bugert P, Corash L, Lin L, Kluter H, Janetzko K. Effect of the psoralen-based photochemical pathogen inactivation on mitochondrial DNA in platelets. Platelets. 2005;16(8):441–445. doi: 10.1080/09537100500129300. [DOI] [PubMed] [Google Scholar]
- 52.Li J, Lockerbie O, de Korte D, Rice J, McLean R, Goodrich RP. Evaluation of platelet mitochondria integrity after treatment with Mirasol pathogen reduction technology. Transfusion. 2005;45(6):920–926. doi: 10.1111/j.1537-2995.2005.04381.x. [DOI] [PubMed] [Google Scholar]
- 53.Matarrese P, Straface E, Palumbo G, Anselmi M, Gambardella L, Ascione B, Del Principe D, Malorni W. Mitochondria regulate platelet metamorphosis induced by opsonized zymosan A–activation and long-term commitment to cell death. FEBS J. 2009;276(3):845–856. doi: 10.1111/j.1742-4658.2008.06829.x. [DOI] [PubMed] [Google Scholar]
- 54.Zharikov S, Shiva S. Platelet mitochondrial function: From regulation of thrombosis to biomarker of disease. Biochem Soc Trans. 2013;41(1):118–123. doi: 10.1042/BST20120327. [DOI] [PubMed] [Google Scholar]
- 55.Keuren JF, Cauwenberghs S, Heeremans J, de Kort W, Heemskerk JW, Curvers J. Platelet ADP response deteriorates in synthetic storage media. Transfusion. 2006;46(2):204–212. doi: 10.1111/j.1537-2995.2006.00702.x. [DOI] [PubMed] [Google Scholar]
- 56.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


