Abstract
Obesity, type 2 diabetes, and HIV-associated lipodystrophy are associated with abnormalities in adipocyte growth and differentiation. In persons with these conditions, adipose depots contain increased numbers of macrophages, but the origins of these cells and their specific effects are uncertain. Peripheral blood mononuclear cells (PBMC)-derived monocytes, but not T cells, cocultured via transwells with primary subcutaneous preadipocytes, increased proliferation (approximately twofold) and reduced differentiation (~50%) of preadipocytes. Gene expression analyses in proliferating preadipocytes (i.e., prior to hormonal induction of terminal differentiation) revealed that monocytes down-regulated mRNA levels of CCAAT/enhancer binding protein, alpha (C/EBPα) and up-regulated mRNA levels of G0/G1 switch 2 (G0S2) message, genes important for the regulation of adipogenesis and the cell cycle. These data indicate that circulating peripheral blood monocytes can disrupt adipogenesis by interfering with a critical step in C/EBPα and G0S2 transcription required for preadipocytes to make the transition from proliferation to differentiation. Interactions between preadipocytes and monocytes also increased the inflammatory cytokines IL-6 and IL-8, as well as a novel chemotactic cytokine, CXCL1. Additionally, the levels of both IL-6 and CXCL1 were highest when preadipocytes and monocytes were cultured together, compared to each cell in culture alone. Such cross-talk amplifies the production of mediators of tissue inflammation.
Introduction
Immune cells such as macrophages, dendritic cells, T cells, and neutrophils are increased in number within adipose depots in persons with metabolic disorders such as obesity, type 2 diabetes, and HIV-associated lipodystrophy (1–6). The source of the macrophages and dendritic cells is likely to be circulating monocytes that have migrated into the adipose depots and subsequently undergone terminal differentiation (7). Macrophages have profound effects on adipocyte growth and metabolic physiology. For example, activated human macrophages disrupt primary preadipocyte development by increasing their proliferation, inducing them to assume an abnormal fibrotic morphology, and preventing them from differentiating into mature adipocytes (8,9). Human adipose tissue-derived macrophages affect the endocrine physiology of adipocytes by upregulating the production of inflammatory cytokines and chemokines such as IL-6, IL-8, MCP-1, MIP-1β, and TNFα in primary adipocytes (8,10,11). Activated murine RAW264.7 macrophages promote insulin resistance in 3T3-L1 adipocytes by a mechanism involving increased NF-κB activity, enhanced lipolysis and fatty acid release, and downregulation of GLUT4 and IRS-1 (12).
Much less is known about the direct effects on adipose tissues of circulating monocytes, the precursors of tissue macrophages. Understanding the role of monocytes in adipocyte dysfunction is important, since these cells migrate continuously into adipose tissue to replenish macrophage numbers. In addition, monocytes differentially regulate the proliferation and differentiation of a variety of cells. For example, monocytes enhance B cell proliferation, but suppress proliferation of PHA-, CD3-, or antigen-stimulated T cells, as well as that of IL-2-treated natural killer cells (13–17). Monocytes enhance the proliferation of retinal pigment epithelial cells and possibly that of human umbilical vein endothelial cells (18–20). Additionally, monocytes increase fibroblast proliferation via IL-1 production (21,22) and may enhance osteoblastic differentiation of calcifying vascular cells (23). Enterotoxin-B-stimulated human primary monocytes secrete cytokines that alter T cell differentiation (24). Finally, human monocytes support B cell differentiation by increasing the formation of immunoglobulin-secreting cells (25).
Adipocytes develop from a highly regulated process that involves the replication of preadipocytes, followed by their differentiation into triglyceride-filled adipocytes. We hypothesized that monocytes might alter this developmental program by affecting human preadipocyte proliferation or differentiation. To test this hypothesis, human peripheral blood monocytes were cocultured via transwells with primary human subcutaneous preadipocytes, then the preadipocytes were examined for proliferation and differentiation. We found that the monocytes increased preadipocyte proliferation and blunted differentiation. Gene expression studies of preadipocytes cocultured with peripheral blood mononuclear cells (PBMCs) using human genome-wide microarray and pathway analysis revealed networks that were significantly associated with cell cycle, immune-related functions, adipogenesis, and inflammation. Further examination of adipogenic gene expression revealed an intriguing pattern of expression of not only key early adipogenic transcription factors, such as C/EBPβ, CCAAT/enhancer-binding protein-α (C/EBPα), and PPARγ2, but also a later adipogenic gene, G0/G1 switch 2 (G0S2), involved in cell cycle regulation, suggesting that enhanced preadipocyte proliferation is associated with the dysregulation of C/EBPα and G0S2. Finally, monocytes up-regulated CXCL1 (also known as GROα) expression in preadipocytes, suggesting that preadipocytes are a major source of this chemoattractant for macrophages, T cells, and neutrophils. Collectively, these data suggest that circulating monocytes that migrate into adipose tissue are capable of regulating the proliferation and differentiation of preadipocytes and promote a proinflammatory process very early during their infiltration of adipose tissues, prior to their differentiation into macrophages or dendritic cells.
Methods and Procedures
Cells and coculture methods
Cryopreserved human subcutaneous preadipocytes were obtained from Zen-Bio (Research Triangle Park, NC) and cultured in Zen-Bio preadipocyte maintenance medium consisting of DMEM/Ham’s F-12, D-glucose, HEPES (pH 7.4), FBS, penicillin, streptomycin, and amphotericin B. The preadipocytes were acquired from donors undergoing elective surgery or liposuction and pooled before freezing. Cells tested negative for HIV-1, HIV-2, HTLV-I, HTLV-II, HCV, and HBV. The absence of stromal vascular cells including endothelial cells and macrophages was confirmed by vascular endothelial growth factor staining and flow cytometry staining for CD45 and CD14.
PBMCs were obtained from buffy coat preparations (Gulf Coast Regional Blood Center, Houston, TX) by density-gradient separation using Histopaque-1077 (Sigma-Aldrich, St Louis, MO). CD3+ T cells and CD14+ monocytes were purified from PBMCs using EasySep negative selection kits (StemCell Technologies, Vancouver, BC), and the purities were verified by flow cytometry (routinely >95% CD3+ for T cells and >85% CD14+ for monocytes). PBMCs, T cells, or monocytes were then placed in transwells for coculture experiments as described below.
Prior to the start of the coculture experiments, ~2 × 104 preadipocytes were placed in Costar 6-well plates (Corning, Lowell, MA) and cultured in preadipocyte maintenance medium overnight. Preadipocyte confluency was no greater than ~30% after the overnight incubation and prior to the start of cocultures. Transwells of 0.4 µm pore size (Costar) containing PBMCs, T cells, or monocytes in preadipocyte maintenance media were then added to the preadipocytes, and cells were cultured at the indicated times.
Preadipocyte flow cytometry cell cycle measurements
At the appropriate coculture time points, the preadipocytes were trypsinized, washed, and fixed with 70% ethanol. The nucleic acid dye propidium iodide, PI (Sigma-Aldrich), was then added at 50 µg/ml, and RNase A (Sigma-Aldrich) was added at 100 µg/ml to degrade RNA. Cell cycle data were acquired with a Beckman-Coulter EPICS XL-MCL Flow Cytometer (Fullerton, CA). Doublet discrimination and debris were excluded from analysis by plotting the linear PI signal vs. the signal peak, and then gating on the single cells. Data analysis was performed on at least 5,000 cells using FlowJo software (Tree Star, Ashland, OR). Percentages of cells in each phase of the cell cycle (G1, S, and G2/M) were resolved using the FlowJo Watson-Pragmatic cell cycle modeling algorithm. Proliferation of preadipocytes was determined by the addition of S and G2/M phases.
Preadipocyte differentiation
Following 5- or 6-day coculture periods, transwells containing leukocytes were removed and the preadipocytes were gently rinsed three times with phosphate buffered saline. Differentiation was then induced using Zen-Bio proprietary Differentiation Media (DM) consisting of DMEM/Ham’s F-12, HEPES (pH 7.4), FBS, biotin, pantothenate, human insulin, dexamethasone, IBMX, PPARγ agonist (non-TZD), penicillin, streptomycin, and amphotericin B. The preadipocytes were cultured in this media for seven days before changing to Zen-Bio Adipocyte Maintenance (AM) medium, which contained the same ingredients as the DM medium but excluding IBMX and PPARγ agonist. The AM medium was changed every 3 days for another week. After 2 weeks of differentiation, lipid accumulation was assessed by cell fixation and staining with the lipid dye Oil Red O (Sigma-Aldrich). Differentiation was quantified by phase contrast microscopy and recording four random fields of view from each condition at ×40 magnification. Oil Red O-stained areas were then analyzed with Adobe Photoshop version CS2 software (Adobe, San Jose, CA). Normal differentiation with control media was established as 100% differentiation.
Microarray and pathway analysis
Two-color cDNA microarray was performed on preadipocytes with Agilent Whole Human Genome Microarray on four biological replicates by Miltenyi Biotec (Auburn, CA). Briefly, after 3 days of coculture with control media or PBMCs, preadipocytes were lysed and samples shipped in accordance with manufacturer’s instructions. Following verification of cDNA integrity using an Agilent 2100 Bioanalyzer (Santa Clara, CA), cDNA of preadipocytes cultured with control media or PBMCs were labeled with Cy3 or Cy5, respectively, then combined and hybridized overnight to an Agilent 4×44K slide. Signal intensities and ratios, background subtraction, normalization, and statistical P values were calculated using Agilent Feature Extraction Software (Agilent Technologies), and differential gene expression was determined from double-log scatter plots of Cy3 compared to Cy5 signal intensities using Rosetta Resolver Software (Rosetta Biosoftware, Seattle, WA). These data have been submitted to NCBI Gene Expression Omnibus (accession number GSE19324). Genes with minimum fold-changes of two and P values <0.01 on three of four bioreplicates were then uploaded into Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) for pathway analysis.
For network generation, differentially expressed genes were mapped to their corresponding genes in the Ingenuity knowledge base. These “focus genes” were overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. Networks of these focus genes were then algorithmically generated based on their connectivity. Further functional analyses of the networks identified the biological functions and/or diseases of the most significant network genes. Those associated with biological functions and/or diseases in the Ingenuity knowledge base were considered for analysis. Fisher’s exact test was used to calculate the probability that each biological function and/or disease was significant.
Real-time PCR
RNA isolation from preadipocytes was performed using RNeasy Mini Kit (Qiagen, Valencia, CA); 0.5 µg of RNA was reverse-transcribed into cDNA using Applied Biosystems High Capacity RNA-to-cDNA Kit (Carlsbad, CA). From these reactions, 2 µl of template was then used for real-time PCR using the Applied Biosystems Prism 7000 Sequence Detection System. Applied Biosystems Taqman Gene Expression Assays were used for real-time PCR detection of C/EBPβ (Hs00270923_s1), PPARγ2 (Hs00234592_m1), C/EBPα (Hs00269972_ s1), G0S2 (Hs00274783_s1), and CXCL1 (Hs00236937_m1). GAPDH (Hs00266705_g1) was used as the internal control, and relative expression was determined using the formula 2−ΔΔCT, in which the fold change in gene expression was determined by comparing preadipocytes cultured with monocytes vs. control media.
Western blot
Preadipocyte whole cell lysates was collected following lysis with RIPA buffer supplemented with Roche Complete Mini protease inhibitor cocktail (Roche, Indianapolis, IN), and protein amounts were determined with Pierce BCA assay (Pierce, Rockford, IL). Equal amounts of protein were loaded into mini- or large-format 12% gels for SDS-PAGE. After overnight transfer to nitrocellulose membranes, gels were blocked with 5% nonfat dry milk in TBST buffer and probed for C/EBPβ, PPARγ2, C/EBPα, or G0S2, and actin was used as the loading control. The following antibodies and dilutions were used: Rabbit C/EBPβ-1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA), Rabbit PPARγ2-1:500 (Cell Signaling Technology, Danvers, MA), Rabbit C/EBPα-1:500 (Cell Signaling Technology), Rabbit G0S2-1:100 (Santa Cruz Biotechnology), and Rabbit β-actin-1:1000 (Santa Cruz Biotechnology). Blots were incubated with primary antibodies overnight and then with GoatxRabbit-HRP secondary antibodies (1:2000) for 1 h. Exposures were done with X-ray film, and densitometric analysis was performed using a Syngene GeneGnome imaging system (Frederick, MD).
Extracellular cytokine measurements
Cell culture supernatants were collected, centrifuged, and stored at −20 °C prior to protein measurement. Concentrations of the inflammatory cytokines IL-1β, IL-6, IL-8, IL-10, IL-12p70, and TNFα were measured using a BD Cytometric Bead Array (CBA) Human Inflammation Kit (BD Biosciences, San Jose, CA). Data were acquired with a Beckman-Coulter EPICS XL-MCL Flow Cytometer and analyzed with BD CBA Software. Concentrations were determined based on a standard curve with a detection range of 20–5,000 pg/ml. Concentrations of CXCL1 were measured using a Human GROα Single Analyte ELISArray Kit (SA Biosciences, Frederick, MD).
Statistics
Data shown are means ± s.d. from at least 3 experiments in duplicate or triplicate, and differences between means were compared using two-tailed Student’s t-test. P < 0.05 was considered significant.
Results
Monocytes increase preadipocyte proliferation
To address the question of whether undifferentiated monocytes influence proliferation or differentiation of preadipocytes, primary subcutaneous preadipocytes were cocultured with PBMCs, negatively selected CD3+ T cells, or negatively selected CD14+ monocytes separated by transwells. The preadipocytes were then harvested, fixed, stained with propidium iodide, and analyzed by flow cytometry and FlowJo software for cell cycle and proliferation measurements.
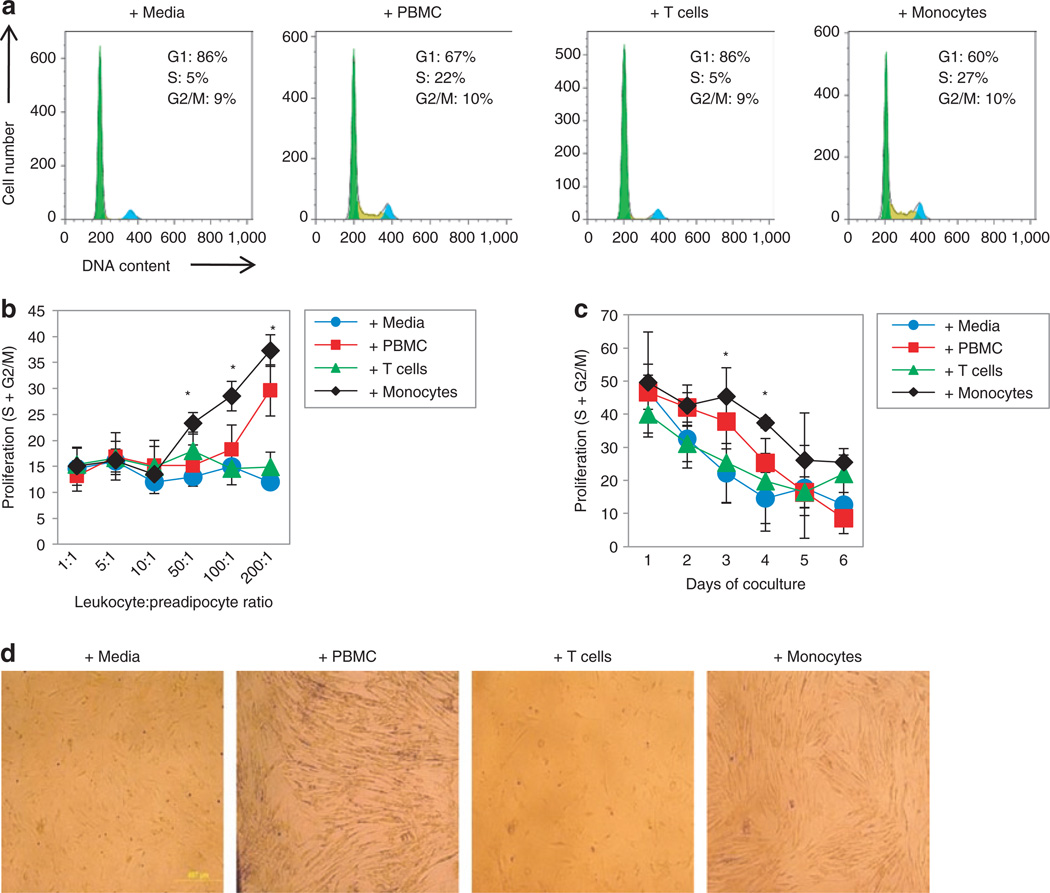
At different preadipocyte:leukocyte ratios, preadipocyte proliferation was significantly and linearly increased by monocytes, beginning at a preadipocyte-to-monocyte ratio of 1:50, and by PBMCs at a ratio of 1:200 (Figure 1b). T cells exerted no effect on preadipocyte proliferation at any ratio tested. Representative cell cycle histograms of preadipocytes cultured with PBMCs, T cells, or monocytes (1:100 ratios) after 3 days are shown in Figure 1a. Additionally, the increase in preadipocyte proliferation caused by PBMCs or monocytes (1:100 ratios) was most pronounced on the third or fourth day of coculture (Figure 1c). At day 3, mean preadipocyte proliferation (S + G2M) was 37.8 ± 8.2% when cocultured with PBMCs compared to 22.1 ± 9.0% (P < 0.05) when cultured with control media. At days 3 and 4, mean preadipocyte proliferation frequencies were 37.4 ± 1.6% and 45.4 ± 8.7%, respectively, when cocultured with monocytes compared to 14.5 ± 9.9% (P < 0.05) and 22.1 ± 9.0% (P < 0.01) when cultured with control media. After 4–5 days of coculture with PBMCs or monocytes, the preadipocytes became densely confluent, whereas culture without added cells or with T cells in transwells resulted in confluency of ~50% (Figure 1d). These data indicate that monocytes, and not T cells, are the PBMC component causing increased preadipocyte proliferation.
Figure 1.
Cell cycle and proliferation of human subcutaneous preadipocytes during coculture with PBMCs, T cells, or monocytes. Preadipocyte cell cycle was measured by flow cytometry after staining with a DNA dye, and the G1, S, and G2/M phases were delineated using FlowJo analysis software. (a) Representative cell cycle histograms of preadipocytes after 3 days of culture with control media, PBMCs, T cells, or monocytes. (b) Proliferation of preadipocytes after 3 days of coculture with increasing numbers of PBMCs, T cells, or monocytes. Proliferation was determined by addition of the S and G2/M phases of the cell cycle. N = 3, error bars are means ± s.d., *P < 0.05 compared to control media. (c) Proliferation of preadipocytes during coculture with PBMCs, T cells, or monocytes for up to 6 days at a 50:1 leukocyte:preadipocyte ratio. N = 3, error bars are means ± s.d., *P < 0.05 compared to control media. (d) Representative phase contrast microscopy images (×40 magnification) of preadipocyte confluency after 6 days of coculture with PBMCs, T cells, or monocytes. PBMC, peripheral blood mononuclear cell.
Increased preadipocyte proliferation by monocytes is associated with reduced differentiation
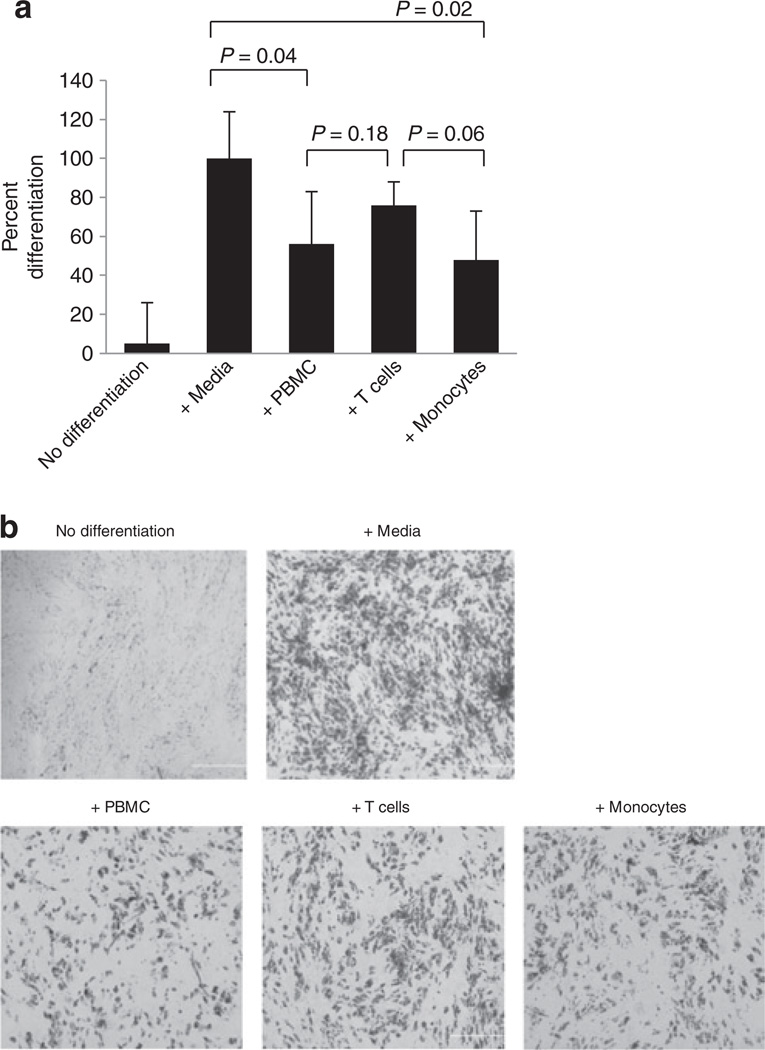
To determine whether the increased preadipocyte proliferation demonstrated in Figure 1 is associated with any effect on subsequent differentiation, preadipocytes were induced to differentiate using medium containing insulin, dexamethasone, IBMX, and PPARγ agonist after 5–6 days of coculture with PBMCs, T cells or monocytes, as described above. After 2 weeks in differentiation media, the adipocytes were fixed and stained with Oil Red O for the presence of triglyceride-containing adipocytes. Exposure to PBMCs and monocytes reduced differentiation, by 56 ± 27% (P < 0.05) and 48 ± 24%, (P < 0.05), respectively (Figure 2a). By contrast, there was no significant reduction of the differentiation of preadipocytes exposed to T cells. Representative images of Oil Red O-stained adipocytes are shown in Figure 2b. These data suggest that PBMCs, specifically monocytes, produce soluble factors that not only cause increased preadipocyte proliferation but also disrupt the ability of preadipocytes to subsequently undergo normal differentiation into mature adipocytes.
Figure 2.
Differentiation of human subcutaneous preadipocytes for 2 weeks following 6 days of coculture with PBMCs, T cells, or monocytes (i.e., after 6 days of coculture, PBMCs, T cells, or monocytes were removed and preadipocytes were differentiated in the absence of these cells). (a) Quantification of preadipocyte differentiation after 2 weeks following hormonal induction. Adipocyte lipids were stained with Oil Red O and four random fields of observation from each condition were recorded. Pixel values of Oil Red O-stained areas were calculated with Adobe Photoshop software and averaged. Normal differentiation with control media was established as 100%. N = 3, error bars are means ± s.d. (b) Representative phase contrast microscopy images (×40) of Oil Red O-stained preadipocytes after 2 weeks of differentiation. PBMC, peripheral blood mononuclear cell.
Increased preadipocyte proliferation alters preadipocyte gene expression of adipogenic proteins, cell cycle proteins, and inflammatory cytokines
To understand better the mechanisms underlying dysregulated preadipocyte proliferation associated with exposure to PBMCs, whole human genome microarray analysis (41,000+ genes) was performed on subcutaneous preadipocytes after coculture with PBMCs (1:200 ratio) or control media for 3 days. Differentially expressed genes were further analyzed using Ingenuity Systems’ Pathway Analysis software (IPA) to place these gene expression changes in the context of specific biological functions, diseases, pathways, or protein interaction networks.
Of the 41,000+ genes examined by microarray, 456 were differentially expressed in the PMBC-exposed preadipocytes. Among these 456 genes, some were identified by the pathway analysis to be members of one of the 14 networks generated by the IPA algorithm, of which the three top-scoring networks are summarized in Table 1. The remaining 11 networks had scores of less than 2 and are not shown. The biological functions most significantly associated with the top-scoring network (score of 14) included cell cycle, cancer, and reproductive system disease, and the differentially expressed genes included in this network included ABCB1, ACTA2, ATM, CCNE2, DZIP3, ESPL1, MXD3, PKN2, PPM1D, PSEN2, SMC1A, and TGFB1. The functions associated with the other two networks (scores of 12) were immunological functions such as antigen presentation and cell-mediated and humoral immune response, and the differentially expressed genes in these networks included CXCL1, CXCL2, DDR1, IL12A, LCP1, NFKBIB, SERPINA3, SERPINB2, SLC1A5, SUV39H1, TFPI2, ABCA1, ANK3, CREM, FAM162A, G0S2, ITGB2, LEP, MN1, TGM2, and TNFRSF11A. The results of these gene expression data suggest that exposure of preadipocytes to PBMCs modulates the expression of cell cycle, adipogenic, and immunological proteins.
Table 1.
Pathway analysis of whole genome microarray data of human subcutaneous preadipocytes after 3 days of coculture with PBMCs
| Top associated network functions |
Network molecules | Score |
|---|---|---|
| Cell cycle, cancer, reproductive system disease | ↓ABCB1, ↓ACTA2, ↑ATM, ↑CCNE2, CD44, CD46, CD59, CDK2, CDKN1A, CDKN1B, CHEK2, CTNNB1, ↓DZIP3, ERBB2, ↑ESPL1, ESR1, JUN, MDM2, MDM4, MRE11A, ↓MXD3, NBN, ↑PKN2, ↓PPM1D, ↓PSEN2, RAD17, RELA, SCD, ↓SMC1A, TERF2, TERT, ↓TGFB1, TGFB3, TP53, YBX1 | 14 |
| Antigen presentation, cell-mediated immune response, humoral immune response | CCL5, CD40LG, CTNNB1, ↑CXCL1, ↑CXCL2, ↓DDR1, ESR1, IFNG, IL4, IL6, IL8, IL10, ↑IL12A, IL17A (includes EG:3605), IL17F, IL1A, IL1B, IL1RN, IL23A, JUN, KLK2, ↓LCP1, MMP2, MMP9, MMP1 (includes EG:4312), ↑NFKBIB, RELA, ↓SERPINA3, ↑SERPINB2, ↓SLC1A5, ↓SUV39H1, TAL1, TERT, ↑TFPI2, TNF | 12 |
| Antigen presentation, cell-mediated immune response, humoral immune response | ↓ABCA1, ↓ANK3, C3, CD40LG, CDC42, CDH1, ↓CREM, ↑CXCL1, ↑FAM162A, FLOT1, FOS, ↓G0S2, ICAM1, IL2, IL4, IL8, IL10, IL13, IL17F, IL1RN, INS, ↓ITGB2, ↓LEP, MMP9, ↓MN1, NCF2, NCF1C, PIGR, RAC1, STAT3, SYK, ↑TGM2, TNF, ↓TNFRSF11A, TNFSF11 | 12 |
Genes whose expression were predetermined to be significantly up- or down-regulated by PBMCs after microarray analysis were uploaded into Ingenuity Pathway Analysis software, of which the three top-scoring networks are shown. Indicated for each network are the top associated biological functions, the molecules comprising the network (molecules with downward and upward arrows indicate downregulation and upregulation, respectively), and a Score (the Score is based on a P value calculation, which calculates the likelihood that the Network Eligible Molecules that are part of a network are found therein by random chance alone. Mathematically, the score is simply the negative exponent of this P value calculation. For example, if the score is 3, then the corresponding P value was 10−3, i.e., there is a 1 in 1,000 chance that the Network Eligible Molecules found in that network appeared there just by chance. In other words, the score is simply a measure of the number of Network Eligible Molecules in a network, and the greater the number of Network Eligible Molecules in a pathway, the higher the score (lower the P value) will be). PBMC, peripheral blood mononuclear cell.
Regulation of preadipocyte proliferation and differentiation by monocytes is associated with suppression of C/EBPα gene expression and induction of G0S2
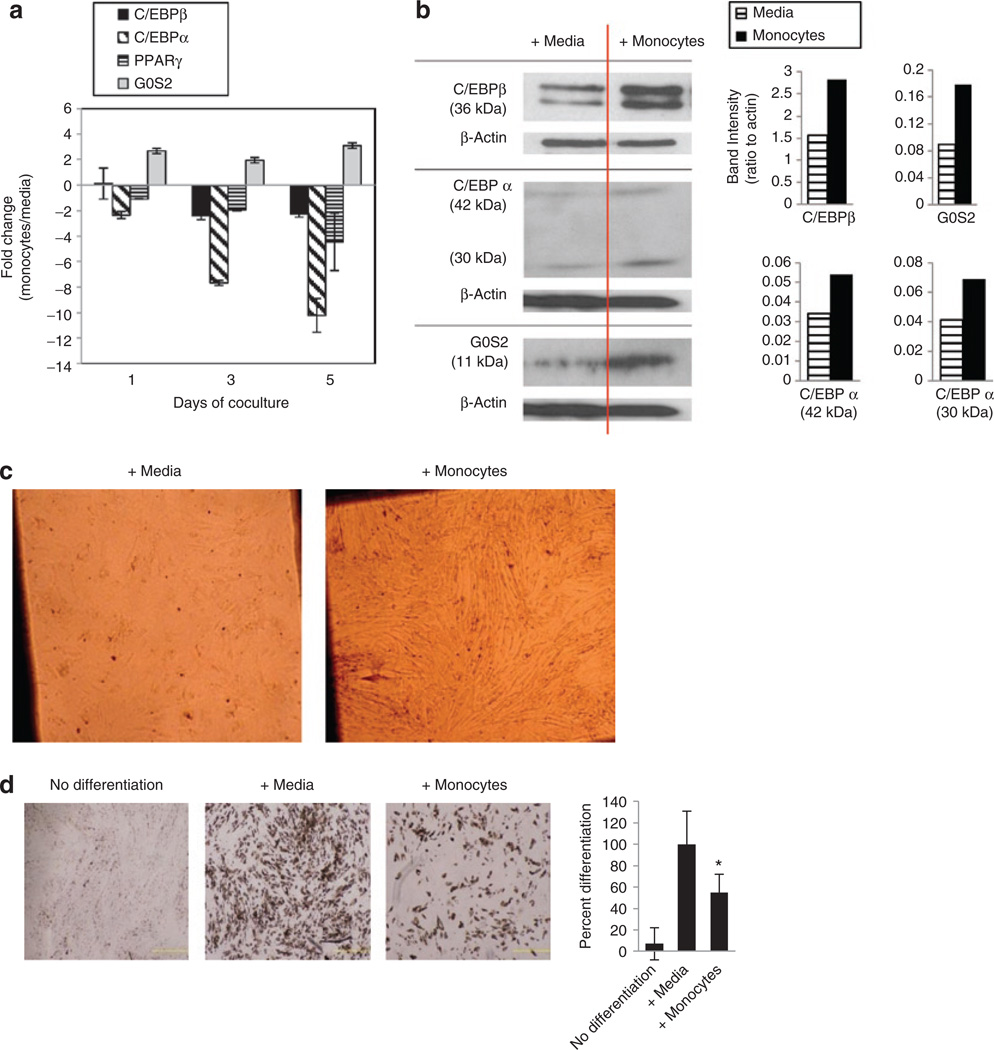
The microarray and pathway analysis indicated that the expression of specific cell cycle regulatory proteins in preadipocytes, such as ATM and G0S2, were affected during coculture with PBMCs. Since the difference was observed prior to hormonal induction and terminal differentiation of preadipocytes, aberrant expression of these genes might be responsible for dysregulation of the proliferative phase of preadipocyte adipogenesis. It was also striking, in the context of adipogenesis, that G0S2 is thought to be a target of PPARγ (26). G0S2 also plays a role in adipocyte lipolysis by regulating adipose triglyceride lipase, a key enzyme involved in triacyglycerol hydrolysis (27). We examined the expression of G0S2 in preadipocytes during coculture with monocytes for up to 5 days, concomitantly with measurement of the mRNA’s of the key early adipogenic proteins CEBPβ, C/EBPα, and PPARγ (Figure 3a). Real-time PCR analysis indicated that C/EBPα was remarkably suppressed in preadipocytes (up to tenfold relative to control media after 5 days) by monocytes at all time points. Interestingly, this suppression of C/EBPα coincided with modest upregulation of G0S2 (up to threefold relative to control media after 5 days). Minor suppression of C/EBPβ (~2-fold after 5 days) and PPARγ (~4-fold after 5 days) was also observed.
Figure 3.
Expression of key adipogenic and cell cycle (C/EBPα and G0S2) proteins in proliferating human subcutaneous preadipocytes during coculture with monocytes and subsequent terminal differentiation. (a) Gene expression of C/EBPβ, C/EBPα, PPARγ, and G0S2 by real-time PCR in preadipocytes during coculture with monocytes for up to 5 days. The fold change expression of each gene in preadipocytes compares coculture with monocytes to culture with control media after normalization to an internal control (GAPDH) using the 2−ΔΔCt calculation for relative expression. Data are from one experiment done in duplicate representative of three independent experiments, error bars are means ± s.e.m. (b) Western blot of C/EBPβ, C/EBPα, and G0S2 in preadipocytes at the 5-day time point during coculture with monocytes. The two bands for C/EBPβ represent the two isoforms for this protein at ~36 kDa. PPARγ was probed for but not detected. Densitometry of blots was normalized to actin loading control. (c) Representative phase contrast microscopy images (×40) of preadipocyte confluency after 5 days of coculture with monocytes just prior to hormonal induction of differentiation. (d) Representative phase contrast microscopy images (×40) of Oil Red O-stained adipocytes after 2 weeks of preadipocyte differentiation following 5 days of preadipocyte coculture with monocytes (i.e., after 5 days of coculture with preadipocytes, monocytes were removed and preadipocytes were differentiated in the absence of monocytes). Error bars are means ± s.d. of pixel intensity quantification of four images per condition from one experiment representative of three. *P < 0.05 compared to control media. C/EBPβ, CCAAT/enhancer binding protein (C/EBP), beta; PPARγ, peroxisome proliferator-activated receptor gamma 2; C/EBPα, CCAAT/enhancer binding protein (C/EBP), alpha; G0S2, G0/G1 switch 2.
The protein levels of CEBPβ, C/EBPα, PPARγ, and G0S2 in preadipocytes cocultured with monocytes were also measured by western blot at the 5-day time point. Protein levels of G0S2 were increased in preadipocytes after 5 days of culture with monocytes (Figure 3b). The level of C/EBPβ protein in preadipocytes was also increased after 5 days of coculture, but protein levels of C/EBPα showed less change. PPARγ protein could not be detected by western blot in our hands, although one factor may be because protein levels of this transcription factor are characteristically very low in preadipocytes prior to hormonal induction of differentiation.
The altered expression patterns of these adipogenic genes were also associated with blunted preadipocyte differentiation (Figure 3d). After 5 days of coculture, the monocytes were removed and the densely confluent preadipocytes (Figure 3c) were induced to differentiate by hormonal induction. Exposure of preadipocytes to monocytes during the proliferative phase of preadipocyte adipogenesis resulted in reduction of subsequent, hormonally induced, terminal differentiation by 45 ± 12% compared to exposure to control media (P < 0.05), as assessed by Oil Red O staining after 14 days of differentiation. These data suggest that altered expression of C/EBPα and G0S2 induced by interaction with monocytes disrupt the orderly progression of preadipocyte differentiation such that the proliferative phase is exaggerated, and subsequent differentiation is compromised.
Levels of specific inflammatory cytokines and chemokines are elevated during preadipocyte coculture with monocytes, but these are not sufficient to induce defects in preadipocyte proliferation and differentiation
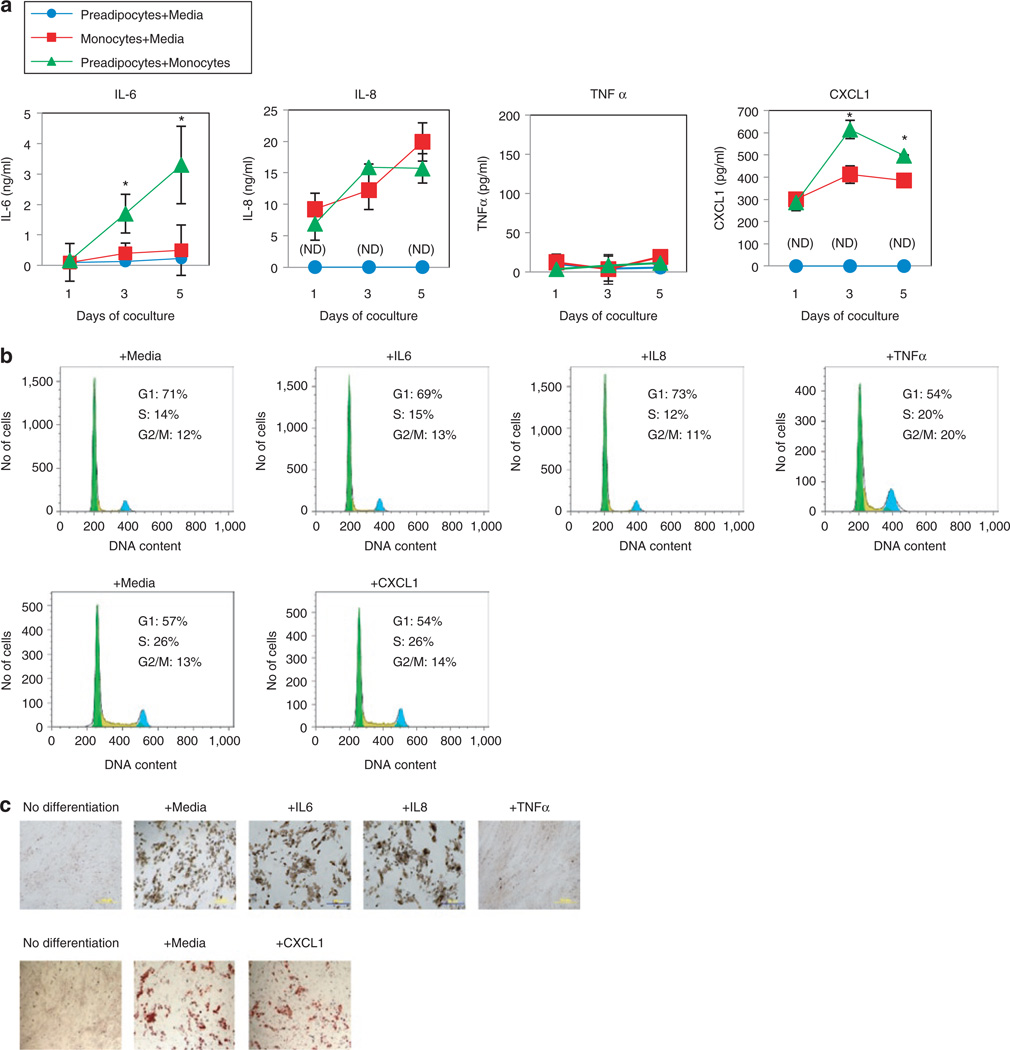
The preadipocyte microarray and pathway analysis indicated that inflammatory cytokines and chemokines result from chemical interactions between preadipocytes and either PBMCs or monocytes. For example, the microarray indicated that the mRNAs for CXCL1 and IL-12 were up-regulated during exposure to PBMCs, and pathway analysis indicated that these were part of a network that also included IL-6, IL-8, IL-10, IL1β, and TNF (Table 1). We measured concentrations of these proteins in supernatants of preadipocytes cocultured with monocytes (Figure 4a).
Figure 4.
Production of inflammatory cytokines during coculture of human subcutaneous preadipocytes with monocytes, and preadipocyte proliferation and differentiation after exogenous treatment with inflammatory cytokines. (a) The inflammatory cytokines IL-6, IL-8, and TNFα were measured in supernatants using a flow cytometry multiplex assay, and CXCL1 was measured with an ELISA kit. Data are from one experiment done in triplicate that is representative of three independent experiments, error bars are means ± s.d. For IL-6 and CXCL1, asterisks indicate P < 0.05 comparing the “preadipocytes + monocytes” condition to either the “preadipocytes + media” or “monocytes + media” condition. (b) Preadipocyte cell cycle after treatment with 100 ng/ml recombinant IL-6, IL-8, TNFα, or CXCL1 for 3 days. CXCL1 is compared with its own media control since that treatment experiment was conducted in a separate experiment with a different lot of preadipocytes. Histograms are representative of two separate experiments. (c) Preadipocyte differentiation after treatment with 100 ng/ml recombinant IL-6, IL-8, TNFα, or CXCL1. Shown are representative images (×40) of Oil Red O-stained adipocytes after 2 weeks of differentiation. ELISA, enzyme-linked immunosorbent assay.
After 3 and 5 days of coculture, production of IL-6 and CXCL1 was markedly elevated in the supernatants from the cocultured cells, compared to preadipocytes or monocytes alone. IL-6 concentration in the preadipocyte + monocyte coculture supernatant was 1.7 ± 0.64 ng/ml on the third day (vs. preadipocytes alone at 0.12 ± 0.05 ng/ml, P < 0.05, or vs. monocytes alone at 0.4 ± 0.33 ng/ml, P < 0.05) and 3.3 ± 1.27 ng/ml on the fifth day (vs. preadipocytes alone at 0.23 ± 0.03 ng/ml, P<0.05, or vs. monocytes alone at 0.5 ± 0.83 ng/ml, P < 0.05). CXCL1 concentration in the preadipocyte + monocyte supernatant was 615.3 ± 41.2 pg/ml on the third day (vs. preadipocytes alone at undetectable levels, or vs. monocytes alone at 412 ± 39.2 pg/ml, P < 0.05) and 497.7 ± 3.5 pg/ml on the fifth day (vs. preadipocytes alone at undetectable levels, or vs. monocytes alone at 385 ± 14.7 pg/ml, P < 0.05). Expression of CXCL1 in preadipocytes during coculture with monocytes was also examined by real-time PCR; consistent with the microarray data, the results revealed substantial upregulation (tenfold or greater) compared to preadipocytes cultured alone (Supplementary Figure S1 online), suggesting that much of the CXCL1 in the preadipocyte + monocyte supernatant originates from preadipocytes. IL-8 levels were elevated comparably in supernatants from both preadipocytes + monocytes and monocytes alone conditions. TNFα, IL-1β, IL-10, and IL-12 proteins were undetectable in all culture conditions (i.e., preadipocytes + monocytes, preadipocytes alone, or monocytes alone).
To investigate the possible role of IL-6, IL-8, or CXCL1 in the dysregulation of preadipocyte proliferation or differentiation, preadipocytes were treated with up to 100 ng/ml recombinant IL-6, IL-8, CXCL1, or TNFα for 3 days, and proliferation by cell cycle analysis was examined. Although TNFα was undetectable in any coculture conditions, TNFα has been reported to increase preadipocyte proliferation and block differentiation (28), and consistent with these reports, we observed that 100 ng/ml TNFα increased preadipocyte proliferation and potently blocked differentiation (Figure 4b,c). However, preadipocyte proliferation and differentiation was unaffected by treatment with 100 ng/ml IL-6, IL-8, or CXCL1 (Figure 4b,c). Additionally, the use of neutralizing antibodies against IL-6, IL-8, or CXCL1, or against their receptors, did not mitigate the effects of monocytes on preadipocyte proliferation and differentiation (data not shown).
These data suggest that the abnormal preadipocyte proliferation and differentiation induced by monocytes is associated with the increased production of inflammatory cytokines such as IL-6, IL-8, and CXCL1 from preadipocytes and monocytes, and that preadipocytes may communicate with monocytes via IL-6 or CXCL1. However, by themselves these cytokines are not sufficient for the induction of aberrant preadipocyte proliferation and differentiation by monocytes.
Discussion
The present data demonstrate that monocytes affect the dynamics of orderly preadipocyte proliferation and differentiation, leading to disruption of adipogenesis. Specifically, they accelerate proliferation of preadipocytes coincident with transcriptional repression of C/EBPα, a critical antimitotic adipogenic transcription factor that has a key role in terminating preadipocyte proliferation prior to subsequent terminal differentiation (although protein levels of C/EBPα were not diminished 5 days after addition of the differentiation cocktail). The increased proliferation of preadipocytes was accompanied by increased mRNA as well as protein expression of G0S2, a cell cycle regulatory protein in preadipocytes that may be positively regulated by PPARγ (26). However, in this case, we found increased G0S2 with no evidence of PPARγ expression. Thus, altered G0S2 expression in the context of preadipocyte–monocyte interaction appears to be dissociated from PPARγ activity, which may be the reason for its dysregulation at this critical juncture of adipocyte development. The underlying mechanisms likely involve soluble factors exchanged between monocytes and preadipocytes, because they are not dependent upon physical contact between these two cell types. Macrophages and T cells affect adipocyte development and function, but a role for monocytes in the regulation of adipogenesis, such as preadipocyte proliferation and terminal differentiation, has not been well defined. Macrophages and T cells reside in the stromal vascular component of adipose tissues, and their numbers increase greatly in persons with obesity or HIV-associated lipodystrophy (1–3). Monocytes regulate the proliferation and differentiation of T cells, B cells, natural killer cells, epithelial cells, endothelial cells, and fibroblasts (13–25). Circulating monocytes could also infiltrate fat depots and exacerbate an inimical inflammatory response distinct from effects that occur after they differentiate into tissue macrophages (5,7).
PBMCs and monocytes increased the proliferation of subcutaneous preadipocytes after 2–3 days of coculture via transwells (Figure 1). The increased proliferation of preadipocytes by PBMCs and monocytes was also associated with reduced preadipocyte terminal differentiation as assessed by lipid accumulation for 14 days following addition of a hormonal cocktail to induce differentiation (Figure 2). In these experiments, the differentiation cocktail was added after 5 or 6 days of preadipocyte coculture with PBMCs or monocytes, and after the PBMCs or monocytes had been removed, suggesting that the mechanism of the differentiation block likely occurred during the proliferative phase of adipogenesis. Although the effect on preadipocyte differentiation was more subtle than that of proliferation, this may be because differentiation was occurring in the absence of PBMCs or monocytes and perhaps continuation of coculture and persistent interaction during differentiation would have resulted in more significant reduction of differentiation. The purified CD3+ T cells, which were quiescent and not exogenously stimulated, did not significantly affect preadipocyte proliferation or differentiation. Although activated T cells drastically affect adipogenesis (2), the T cells used in these experiments were not in a state of activation (as determined by lack of surface expression of CD69 or CD25 by flow cytometry). Thus, peripheral blood monocytes affect adipogenesis at an early stage.
Pathway analysis based on a comprehensive human genome microarray pointed to protein interaction networks of cell cycle, adipogenic, and inflammatory factors in the regulation of preadipocyte proliferation and differentiation by PBMCs (Table 1). Microarray revealed that preadipocyte expression of cell cycle regulatory proteins such as G0S2 and ATM is altered during exposure to PBMCs. Real-time PCR indicated that suppression of C/EBPβ, C/EBPα, and PPARγ2 occurred concomitantly with modest G0S2 induction (Figure 3a). Interestingly, G0S2 has also been characterized as a PPARγ2 target gene (26), and the differential expression of G0S2 and other PPARγ2 target genes such as leptin (LEP) and cAMP responsive element modulator (CREM), suggests that PBMC exposure affects the expression of C/EBPβ, C/EBPα, and PPARγ2 during the proliferative phase of preadipocyte adipogenesis (prior to hormonal induction of terminal differentiation).
The suppression of C/EBPα gene expression in association with increased proliferation of preadipocytes raises the possibility that the latter occurred because of the lack of antimitotic function of C/EBPα. The mechanism of suppression of C/EBPα is unclear, although other reports have investigated mechanisms of C/EBPα downregulation. IL-6 has been reported to suppress C/EBPα expression in hepatoma cells (29). C/EBPα expression can also be down-regulated during hypoxic conditions in cancer cells by hypoxia-inducible factor-1α, a transcription factor that abolishes transcriptional activity by binding to HRE sequences in the C/EBPα promoter (30), and this suppression of C/EBPα is also associated with increased proportions of the S and G2 phases of cell cycle and increased proliferation. The loss of other mechanisms of negative regulation of preadipocyte proliferation also may have been involved. For example, SH2 domain-containing inositol 5-phosphatase 2 inhibits preadipocyte proliferation induced by platelet-derived growth factor by mediating degradation of the platelet-derived growth factor receptor (31), and macrophages are a source of platelet-derived growth factor in adipose tissue (32). However, our microarray indicated that gene expression of SH2 domain-containing inositol 5-phosphatase 2 or platelet-derived growth factor receptor in preadipocytes was unaffected during coculture.
At the protein level, the abundance of C/EBPβ was increased and that of C/EBPα was less affected compared to preadipocytes cultured in control media (Figure 3b). The discordance between the mRNA and protein concentration data may be because of altered translational processes that regulate the synthesis and degradation of these proteins during cellular stress (33,34), but the higher protein levels of C/EBPβ in preadipocytes during coculture with monocytes was associated with reduction of terminal differentiation, rather than expected enhancement of differentiation. However, the role of C/EBPβ during adipogenesis has mostly been studied after preadipocytes have been induced to differentiate, and less is known regarding the influence of C/EBPβ prior to hormonal induction and in the absence of exogenous induction agents. It could be that the protein is bound in preadipocytes to avoid turnover, but because the level of transcript is diminished, insufficient amounts of the protein are present—that is, perhaps the effects of the down-regulated C/EBPβ mRNA are not felt until differentiation is induced and the existing C/EBPβ is used up.
Inflammatory cytokines IL-6, IL-8, and CXCL1 were present in high amounts in supernatants of preadipocytes cocultured with monocytes (Figure 4). The higher amounts of IL-6 and CXCL1 in the supernatants of preadipocytes cocultured with monocytes than in the supernatants of either of the two cell types cultured alone suggest that these cytokines were produced as a result of cross-talk between preadipocytes and monocytes. IL-6 was recently shown to be a cross-talk cytokine in a coculture system between U937 monocytes and mature adipocytes, in which enhanced production of osteopontin by monocytes was observed during coculture with adipocytes (35). This upregulation of osteopontin was mediated by IL-6, which was mainly produced by adipocytes, but similar to our data, the levels of IL-6 were highest when the monocytes and adipocytes were cultured together. CXCL1 is expressed in 3T3-L1 cells under inflammatory conditions and in adipose tissues of obese mice and humans (36–38), but induction of CXCL1 expression specifically in primary preadipocytes has not been reported. We confirmed by quantitative reverse transcription-PCR that CXCL1 expression in human subcutaneous preadipocytes was up-regulated by monocytes (Supplementary Figure S1 online). Thus, in addition to known chemokines such as MCP-1, IL-8, and MIP-1α (39), CXCL1 is produced by primary human preadipocytes, which presumably targets CXCR2 (CXCL1 receptor)-expressing leukocytes such as T cells, monocytes, and neutrophils. However, addition of recombinant IL-6, IL-8, or CXCL1 to preadipocytes had no effect on their proliferation and differentiation (Figure 4b,c), nor did blocking antibodies against these cytokines mitigate the increased proliferation or reduced differentiation of preadipocytes by monocytes (data not shown). This suggests the need for other factors that are necessary for the observed effects of monocytes upon preadipocytes. It is also possible that pretreatment of monocytes with these cytokines could induce production of the necessary factors that result in abnormal preadipocyte growth.
In summary, circulating monocytes produce soluble molecules that increase primary preadipocyte proliferation and block differentiation, and the dysregulation of this early phase of adipogenesis is associated with reduced C/EBPα gene expression and modest upregulation of G0S2. Further studies may establish a more important role for G0S2 in linking preadipocyte cell cycle control with adipogenesis. Additionally, subcutaneous preadipocytes can be a significant source of IL-6 and CXCL1 via interactions with monocytes, the consequences of which are currently being explored. These findings highlight novel effects of circulating monocytes as a key component of immune mechanisms that disrupt the functions of adipose tissue.
Supplementary Material
Acknowledgments
We thank Miguel Medina, Aaron Orozco, Manisha Singh, Cassandra Horne, Alexander Hutchison, John Rodgers, Neeti Agarwal, Toni Oplt, Alejandro Castellanos, and Scott Anthony for countless helpful comments and ideas. This work was supported by a pilot and feasibility grant from the Baylor College of Medicine Diabetes/Endocrinology Research Center (DERC, NIH P30-DK079638), a grant from the Baylor College of Medicine Center for AIDS Research (CFAR, NIH 5P30AI036211-15 to D.E.L.) and grant R01-DK081553 (to A.B.).
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
Disclosure
The authors declared no conflict of interest.
REFERENCES
- 1.Weisberg SP, McCann D, Desai M, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu H, Ghosh S, Perrard XD, et al. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007;115:1029–1038. doi: 10.1161/CIRCULATIONAHA.106.638379. [DOI] [PubMed] [Google Scholar]
- 3.Jan V, Cervera P, Maachi M, et al. Altered fat differentiation and adipocytokine expression are inter-related and linked to morphological changes and insulin resistance in HIV-1-infected lipodystrophic patients. Antivir Ther (Lond) 2004;9:555–564. [PubMed] [Google Scholar]
- 4.Bedford PA, Todorovic V, Westcott ED, et al. Adipose tissue of human omentum is a major source of dendritic cells, which lose MHC Class II and stimulatory function in Crohn’s disease. J Leukoc Biol. 2006;80:546–554. doi: 10.1189/jlb.0905501. [DOI] [PubMed] [Google Scholar]
- 5.Kappert K, Meyborg H, Clemenz M, et al. Insulin facilitates monocyte migration: a possible link to tissue inflammation in insulin-resistance. Biochem Biophys Res Commun. 2008;365:503–508. doi: 10.1016/j.bbrc.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Elgazar-Carmon V, Rudich A, Hadad N, Levy R. Neutrophils transiently infiltrate intra-abdominal fat early in the course of high-fat feeding. J Lipid Res. 2008;49:1894–1903. doi: 10.1194/jlr.M800132-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Curat CA, Miranville A, Sengenès C, et al. From blood monocytes to adipose tissue-resident macrophages: induction of diapedesis by human mature adipocytes. Diabetes. 2004;53:1285–1292. doi: 10.2337/diabetes.53.5.1285. [DOI] [PubMed] [Google Scholar]
- 8.Lacasa D, Taleb S, Keophiphath M, Miranville A, Clement K. Macrophage-secreted factors impair human adipogenesis: involvement of proinflammatory state in preadipocytes. Endocrinology. 2007;148:868–877. doi: 10.1210/en.2006-0687. [DOI] [PubMed] [Google Scholar]
- 9.Keophiphath M, Achard V, Henegar C, et al. Macrophage-secreted factors promote a profibrotic phenotype in human preadipocytes. Mol Endocrinol. 2009;23:11–24. doi: 10.1210/me.2008-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suganami T, Nishida J, Ogawa Y. A paracrine loop between adipocytes and macrophages aggravates inflammatory changes: role of free fatty acids and tumor necrosis factor alpha. Arterioscler Thromb Vasc Biol. 2005;25:2062–2068. doi: 10.1161/01.ATV.0000183883.72263.13. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita A, Soga Y, Iwamoto Y, et al. DNA microarray analyses of genes expressed differentially in 3T3-L1 adipocytes co-cultured with murine macrophage cell line RAW264.7 in the presence of the toll-like receptor 4 ligand bacterial endotoxin. Int J Obes (Lond) 2008;32:1725–1729. doi: 10.1038/ijo.2008.153. [DOI] [PubMed] [Google Scholar]
- 12.Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Diabetes. 2007;56:16–23. doi: 10.1152/ajpendo.00284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passwell JH, Levanon M, Davidsohn J, Kohen F, Ramot B. The effect of human monocytes and macrophages on lymphocyte proliferation. Immunology. 1982;47:175–181. [PMC free article] [PubMed] [Google Scholar]
- 14.Ruggiero G, Racioppi L, Manzo C, et al. HLA class II molecules on monocytes regulate T cell proliferation through physical interaction in the CD3 activation pathway. Eur J Immunol. 1991;21:29–33. doi: 10.1002/eji.1830210106. [DOI] [PubMed] [Google Scholar]
- 15.Mueller CG, Boix C, Kwan WH, et al. Critical role of monocytes to support normal B cell and diffuse large B cell lymphoma survival and proliferation. J Leukoc Biol. 2007;82:567–575. doi: 10.1189/jlb.0706481. [DOI] [PubMed] [Google Scholar]
- 16.Hellstrand K, Hermodsson S. Cell-to-cell mediated inhibition of natural killer cell proliferation by monocytes and its regulation by histamine H2-receptors. Scand J Immunol. 1991;34:741–752. doi: 10.1111/j.1365-3083.1991.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 17.Birdsall HH, Porter WJ, Trial J, Rossen RD. Monocytes stimulated by 110-kDa fibronectin fragments suppress proliferation of anti-CD3-activated T cells. J Immunol. 2005;175:3347–3353. doi: 10.4049/jimmunol.175.5.3347. [DOI] [PubMed] [Google Scholar]
- 18.Schubert SY, Benarroch A, Ostvang J, Edelman ER. Regulation of endothelial cell proliferation by primary monocytes. Arterioscler Thromb Vasc Biol. 2008;28:97–104. doi: 10.1161/ATVBAHA.107.157537. [DOI] [PubMed] [Google Scholar]
- 19.Lioté F, Wautier MP, Kuntz D, Wautier JL. Inhibition of human endothelial cell growth by human monocytes in coculture. Nouv Rev Fr Hematol. 1992;34:183–189. [PubMed] [Google Scholar]
- 20.Osusky R, Ryan SJ. Retinal pigment epithelial cell proliferation: potentiation by monocytes and serum. Graefes Arch Clin Exp Ophthalmol. 1996;234(Suppl 1):S76–S82. doi: 10.1007/BF02343052. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt JA, Oliver CN, Lepe-Zuniga JL, Green I, Gery I. Silica-stimulated monocytes release fibroblast proliferation factors identical to interleukin 1 – A potential role for interleukin 1 in the pathogenesis of silicosis. J Clin Invest. 1984;73:1462–1472. doi: 10.1172/JCI111350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Austgulen R, Hammerstrøm J, Nissen-Meyer J. In vitro cultured human monocytes release fibroblast proliferation factoRs) different from interleukin 1. J Leukoc Biol. 1987;42:1–8. doi: 10.1002/jlb.42.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Tintut Y, Patel J, Territo M, et al. Monocyte/macrophage regulation of vascular calcification in vitro. Circulation. 2002;105:650–655. doi: 10.1161/hc0502.102969. [DOI] [PubMed] [Google Scholar]
- 24.Turcanu V, Hirst TR, Williams NA. Modulation of human monocytes by Escherichia coli heat-labile enterotoxin B-subunit; altered cytokine production and its functional consequences. Immunology. 2002;106:316–325. doi: 10.1046/j.1365-2567.2002.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg SA, Lipsky PE. The role of monocytes in pokeweed mitogen-stimulated human B cell activation: separate requirements for intact monocytes and a soluble monocyte factor. J Immunol. 1981;126:1341–1345. [PubMed] [Google Scholar]
- 26.Zandbergen F, Mandard S, Escher P, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Lu X, Lombès M, et al. The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metabol. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isakson P, Hammarstedt A, Gustafson B, Smith U. Impaired preadipocyte differentiation in human abdominal obesity: role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes. 2009;58:1550–1557. doi: 10.2337/db08-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foka P, Irvine SA, Kockar F, Ramji DP. Interleukin-6 represses the transcription of the CCAAT/enhancer binding protein-alpha gene in hepatoma cells by inhibiting its ability to autoactivate the proximal promoter region. Nucleic Acids Res. 2003;31:6722–6732. doi: 10.1093/nar/gkg861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seifeddine R, Dreiem A, Blanc E, et al. Hypoxia down-regulates CCAAT/enhancer binding protein-alpha expression in breast cancer cells. Cancer Res. 2008;68:2158–2165. doi: 10.1158/0008-5472.CAN-07-1190. [DOI] [PubMed] [Google Scholar]
- 31.Artemenko Y, Gagnon A, Sorisky A. Catalytically inactive SHIP2 inhibits proliferation by attenuating PDGF signaling in 3T3-L1 preadipocytes. J Cell Physiol. 2009;218:228–236. doi: 10.1002/jcp.21595. [DOI] [PubMed] [Google Scholar]
- 32.Pang C, Gao Z, Yin J, et al. Macrophage infiltration into adipose tissue may promote angiogenesis for adipose tissue remodeling in obesity. Am J Physiol Endocrinol Metab. 2008;295:E313–E322. doi: 10.1152/ajpendo.90296.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Bevilacqua E, Chiribau CB, et al. Differential control of the CCAAT/enhancer-binding protein beta (C/EBPbeta) products liver-enriched transcriptional activating protein (LAP) and liver-enriched transcriptional inhibitory protein (LIP) and the regulation of gene expression during the response to endoplasmic reticulum stress. J Biol Chem. 2008;283:22443–22456. doi: 10.1074/jbc.M801046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawagishi H, Wakoh T, Uno H, et al. Hzf regulates adipogenesis through translational control of C/EBPalpha. EMBO J. 2008;27:1481–1490. doi: 10.1038/emboj.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuvel DJ, Sundararaj KP, Li Y, Lopes-Virella MF, Huang Y. Adipocyte-mononuclear cell interaction, Toll-like receptor 4 activation, and high glucose synergistically up-regulate osteopontin expression via an interleukin 6-mediated mechanism. J Biol Chem. 2010;285:3916–3927. doi: 10.1074/jbc.M109.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee YH, Nair S, Rousseau E, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gülden E, Mollérus S, Brüggemann J, Burkart V, Habich C. Heat shock protein 60 induces inflammatory mediators in mouse adipocytes. FEBS Lett. 2008;582:2731–2736. doi: 10.1016/j.febslet.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Nair S, Lee YH, Rousseau E, et al. Increased expression of inflammation-related genes in cultured preadipocytes/stromal vascular cells from obese compared with nonobese Pima Indians. Diabetologia. 2005;48:1784–1788. doi: 10.1007/s00125-005-1868-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerhardt CC, Romero IA, Cancello R, Camoin L, Strosberg AD. Chemokines control fat accumulation and leptin secretion by cultured human adipocytes. Mol Cell Endocrinol. 2001;175:81–92. doi: 10.1016/s0303-7207(01)00394-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.