Summary
A short conserved motif located at the carboxy terminus of FtsZ, referred to here as the CCTP (conserved C-terminal peptide), is required for the interaction of FtsZ with many of its partners. In E. coli interaction of FtsZ with its membrane anchors, ZipA and FtsA, as well as the spatial regulators of Z-ring formation, MinC and SlmA, requires the CCTP. ZipA interacts with FtsZ with high affinity and interacts with the CCTP with low affinity, but the reason for this difference is not clear. In this study we show that this difference is due to the oligomerization of FtsZ converting the CCTP to a multivalent ligand that binds multiple ZipAs bound to a surface with high avidity. Artificial dimerization of the CCTP is sufficient to increase the affinity for ZipA in vitro. Similar principles apply to the interaction of FtsZ with SlmA. Although done in vitro these results have implications for the recruitment of FtsZ to the membrane in vivo, the interaction of FtsZ with spatial regulators and the reconstitution of FtsZ systems in vitro.
Introduction
The first generally recognized step in bacterial cell division in E. coli is the formation of the Z ring, which is under spatial and temporal regulation so that it forms at the proper time and place in the cell (Bi & Lutkenhaus, 1991, Lutkenhaus et al., 2012, Lutkenhaus, 2007). The Z ring is about 100 nm in width and accumulating evidence indicates it consists of a band of short, loosely arranged FtsZ filaments that are attached to the membrane through the membrane anchors ZipA and FtsA (Pichoff & Lutkenhaus, 2002, Fu et al., 2010, Strauss et al., 2012). These filaments arise from the GTP dependent polymerization of FtsZ and are extremely dynamic due to the subunit turnover associated with FtsZ’s GTPase activity (Stricker et al., 2002, Mukherjee & Lutkenhaus, 1998).
The FtsZ protein can be divided into distinct functional domains; an N-terminal globular domain with a tubulin fold, which binds GTP and is responsible for polymerization and GTP hydrolysis, is connected by a flexible linker to a short conserved carboxy-terminal peptide (CCTP) that mediates interaction with a number of FtsZ interacting proteins (Erickson et al., 2010, Shen & Lutkenhaus, 2009, Mosyak et al., 2000, Haney et al., 2001, Szwedziak et al., 2012, Du & Lutkenhaus, 2014). The connecting linker is intrinsically disordered and about 50 amino acids in length in E. coli and B. subtilis (Gardner et al., 2013, Buske & Levin, 2013). The sequence of the linker is not important, only that it be intrinsically disordered, and for E. coli FtsZ, that it be longer than about 43 residues and shorter than 95 (Buske & Levin, 2013, Gardner et al., 2013). In some bacteria the linker is much longer, up to 257 residues, but such long linkers cannot functionally replace the linker in the E. coli FtsZ (Gardner et al., 2013).
FtsZ’s CCTP is required for interaction of FtsZ with a number of proteins, including ZipA and FtsA, the membrane anchors for FtsZ filaments in E. coli (Pichoff & Lutkenhaus, 2002, Haney et al., 2001, Mosyak et al., 2000, Szwedziak et al., 2012, Ma & Margolin, 1999). The CCTP is also required for the action of the spatial regulators of the Z ring, MinC and SlmA. MinC, an antagonist of FtsZ assembly, uses a two-pronged mechanism to interact with FtsZ (Shen & Lutkenhaus, 2009, Shen & Lutkenhaus, 2010). It contacts the CCTP through its C-terminal domain and contacts the globular domain through its N-terminal domain (Shen & Lutkenhaus, 2009, Shen & Lutkenhaus, 2010). SlmA, the mediator of nucleoid occlusion, also appears to use a two-pronged mechanism to interact with FtsZ (Du & Lutkenhaus, 2014). It requires interaction with the CCTP to depolymerize FtsZ filaments and genetic as well as biochemical evidences suggests that SlmA contacts the globular domain in addition to the CCTP (Du & Lutkenhaus, 2014, Cho et al., 2011, Tonthat et al., 2011). In addition, the CCTP is required for the interaction of FtsZ with ZapD and ClpXP (Durand-Heredia et al., 2012, Camberg et al., 2009, Camberg et al., 2014). In other organisms the CCTP is involved in binding additional proteins involved in regulating or anchoring the Z ring to the membrane. In addition to FtsA in B. subtilis these include SepF, which can serve as a membrane anchor in this organism, and EzrA, which contributes to the spatial regulation of Z ring assembly (Krol et al., 2012, Duman et al., 2013, Singh et al., 2007, Singh et al., 2008).
The structure of the CCTP bound to ZipA and FtsA is similar but not identical (Szwedziak et al., 2012, Mosyak et al., 2000). The former is from E. coli and the latter from T. maritima. In each case the CCTP has a helical region starting at a conserved proline. The helical region in the CCTP bound to FtsA has a kink, but this occurs at a glycine residue not present in the E. coli CCTP (Szwedziak et al., 2012). The interaction between CCTP and ZipA is largely hydrophobic whereas the interaction between FtsA and CCTP is largely electrostatic (Szwedziak et al., 2012, Mosyak et al., 2000). In both cases the interaction is rather weak with Kds estimated in the 10–50 µmolar range.
Although the interaction between ZipA and the CCTP is rather weak, the interaction between ZipA and full length FtsZ is reported to be about 100 fold stronger (Mosyak et al., 2000, Haney et al., 2001). Two possible explanations for this higher affinity were proposed. One was that the presence of the globular domain influenced the conformation of the CCTP, which favored binding. The second was that ZipA made two contacts with FtsZ, one through the CCTP and the other through the globular domain (Mosyak et al., 2000). This latter possibility seems likely since monomeric ZipA has been reported to bundle FtsZ filaments (Mateos-Gil et al., 2012, Hale et al., 2000, RayChaudhuri, 1999, Kuchibhatla et al., 2011), which would not be expected if ZipA only bound to the CCTP. However, additional studies report that ZipA interacts with FtsZ monomers or oligomers with moderate affinity (micromolar range) and ZipA bundling of FtsZ filaments is salt sensitive (Martos et al., 2010, Hernandez-Rocamora et al., 2012b). We undertook a series of experiments to understand this difference in affinities between ZipA binding to full length FtsZ versus a peptide corresponding to the CCTP, as well as the effect of ZipA on bundling. We also analyze the interaction of SlmA and FtsZ, which has similar characteristics to the interaction of ZipA with FtsZ (Du and Lutkenhaus, 2014). The results have implications for the binding of filaments to the membrane in vivo, the interaction of spatial regulators with FtsZ filaments and in vitro reconstitution experiments.
Results
Differential affinity of ZipA for FtsZ and the CCTP
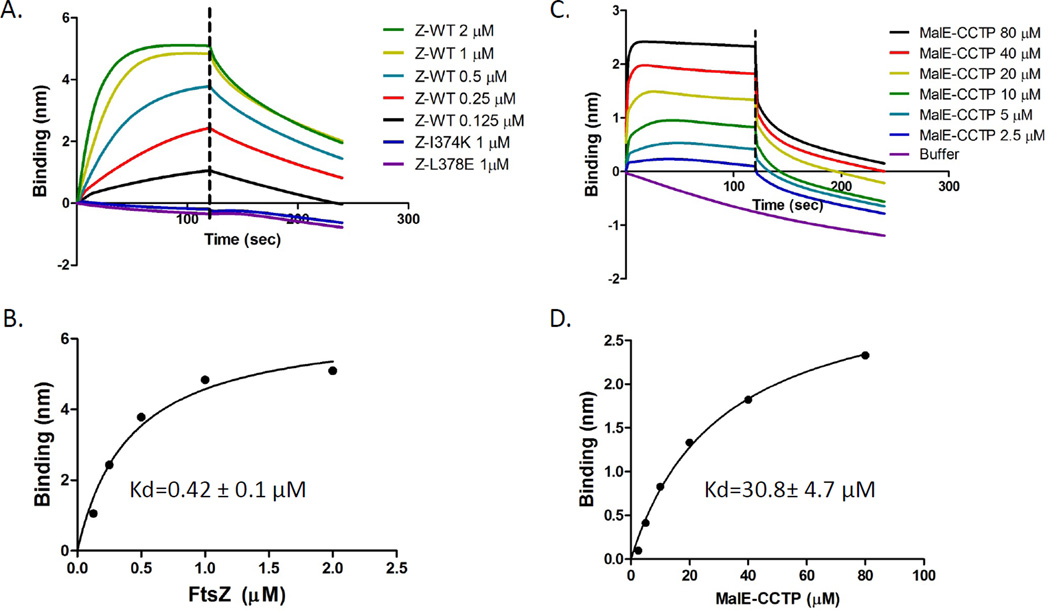
Initially we set out to determine if we could reproduce the different reported affinities of ZipA for FtsZ and the CCTP using a biosensor assay (see Experimental Procedures for the principles and details of the assay). Previous studies have found that the structured domain of ZipA, ZipA185–328, is necessary and sufficient to interact with FtsZ (Mosyak et al., 2000, Haney et al., 2001), so an N-terminally His-tag ZipA185–328 was purified for our study. In this assay 250 µl of His-ZipA185–328 (1 µM) was loaded onto Ni-NTA biosensor pins (ForteBio Inc.). These pins were then incubated with various concentrations of FtsZ or a fusion protein containing the last 17 amino acids of FtsZ (CCTP- residues 367–383) fused to the maltose binding protein (MalE-CCTP). FtsZ bound to the His- ZipA185–328 with an apparent Kd calculated to be 0.42±0.1 µM, which is close to previous reports (Mosyak et al., 2000, Haney et al., 2001)(Fig. 1A &B; Table S1). To ensure that the binding affinity we observed for the full length FtsZ required the CCTP of FtsZ we tested FtsZ mutants containing amino acid substitutions in the CCTP. Neither of the two point mutants (FtsZ-I374K and FtsZ-L378E) displayed an appreciable binding signal when added at 1 µM (Fig. 1A).
Fig. 1.
Differential affinity of His-ZipA185–328 for FtsZ and MalE-CCTP. A&B) Kinetic and equilibrium binding analysis of FtsZ binding to His-ZipA185–328. His-ZipA185–328 was loaded onto Ni-NTA tips by dipping the tips into a 250 µl solution of 1 µM His-ZipA185–328 (50 mM HEPES-NaOH [pH 6.8], 50 mM KCl, 10 mM MgCl2). The tips were rinsed in buffer and dipped in tubes containing various concentrations of FtsZ in the same buffer and the association monitored. After 2 minutes the tips were placed in buffer and disassociation monitored. The association of two FtsZ tail mutants was also examined at 1 µM (FtsZ-I374K and FtsZ-L378E). B) Binding signal at the end of association was plotted against the concentration of FtsZ in GraphPad Prism 5.0 and the apparent Kd value was obtained with nonlinear fitting. C and D) are similar to A) and B) except that MalE-CCTP was used instead of FtsZ.
Next we measured the apparent Kd for the interaction between MalE-CCTP and His- ZipA185–328 and determined that it was 30.8±4.7 µM (Fig. 1C & D). This is similar to the previously reported affinity for a 17 amino acid peptide corresponding to the CCTP (21.6 & 35 µM; Table S1) (Mosyak et al., 2000). We also examined the binding affinity of a 14 amino acid peptide corresponding to residues 370–383 of FtsZ. The affinity of ZipA for this peptide had an apparent Kd of 140±22 µM (Fig. S1 and Table 1). It is likely that this reduced affinity compared to that reported previously for the CCTP is due to the slightly shorter peptide (14 versus 17 amino acids) that we used. Nonetheless, this system reproduces the large differences in affinities between the full length FtsZ and the CCTP reported earlier.
Table 1.
Affinities of proteins in solution for proteins attached to a biosensor1
| Binding affinities of various proteins or peptide to 6×His-ZipA185–328 | |||
|---|---|---|---|
| Apparent Kd (µM) | Bmax (nm)2 | R2(3) | |
| FtsZ-WT | 0.42±0.1 | 6.5±0.6 | 0.97 |
| FtsZ-WT (pH 8.0, 200 mM KCl) | 0.77±0.1 | 4.1±0.2 | 0.99 |
| MalE-CCTP | 30.8±4.7 | 3.2±0.2 | 0.99 |
| FtsZ-L178E | 5.3±0.7 | 4.6±0.2 | 0.99 |
| MinDΔ10-linker-CCTP + ADP | 11.7±5.2 | 4.1±0.7 | 0.94 |
| MinDΔ10-linker-CCTP + ATP | 1.4±0.2 | 4.2±0.2 | 0.99 |
| CCTP (14 amino acids) | 140±22 | 1.0±0.07 | 0.99 |
| Binding affinity of various proteins to 6×His-FtsZ | |||
| ZipA185–328 | 8.1±1.3 | 2.8±0.1 | 0.98 |
| MinDΔ10-ZipA185–328 + ADP | 8.1±2.5 | 5.0±0.7 | 0.97 |
| MinDΔ10-ZipA185–328 + ATP | 1.3±0.1 | 3.4±0.1 | 1.00 |
| Binding affinity of FtsZ360 to 6×His-SlmA-SBS | |||
| FtsZ-360 (pH 8.0, 200 mM KCl) | 82±35 | 0.6±0.2 | 0.99 |
All the measurements were performed in buffer (50 mM HEPES-NaOH pH 6.8, 50 mM KCl, 10 mM MgCl2) unless otherwise indicated.
Bmax is the best-fit value for maximum specific binding in the same unit as Y (nm) in the plot.
R2 indicates the goodness of the fitting curve.
Failure to find support for the two postulated models for the differential affinity
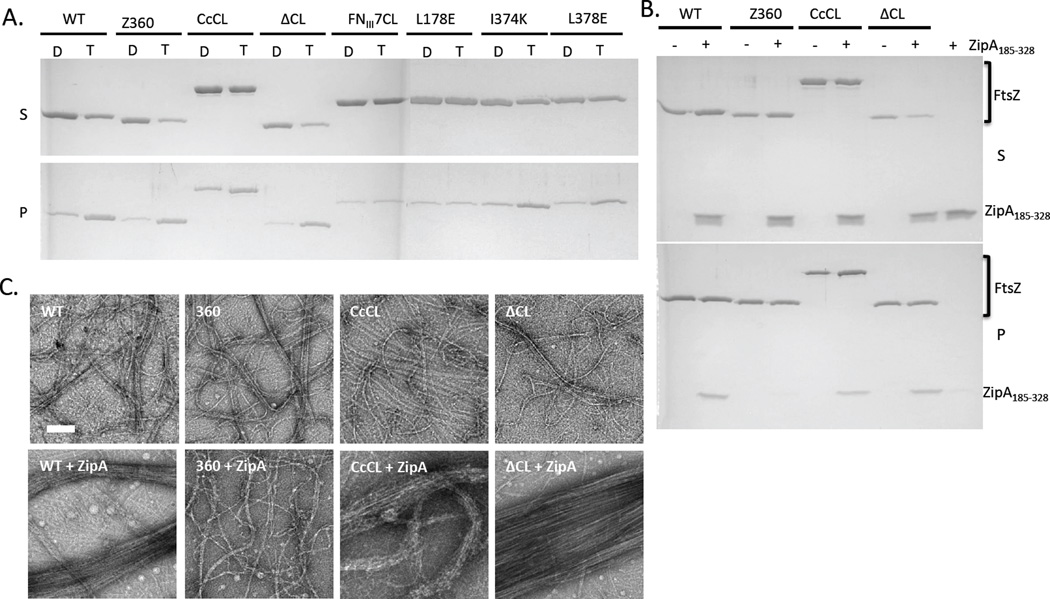
Having verified the differential binding affinities of ZipA for FtsZ and the CCTP we set out to test the two proposed possibilities for the higher affinity of ZipA for FtsZ than the CCTP. One of the explanations was that the globular domain influenced the conformation of the CCTP, which enhanced binding (Mosyak et al., 2000). To test this we exploited constructs from the Erickson lab in which various alterations were made to the linker region (Gardner et al., 2013). We assumed that varying the linker length and/or flexibility would minimize the influence of the globular domain on the CCTP. One construct contained a deletion of the linker region (FtsZΔCL), another contained a much longer linker (linker from Caulobacter crescentus FtsZ, FtsZ-CcCL) and in a third, the unstructured linker was replaced with a structured domain (linker replaced with fibronectin type III domain 7, FtsZ-FNIII7CL). Sedimentation assays and electron microscopy showed that FtsZΔCL and FtsZ-CcCL, polymerized in the presence of GTP, consistent with the previous finding that the linker region is not necessary for polymerization (Fig. 2A) (Gardner et al., 2013, Wang et al., 1997). Surprisingly, FtsZ-FNIII7CL did not polymerize, however, this is consistent with the previous observation that it did not complement an FtsZ depletion strain and the finding that B. subtilis FtsZ with a structured linker polymerizes poorly (Gardner et al., 2013, Buske & Levin, 2013).
Fig. 2.
Interaction of FtsZ mutants with His-ZipA185–328 in solution. A) Polymerization of FtsZ and FtsZ mutants assessed by sedimentation. FtsZ or various mutants (5 µM) was incubated at room temperature for 5 minutes and then GDP or GTP (1 mM) was added. After 5 additional minutes at room temperature the reactions were subjected to ultracentrifugation for 15 minutes and the supernatants and pellets analyzed by SDS-PAGE (D=GDP, T=GTP). The various FtsZ constructs are indicated at the top. B) His-ZipA185–328 co-sediments with FtsZ and FtsZ mutants with modified linkers. Reactions were performed as (A) except that His-ZipA185–328 was included (5 µM). C) EM analysis of effect of His-ZipA185–328 on the bundling of FtsZ and FtsZ mutant polymers. FtsZ and its mutants with or without His-ZipA185–328 (5 µM) were incubated at room temperature for 5 minutes followed by the addition of GTP (1 mM) to initiate polymerization. After 5 minutes 15 µl of each sample was spotted on pre-discharged EM grids and stained with uranyl acetate.
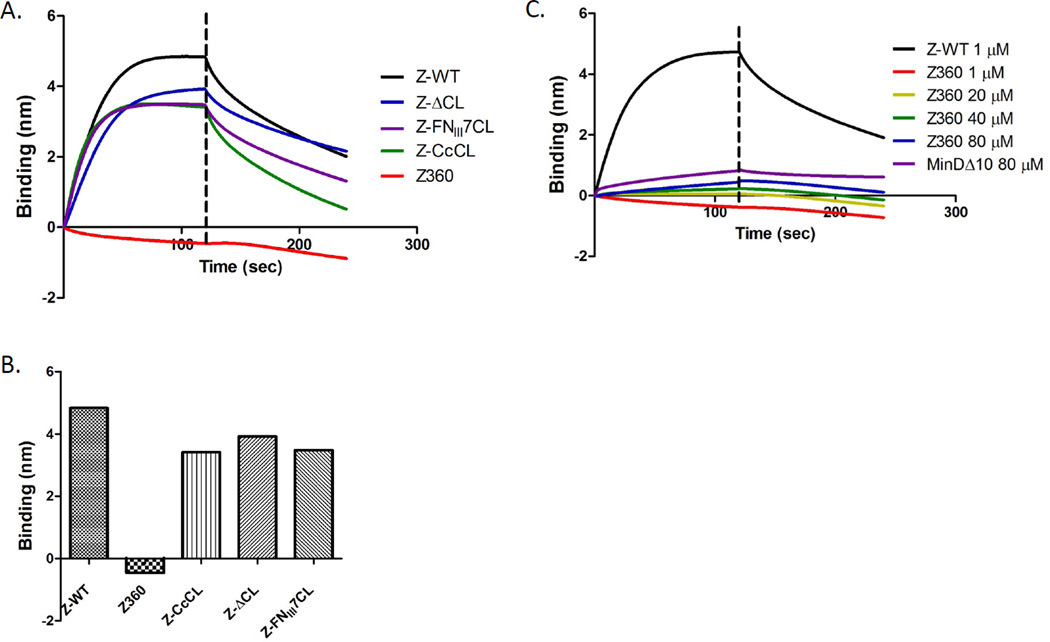
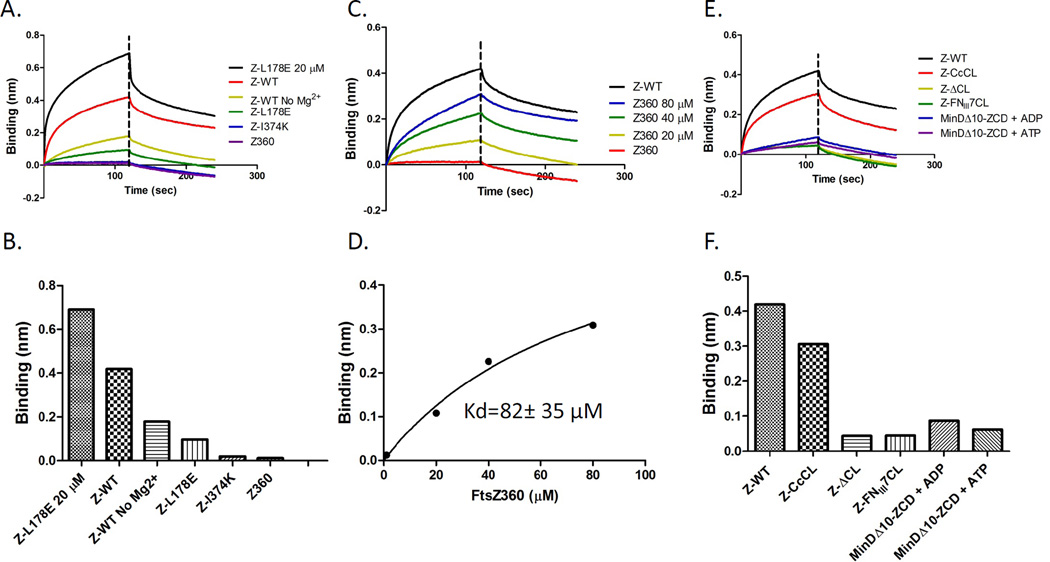
The binding of these three FtsZ constructs with altered linker regions to His-ZipA185–328 in the biosensor assay was fairly similar to native FtsZ with the FtsZΔCL construct having slightly slower kinetics than the other two (Fig. 3A & B). Since the binding was so similar to WT FtsZ, we think it is unlikely that the globular domain of FtsZ influences the conformation of the CCTP resulting in the higher affinity. We therefore tested the possibility that ZipA makes a second contact with FtsZ (Mosyak et al., 2000). To test this we used an FtsZ truncated after amino acid 360, which lacks the CCTP (FtsZ-360). Increasing the concentration of FtsZ-360 from 1 to 80 µM did not result in a significant binding signal (Fig. 3C). Even at 80 µM the binding signal was much lower than the binding signal seen with 1 µM FtsZ and lower than the signal observed with a nonspecific protein, MinDΔ10. Although we cannot exclude the possibility that a weak interaction between ZipA and the globular domain of FtsZ may be dependent on the initial contact between ZipA and the CCTP, our data suggest that the CCTP is the only region of FtsZ important for binding to ZipA in the biosensor assay.
Fig. 3.
Effect of the linker region and CCTP on FtsZ binding to His-ZipA185–328. A) Influence of the linker region on FtsZ binding to His-ZipA185–328. Reactions were performed as Fig. 1A. The various FtsZ constructs contain an altered linker region as described in the text. B) Comparison of the binding signal of His-ZipA185–328 and various FtsZ mutants with altered linker regions. FtsZ and FtsZ-360 (deleted for the CCTP) were run as controls. C) FtsZ-360 does not bind His-ZipA185–328. Reactions were performed as in Fig. 1A. The FtsZ-360 concentration varied from 1–80 µM. MinDΔ10 was run at 80 µM as a control.
Consistent with the results above, His-ZipA185–328 did not sediment with polymers of FtsZ-360 in the presence of GTP, but it did sediment with polymers of FtsZ, FtsZΔCL and FtsZ-CcCL (Fig. 2B). In addition, His-ZipA185–328 bundled FtsZ polymers as reported and elimination of the linker region or increasing the length of the linker did not affect the bundling (Fig. 2C). As expected, His-ZipA185–328 did not bundle FtsZ-360. Since these drastic alterations of the linker region did not affect FtsZ binding to His-ZipA185–328, whether the reaction was performed on the biosensor tips or in solution, we conclude that the CCTP is probably the sole binding site for ZipA.
Evidence that oligomerization is responsible for the differential affinity
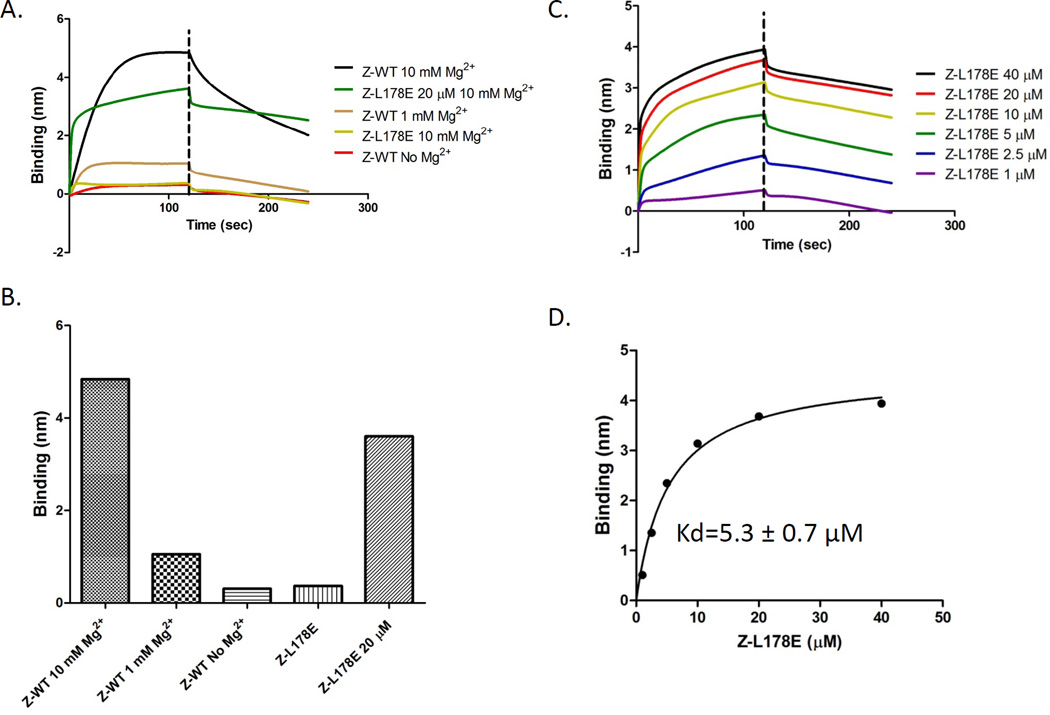
Having found no evidence for the proposed explanations for the different affinities of ZipA for FtsZ and the CCTP we tested the possibility that the higher affinity of ZipA for FtsZ was due to the oligomerization of FtsZ. In this scenario oligomers of FtsZ in solution interact with multiple ZipAs present on the biosensor tip and the higher affinity displayed by FtsZ over the CCTP is due to avidity arising from multiple CCTPs contacting multiple ZipAs on the tip. This possibility may actually reflect the in vivo and in vitro situation in which polymers of FtsZ in the cytoplasm are recruited to a membrane by interaction with multiple ZipAs present in the membrane. Our studies so far were done in the presence of 10 mM Mg2+, which is known to promote the formation of FtsZ oligomers (Sossong et al., 1999, Martos et al., 2010). Although GDP is also present in the buffer, which favors FtsZ monomers, this effect is overridden by Mg2+. (Sossong et al., 1999, Martos et al., 2010). Reducing the Mg2+ concentration from 10 mM to 1 mM dramatically reduced the binding signal and eliminating the Mg2+ reduced the signal further (Fig. 4A). Size exclusion chromatography confirmed that FtsZ exhibited a higher oligomerization state in the presence of 10 mM Mg2+ than in its absence (Fig. S2).
Fig. 4.
Oligomerization of FtsZ enhances binding to His-ZipA185–328 present on the biosensor. A) Influence of Mg2+ concentration and FtsZ oligomerization on FtsZ binding to His-ZipA185–328. Reactions were performed as in Fig. 1A. The Mg2+ concentration in the buffer is indicated and protein concentration was 1 µM unless otherwise indicated. B) Comparison of the binding signal between FtsZ or FtsZ-L178E and His-ZipA185–328 at different Mg2+ or protein concentrations. C&D) Kinetic and equilibrium binding analysis of FtsZ-L178E (concentration indicated in panel) binding to His-ZipA185–328 Reactions were performed as in Fig. 1A and data were plotted as in Fig. 1B.
It was possible, however, that Mg2+ affected the interaction between FtsZ and ZipA directly so we looked for additional support for the oligomerization model. Ideally we wanted an FtsZ mutant that would remain a monomer under all conditions. Recently, Li et al. reported the structure of FtsZ from Mycobacterium tuberculosis in the presence of GDP (Li et al., 2013). FtsZ was present in a twisted filament and a variety of mutations that altered key contact residues between subunits in the structure were made that supported the physiological relevance of the filament structure (Li et al., 2013). Based on their work and with the goal of obtaining a mutant that would be unable to oligomerize we made several E. coli FtsZ mutants corresponding to the mutants which were reported to be unable to polymerize with GTP or assemble into short oligomers at higher concentrations with GDP. One of these, FtsZ-L178E, behaved as a monomer in size exclusion chromatography (Fig. S2). It ran as a monomer with or without GDP and in the presence or absence of 10 mM Mg2+. A sedimentation assay revealed that it also failed to polymerize with GTP (Fig. 2A). Since FtsZ-L178E was a monomer in solution under all conditions tested, it was used to analyze the binding of monomeric FtsZ to His- ZipA185–328.
In the presence of 10 mM Mg2+ FtsZ-L178E (1 µM) produced a very weak binding signal relative to WT FtsZ (Fig. 4A & B). By measuring the binding signal with increasing concentrations of FtsZ-L178E, the apparent Kd of FtsZ-L178E for His-ZipA185–328 was calculated to be 5.3±0.7 µM (Fig. 4C & D). This value is higher than the Kd value for MalE-CCTP (30.8±4.7 µM), but it is still 10 fold weaker than the Kd value with FtsZ-WT. It is possible that the higher value is due to FtsZ-L178E self interacting at higher concentrations since it assembled into filaments when crystallized (data not shown). Together, we conclude that the oligomerization of FtsZ in solution significantly enhances its binding to His-ZipA185–328 located on the biosensor tip.
Confirmation of the oligomerization model by construction of a chimera
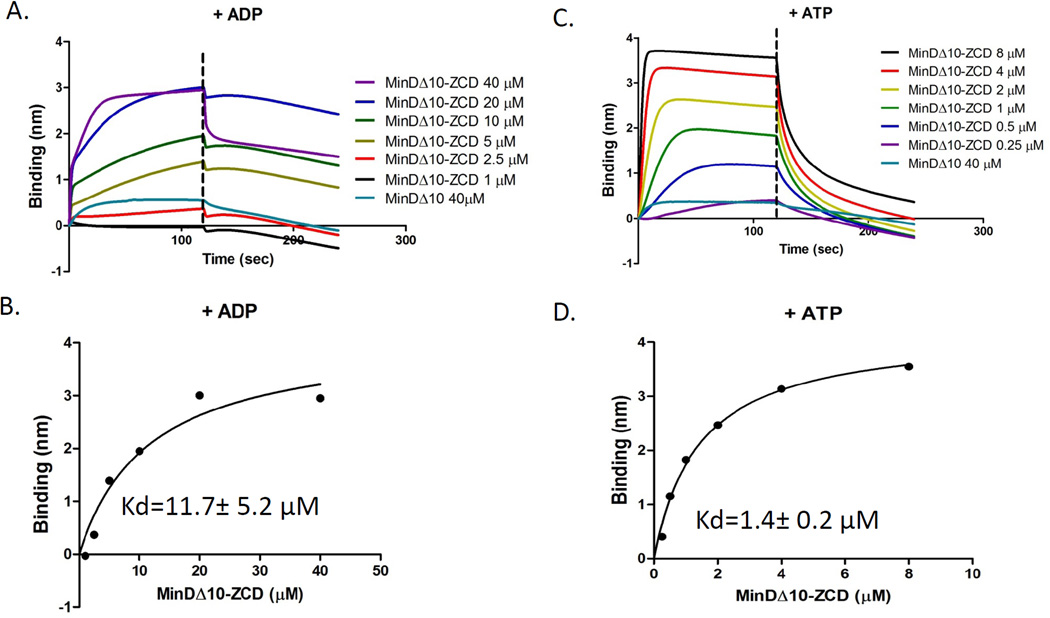
As an additional approach to examine the effect of oligomerization of the protein in the solution on the affinity for the protein on the biosensor pin we constructed a chimera in which the linker and CCTP of FtsZ were fused to the C-terminus of MinDΔ10, essentially replacing the amphipathic tail of MinD (last 10 amino acids) with the linker and CCTP of FtsZ. MinD was chosen since dimerization of MinD can be induced by the addition of ATP (Hu et al., 2003). Only a weak signal was observed when a biosensor pin loaded with His- ZipA185–328 was dipped in a 1 µM solution of MinDΔ10-linker-CCTP in the presence of ADP (Fig. 5A, [MinDΔ10-linker-CCTP is abbreviated as MinDΔ10-ZCD in the figure]). In the presence of ATP, however, a significant signal was observed indicating that dimerization of the chimera significantly increased the affinity (Fig. 5B). To quantify the binding signal, we incubated the His- ZipA185–328 coated pins with increasing concentrations of MinDΔ10-linker-CCTP in the presence of ADP or ATP. The fitting curves of the binding signal plotted versus the concentration of MinDΔ10-linker-CCTP showed that in the presence of ADP, MinDΔ10-linker-CCTP bound to His-ZipA185–328 with an apparent Kd of 11.7±5.2 µM (Fig. 5A&B, Table 1), while in the presence of ATP, the apparent Kd was 1.4±0.2 µM (Fig. 5C &D, Table 1). Together, these results demonstrate that oligomerization of the CCTP significantly enhances its binding to His- ZipA185–328 located on the tip.
Fig. 5.
Dimerization of the CCTP increases its affinity for His-ZipA185–328 loaded on the biosensor. Kinetic and equilibrium analysis of MinDΔ10-linker-CCTP (indicated as MinDΔ10-ZCD in the figure) binding to His-ZipA185–328 in the presence of ADP (A&B) or ATP (C&D). Reactions were performed as in Fig. 1A and data were plotted as in Fig. 1B.
The effect of reversing the components in the reaction
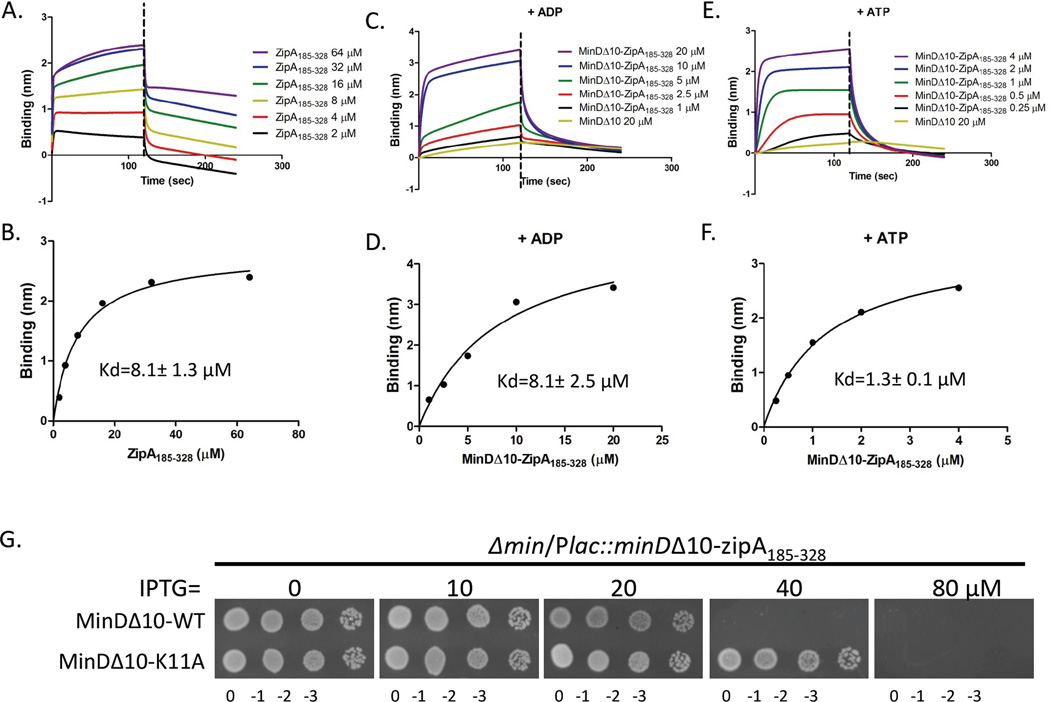
In the above experiments we provide strong evidence that oligomerization of FtsZ in solution augments binding to ZipA on the biosensor tip. If, however, the components in the reaction are reversed (FtsZ attached to the biosensor and ZipA in solution) only weak binding should be observed, since ZipA185–328 does not oligomerize (Mosyak et al., 2000, Martos et al., 2010). Size exclusion chromatography of ZipA185–328 confirmed that it was a monomer as previously reported (Fig. S3). The apparent Kd of ZipA185–328 (untagged) for His-FtsZ attached to the pin was determined to be 8.1±1.3 µM (Fig. 6A & B). To see if dimerization of ZipA would enhance its affinity for FtsZ attached to the biosensor pin we constructed a MinDΔ10-ZipA185–328 fusion and tested it for binding to His-FtsZ loaded onto the biosensor pin in the presence of ADP and ATP. As shown in Fig. 6C–F the apparent Kds for MinDΔ10-ZipA185–328 binding to His-FtsZ in the presence of ADP and ATP were 8.1±2.5 µM and 1.3±0.1 µM, respectively. These results further demonstrate that oligomerization of the component in solution results in a higher affinity for the component attached to the biosensor pin.
Fig. 6.
Dimerization of ZipA185–328 enhances its binding to His-FtsZ. A&B) ZipA185–328 binds to His-FtsZ on the biosensor with moderate affinity. Reactions were performed as Fig. 1A but the orientation of proteins was reversed. His-FtsZ was loaded onto the Ni-NTA tips by incubating the tips with 250 µl solution containing 1 µM His-FtsZ. The tips were then washed and incubated with increasing concentrations of ZipA185–328 (untagged) for 2 minutes followed by a wash and a dissociation phase. Data were plotted as in Fig. 1B. C–F) Kinetic and equilibrium analysis of MinDΔ10-ZipA185–328 binding to His-FtsZ in the presence of ADP (C&D) or ATP (E&F). Reactions were performed as in Fig. 1A and data were plotted as in Fig. 1B. G) Dimerization of MinDΔ10-ZipA185–328 increases toxicity. Single colonies of strain S4/pSD198 (Δmin/Plac minDΔ10-zipA185–328) and S4/pSD198-K11A (Δmin/Plac minDΔ10(K11A)-zipA185–328) were resuspended in 1 ml of LB medium and serially diluted by 10. Three µl of each dilution was spotted on plates containing appropriate antibiotics and IPTG, grown overnight at 37°C and photographed.
In vivo effect of dimerization of ZipA185–328
Overexpression of the soluble ZipA185–328 is toxic to E. coli and inhibits cell division, presumably by disrupting the Z ring (Hale et al., 2000). This toxicity likely results from competition between this soluble ZipA185–328 and FtsZ’s membrane anchors, dislodging FtsZ filaments from the membrane. Based upon our in vitro results a soluble dimeric ZipA185–328 should be more toxic as it would be a better competitor since it binds oligomeric FtsZ with higher affinity. A monomeric MinDΔ10-ZipA185–328 fusion was made by introduction of the minD-K11A mutation into MinDΔ10-ZipA185–328. The toxicity was examined in a Δmin strain so there would be no contribution from interaction with the WT Min proteins. Consistent with expectations MinDΔ10-K11A–ZipA185–328 was less toxic than the wild type MinDΔ10-ZipA185–328 (Fig. 6G). Quantitative immunoblotting indicated that the monomeric mutant was stable and ∼6 fold less toxic than the WT (Fig. S4).
ZipA bundling of FtsZ filaments
Although ZipA has the ability to bundle FtsZ filaments, the basis for this activity is not clear. We only found that ZipA185–328 bound to the CCTP, and since it is monomeric, one would not expect it to promote bundling. However, the bundling of FtsZ filaments by ZipA has been observed repeatedly (Hale et al., 2000, Kuchibhatla et al., 2011, RayChaudhuri, 1999, Mateos-Gil et al., 2012). In some cases it could be explained by the use of full length ZipA, which forms large oligomers due to the presence of the transmembrane region (RayChaudhuri, 1999, Kuchibhatla et al., 2011), but in other cases only the soluble region of ZipA was used (Fig. 2) (Hale et al., 2000). Also, bundling of FtsZ filaments by full length ZipA was observed at low salt (0.05 mM KCl) but was not observed at a higher salt concentration (0.5 mM KCl)(Hernandez-Rocamora et al., 2012a, Martos et al., 2010). This result indicates some step in the ZipA induced bundling is sensitive to the ionic strength so we decided to examine this.
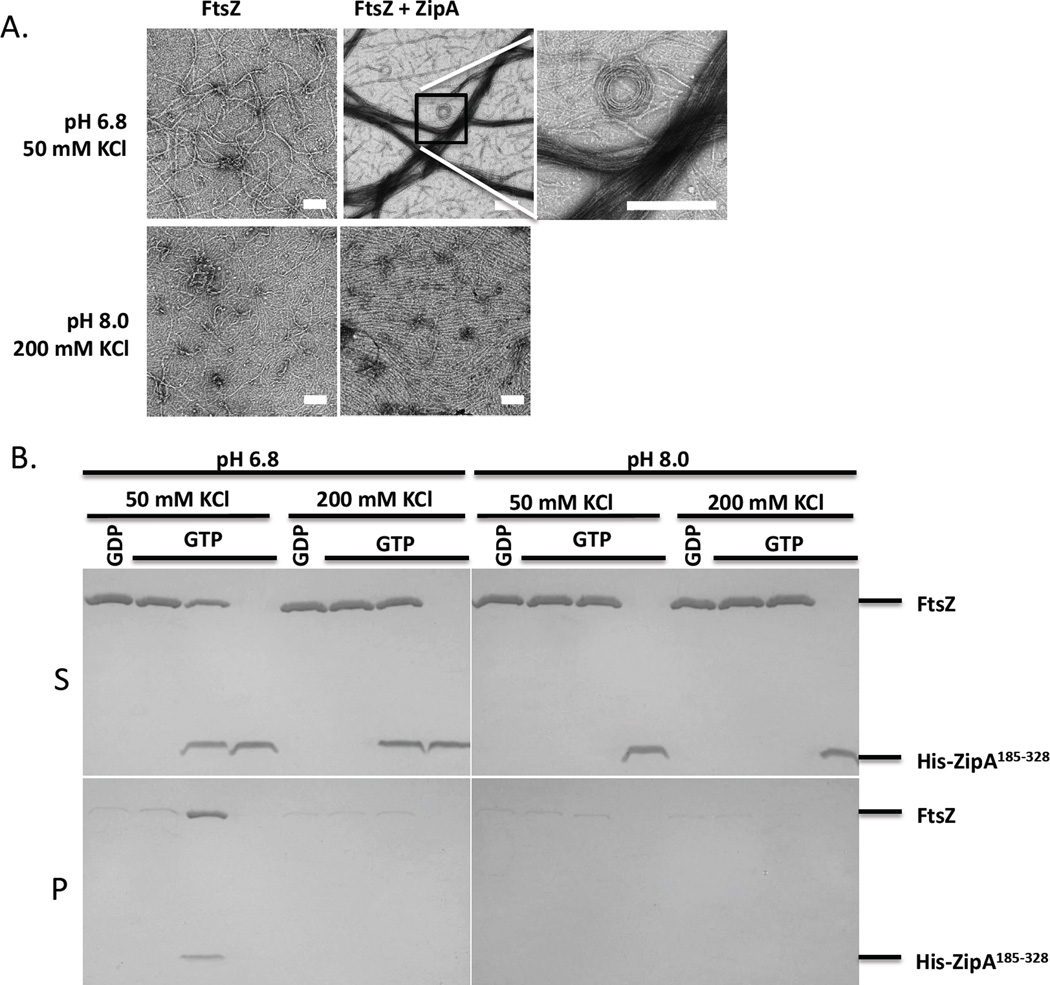
To assess the effect of in vitro buffer conditions on bundling of FtsZ filaments by ZipA we examined the effect of increasing the salt and pH on the bundling of FtsZ filaments by His-ZipA185–328. Under our standard polymerization buffer conditions (50 mM HEPES-NaOH [pH 6.8], 10 mM MgCl2, 50 mM KCl) His-ZipA185–328 induced dramatic bundling of FtsZ filaments as reported by others (Fig. 2 and Fig. 7A). At higher pH and salt concentration (50 mM HEPES-NaOH [pH 8.0], 10 mM MgCl2, 200 mM KCl), however, the dramatic bundling was virtually eliminated. We also verified that the interaction of His-ZipA185–328 and unpolymerized FtsZ was not significantly affected by 200 mM KCl and the higher pH (Fig. S5, Table 1). To quantify the effect of pH and salt concentration on FtsZ bundling caused by His-ZipA185–328, we used low speed centrifugation in which FtsZ filaments bundled by ZipA, but not unbundled filaments, appear in the pellet. As shown in Fig. 7B, FtsZ and His-ZipA185–328 only appeared in the pellet with the low salt low pH buffer (50 mM KCl, pH 6.8), consistent with the EM results. Increasing the pH from 6.8 to 8.0 without changing the salt concentration (50 mM KCl, pH 8.0) or increasing the salt concentration from 50 mM to 200 mM without changing the pH (200 mM KCl, pH 6.8) prevented the pelleting of FtsZ and His-ZipA185–328. Thus, the results indicate the bundling induced by ZipA185–328 in vitro is likely due to electrostatic interactions between ZipA185–328 and FtsZ that occur in addition to the binding to the CCTP.
Fig. 7.
Bundling of FtsZ filaments by ZipA is sensitive to buffer conditions. A) EM analysis of the effects of salt and pH on FtsZ bundling by His-ZipA185–328. FtsZ (5 µM) with or without His-ZipA185–328 (5 µM) was incubated at room temperature for 5 minutes and then GTP (1 mM) was added to initiate polymerization. Two different buffers were used, one with low salt and low pH (50 mM HEPES-NaOH [pH 6.8], 50 mM KCl and 10 mM MgCl2) as in Fig. 2 and the other with high pH and high salt (50 mM Tris-HCl [pH 8.0], 200 mM KCl and 10 mM MgCl2). After 5 minutes 15 µl of each sample was spotted on pre-discharged EM grids and stained with uranyl acetate. The scale bar indicates 100 nm. B) Sedimentation analysis of the effects of salt and pH on FtsZ bundling by His-ZipA185–328. Samples prepared as in panel A were centrifuged at 14K and the supernatants (S) and pellets (P) were analyzed by SDS-PAGE. Lanes 1–4 were in low salt low pH buffer (50 mM KCl and pH 6.8), lanes 5–8 were in high salt low pH buffer (200 mM KCl and pH 6.8), lanes 9–12 were in low salt high pH buffer (50 mM KCl and pH 8.0) and lanes 13–16 were in high salt high pH buffer (200 mM KCl and pH 8.0). Note: lanes 1–2, 5–6, 9–10 and 13–14 contain only FtsZ while lanes 4, 8, 12 and 16 contain only His- ZipA185–328.
FtsZ interaction with SlmA-SBS
The differential affinity ZipA displayed for FtsZ and the CCTP is similar to what we observed in examining the binding of SlmA/SBS to FtsZ and CCTP (Du and Lutkenhaus, 2014). In that study we observed FtsZ bound to SlmA/SBS with a high affinity similar to that reported by Tonthat et al., and the CCTP bound with a weak affinity similar to what we observed between the CCTP and ZipA (Du & Lutkenhaus, 2014, Tonthat et al., 2011). Since the differential affinity between soluble FtsZ and His-ZipA185–328 attached to the biosensor was largely due to the oligomerization state of FtsZ, we speculated that this was also the case for the FtsZ-SlmA interaction. To explore this, we examined the effect of Mg2+ and the FtsZ-L178E substitution on the interaction between FtsZ and His-SlmA/SBS (SBS17, a SlmA binding site with high affinity for SlmA was used)(Cho et al., 2011). Since the interaction between FtsZ and SlmA is dependent upon SlmA bound to DNA, His-SlmA/SBS complexes were loaded on a biosensor pin, which was then dipped in a solution containing FtsZ. Elimination of Mg2+ from the buffer significantly reduced the binding signal between FtsZ and His-SlmA/SBS, although the reduction was not as dramatic as that observed with ZipA (Fig. 8A, olive green line). At 1 µM FtsZ-L178E displayed a weak signal with His-SlmA/SBS, which increased when the concentration was increased to 20 µM indicating it had reduced affinity for His-SlmA/SBS.
Fig. 8.
Interaction of FtsZ with His-SlmA/SBS loaded on the biosensor. A&B) Influence of oligomerization of FtsZ on the interaction with SBS-His-SlmA loaded on the biosensor. SBS-His/SlmA was loaded on the Ni-NTA tips by incubating the tips with 250 µl of solution containing 1 µM His-SlmA and 1 µM SBS DNA. After a 10 second wash, association of FtsZ to SBS-His/SlmA was initiated by incubating the tips with solutions containing the indicated proteins (at 1 µM FtsZ [WT or mutant] and 10 mM Mg2+ unless otherwise indicated). After 2 minutes the tips were incubated with buffer to monitor the dissociation of the protein complexes. Binding signals at the end of association in panel A are plotted in panel B. C&D) Kinetic and equilibrium analysis of FtsZ-360 binding to SBS/His-SlmA on the biosensor. Reactions were performed as in panel A and the binding signals at the end of association were plotted against the protein concentration. An apparent Kd value was obtained by nonlinear fitting in GraphPad Prism 5.0. (E&F) Dimerization of the CCTP and influence of the linker region on FtsZ’s interaction with SBS/His-SlmA loaded on the biosensor. Reactions were performed as in panel A and data were compared as panel B. The protein concentrations were 1 µM and nucleotides were added at 1 mM.
To further explore the interaction between FtsZ and His-SlmA/SBS we examined the various FtsZ constructs with alterations to the CCTP. Deletion of the CCTP (FtsZ-360) or substitution of key residues within the CCTP eliminated the binding signal for His-SlmA/SBS at low concentration (1 µM, Fig. 8C & D), as previously reported (Du & Lutkenhaus, 2014). Interestingly, increasing the FtsZ-360 concentration resulted in increased signal suggesting a second weak interaction outside of the CCTP, something that was not observed with ZipA (Fig. 8C versus Fig. 3C). The apparent Kd for this association was estimated to be 82±35 µM, similar to what we observed for the CCTP (Fig. 8D, Table 1) (Du & Lutkenhaus, 2014). A weak interaction between FtsZ lacking the CCTP and SlmA was observed previously (Tonthat et al., 2011).
We next explored the effect of the linker region on the FtsZ-SlmA/SBS interaction. In the FtsZ-ZipA binding experiments the linker region did not seem important and the various constructs with the linker region deleted or replaced with a longer or structured linker, behaved similar to FtsZ with the WT linker. In contrast to the results with ZipA, the linker impacted the binding of FtsZ to His-SlmA/SBS as only the construct with the long Caulobacter crescentus linker (FtsZ-CcCL) displayed strong binding (Fig. 8E & F). The other two constructs, including the one with the linker deleted (FtsZΔCL) and one with the structured linker (FNIII7CL), displayed very weak binding. These results suggest that a long flexible linker is required for high affinity binding to SlmA/SBS. A long flexible linker must be required for FtsZ to interact with the two binding sites on one SlmA monomer or to bind simultaneously to binding sites on adjacent dimers, since SlmA exists as a dimer of dimers on the SBS DNA. Furthermore, we found that addition of ATP did not augment the binding of MinDΔ10-linker-CCTP to His-SlmA/SBS on the biosensor. The signal was weak in the presence of ADP or ATP (Fig 8E & F). Taken together, these results suggest that oligomerization of FtsZ enhances the binding of FtsZ to SlmA/SBS significantly but has more stringent requirements than we observed with ZipA.
Discussion
Protein-protein interactions driven by short linear peptide sequences embedded within or at the end of a region of predicted disorder is quite common in eukaryotes (Uversky, 2013). An example is the intrinsically disordered C-terminal regulatory domain of p53, which binds many different partners (Oldfield et al., 2008). Although less common in prokaryotes, a few examples have emerged over the last decade. The single-stranded DNA binding protein (SSB) of E. coli, which is required for DNA replication, recombination and repair, contains an intrinsically disordered C-terminal tail that is required for SSB binding to more than 10 of its partners (Shereda et al., 2008). The CCTP of FtsZ is another example of a short peptide in prokaryotes that mediates the interaction of FtsZ with many proteins, including essential membrane tethers and the spatial regulators that help position the Z ring. Here, we show that the highly conserved CCTP binds to FtsZ’s partners with high specificity and low affinity, and the oligomerization of FtsZ generates a multivalent CCTP that binds cooperatively to these partners with high avidity. This kind of behavior favors polymers of FtsZ over monomers interacting with FtsZ’s partners. Thus, it is likely that only polymers of FtsZ are attached to the membrane and that spatial regulators of FtsZ interact primarily with FtsZ polymers. Although the system we studied is artificial it likely has characteristics that mimic the in vivo situation. In particular, it is likely that attaching a protein to a chip is somewhat akin to having a protein attached to a membrane (although it can not diffuse) allowing an estimation of the impact of oligomerization of the protein in solution on its recruitment to the membrane.
The importance of oligomerization for high affinity binding between ZipA and FtsZ
In this study we set out to understand the differential affinity of ZipA for FtsZ and the CCTP. In our experimental system ZipA is attached to a biosensor tip and the FtsZ ligand is in solution. This setup reproduced the reported ∼100 fold difference in affinity that ZipA displays for FtsZ and the CCTP. Our conclusion that higher affinity binding is due to oligomerization that produces a multivalent CCTP binding to ZipA on the biosensor is based upon results from several approaches. First, conditions that reduced oligomerization (lower Mg2+ or the ftsZ-L178E mutation) reduced the affinity close to that observed with the CCTP attached to a monomeric carrier protein (MalE-CCTP or MinDΔ10-linker-CCTP). Second, dimerization of a monomeric carrier protein (MinDΔ10-linker-CCTP) was sufficient to increase the affinity ∼10 fold. Third, we demonstrated the importance of oligomerization of the protein in solution by reversing the components in the reaction. The monomeric C-terminal domain of ZipA alone or fused to MinDΔ10, had low affinity for FtsZ on the biosensor, however, dimerization of MinDΔ10-ZipA led to a ∼6 fold increase in affinity. This result is consistent with the previous finding that ZipA interacts with FtsZ-GTP polymers with moderate affinity when monomeric ZipA was immobilized on a nanodisc (apparent Kd ∼ 40 µM, each nanodisc contains one ZipA molecule)(Hernandez-Rocamora et al., 2012b). Taken together, these results demonstrate that interaction between the CCTP and ZipA has low affinity if the protein in solution is monomeric, but high affinity if the protein is converted to a multimer. It should be noted that in earlier studies in which high affinity between FtsZ and ZipA were observed the measurements were done in the presence of Mg2+ which favors the oligomerization of FtsZ (Hale et al., 2000, Mosyak et al., 2000).
The linker region of FtsZ did not play a significant role in the binding affinity to ZipA under these experimental conditions. Even deletion of the linker region did not attenuate the binding indicating that FtsZΔCL multimers are still able to contact multiple ZipAs on the biosenor. It should be noted that the FtsZΔCL construct still contains a 6 amino acid linker (GRSGLE) due to the way it was constructed.
ZipA bundling of FtsZ filaments
Although ZipA has been repeatedly observed to bundle FtsZ filaments the significance is not clear. Our interest was in the mechanism by which monomeric ZipA could cause bundling; other bundling proteins are at least dimeric. Our results show that ZipA does indeed bundle FtsZ filaments under some buffer conditions, however, this bundling effect of ZipA was eliminated by increasing the pH and salt concentration of the buffer. Hale et al. also mentioned that increasing the pH and salt concentration significantly reduced the bundling of FtsZ filaments caused by ZipA (Hale et al., 2000). This raises the possibility that ZipA bound to the CCTP in one FtsZ filament interacts electrostatically with another FtsZ filament, essentially bridging two filaments. Such an effect may be similar to adding the extreme carboxy 6 residues of the B. subtilis FtsZ to the end of the E. coli FtsZ (Buske & Levin, 2012). The presence of these extra residues, three of which carry a positive charge, is enough to cause the filaments to assemble into sheets and bundles at low salt concentration (50 mM MES, pH 6.5, 50 mM KCl, 2.5 mM MgCl2, 1 mM EGTA)(Buske & Levin, 2012). Consistent with this, the packing of FtsZ filaments appears irregular unlike FtsZ filaments bundled by ZapA or the packing of tubulin protofilaments in the wall of a microtubule (Mohammadi et al., 2009). Even though we raise doubts about the significance of ZipA bundling observed in vitro it is possible that ZipA could bundle polymers in vivo if it dimerizes through its N-terminal domain (Skoog & Daley, 2012).
FtsZ and SlmA
SlmA displays differential affinity for FtsZ and the CCTP that is similar to the interaction between ZipA and FtsZ, however, the interaction displays several differences. Conditions that favor FtsZ monomers (low Mg2+ or the ftsZ-L178E mutation) decrease the binding between FtsZ and SlmA/SBS as it did between FtsZ and ZipA. This is consistent with the observation by Cho et al. who observed that SBS/SlmA interacted with stable FtsZ polymers (FtsZ-D212N–GTP polymers) but not with FtsZ in the presence of GDP in a pull down assay (Cho et al., 2011). In contrast to our results with ZipA, however, we were unable to recapitulate high affinity binding by dimerization of MinDΔ10-linker-CCTP. In addition, only one of the constructs with an altered linker, FtsZ-CcCL displayed high affinity binding, the ones with the deleted linker or a structured linker did not. This suggests that the interaction between FtsZ and SlmA/SBS is less permissive than the FtsZ-ZipA interaction and requires a long, flexible linker.
Another difference between the FtsZ-SlmA interaction and the FtsZ-ZipA interaction was that FtsZ interacted with SlmA through a second site. This interaction is weak similar to the CCTP-SlmA/SBS interaction. The possibility of an interaction between SlmA and FtsZ outside of the CCTP was suggested from in vivo and in vitro work. As the CCTP is not necessary for polymerization, SBS-bound SlmA must also bind to the globular domain of FtsZ to antagonize FtsZ polymerization (Cho et al., 2011, Du & Lutkenhaus, 2014). Consistent with this, mutations in the globular domain of FtsZ confer resistance to SlmA/SBS and a SlmA mutant unable to bind DNA (which is necessary to bind the CCTP) inhibits division inefficiently, but genetic evidence indicates it does so through FtsZ (Du & Lutkenhaus, 2014, Cho et al., 2011). In addition, Tonthat et al. found that a SlmA dimer forms a complex with FtsZ316-GFP (the linker and CCTP replaced with GFP), indicating a weak interaction with the globular domain of FtsZ (Tonthat et al., 2011). Surprisingly, even though we found evidence for FtsZ interacting with SlmA through two different sites, the CCTP and somewhere else, our results indicate that a monomer of FtsZ binds only weakly to SlmA/SBS on the biosensor. This suggests that the two interaction sites on one FtsZ monomer are probably unable to both interact with the same SlmA monomer or even the combination of these two bindings is still weak.
Implications of this work
The results reported here suggest that studying the interaction between FtsZ and its interacting partners attached to a biosensor may be quite useful when that interaction depends upon the CCTP of FtsZ. Since the affinity of the CCTP for its interacting partner is weak, it is likely that the interaction of FtsZ and its partners involves short polymers of FtsZ and not monomers. Monomeric FtsZ-GDP may only exist transiently in the cytolplasm as conditions in the cell favor formation of FtsZ-GTP oligomers, which are quickly recruited to the membrane (Fig. 9A.). The weak affinity of the CCTP for ZipA suggests that FtsZ monomers detach from the membrane when they are released from the membrane-localized polymer, whether or not they directly interact with ZipA (Fig. 9A.) This is likely the case for FtsZ polymers recruited by FtsA as well. In a recent in vitro study the interaction of FtsZ with a supported membrane layer containing ZipA or FtsA was examined (Loose & Mitchison, 2014). One of the findings was that the FtsZ pattern was different depending upon whether the membrane anchor was ZipA or FtsA. One possible explanation proposed for the difference was that ZipA but not FtsA could recruit FtsZ monomers to the membrane and nucleate FtsZ polymers (Loose & Mitchison, 2014). However, because of the Mg2+ concentration used in that study (5 mM) they were likely observing the recruitment of FtsZ multimers to the membrane by ZipA, something not observed with FtsA.
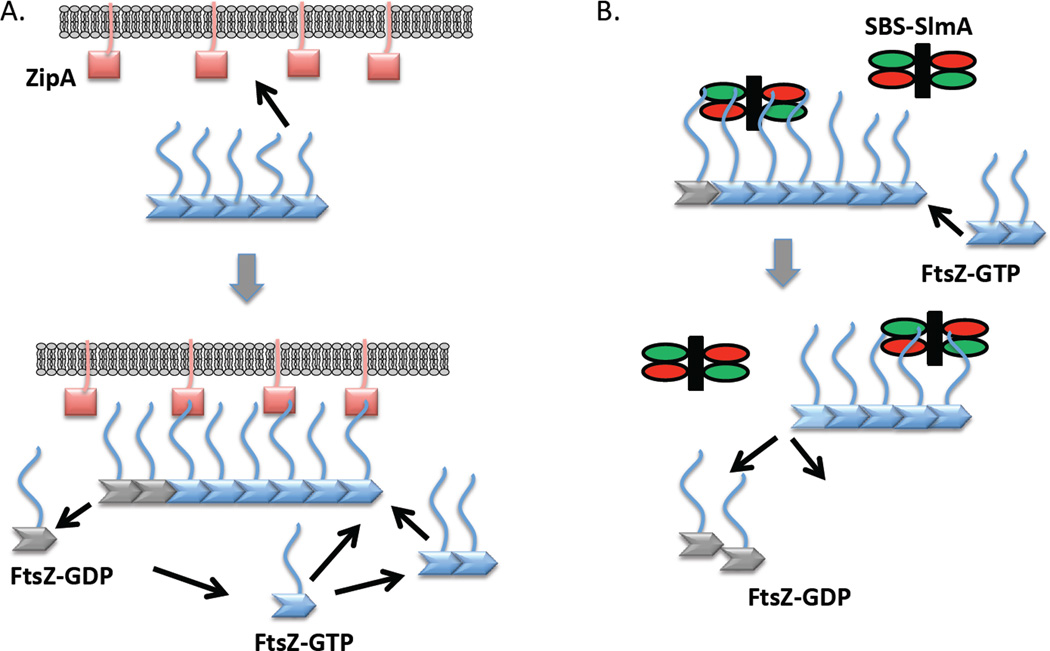
Fig. 9.
How the weak affinity of the CCTP of FtsZ for partners such as ZipA contributes to FtsZ polymer dynamics at the membrane. (A) Oligomers of FtsZ in solution bind to the membrane (or biosensor) through multiple ZipAs binding multiple CCTPs. As new FtsZ subunits (monomers or short polymers) are added GTP hydrolysis leads to release of FtsZ monomers. Even though such monomers may be bound to ZipA through the CCTP, the weak affinity results in release of the FtsZ monomer from the membrane. (B) SBS-SlmA complex interacts with FtsZ polymers by binding to the CCTP. A second interaction (not depicted) between SBS-SlmA and FtsZ filament (globular domain of FtsZ) leads to breakage of the filament. After severing the FtsZ polymers, SBS-SlmA quickly dissociates from the bound FtsZ monomers and binds to the next available FtsZ polymer to initiate another round of breakdown of FtsZ polymers.
Previously, it was suggested that SlmA interacts with preformed polymers, in part, because an interaction was detected between SlmA and polymerized FtsZ and not between SlmA and FtsZ monomers in a pull down assay (Cho et al., 2011). This inability to observe monomers in the pull down assay is due to the weak affinity of FtsZ monomers for SlmA compared to the affinity for FtsZ oligomers. This is similar to the results with MinCC and FtsZ where stable FtsZ polymers but not monomers are recruited to vesicles in the presence of MinD and ATP suggesting that MinCC also has higher affinity for FtsZ polymers (Shen & Lutkenhaus, 2009). This higher affinity of spatial regulators for FtsZ oligomers ensures that they interact with FtsZ polymers, and that monomers do not compete, and is consistent with the proposal that these spatial regulators function by attacking FtsZ polymers (Shen & Lutkenhaus, 2010, Cho et al., 2011, Du & Lutkenhaus, 2014, Hu et al., 1999). FtsZ monomers bound to an SBS-SlmA or MinCD complex would quickly dissociate from the SBS-SlmA or MinCD complex once the polymer has been severed (Fig. 9B). In this way, the SBS-SlmA or MinCD complex avoids saturation by FtsZ monomers and can work efficiently at a much lower concentration than the level of FtsZ. Thus, our finding that oligomerization of FtsZ is responsible for the high affinity of FtsZ for ligands that bind the CCTP is useful to discern the different affinity FtsZ monomers and oligomers have for FtsZ ligands.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Strains and plasmids used in this study are listed in Table 2. Strains were grown in LB medium at 37°C unless otherwise indicated. When needed, antibiotics were used at the following concentrations: ampicillin=100 µg/ml; kanamycin=25 µg/ml; and chloramphenicol=20 µg/ml.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain | Genotype | Source/reference | |
|---|---|---|---|
| BL21 (λDE3) | ompT rB– mB (PlacUV5::T7gene1) | Novagen | |
| JS238 | MC1061 malPp::lacIq srlC::Tn10 recA1 | Lab collection | |
| S4 | W3110 min::kan | (Shen & Lutkenhaus, 2009) | |
| Plasmid | Genotype | Origin | Source/reference |
| pET11B–FRCCc | pET11B PT7::FtsZ-CcCL | ColE1 | (Gardner et al., 2013) |
| pJF118EH-MinDΔ10_L5 | Ptac::minDΔ10-L5 | ColE1 | KT Park |
| pJSB-FNIII7 | pBAD18 Para::FtsZ-FNIII7 Ampr | ColE1 | (Gardner et al., 2013) |
| pJSB-ΔCLa | pBAD18 Para::FtsZ-FNIII7 Ampr | ColE1 | (Gardner et al., 2013) |
| pMT219b | pET21(a) PT7::CcftsZ Ampr | ColE1 | (Thanbichler & Shapiro, 2006) |
| pSEB160 | pBAD18, Para::ftsZ, Ampr | ColE1 | This study |
| pSD119 | pBAD18, Plac::6×his-ftsZ, Ampr | ColE1 | (Du & Lutkenhaus, 2014) |
| pSD197 | pMAL-c2 Ptac::malE-ftsZ367–383, Ampr | ColE1 | This study |
| pSD198 | pQE80 Plac::6×his-zipA185–328Ampr | ColE1 | (Du & Lutkenhaus, 2014) |
| pSD207 | pET21(a) PT7::ftsZΔCL Ampr | ColE1 | This study |
| pSD208 | pET21(a) PT7::ftsZ-FNIII7C L Ampr | ColE1 | This study |
| pSD210 | pBAD18, Para:: ftsZΔCL, Ampr | ColE1 | This study |
| pSD211 | pBAD18, Para:: ftsZ-FNIII7CL, Ampr | ColE1 | This study |
| pSD212 | pJF118EH Ptac::minDΔ10-linker-CCTP | ColE1 | This study |
| pSD217 | pE-SUMO PT7::sumo-zipA185–328Ampr | ColE1 | This study |
| pSD218 | pQE80 Plac::minDΔ10-zipA185–328Ampr | ColE1 | This study |
The original pJSB-ΔCL contains one base insertion (Cytosine) after the codon for residue I374 due to the designed primer. This insertion causes frame shift and produces a protein without the CCTP of FtsZ. We corrected this mutation by replacing the FtsZ-FNIII7CL fragment of pJSB-FNIII7 with the FtsZΔCL (BamHI/XhoI digested) fragment from pJSB-ΔCL. Note that the fragment of FtsZΔCL does not contain the CCTP of FtsZ.
CcftsZ stands for the Caulobacter crescentus FtsZ
Construction of plasmids
Various constructs expressing FtsZs with variations in the linker region were made. A plasmid pJSB-ΔCL (Gardner et al., 2013) from the Erickson lab contained a single base insertion after codon 374 in ftsZ. This mutation causes a frame shift that produced a protein without the last 10 amino acids in CCTP of FtsZ. To correct the mutation we digested pJSB-FNIII7CL and pJSB-ΔCL with BamHI/XhoI and ligated the ftsZ-ΔCL (without CCTP) fragment from pJSB-ΔCL to the digested pJSB-FNIII7CL. As the sequence of pJSB-FNIII7CL was correct, this ligation resulted in the correct plasmid that can express ftsZ without the linker region but still contain the CCTP, FtsZΔCL.
pSD197, which expresses MalE fused to the CCTP of FtsZ, was constructed by inserting an EcoRI and HindIII digested fragment encoding ftsZ367–383 into the EcoRI and HindIII double digested pMAL-c2 plasmid. The fragment was amplified from pBANG112 using primer pairs Ztail-5’-EcoRI: 5’- GACTGAATTCAAAGAGCCGGATTATCTGG-3’ and Ztail-3’-HindIII: 5’- GACTAAGCTTGCCGGGAAATCTACCGGTG-3’.
Plasmids pSD207 and pSD208 were constructed by replacing the CcftsZ coding sequence of pMT219 (Thanbichler & Shapiro, 2006) with sequences coding for ftsZ-ΔCL-CCTP and ftsZ-FNIII7CL-CCTP amplified from pJSB-ΔCL and pJSB-FNIII7CL using primers EcZ-NdeI-F: 5’-GACTCATATGTTTGAACCAATGGAACT-3’ and EcZ-HindIII-R: 5’-GCCAAGCTTGCATGCCTGCAG-3’. The amplified fragments were digested with NdeI and HindIII and ligated to pMT219 digested with the same enzymes. Although pSD207 and pSD208 could be transformed into nonexpression strains easily, they could not be transformed into the BL21/pLysS strain, presumably due to the toxicity associated with the leaky expression of the mutant proteins.
Plasmid pSD210 and pSD211 were constructed by replacing the ftsZ coding sequence of pSEB160 with sequences coding for ftsZ-ΔCL-CCTP and ftsZ-FNIII7CL-CCTP. As there is an EcoRI site at the beginning of ftsZ, we cut pSD207 and pSD208 with EcoRI and HindIII to obtain the fragments coding for ftsZ-ΔCL-CCTP and ftsZ-FNIII7CL-CCTP. The digested fragments were ligated into pSEB160 cut with the same enzymes. The resulting plasmid did not cause any filamentation in JS238 in the absence or presence of glucose due to the tightly controlled Para promoter.
Plasmid pSD212 was created by inserting a BamHI and HindIII digested fragment encoding ftsZ321–383 into the BamHI and HindIII double digested pJF118EH-minDΔ10-L5 constructed by KT ParK. The fragment was amplified from plasmid pSEB160 using primers Z321-BamHI-F: 5’-GATCGGATCCCCTGAAATCACTCTGGTGAC-3’ and EcZ-HindIII-R.
Plasmid pSD217 was constructed by ligation of an of a BsaI-XbaI fragment containing the ZipA185–328 coding sequence and pE-SUMO-amp (LifeSensors) cut with the same enzymes. The fragment was amplified from plasmid pSEB370 using primers zipA185-BsaI-F: 5’-CAGGTCTCTAGGTGATAAACCGAAGCGCAAAG-3’ and zipA-XbaI-R: 5’-GTCATCTAGATCAGGCGTTGGCGTCTTTG-3’.
pSD218 was made by ligation of a BamHI/EcoRI double digested fragment containing coding sequence of minDΔ10 to pSD198 (Du & Lutkenhaus, 2014) digested with the same enzymes.
Protein purification
FtsZ-WT, FtsZ-360, FtsZΔCL, FtsZ-FNIII7CL, FtsZ-L178E, FtsZ-I374K and FtsZ-L378E were purified as previously described (Du & Lutkenhaus, 2014). FtsZ-CcCL was purified similarly except that the protein was expressed in strain BL21/pLysS.
Purification of His-FtsZ, His-SlmA and His-ZipA185–328 were previously described using affinity chromotography (Du & Lutkenhaus, 2014).
MinDΔ10, MinDΔ10-linker-CCTP, MinDΔ10-ZipA185–328 were purified from JS238 containing pJF118EH-minDΔ10-L5, pSD212 and pSD218 following the procedure of MinD purification with a few modifications (de Boer et al., 1991). Cells expressing the indicated proteins were collected from 1L of culture grown in LB with 1 mM IPTG for 3 hours. Collected cells were resuspended in buffer A (25 mM Tris-HCl [pH 7.5], 20 mM NaCl, 1 mM EDTA, 1 mM DTT and 10% glycerol) and lysed by passing through the French press twice. The lysates were centrifuged at 12,000 rpm for 15 min at 4°C to remove cell debris. The supernatants were removed and loaded onto a pre-equilibrated DEAE column and then washed once with 50 mL buffer A. MinDΔ10, MinDΔ10-linker-CCTP, MinDΔ10-ZipA185–328 proteins were eluted with a 20–200 mM NaCl gradient in buffer A. The peak fractions were pooled and ran over a HiLoad Superdex 200 column in buffer A. The peak fractions were collected and concentrated with Amicon Ultra Centrifugal Filters.
Purification of ZipA185–328 from BL21/pLysS cells containing pSD217 was similar to purification of SlmA described previously (Du & Lutkenhaus, 2014). Cell pellets were resuspended in 20 ml lysis buffer (20 mM Tris-HCl [pH7.9], 70 mM NaCl and 20 mM imidazole) and lysed by French press. The clarified supernatant was loaded onto a pre-equilibrated Ni-NTA resin and washed twice with high salt wash buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl and 20 mM imidazole/50 mM imidazole). The bound 6×His-SUMO-ZipA185–328 protein was eluted with elution buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl and 250 mM imidazole). The peak fractions were dialyzed against the storage buffer (25 mM Tris-HCl [pH 7.9], 200 mM KCl, 1 mM EDTA and 10% glycerol) overnight. After dialysis, the 6×His-SUMO tag was cleaved with purified 6×His-tagged SUMO protease (Ulp1) for 4 hour on ice in the protein storage buffer. The released tag and protease were removed by passing it through the pre-equilibrated Ni-NTA resin. Untagged ZipA185–328 was collected in the flow through and wash fractions, concentrated and stored at −80°C.
MalE-CCTP was purified from JS238 containing pSD197 (pMAL-c2-ftsZ367–383) following the protocol of purification of maltose fusion protein from New England Biolabs.
Sedimentation assay and electron microscopy
FtsZ polymerization reactions were performed in 50 µL Pol buffer (50 mM HEPES-NaOH pH 6.8, 50 mM KCl and 10 mM MgCl2 for His-ZipA185–328, and 50 mM HEPES-NaOH pH 8.0, 200 mM KCl and 10 mM MgCl2 for reactions with SBS-SlmA) at room temperature. FtsZ or its mutants (5µM) was incubated with or without equal molar His-ZipA185–328 at room temperature for 5 min. After the addition of 1 mM GDP or GTP, the reactions were subjected to ultracentrifugation at 80,000 rpm for 15 min at 25°C in TLA100.2 rotor and a Beckman TL-100 centrifuge. Supernatants and pellets were then analyzed by SDS-PAGE. Low speed centrifugation experiments were performed similarly except that the buffers varied in pH or KCl concentration.
To visualize the effect of His-ZipA185–328 on FtsZ polymerization, 5 minutes after addition of GTP, 15 µL reaction samples were loaded onto glow discharged grids. 15 µl of 1% uranyl acetate was spotted on the grid for 1 min and blotted away. The grids were air-dried and imaged with a JEOL-JEM-1400 transmission electron microscope.
Biolayer interferometry assays
The BLItz system developed by Fortebio employs a label free, optical analytical technology to measure protein interactions based on Biolayer Interferometry. In these assays, a protein is immobilized on the biosensor surface and its binding partner is provided in solution. Any interaction between the immobilized protein with its binding partners in solution increases the optical thickness of the biolayer in the biosensor, resulting in a wavelength shift (Δλ) that is reported with nm as units. In our study, the assays with His-ZipA185–328 were performed in 250 µl of 1× Pol buffer (50 mM HEPES-NaOH [pH 6.8], 50 mM KCl, and 0, 1 or 10 mM MgCl2) with the BLItz™ system (FortéBio) at room temperature. 250 µL of His-ZipA185–328 (1 µM) was incubated with pre-equilibrated Ni-NTA biosensors for 3 minutes to immobilize His-ZipA185–328 on the surface of the biosensor and then washed 10 seconds to remove any loosely bound protein. Binding of FtsZ, FtsZ mutants, MalE-CCTP or MinDΔ10-linker-CCTP to the immobilized His-ZipA185–328 was monitored for 2 minutes with agitation at 2,200 rpm followed by dissociation in the same buffer without proteins for 2 minutes. Data were automatically collected by the BLItz system and analyzed with GraphPad Prism 5. To obtain the apparent Kd, the binding signals at the end of the association step were plotted against the protein concentrations using a one-site specific nonlinear regression fitting.
In the reciprocal approach, His-FtsZ was immobilized at the surface of Ni-NTA biosensors and untagged ZipA185–328, or MinDΔ10-ZipA185–328 in the presence of ADP or ATP, was tested for binding to His-FtsZ. In these assays, 250 µL of 1× Pol buffer containing 1 µM His-FtsZ was incubated with the biosensor tips with shaking at 2,200 r.p.m for 3 minutes. After the immobilization, the biosensor tips were washed with 1× Pol buffer for 10 seconds. Association of untagged ZipA185–328, MinDΔ10-ZipA185–328 in the presence of ADP or ATP was monitored for 2 minutes and dissociation was initiated by dipping the biosensor tips into 250 µl of 1× Pol buffer. Data were generated automatically by the BLItz™ User Software version and were subsequently analyzed by global fitting using the GraphPad Prism 5 software.
The assays with His-SlmA were performed similarly but in a different 1× Pol buffer (50 mM HEPES-NaOH [pH 8.0], 200 mM KCl, 0 or 10 mM MgCl2). His-SlmA (1 µM) was incubated with 1 µM of SBS17 at room temperature for 10 minutes before its association to Ni-NTA biosensors. The His-SlmA/SBS complexes was immobilized on the surface of the biosensors for 3 minutes and then washed for 10 seconds to remove the loosely bound complexes. Binding of FtsZ, FtsZ mutants, or MinDΔ10-linker-CCTP with ADP or ATP to the immobilized His-SlmA/SBS complexes was monitored for 2 minutes with agitation at 2,200 rpm. Dissociation was initiated by moving the tips into the same buffer without proteins and monitored for 2 minutes. Data were collected and analyzed with GraphPad Prism 5.
Supplementary Material
Acknowledgements
We thank Harold Erickson (Duke University) for supplying strains and plasmids. This work was supported by the National Institutes of Health with the Grant GM 29764.
References
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. Journal of bacteriology. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske PJ, Levin PA. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. The Journal of biological chemistry. 2012;287:10945–10957. doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske PJ, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Molecular microbiology. 2013;89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Viola MG, Rea L, Hoskins JR, Wickner S. Location of dual sites in E. coli FtsZ important for degradation by ClpXP; one at the C-terminus and one in the disordered linker. PloS one. 2014;9:e94964. doi: 10.1371/journal.pone.0094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Hand AR, Rothfield LI. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. The EMBO journal. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Lutkenhaus J. SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD. PLoS genetics. 2014;10:e1004460. doi: 10.1371/journal.pgen.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R, Ishikawa S, Celik I, Strahl H, Ogasawara N, Troc P, Lowe J, Hamoen LW. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4601–E4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. Journal of bacteriology. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and molecular biology reviews : MMBR. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PloS one. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KA, Moore DA, Erickson HP. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Molecular microbiology. 2013;89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. Journal of bacteriology. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. The Journal of biological chemistry. 2001;276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rocamora VM, Garcia-Montanes C, Rivas G, Llorca O. Reconstitution of the Escherichia coli cell division ZipA-FtsZ complexes in nanodiscs as revealed by electron microscopy. Journal of structural biology. 2012a;180:531–538. doi: 10.1016/j.jsb.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rocamora VM, Reija B, Garcia C, Natale P, Alfonso C, Minton AP, Zorrilla S, Rivas G, Vicente M. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. The Journal of biological chemistry. 2012b;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. Journal of bacteriology. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, van Kessel SP, van Bezouwen LS, Kumar N, Boekema EJ, Scheffers DJ. Bacillus subtilis SepF binds to the C-terminus of FtsZ. PloS one. 2012;7:e43293. doi: 10.1371/journal.pone.0043293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchibhatla A, Bhattacharya A, Panda D. ZipA binds to FtsZ with high affinity and enhances the stability of FtsZ protofilaments. PloS one. 2011;6:e28262. doi: 10.1371/journal.pone.0028262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, Wang HW, Ye S. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Mitchison TJ. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nature cell biology. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annual review of biochemistry. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. Journal of bacteriology. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martos A, Alfonso C, Lopez-Navajas P, Ahijado-Guzmn R, Mingorance J, Minton AP, Rivas G. Characterization of self-association and heteroassociation of bacterial cell division proteins FtsZ and ZipA in solution by composition gradient-static light scattering. Biochemistry. 2010;49:10780–10787. doi: 10.1021/bi101495x. [DOI] [PubMed] [Google Scholar]
- Mateos-Gil P, Marquez I, Lopez-Navajas P, Jimenez M, Vicente M, Mingorance J, Rivas G, Velez M. FtsZ polymers bound to lipid bilayers through ZipA form dynamic two dimensional networks. Biochimica et biophysica acta. 2012;1818:806–813. doi: 10.1016/j.bbamem.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, Martos A, Alfonso C, van Marle J, Rivas G, den Blaauwen T. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry. 2009;48:11056–11066. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. The EMBO journal. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. The EMBO journal. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. The EMBO journal. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. The EMBO journal. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Molecular microbiology. 2009;72:410–424. doi: 10.1111/j.1365-2958.2009.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Lutkenhaus J. Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Molecular microbiology. 2010;75:1285–1298. doi: 10.1111/j.1365-2958.2010.07055.x. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Critical reviews in biochemistry and molecular biology. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh JK, Makde RD, Kumar V, Panda D. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry. 2007;46:11013–11022. doi: 10.1021/bi700710j. [DOI] [PubMed] [Google Scholar]
- Singh JK, Makde RD, Kumar V, Panda D. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. The Journal of biological chemistry. 2008;283:31116–31124. doi: 10.1074/jbc.M805910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog K, Daley DO. The Escherichia coli cell division protein ZipA forms homodimers prior to association with FtsZ. Biochemistry. 2012;51:1407–1415. doi: 10.1021/bi2015647. [DOI] [PubMed] [Google Scholar]
- Sossong TM, Jr, Brigham-Burke MR, Hensley P, Pearce KH., Jr Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D–SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS biology. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. The EMBO journal. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. The EMBO journal. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. The most important thing is the tail: multitudinous functionalities of intrinsically disordered protein termini. FEBS letters. 2013;587:1891–1901. doi: 10.1016/j.febslet.2013.04.042. [DOI] [PubMed] [Google Scholar]
- Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. Journal of bacteriology. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.