Abstract
Cell-surface markers and transcription factors are being used in the assessment of stem cell fate and therapeutic safety, but display significant variability in stem cell cultures. We assessed nuclear patterns of 5-hydroxymethylcytosine (5hmC, associated with pluripotency), a second important epigenetic mark, and its combination with 5-methylcytosine (5mC, associated with differentiation), also in comparison to more established markers of pluripotency (Oct-4) and endodermal differentiation (FoxA2, Sox17) in mouse embryonic stem cells (mESC) over a ten-day differentiation course in vitro: by means of confocal and super-resolution imaging together with high-content analysis, an essential tool in single-cell screening. In summary: 1) We did not measure any significant correlation of putative markers with global 5mC or 5hmC. 2) While average Oct-4 levels stagnated on a cell-population base (0.015 lnIU per day), Sox17 and FoxA2 increased 22-fold and 3-fold faster, respectively (Sox17:0.343 lnIU/day; FoxA2: 0.046 lnIU/day). In comparison, DNA global methylation levels increased 4-fold faster (0.068 lnIU/day), and global hydroxymethylation declined at 0.046 lnIU/day, both with a better explanation of the temporal profile. 3) This progression was concomitant with the occurrence of distinct nuclear codistribution patterns that represented a heterogeneous spectrum of states in differentiation; converging to three major coexisting 5mC/5hmC phenotypes by day 10: 5hmC+/5mC−, 5hmC+/5mC+, and 5hmC−/5mC+ cells. 4) Using optical nanoscopy we could delineate the respective topologies of 5mC/5hmC colocalization in subregions of nuclear DNA: in the majority of 5hmC+/5mC+ cells 5hmC and 5mC predominantly occupied mutually exclusive territories resembling euchromatic and heterochromatic regions, respectively. Simultaneously, in a smaller subset of cells we observed a tighter colocalization of the two cytosine variants, presumably delineating chromatin domains in remodeling. We conclude that 1) 5mC emerges as the most differential marker in our model system. 2) However, the combined enrollment of the two DNA modifications provided higher-definition screening and lead to the identification of cell subpopulations based on differential 5hmC/5mC phenotypes corresponding to different 5hmC/5mC ratios. The results encourage: a) assessing the regenerative potential of early-endodermal cells enriched for the three DNA methylation/hydroxymethylation categories, and b) exploring the universality of this type of epigenetic phenotyping across other lineage-specific differentiations.
Keywords: DNA methylation, hydroxymethylcytosine, stem cell heterogeneity, super-resolution imaging, 3D high-content analysis, epigenetic phenotyping
Introduction
Pluripotent stem cells such as ESC and induced pluripotent stem cells (iPSC) provide an unlimited source of cells for disease modeling, and therapeutic intervention (regenerative medicine) of damaged tissue by cell transplantation and/or pharmaceutical approaches [1–3]. The assessment of stem cell fate during in vitro propagation and differentiation is an important factor in the evaluation of such biomedical operations, also in terms of therapeutic safety [4]. Molecular markers derived from cell phenotyping and transcriptional profiling are amongst the most established indicators in this field. However, much evidence has brought to light that pluripotent cell populations in the embryo or in ESC cultures display significant heterogeneity at the molecular level that could be associated with the probabilistic nature of fate determination [5, 6]. This molecular heterogeneity is reflected by the variability in expression of cell surface antigens and also more cell-interior early-lineage differentiation markers, such as the nuclear DNA-binding protein forkhead box A2 (FoxA2) and the transcription factor sex determining region Y (SRY)-related HMG-box (Sox17), that we recently confirmed on a cell-by-cell resolution using multi-parametric high-content analysis (HCA) of mouse ESC [7]. In the search for additional and more deterministic indicators of stem cell differentiation we assessed the utility of global DNA methylation, an epigenetic key regulator of chromatin structure and relevant DNA expression in cellular differentiation and functionality [8]. In comparison, we had found that during the six days of early endodermal lineage commitment global DNA methylation increased in a linear fashion. As a result of this development we had observed distinct DNA methylation phenotypes that did not correlate with the expression of the two differentiation markers, as well as with other proteins that we had covisualized and analyzed together with 5-methylcytosine: including the canonical pluripotency marker octamer-binding transcription factor 4 (Oct-4), the cell-cell adhesion molecule E-cadherin-1 (CDH1), and the insulin-like growth factor 1 receptor (IGFR) involved in cell transformation, which are respective indicators of the epithelial and mesenchymal phenotypes of cells. By tracking nuclear load and spatial distribution patterns of 5mC we found that changes in these parameters seem to follow more deterministic cues as opposed to the covisualized protein markers. In fact, we were able to identify progressive 5mC-based chromatin texture patterns indicative of the very early stages of lineage commitment: with the ultimate notion of finding in situ patterns of epigenetic marks as signatures for the selection of multipotent (non-pluripotent) cell phenotypes for regenerative medicine applications. Nevertheless, the field of hematology/immunology, in which cell phenotyping is routinely applied for quantifying the different cellular constituents in blood, has established the use of more than one marker. This screening practice can lead to the identification of cellular subpopulations and subsequently to a higher-definition phenotyping in blood testing. In order to emulate this successful model we were tempted to recruit additional DNA-specific epigenetic modifications to be tested in combination with 5mC as biomarkers towards a more detailed characterization of mouse ESC cultures. For this purpose, we focused on 5-hydroxymethylcytosine, a chemical modification of 5mC, that had been originally discovered in bacteriophages [9], and is gaining much attention in biology and the biomedical space since its recent re-encounters in human and mouse brain cells [10]. In mammals, it appears as a product of 5mC oxidation by ten-eleven translocation (TET) family of enzymes [11]. In human and mouse embryonic stem cells, 5hmC is not as abundant as in Purkinje neurons but still at significant levels [12–15] and highly enriched in primordial germ cells [16], therefore seemingly playing a role in the gender-specific resetting of genome-wide imprints with importance for embryonic development [17] and chromatin states related to pluripotency [18]. Genome-wide analyses using chromatin immunoprecipitation with antibodies specific to 5mC and 5hmC have revealed that the two cytosine variants occupy mutually exclusive sites within the genome, and that 5hmC decreases as a result of down-regulation of TET enzymes, concomitant with the down-regulation of pluripotency-related genes and an increase in global DNA methylation, in parallel to elevated expression of lineage-specific markers [14]. Considering the existing knowledge, here we report on the recruit of 5hmC as a second DNA-specific epigenetic imprint and a putative marker for pluripotency for the higher-definition phenotyping of ESC cultures in early endodermal differentiation. The novelty of the work is a) in part based on the application of confocal and cutting-edge super-resolution imaging in combination with highly-parallel cell-by-cell data extraction, and b) in part based on the utility of bigdata analysis principles that lend themselves to time-saving higher-throughput stem cell screening. Thereby the notion was to reveal the parallel dynamics in the levels and phenotypes (spatial distributions) of 5hmC and 5mC and their respective cell population using mouse early endodermal differentiation as a model.
Materials and methods
Stem-cell culture and endodermal differentiation
We used mouse ES cell line R1 (SCRC-1011, American Type Tissue Collection, Manassas, VA). For maintenance culture, ES cells were kept on a mitotically inactivated feeder-layer of mouse embryonic fibroblasts in Dulbecco’s Modified Eagle’s Medium (DMEM) high glucose (Life Technologies, Carlsbad, CA) supplemented with 15% ES-qualified fetal bovine serum (FBS) (Atlanta Biotech), 10 mM of 2-mercaptoethanol (Sigma, St. Louis, MO), 2 mM L-glutamine (Invitrogen), and 10 ng/ml of leukemia inhibitory factor (LIF) (Life Technologies). The cells were initially divided into subpopulations that were cultured on 18 mm round glass coverslips (Fisher Scientific) —that were placed into a 12-well microplate and coated with Type I collagen (Sigma)— for immunofluorescence (IF) assays. ES cells were removed from culture wells and seeded at a density of 8,000–12,000 cells/cm2 for the naïve control population and Day 1–3 propagation, and 4,000–6,000 cells/cm2 for Day 5–10 propagation in the abovementioned medium but without LIF and substituted with heat-inactivated FBS and 100 ng/ml aFGF (Sigma). Parallel cultures were allowed to grow up to six different time points: 24 hours = day 1, day 3, day 5, day 7, and day 10. At each time point cells were fixed for 30 minutes in 4% paraformaldehyde (Sigma) before cytochemical processing.
Immunofluorescence
Immunofluorescence was performed according to previously established protocols [7, 19]. Additional primary and secondary antibody sets included unconjugated monoclonal mouse anti-5-methylcytosine monoclonal antibody (Cat. No. AMM99021, Aviva Systems Biology, San Diego, CA) and rabbit anti-hydroxymethylcytosine, polyclonal IgG (Cat. No. 39791, Active Motif, Carlsbad, CA), both at the concentration of 1 µg/ml, as well as secondary Alexa488-linked donkey anti-mouse IgG (Cat. No. A21202, Life Technologies, Carlsbad, CA) and Alexa 647-conjugated chicken anti-rabbit IgG (Cat. No. A21443, Life Technologies), respectively at 5 µg/ml final concentration. The specificity/dynamic range of the antibodies was tested as previously reported in [20] (data not shown in here).
Confocal and super-resolution imaging, 3-D reconstruction, analysis and animation
Confocal microscopy and three-dimensional (3-D) image analysis were performed as previously described in [7]. 3-D reconstructions of confocal images were performed with the CytoFx software (Illucida, LLC). The process used maximum intensity projection combined with shadow projection [21]. Channels were shown in different combinations. For super-resolution imaging we applied 3-D localization by ground state depletion (3D-GSD) using a Leica SR GSD 3-D nanoscope (Leica Microsystems, Mannheim, Germany). The system has a 20 nm lateral resolution and an improved axial resolution of 50 nm. 3-D reconstructions of super-resolution images were performed with the Leica Application Suite Advanced Fluorescence (LAS AF).
Statistical data analysis
Analysis of obtained imaging data was performed with the statistical package STATA 12 (StataCorp, College Station, TX). We conducted the statistical analysis in two stages. In the first part we have analyzed the data using Principle Component Analysis (PCA), one of the oldest multivariate exploratory statistical analyses that are still in use [22]. The purpose of these exploratory analyses was to determine the number and types of groups of cells that can be identified based on the six biomarkers utilized in this study and across a period of 10 days of incubation. In the second and final step we utilized weighted least-squares regression analysis with the weights estimated by an iterative process called robust regression. The process is an alternative to ordinary linear regression in cases where the observed data is contaminated with outliers or influential observations. It can also be used for the purpose of detecting influential observations.
Results
Spatiotemporal changes in the distribution of cell phenotypes during early differentiation
The objective of our study was to characterize mouse ESC during early in vitro differentiation towards definitive endoderm (day 0 – day10) regarding their global nuclear DNA methylation and hydroxymethylation patterns. Lineage-specific differentiation was induced by culturing ESC in the presence of acidic fibroblast growth factor 1 (aFGF), as previously reported [7, 23]. Subpopulations of the cells deriving from one initially undifferentiated batch were cultured in parallel and fixed at day 0 (after the cells had attached to the culture dish surface), and subsequently at 24 hours post induction of differentiation (day 1), as well as at days 3, 5, 7 and 10. The analyzed nuclear targets besides 5mC and 5hmC included Oct-4, FoxA2, Sox17, and global DNA (gDNA) visualized by 4',6-diamidino-2-phenylindole (DAPI). The high-content assay and analysis was performed on the three-dimensional quantitative DNA methylation imaging (3D-qDMI) platform that we had previously introduced [19, 24–26]. The technology constitutes an image-cytometric approach, by which fluorescence signals of nuclear targets —generated through established immunocytochemistry and light microscopy— are extracted from 3-D images to visualize and measure changes in global DNA methylation and hydroxymethylation in nuclei of thousands of cells in parallel. This capability to analyze cell populations on a per-cell basis is a powerful means in addressing ESC population heterogeneity that usually display a high level of spatiotemporal complexity [27,28] in the regulation of pluripotency maintenance and lineage commitment, in an intricate network of stochastic and binary signaling cues. Thereby fluctuations at the single-cell level can often also shape changes in the development of cell populations [29]. Following the in situ visualization of cells, the subsequent quantitative analyses were based on the three extracted features: 1) the overall abundance level of each target measured as the respective mean nuclear intensity, and 2) the codistributions of 5mC, 5hmC, and gDNA (DAPI); both parameters were used for phenotyping of cells. 3) As an additional parameter, we determined the heterogeneity of the two first features in interrogated cell populations. For each day and combination of targets we were able to image multiple ES cell colonies containing a representative subset of ~2,000 propagated cells for statistical evaluation.
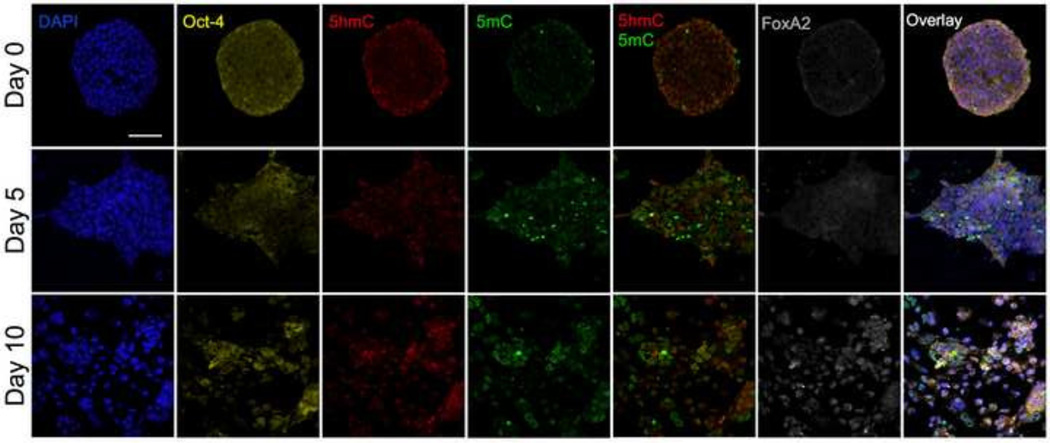
Fig. 1 shows sample cell colonies on the three days 0, 5, and 10, which in comparison revealed more significant changes in cell phenotypes and colony structure than the days in between (day 3 and day 7). Fig. 2 illustrates the 3-D reconstruction of the same cell populations for an enhanced visual impression of the topographical and subcellular topological (tectonic) changes of the visualized markers. Both animations clearly demonstrate that originally more spherical colonies grew into diverse directions (as seen at day 5), before they seemingly segregated into smaller cores as observed on day 7 (not shown in here) and day 10. These cores became surrounded by a large number of cells including a majority of differentiating cells as judged by their enlarged and flatter morphology in conjunction with comparatively increased levels of 5mC signal. The outbound migration of these derivative cells from the cores then formed a tighter meshwork of valley cells in between the remaining slightly elevated cores, as can be discerned from the 3D reconstructed images in Fig. 2.
Fig. 1.
Structure and cellular heterogeneity of mESC colonies regarding five visualized nuclear markers by confocal scanning microscopy: gDNA (represented by DAPI), Oct-4, 5hmC, 5mC, and FoxA2. Each marker (false-colored) was recorded in a separate channel. For each colony (day) all channels —including the five-color overlay image— are presented from the same focal plane (midsection). The aerial views depict changes in overall structure and marker heterogeneity during the 10-day period of early endodermal differentiation. At day 5, the colonies reached their maximal size before they transitioned into smaller core colonies with more Oct-4 positive (pluripotent) cells surrounded by a mesh of detached cells that appear to be in the various stages of differentiation. In addition, Oct-4-positive regions also include cells with strong 5hmC and low 5mC signals. The 5mC channel reveals a number of high-intensity objects that may represent non-viable cells or larger cell debris from dying cells. White bar is 20 µm.
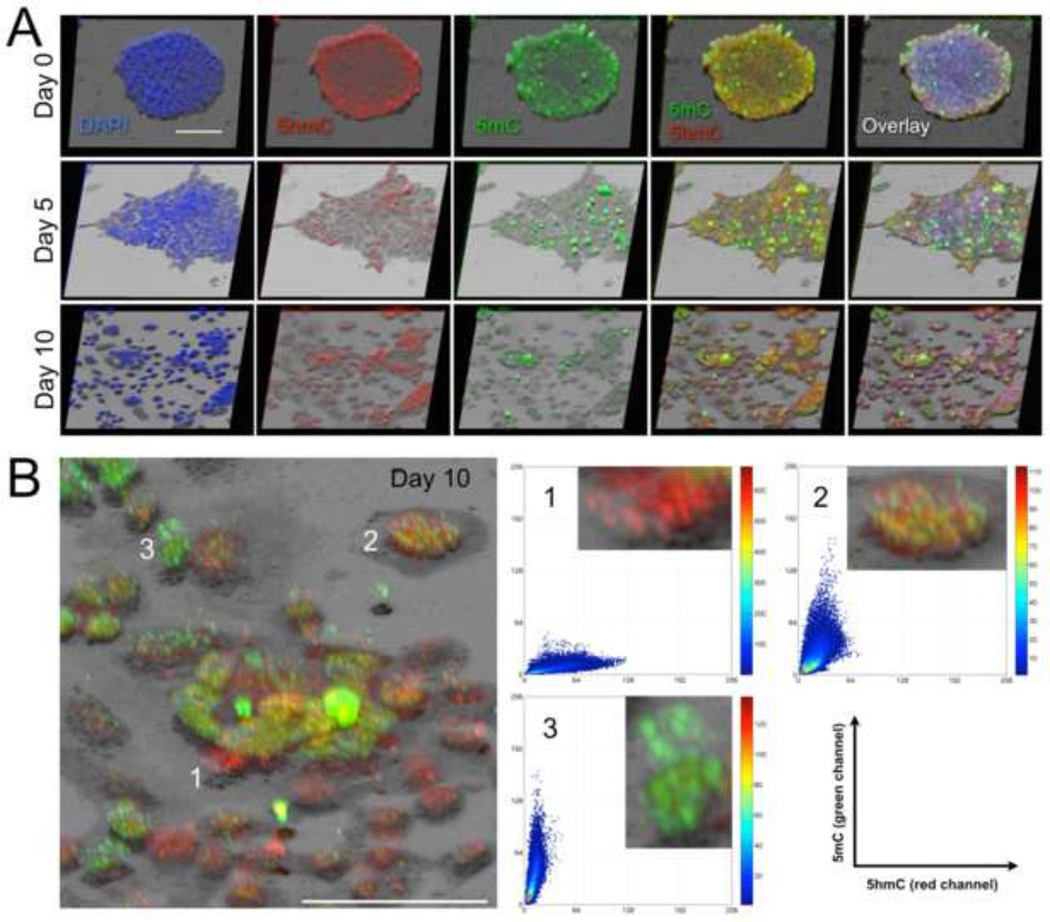
Fig. 2.
3-D reconstruction of mESC colonies presented in Fig. 1. Rendering was performed with the respective stacks of confocal 2-D images. The volume reconstruction used maximum intensity projection combined with shadow projection in order to augment the structural and subcellular spatiotemporal changes that occurred during differentiation. (A) The additional shadow projection allows for a better perception of the regional thicknesses of and within the cell clusters. Regions with a higher number of strong 5hmC-positive cells (Oct-4 positive as seen in Fig. 1) are more elevated and resemble the pluripotent cores of the cluster(s), whereas strong 5mC-positive cells (that are 5hmC+ or 5hmC−) appear relatively flat. (B) Magnification of a subregion of the day-10 sample image (5hmC/5mC overlay) discloses three major cell subtypes based on the abundance of the two epigenetic markers: 5hmC+/5mC− cells, 5hmC+/5mC+ cells, and 5hmC−/5mC+ cells. The three phenotypes supposedly correspond to cells at various differentiation stages. The corresponding quantitative scatter plots indicate that in type-2 cells, which display both markers (at near-equivalent ratios), 5hmC and 5mC occupy distinct nuclear territories but also tightly colocalize in nuclear subregions. Bars are 20 µm.
In summary, the confocal images from the individual channels and the composite images in Fig. 1 provide an overall impression that agrees with previous observations that we and other investigators had experienced. As differentiation progressed we found an increasing number of cells with lower abundance of markers associated with pluripotency, i.e. Oct-4 and 5hmC. Simultaneously, we observed a growth in the number of cells with increasing levels of differentiation markers such as FoxA2, Sox17 (not shown in here) and 5mC. However, from a visual perspective it is often challenging to estimate the proportions of the individual markers in cells. Nevertheless, a topological evolution regarding the composition of cells with differential marker presentation (cellular heterogeneity) can be perceived along differentiation. We assume that day 5 may have marked the start of a transition interval, at which we observed the highest degree of cellular heterogeneity, before cell populations became regionally diverged with pockets that resemble more the original pluripotent day 0-population and areas with more heterogeneous phenotypes. This is concordant with the fact that ESC differentiation (also in vitro) is an asymmetric process resulting in a remainder of pluripotent cells that steadily give rise to new differentiating cells. Therefore, even advanced cell populations still harbor cells that represent the various differentiation stages. An example is provided in Fig. 2B for a day 10-population: showing cells with three major phenotypes (codistribution signatures) of the two cytosine modifications, including 5hmC+/5mC− cells, 5hmC+/5mC+ cells, and 5hmC−/5mC+ cells representing consecutive stages in ESC differentiation. Interestingly, in cells displaying both markers, 5hmC and 5mC occupied distinct nuclear territories but also partially overlapped in localization. We then performed super-resolution microscopy using 3-D localization by ground state depletion (3D-GSD) that had been first experimentally demonstrated in 2007 [30], and has produced images of nuclear patterns similar to direct stochastic optical reconstruction microscopy (dSTORM) [31]. Utilizing this technology, we were able to retrieve images from day 10 cells at single-antibody resolution, and to make the following observations (displayed in Fig. 3): (i) the overwhelming majority of 5mC signals were concentrated in smaller to larger foci, whereas 5hmC signals exhibited a more punctate pattern throughout the nucleus. (ii) The two signals largely occupied exclusive regions. However, in a smaller subset of cells we could find a few areas in which 5hmC colocalized with 5hmC signals. The two cytosine analogs either a) formed mixed-sprinkled hubs (Fig. 3C) or b) smaller 5hmC foci were found next to larger 5mC hubs, some of them even engulfed by a 5mC focus (Fig. 3D). Our interpretation of these observations is that 5mC is more prevalent in high-density heterochromatic regions of the genome, whereas the majority of 5hmC signals could be retrieved from the more scattered euchromatin compartment. The visual impressions very much complemented the respective association and putative roles of the two molecules in transcriptional silencing and compaction of DNA versus maintenance of the permissive state of DNA in connection with a more open chromatin structure. Therefore, we speculate that colocalized foci of both cytosine modifications represented proximal loci that needed to be regulated (and occasionally spatially separated) for the two opposite transcriptional states. In this scenario, the different types of colocalized 5hmC signals may have constituted sites of ongoing DNA demethylation by oxidation.
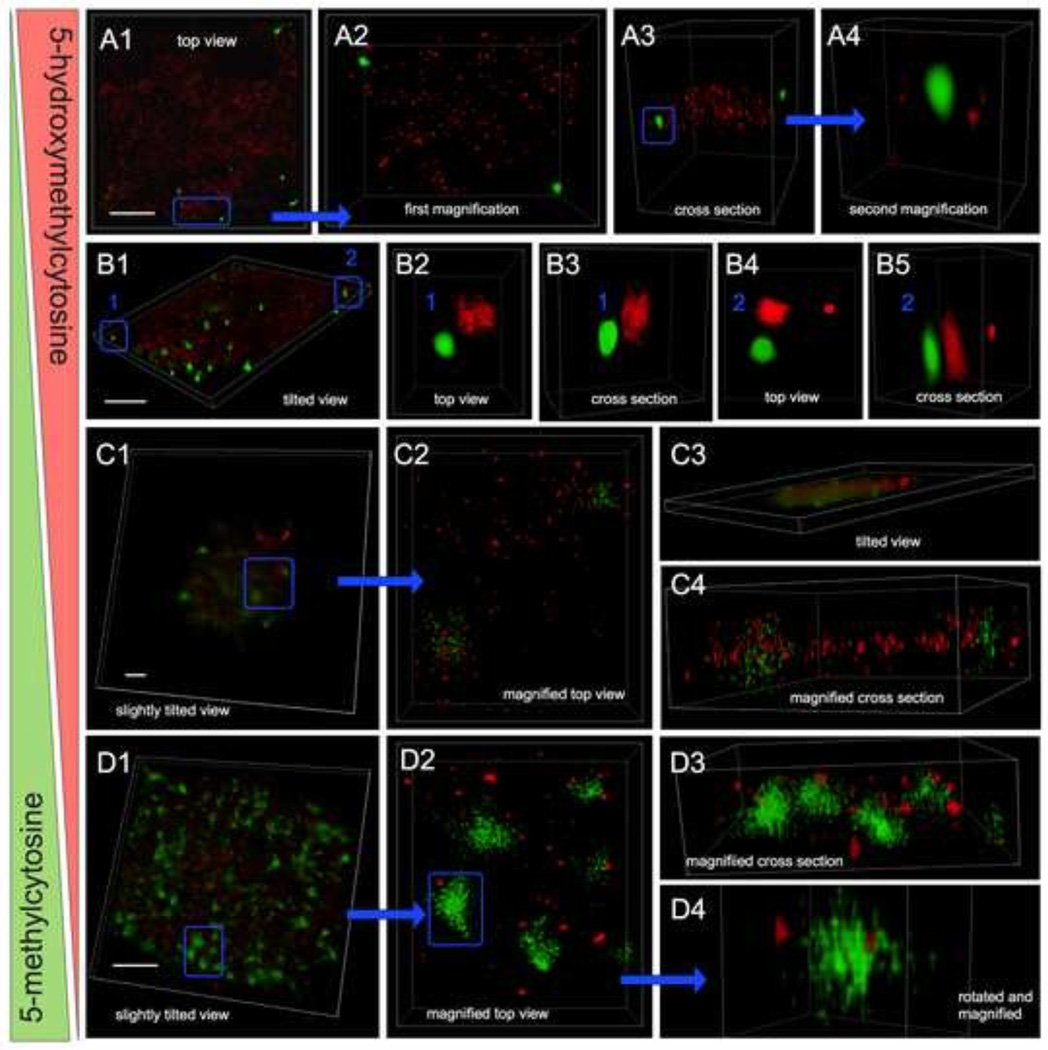
Fig. 3.
Dynamic nuclear topology of 5mC and 5hmC of day-10 cells as disclosed by 3-D superresolution localization using ground state depletion nanoscopy. Images were taken from midsections of cells that had been dual-labeled for 5hmC (red) and 5mC (green). No DAPI counterstaining was used, as it would have caused bleed-through of the DAPI signal into all other channels, due to the use of high-power lasers necessary for GSD. Panels A–D: 3-D reconstructions revealed nuclear abundance and spatial distribution of both cytosine variants within nuclear DNA and the degree of colocalization in sub-genomic regions. Each panel represents one nucleus, with sub-regions (framed in blue) being magnified and displayed at various angles (top views and cross sections). 5hmC showed a more punctate and dispersed pattern, whereas 5mC appeared more concentrated in larger foci. In nuclei with higher 5hmC and lower 5mC content (supposedly in pluripotent cells) the two types of molecules occupied separate areas, even at closer proximity (A4 and B2–B5). In contrast, in nuclei of transitioning cells with approximately equal amounts of both cytosine variants, the two analogs colocalized in less-condensed foci (C2 and C4). Finally in cells with an overwhelming 5mC pattern and relatively minimal 5hmC signals (assumingly representing early-differentiated cells), the two cytosine variants were seen more apart similar to pluripotent cells; with a few larger 5mC foci harboring 5hmC sites as seen in Fig. D4. White bars are 2 µm.
Our observations were largely concordant with earlier reports based on immunofluorescence and immunoprecipitation (IP) of methylated and hydroxymethylated DNA from mESCs (using target-specific antibodies of the same clone as used in our study) followed by massively parallel sequencing (IP-seq) [14,32]. The IP-seq based investigation had unveiled the existence of genomic regions that are enriched for either cytosine modification but also ambiguous regions with enrichment (at different ratios) for both variants. Though IP-seq experiments provide information at the single-nucleotide resolution, it is often difficult to interpret their average calls across a large number of cells in terms of cell-to-cell variability. The single-molecule resolution imaging and the single-cell 3-D image analysis used in our study confirmed that this ambiguity can indeed be attributed to the coexistence of the two epigenetic marks within tight genomic regions and was not only due to the possible heterogeneity of marker localization among different cells in the tested populations. Certainly, single-cell sequencing could ultimately elucidate the highest precision in genomic localization and proximity of 5hmC/5mC overlap.
Measuring phenotypic heterogeneity in differentiating stem cell populations
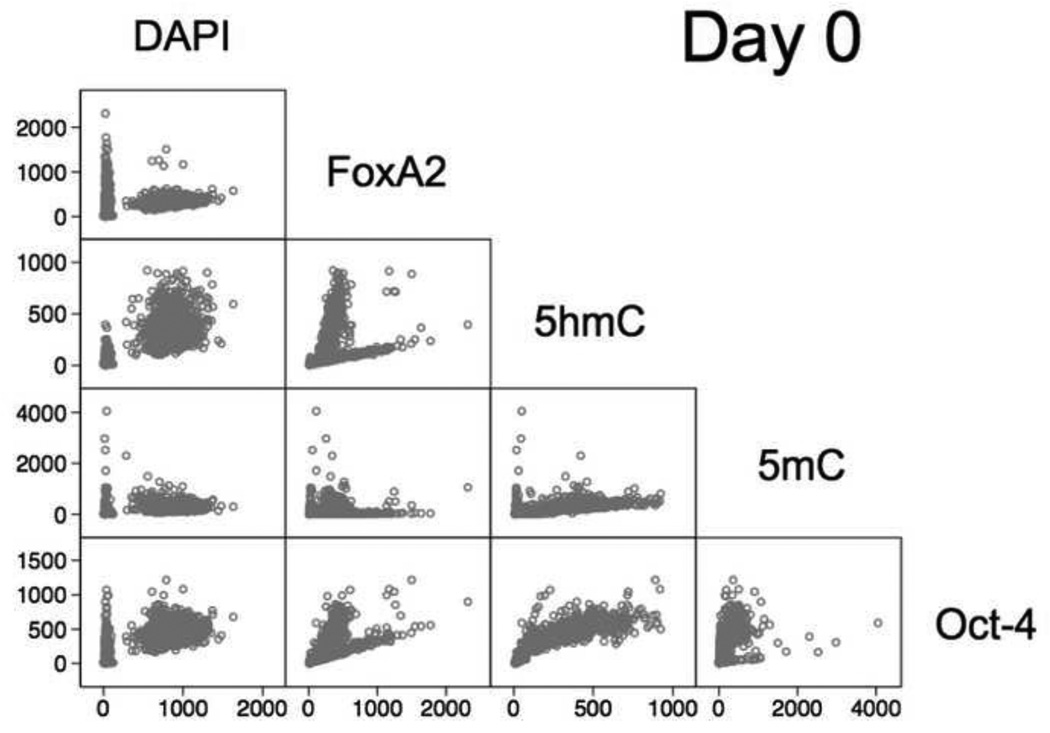
In this study our second aim was to characterize stem cell population heterogeneity based on determined cell phenotypes. Therefore, although the generated data has been derived from imaging signals as previously described [7], in here we report a different strategy for data analysis that would be more compatible with higher throughput and even real-time image analysis, which constitutes a desired application mode of HCA in the assessment of stem cells for therapeutic screening [33]. Hence, data analysis was conducted in two steps. In an initial step, we explored the covariance/correlation of the pairwise relationship between the six markers assessed in this study. Since Sox17 and FoxA2 were non-simultaneously visualized in different groups of cells, we evaluated the overall generated data in two batches, one including FoxA2 parameters and one analogously containing Sox17 features. For simplicity we have displayed a sample matrix of pairwise two-dimensional (2-D) plots for day 0 (Fig. 4). The complete 2D-plot matrices for all six days (time points) can be found in the Supplementary Fig. S1.
Fig. 4.
Graphical summary of the correlation of the overall intensities of the five markers (signal channels), i.e. DAPI, FoxA2, 5hmC, 5mC, and Oct-4 and display as a matrix of 2-D scatterplots for day 0: whereby each plot represents the pairwise values of two channels per cell/nucleus (hollow dot). The individual plots indicate the coexistence of different subgroups. Particularly, in the DAPI-specific plots we observed two cell fractions, with one fraction having very low DAPI intensities and low to high intensities for the other channels (markers), therefore suspected to represent perishing cells and cell debris.
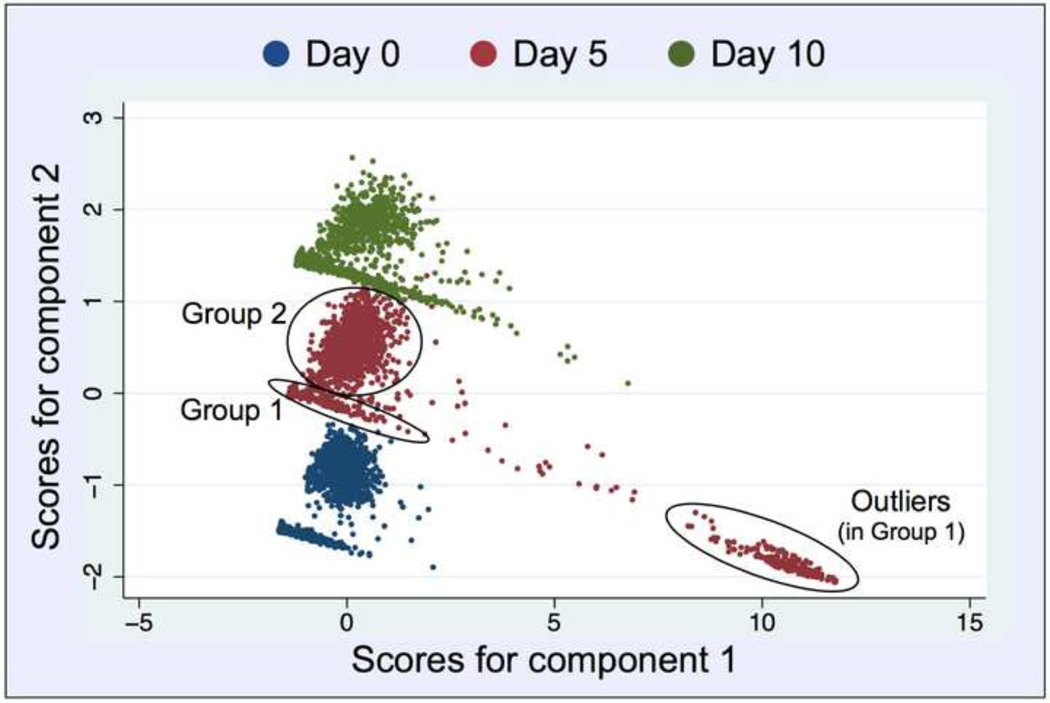
Following this procedure it became apparent that the pairwise relationship between the various markers was more complex than just calculating a simple covariance or the lack of it. By 2D-plotting we observed graphical clustering of the cells, with each cluster showing a different (specific) covariance. Furthermore, because the data had been derived from different batches of cells —cultured in parallel and frozen in time at the six different days— it was not possible to conduct a continuous assessment of covariance over time, i.e. for the ten days of differentiation. Therefore, we needed to apply a comprehensive exploratory statistical approach that enables the reduction of observed variables, while preserving a large portion of the variance in the data. Using PCA, the new produced variables were derived in decreasing order of importance pertaining to the amount of variation they explain of the original variables. For example, principle component 1 explains for as much of the variability of the original data as possible. The second principle component explains as much of the remaining variance as possible under assertion that it is not correlated to principle component 1, etc. This reduction is useful as our data could be graphically summarized, which simplified the further analysis that we based on Fig. 5. An informal rule for the selection of principle components is to consider components, where the eigenvalues are greater than 1, which in our case was true for two principle components. Also, the initial Scree-Plot (not shown in here) that we generated for estimation of components justified the use of two principle components separately for both, the FoxA2 and Sox17 data sets. The first two components explained 75% (60%) of the variance in case of FoxA2 (Sox17), as displayed in the Supplementary Table S1. Ultimately, we decided to further analyze the FoxA2-data due to its better graphical segregation. Furthermore, the loading matrix (Table S1) showed that principle component 1 (PC1) is an average of the 4 markers applying equal weight to 5mC, 5hmC, Oct-4 and FoxA2, while principle component 2 (PC2) is dominated by day and DAPI variables. For improved visualization, Fig. 5 shows a reduced representation of the results for the selected days 0, 5 and 10 as they appeared almost equidistant from each other exhibiting a less crowded segregation. The scatter plots of the predictions of PC2 versus PC1 revealed a repetitive pattern between the different days, in which two clusters of cells can be identified for each day: one that forms a diagonal line and another cluster of data resembling a cloud.
Fig. 5.
Principle Component Analysis (with two components) of the entire data (mean nuclear intensities of the five targets). For each day, the data segregates into two distinct populations (groups) as pointed out by circles for day 1: Group 1 (arm) presumably a mixture of haploid G1-cells and nonviable cells and debris, and Group 2 (cloud) more likely a collection of viable cells in various cell cycle interphases (G1 and S/G2), as judged by their respective DAPI signals (gDNA content) listed in Tables 1 and 2. Each dot corresponds to one object (intended to be a segmented nucleus). On day 5, Group 1 is divided into a main group (arm) and a separate outlier group of objects (arm extension).
Based on the cutoff values for PC2, the two groups of cells showed distinct mean intensity values in their DAPI channels: Group 1 with 50–200 intensity units (IU) and Group 2 with 650–900 IU, respectively (Tables 1 and 2).
Table 1.
Mean nuclear intensity (mean) and standard deviations (std. dev.) of objects in Group 1.
| Day 0 (n = 1000) | Day 5 (n = 400) | Day 10 (n = 700) | ||||
|---|---|---|---|---|---|---|
| Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | |
| DAPI | 44.86 | 70.81 | 175.37 | 80.40 | 140.65 | 44.37 |
| Oct-4 | 95.30 | 150.17 | 2035.82 | 1723.83 | 355.98 | 414.82 |
| 5hmC | 47.07 | 86.46 | 1831.40 | 1740.18 | 174.12 | 238.00 |
| 5mC | 66.70 | 229.07 | 2020.36 | 1795.05 | 210.47 | 558.89 |
| FoxA2 | 168.14 | 270.69 | 1867.11 | 1529.25 | 574.39 | 694.16 |
Table 2.
Mean nuclear intensity and standard deviations of objects in Group 2.
| Day 0 (n = 800) | Day 5 (n = 1800) | Day 10 (n = 700) | ||||
|---|---|---|---|---|---|---|
| Mean | Std. Dev. | Mean | Std. Dev. | Mean | Std. Dev. | |
| DAPI | 884.91 | 181.39 | 755.78 | 242.83 | 684.30 | 229.49 |
| Oct-4 | 466.10 | 134.94 | 441.35 | 154.02 | 472.04 | 232.26 |
| 5hmC | 376.63 | 154.62 | 318.08 | 152.56 | 270.68 | 132.68 |
| 5mC | 307.35 | 157.60 | 461.83 | 253.60 | 533.49 | 394.33 |
| FoxA2 | 333.14 | 87.84 | 361.16 | 129.45 | 427.04 | 232.26 |
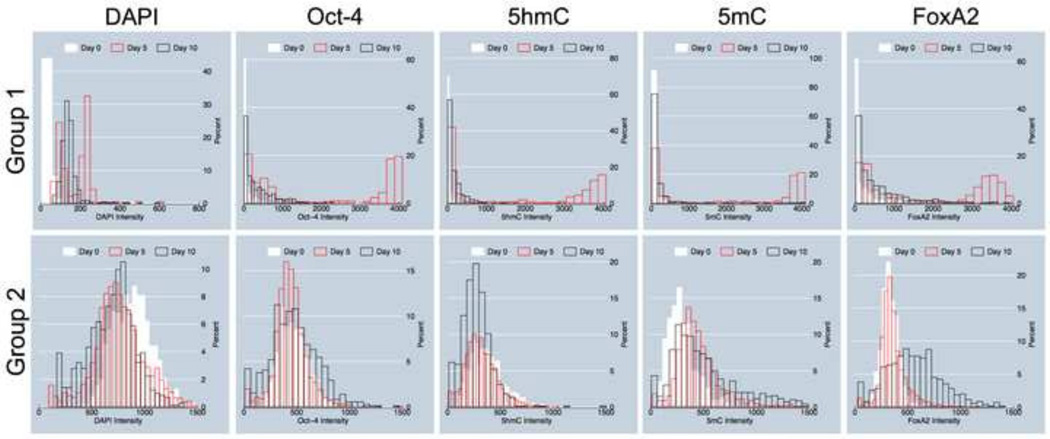
We assumed that probably Group 1 objects in the diagonal line cluster included cells in G1-phase of the cell cycle but also to a large extent outlier objects with low DAPI and extremely high values for the other four markers: 5mC, 5hmC, Oct-4, and FoxA2 (see Fig. 5). These objects may represent non-viable cells and larger cell debris (especially in day 5 cell colonies as seen in Figs. 1 and 2A) that might have been slipped through size exclusion during automated image processing; whereas Group 2 comprised seemingly healthy cells in all cell cycle interphases (G1 and S/G2). In order to assess the influence of 5hmC on the outcome of PCA, we performed the same analyses without the input of 5mC-specific data. We observed a significant reduction in data segregation (data not shown). We then decided to separately analyze the signal distribution of each marker across the reported days for the two groups, since Group1 objects showed extreme variability and consisted of outliers with intensity properties that were difficult to interpret. The results are presented in Fig. 6: Group 1 cells start off as a single distribution on day 0 with low levels of all targets including DAPI signals (gDNA content). Then, on day 5 a dichotomous population appears, harboring partially cells that resembled day 0 values and a second cohort of objects with DAPI signals in the range of (200–300 IU) and extreme high intensities (3000–4000 IU) for all other targets. We assume that these signals partially derive from perishing (apoptotic) cells, which is a frequent observation in in vitro propagated stem-cell colonies beyond day 3 in early differentiation that we also previously experienced [7]. Interestingly, on day 10 we again observed single populations with values similar to day 0. As mentioned above, Group 1 cells seemed less consistent in normal distribution and therefore less predictable for modeling of differentiation, thus had been omitted from further analyses. In contrast, histograms derived from Group 2 cells showed single Gaussian distributions for all nuclear targets across all days, which explains that the cells within a population acted more uniformly in their early developmental path, and may had not yet branched into the different lineage-specific cell types. A more precise observation of the histograms lead to the general impression across all markers that the Group 2 cells had a narrower bandwidth on day 0 compared to the following days, and that by time the curves developed tails. For Oct-4 the maximum did not change but tail development was symmetrical, indicating that cells (~15% for each category) had evolved with either reduced or increased levels of the pluripotency marker. In the case of 5hmC the maximum changed from ~400 IU to ~250 IU with an increasing tail of cells (~25%) that exhibited DNA hydroxymethylation levels below 200 IU. For 5mC the maximum did not change significantly (from ~400 IU to ~500 IU). However, a tail occurred comprising ~20% cells that had doubled to quadrupled levels of global DNA methylation (1000–2000 IU), especially on day 10. Along the same lines, the FoxA2 maximum slightly shifted from ~300 IU to ~500 IU, but at the same time a majority (~65%) of the cells were found to have FoxA2 levels between 500 IU and 1300 IU. The shift in the latter two markers must have happened between days 5 and 10, which are indications for day 5 to possibly be a key day for endodermal commitment of cells in vitro. The observations were consistent with previous experimental results that had been reported in [7].
Fig. 6.
Intensity distribution among sampled cells. The histograms show dynamic alterations in marker heterogeneity in cell populations during differentiation. Group 1-objects and Group 2-objects are described in Fig. 5. In Group 1-objects, red histograms represent outlier objects with extreme intensities in the channels for Oct-4, 5hmC, 5mC, and FoxA2, but low DAPI values.
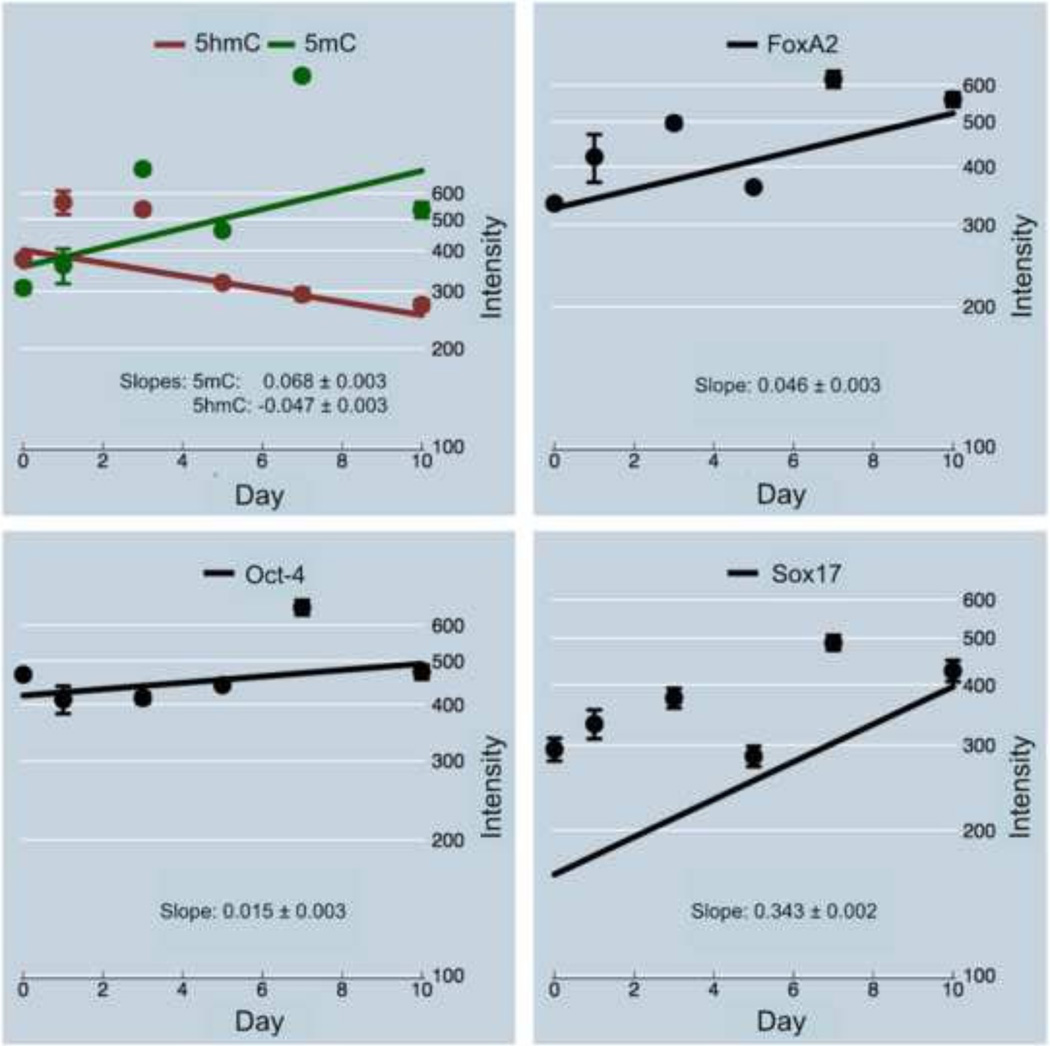
In summary, we would like to highlight the following crystalizing aspects. PCA analysis reported the existence of two groups of cells, where Group 2 most likely consisted of healthy cells. All markers displayed heterogeneity across all time points. 5mC and FoxA2 initially had significantly tighter distributions —equivalent to a smaller standard deviation— underlining their lower heterogeneity in abundance within cell populations, as compared with 5hmC and Oct-4. However, FoxA2 distribution radically increased beyond day 5 with extreme cellular heterogeneity, emphasizing the fact that the endodermal marker is expressed at different levels in differentiating cells. This could have been due to either the fact that the cells in cultured populations were not synchronized in differentiation or that generally differentiating cells may exert stochasticity of the marker in the early phases of lineage commitment. In contrast, 5mC largely maintained its major bandwidth, although with a steady increase in the number of hypermethylated cells (>500 IU) towards day 10. While the markers successfully segregated the cells into two major populations, the intensity distribution of each of the markers (presented as histograms) for the two groups of cells over time revealed more details during developmental changes. This was most obvious for Group 2 cells, as they resembled Gaussian distribution. In the second step of statistical analyses, we were trying to quantify the abovementioned changes in marker levels for Group 2 cells on per day basis utilizing robust regression analysis. Based on derived normal-quantile plots for each biomarker (Fig. S3) we identified high- and low-value outliers for all markers in Group 2 cells. Log-transformation was used to normalize the data. Furthermore, in order to reduce the effect of outliers on the data we applied robust regression methodology. A reciprocal (to 5mC) but weaker shift can be gleaned for 5hmC. Fig. 7 illustrates the global linear trends of the five biomarkers 5mC, 5hmC, Oct-4, FoxA2 and Sox17: represented by the regression-line slopes of the respective overall mean intensities across imaged cell populations over a ten-day inspection (six time points).
Fig. 7.
Regression plots of mean nuclear intensities of the five markers Oct 4, Sox17, FoxA2, as well as 5mC, and 5hmC (co-plotted for direct comparison).
With the exception of 5hmC, the other four markers showed their highest average at day 7, and declined to ~400 500 IU on day 10. We assume that somewhere around day 7 another significant change in the differentiation path of the original stem-cell clusters occurred, which coincided with alterations in the spatial organization of these colonies: as mentioned above on day 7 we observed the formation of smaller cell foci around assumingly poorly differentiated Oct-4-positive cells (not shown in here). This segregation became more distinct on day 10 (as seen in Fig. 1). Hence we focused on the estimation including all days. The pluripotency marker Oct-4 showed a relatively weak increase (0.015 lnIU per day, P<0.0001), almost resembling stagnation. This may partially be due to the asymmetric stem-cell division that maintains a certain contingent of highly Oct-4 expressing (pluripotent) cells, even in vitro. Furthermore, both epigenetic markers had close average starting levels before initiation of differentiation (day 0) with 5hmC (~400 IU) exceeding 5mC by only ~20% (~350 IU). Also, the endodermal markers FoxA2 and Sox17 showed comparable starting volumes of ~350 IU and ~150 IU, respectively. A possible explanation could be the uncontrolled spontaneous and rapid differentiation of cells during in vitro maintenance culturing of cells. Concisely, only 5hmC presented a statistically significant decrease (−0.047 lnIU per day, P<0.0001). In contrast, among those recruited markers that are known to increase during the course of early differentiation in mouse cells, 5mC showed the second highest slope, which was significantly different than the slope of all other targets (P<0.0001). The highest estimated slope and therefore the most rapid increase was extracted for Sox17 (0.34 lnIU daily, P<0.0001). However, Sox17 faired the weakest match, where only two points lined up with the model: an indication that the simple univariate regression model based on day as independent variable could not adequately explain the temporal pattern of Sox17. In stark contrast to Sox17, prediction curves for 5mC and 5hmC were close to the majority of mean day estimates (regression line) signifying compatibility between observed data and the simple regression model.
Discussion
Our approach to evaluate the co-evolution of the two cytosine modifications 5mC and 5hmC in differentiating pluripotent stem cells of the mouse system included three novelties: 1) the use of PCA for the high-throughput analysis of all imaged cells within cultured cell populations without any time and labor-consuming pre-selection of cells, 2) the quantitative assessment of changes in 5mC/5hmC load heterogeneity within differentiating cell populations, and 3) the identification of differential 5mC/5hmC colocalization patterns using optical nanoscopy. We conclude that based on our findings all studied biomarkers with the exception of Oct-4 showed anticipated trends during induced early cellular differentiation and commitment, in our experiments. The average Oct-4 levels almost stagnated, although we observed an increase of cells with lower Oct-4 levels. Hence we assume that the asymmetric division of stem cells may have resulted in the co-existence of low-expressing Oct-4 cells and a remainder of pluripotent cells with extremely high Oct-4 volumes that balances the average population value. In contrast, 5mC revealed the second highest average increase (slope) among the tested markers up to day 10. By that we found that 5mC, also in comparison to 5hmC, provides the most differential marker in in vitro cell development. Furthermore, the addition of 5hmC as a second imprint led to the identification of distinct cellular subtypes that represent and correlate with different levels and codistribution patterns of the two cytosine modifications. It is conceivable that the initial quantities of the two epigenetic markers may vary depending on the degree of spontaneous differentiation. From a population perspective, day 3 seems to be a time point by which a vast number of cells enter endodermal commitment. This number continues to grow until day 7. For that day we calculated an approximate quadrupling of the cells’ 5mC content, concomitant with an average decline of ~24% in 5hmC content compared with the initial day-0 values. The disproportional changes in the two markers may be explained by two proven facts: (i) that alterations in DNA methylation on the gene-specific as well as the global level are important in driving stem-cell differentiation [34–37], and (ii) the recent finding that 5hmC is predominantly present in gene-rich euchromatic regions that harbor CpG islands, which comprise only up to ~2% of the mammalian genome and being gradually depleted during ESC differentiation [14,31]. In contrast, the bulk of de novo DNA methylation during differentiation occurs at repeat sequences within heterochromatic regions representing a substantial portion (~45%) of the genome that appear as DAPI-dense nuclear areas in fluorescence microscopy [11, 38–40]. This fact may have contributed to the relatively more massive increase in 5mC. Conversely, the induction of pluripotent stem cells from terminally differentiated cells has revealed the exact opposite mechanisms during cellular dedifferentiation, namely a decrease in global 5mC levels accompanied by an increase in global hydroxymethylation [15], partially due to the conversion of 5mC to 5hmC in CpG islands of genes by the TET family of enzymes. Beyond these more global cell population-specific observations we gleaned subcellular topological information on the codistribution of 5hmC and 5mC sites, previously not reported. The novel application of single-molecule imaging visually uncovered genomic regions, at which the two cytosine variants could be found in close spatial proximity. These areas that are below the resolution of conventional confocal microscopy (~100 nm axial and up to ~500 nm vertical) may present captured 5mC-rich sites that were possibly undergoing active DNA demethylation through oxidation and conversion to 5hmC. Several lines of evidence exist that describe DNA methylation and demethylation as dynamic processes, with hypomethylation occurring during the embryonic blastocyst stage and also lasting through differentiation [41,42]. Previously we had observed that up to the sixth day in early differentiation the mouse genome becomes predominantly hypermethylated, first in its euchromatic compartments followed by heterochromatin [7]. However, differentiated mouse cells show a distinct 5mC phenotype, in which only the heterochromatic regions (chromocenters) appear as hypermethylated foci. These observations imply additional demethylation steps in the following period that would be necessary to create the more mature DNA methylation profiles and relevant nuclear patterns in differentiated cells. As seen in day-10 cell nuclei (Fig. 3, panel D), the type of optical nanoscopy we applied in here revealed conceivable sites of demethylation in both genomic domains, providing a visual snapshot of chromatin areas under remodeling. These visual evidences speak in favor of DNA methylation and demethylation via hydroxymethylation as iterative epigenetic processes for fine-tuning the genome during cellular differentiation.
Conclusion
Based on the abovementioned findings and the results of our study, we derive that DNA methylation/hydroxymethylation phenotyping —i.e. tracking the ratio of the two epigenetic marks and their codistribution patterns as the cytosine-modification index (CMI) in cells using 3D high-content screening— might have the potential to serve as an indicator of either cellular fate (directions), at least for endodermal differentiation of ESCs. The results encourage the appraisal of functional implications of the CMI: by using our cytometric approach in the selection of propagated stem cells for engraftment studies and evaluating the regenerative potential of enriched early-endodermal cells based on differential 5mC/5hmC phenotypes. In parallel, it would be worth to test the universality of our hypothesis in other types of lineage-specific differentiations derived from diverse types of pluripotent stem cells. As cytosine-modification indexing is compatible with high-throughput automation —an essential tool in the in vitro characterization of cell populations at various scales— feasible biomedical applications are: the quality assessment of stem cell differentiation, also involving patient-derived iPSC, that are increasingly utilized as autologous sources towards cellular therapies and personalized cell models for drug screening [1, 32].
Supplementary Material
Highlights.
First reported single-molecule super-resolution 3D-visualization of 5mC/5hmC sites in cells.
Identification of cells with differential 5mC/5hmC nuclear codistribution phenotypes.
Application of principle component and robust regression analyses for evaluation of 3D high-content-screening data.
Global 5mC constitutes a highly differential spatiotemporal in situ marker in early endodermal cell differentiation.
Acknowledgments
The authors thank Carlos Alonso and Jason Massey (Leica Microsystems Inc.) for helping with 3DGSD imaging, and Arkadiusz Gertych (CSMC) with image analysis support. This study was made possible by institutional support from the Department of Surgery at CSMC and the contract award W81XWH-10-1-0939 from the United States Department of Defense (to JT), as well as the NIH research grant R01HL082606 (to JHF).
Abbreviations
- ESC
embryonic stem cell
- iPSC
induced pluripotent stem cell
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- FoxA2
forkhead box A2
- Sox17
sex determining region Y box 17
- Oct-4
octamer-binding transcription factor 4
- CDH1
E-cadherin-1
- IGFR
insulin-like growth factor 1 receptor
- TET
ten-eleven translocation
- LIF
leukemia inhibitory factor
- FBS
fetal bovine serum
- aFGF
acidic fibroblast growth factor
- DMEM
Dulbecco’s Modified Eagle’s Medium
- 3-D or 3D
three-dimensional
- gDNA
global DNA
- DAPI
4',6-diamidino-2-phenylindole
- dSTORM
direct stochastic optical reconstruction microscopy
- 3D-GSD microscopy
three-dimensional ground state depletion microscopy
- HCA
high-content analysis
- PCA
principle component analysis
- IU
intensity unit
- lnIU
natural logarithm of the intensity unit
- CMI
cytosine-modification index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that they have no conflict of interest.
References
- 1.Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin. Pharmacol. Ther. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- 2.Ben-David U, Kopper O, Benvenisty N. Expanding the boundaries of embryonic stem cells. Cell Stem Cell. 2012;10:666–677. doi: 10.1016/j.stem.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Klimanskaya I, Rosenthal N, Lanza R. Derive and conquer: sourcing and differentiating stem cells for therapeutic applications. Nat. Rev. Drug Discov. 2008;7:131–142. doi: 10.1038/nrd2403. [DOI] [PubMed] [Google Scholar]
- 4.Goldring CE, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, et al. Assessing the safety of stem cell therapeutics. Cell Stem Cell. 2011;8:618–628. doi: 10.1016/j.stem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Enver T, Pera MF, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tajbakhsh J, Gertych A, Fagg WS, Hatada S, Fair JH. Early in vitro differentiation of mouse definitive endoderm is not correlated with progressive maturation of nuclear DNA methylation patterns. PLoS One. 2011;6:e21861. doi: 10.1371/journal.pone.0021861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–568. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170:1072–1073. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- 10.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 15.Ruzov A, Tsenkina Y, Serio A, Dudnakova T, Fletcher J, Bai Y, Chebotareva T, Pells S, Hannoun Z, Sullivan G, Chandran S, Hay DC, Bradley M, Wilmut I, De Sousa P. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 18.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tajbakhsh J. Covisualization of methylcytosine, global DNA, and protein biomarkers for in situ 3D DNA methylation phenotyping of stem cells. Methods Mol. Biol. 2013;1052:77–88. doi: 10.1007/7651_2013_18. [DOI] [PubMed] [Google Scholar]
- 20.Gertych A, Oh JH, Wawrowsky KA, Weisenberger DJ, Tajbakhsh J. 3-D DNA methylation phenotypes correlate with cytotoxicity levels in prostate and liver cancer cell models. BMC Pharmacol. Toxicol. 2013;14:11. doi: 10.1186/2050-6511-14-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barcia C, Wawrowsky K, Barrett RJ, Liu C, Castro MG, Lowenstein PR. In vivo polarization of IFN-gamma at Kupfer and non-Kupfer immunological synapses during the clearance of virally infected brain cells. J. Immunol. 2008;180:1344–1352. doi: 10.4049/jimmunol.180.3.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everitt BS, Rabe-Hesketh S. Handbook of Statistical Analyses Using Stata. fourth edition. CRC Press; 2006. [Google Scholar]
- 23.Fair JH, Cairns BA, Lapaglia MA, Caballero M, Pleasant WA, Hatada S, et al. Correction of factor IX deficiency in mice by embryonic stem cells differentiated in vitro. Proc. Natl. Acad. Sci. U S A. 2005;102:2958–2963. doi: 10.1073/pnas.0409840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gertych A, Wawrowsky KA, Vishnewsky E, Lindsley EH, Farkas DL, Tajbakhsh J. Automated quantification of DNA demethylation effects in cells via 3D mapping of nuclear signatures and population homogeneity assessment. Cytometry A. 2009;75:559–583. doi: 10.1002/cyto.a.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gertych A, Farkas DL, Tajbakhsh J. Measuring topology of low-intensity DNA methylation sites for high-throughput assessment of epigenetic drug-induced effects in cancer cells. Exp. Cell Res. 2010;316:3150–3160. doi: 10.1016/j.yexcr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertych A, Tajbakhsh J. Homogeneity assessment of cell populations for high content screening platforms. In: Pietka E, Kawa J, editors. Advances in Soft Computing, Information Technologies in Biomedicine. Vol. 69. Heidelberg, Germany: Springer Berlin; 2010. pp. 309–319. [Google Scholar]
- 27.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canham MA, Sharov AA, Ko MS, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bretschneider S, Eggeling C, Hell SW. Breaking the diffraction barrier in fluorescence microscopy by optical shelving. Phys. Rev. Lett. 2007;98:218103. doi: 10.1103/PhysRevLett.98.218103. [DOI] [PubMed] [Google Scholar]
- 31.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 32.Alioui A, Wheldon LM, Abakir A, Ferjentsik Z, Johnson AD, Ruzov A. 5-Carboxylcytosine is localized to euchromatic regions in the nuclei of follicular cells in axolotl ovary. Nucleus. 2012;3:565–569. doi: 10.4161/nucl.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbaric I, Gokhale PJ, Andrews PW. High-content screening of small compounds on human embryonic stem cells. Biochem. Soc. Trans. 2010;38:1046–1050. doi: 10.1042/BST0381046. [DOI] [PubMed] [Google Scholar]
- 34.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 35.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunner AL, Johnson DS, Kim SW, Valouev A, Reddy TE, Neff NF, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Research. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilchrist S, Gilbert N, Perry P, Bickmore WA. Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 2004;12:505–516. doi: 10.1023/B:CHRO.0000034892.64739.ff. [DOI] [PubMed] [Google Scholar]
- 39.Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 40.Tajbakhsh J, Wawrowsky KA, Gertych A, Bar-Nur O, Vishnevsky E, Lindsley EH, et al. Characterization of tumor cells and stem cells by differential nuclear methylation imaging. In: Farkas DL, Nicolau DV, Leif RC, editors. Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues, Proceedings of the SPIE. Vol. 6859. San Jose, CA: 2008. pp. F1–F10. [Google Scholar]
- 41.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Montera B, Fournier E, Shojaei Saadi HA, Gagné D, Laflamme I, Blondin P, Sirard MA, Robert C. Combined methylation mapping of 5mC and 5hmC during early embryonic stages in bovine. BMC Genomics. 2013;14:406. doi: 10.1186/1471-2164-14-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.