Abstract
Pyruvate dehydrogenase E1 alpha (PDHE1α or PDHA1) is the first component enzyme of the pyruvate dehydrogenase (PDH) complex (PDC) that transforms pyruvate, via pyruvate decarboxylation, into acetyl-CoA that is subsequently used by both the citric acid cycle and oxidative phosphorylation to generate ATP. As such, PDH links glycolysis and oxidative phosphorylation in normal as well as cancer cells. Herein we report that SIRT3 interacts with PDHA1 and directs its enzymatic activity via changes in protein acetylation. SIRT3 deacetylates PDHA1 lysine 321 (K321) and a PDHA1 mutant, mimicking a deacetylated lysine (PDHA1K321R) increases in PDH activity, as compared to the K321 acetylation mimic (PDHA1K321Q) or wild-type PDHA1. Finally, PDHA1K321Q exhibited a more transformed in vitro cellular phenotype as compared to PDHA1K321R. These results suggest that the acetylation of PDHA1 provides another layer of enzymatic regulation, in addition to phosphorylation, involving a reversible acetyl-lysine suggesting that the acetylome, as well as the kinome, links glycolysis to respiration.
Keywords: Pyruvate dehydrogenase, SIRT3, PDHA1, Acetylation, Carcinogenesis, Warburg
INTRODUCTION
Pyruvate dehydrogenase is a gatekeeper enzyme complex for carbohydrate and fat metabolisms by catalyzing irreversible decarboxylation of pyruvate into acetyl-CoA, which is one of the key molecules in various biological reactions [1]. PDC is a multi-enzyme complex comprised of many copies of each of three enzymes: (1) E1α or PDHA1 that uses thiamine pyrophosphate (TPP) as its prosthetic group; (2) E2 or dihydrolipoyl transacetylase that uses lipoamide and coenzyme A as its prosthetic groups; and (3) E3 or dihydrolipoyl dehydrogenase that has flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide (NAD+) as its cofactors [2-5]. PDC is located in the mitochondrial matrix of eukaryotes and consists of a total of 96 subunits that transforms pyruvate into acetyl-CoA via pyruvate decarboxylation. Acetyl- CoA is subsequently used by the citric acid cycle, via oxidative phosphorylation, to carry out cellular respiration [2-6]. Thus PDH, which is the main target of upstream signaling factors that direct PDC enzymatic activity, links the glycolysis metabolic pathway to the citric acid cycle and ultimately cellular respiration.
PDH is regulated by phosphorylation of the E1α subunit in response to specific physiological conditions in a tissue-specific manner [7,8]. In the case of scarce oxygen or cellular nutrient status, phosphorylation of PDHA1 by pyruvate dehydrogenase kinases (PDK1-4) on serine 293, 300, and 232 residues inhibits the PDH activity [4,7]. In conditions of sufficient nutrients, dephosphorylation by pyruvate dehydrogenase phosphatases (PDP1 and 2) restores complex activity, ultimately leading to increased oxidative phosphorylation and lower glycolytic rates [8,9]. As such, it seems logical that mutations found in the PDH complex have been associated with various metabolic dysfunctions as well as age-associated diseases, e.g. neurodegenerative diseases, cardiovascular disease, glucose intolerance, and cancer [10].
In numerous cancer cells, aerobic glycolysis is preferentially used rather than oxidative phosphorylation, even in the presence of oxygen, to provide cellular energy resources. When this is biochemically connected to carcinogenesis, this process is often referred to as Warburg effect [11-13]. Transformed cells seem to prefer using glycolysis since it is thought that the Warburg effect favors growth by not only supplying ATP, but also raw materials for high macromolecule synthesis demands. It is also believed that pyruvate is preferentially converted into lactate in cancer cells, partly due to the inhibition of the PDH complex as well as mitochondrial respiration [10-13].
SIRT3 is the primary mitochondrial acetyl-lysine deacetylase [14] as well as a tumor suppressor in the mitochondria [15]. SIRT3 is associated with the regulation of many metabolic enzymes, such as acetyl-CoA synthetase 2 (AceCS2), glutamate dehydrogenase (GDH), long-chain acyl-CoA dehydrogenase (LCAD), and isocitrate dehydrogenase 2 (IDH2) [16-20]. Deletion of Sirt3 increases the rate of tumor formation in various tissues including mammary tumorigenesis [15]. At least one copy of SIRT3 is deleted in 40% of breast and ovarian cancers [21,22]. While the mechanism(s) by which loss of Sirt3 results in a tumor permissive phenotype is complex, one interesting, and perhaps informative, observation is that mice lacking Sirt3 have biochemical features similar to the Warburg effect [21]. For example, Sirt3−/− mouse embryonic fibroblasts (MEFs) consumed more glucose and produced more lactate than wild type cells [15,17,18,21,23]. In addition, overexpression of SIRT3 in vitro was sufficient to reverse this metabolic shift [24].
The mechanisms underlying the role of SIRT3 in reversing the Warburg effect and protecting against cancer formation are complex and poorly understood. SIRT3 has been proposed to have protective roles by decreasing the levels of reactive oxygen species, maintaining genomic stability, cell survival, and regulating metabolism [15,25]. In this work, experiments are presented showing that the PDHA1 subunit of PDC can be acetylated in vitro and in vivo and there is a physical interaction between SIRT3 and PDH. SIRT3 mediated changes in the acetylation status of PDHA1 altering the activity of PDH as well as in vitro tumor cell metabolism. Finally, both mass spectrometry and in vitro deacetylation assays showed: (1) SIRT3 deacetylates PDHA1 K321 and enforced expression of PDHA1K321R, a deacetylation lysine mutant; (2) increased PDH activity; and (3) these cells exhibited a decreased in vitro transformed cell phenotype. These results suggest that in addition to PDHA1 lysine 336 [26], it also appears that lysine 321 acetylation directs the activity of PDC.
MATERIALS AND METHODS
Cell culture and transfection
293T, HCT116, HeLa, T47D, MMT, and MCF7 cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% FBS and were seeded in 10 cm dishes at a density of 2.5 × 106 in 10 mL culture medium, and transiently transfected by Fugene6 (Roche Applied Science) or PEI (polyethylenimine; Polysciences, Inc.) according to manufacturer's recommendation and grown in a CO2 incubator maintained at atmospheric oxygen levels and 5% CO2. Only MEFs were maintained in DMEM containing 15% FBS and grown in 5% oxygen. For the in vitro deacetylase assay, Flag-tagged SIRT3 and Flag-tagged PDHA1 were used. For in vivo deacetylation assay, HA-tagged wild type and mutant SIRT3 were utilized. Endogenous PDHA1 was knocked down by designing various shRNA oligos against 5’ and 3’ of UTR of PDHA1. After annealing forward and reverse oligos to form hairpin structures, it was cloned into pLKO.1. 293T and MCF7 cells were infected with the virus containing this plasmid. Selection was performed adding 2 μg/mL puromycin in the culture medium. The most efficient silencing construct was chosen (>70%) among different shRNA constructs, and used for the subsequent assays. Lenti Flag-PDHA1K321, Flag-PDHA1K321Q, and Flag-PDHA1K321R were infected into MCF7 cells constitutively expressing shPDHA1, which stably knocked down endogenous PDHA1. Positive clones were isolated, grown, and used for subsequent assays.
Immunoprecipitation of flag-tagged SIRT3 and PDHA1
293T cells, transiently transfected with Flag-SIRT3 or Flag-PDHA1 by PEI were lysed with IP buffer (10 mM HEPES, pH 7.9, 180 mM KCl, 1.5 mM MgCl2, 0.1% NP-40, 1 mM EDTA, 0.1 mM PMSF), including protease and phosphatase inhibitors. Transfection efficiency of 293T cells was approximately 70% as determined using a GFP vector. Total cell extracts were incubated with anti-Flag M2 agarose (Sigma) for 16 h at 4 °C. After washing five times with IP buffer, bound proteins were eluted using 0.25 mg/mL Flag peptide (Sigma), concentrated using a 10,000 MWCO spin column (Millipore), and proteins were resolved via 10% SDS PAGE.
Western blotting
Western blotting on nitrocellulose membrane was performed using iBlot (Invitrogen). Nitrocellulose membranes were incubated with anti-pyruvate dehydrogenase (Cell Signaling), anti-SIRT3 (Cell Signaling), phosphor-PDH S293 (Abcam), and acetyl-lysine (Immunochem and Abcam) primary antibodies for 16 h at 4 °C.
Immunofluorescence
Cells seeded on glass coverslips were fixed in 4% paraformaldehyde and then blocked with 1% BSA in PBS. Cells were incubated with anti-PDHA1 (Cell Signaling) or anti-SIRT3 (Cell Signaling) antibody in PBS followed by incubation with goat-rabbit IgG conjugated with Alexa Fluor 488 and 594 (Invitrogen) in PBS with 5% goat serum. Cells were washed in PBS, mounted, and imaged on a fluorescence microscope.
In vitro deacetylation assay
293T cells were transiently transfected with Flag-PDHA1 with PEI. 24 h after transfection, cells were exposed to trichostatin A (TSA) and nicotinamide (NAM) for 10 h to hyper-acetylate proteins. Next, cells were harvested and lysed with IP buffer. Proteins were purified as explained above. For in vitro deacetylation assay, after isolation of flag-tagged PDHA1 protein, an in vitro deacetylation reaction in a 1.5 mL Eppendorph tube was set up. Briefly, commercially available recombinant SIRT3 enzyme, hyperacetylated purified PDHA1 protein as a substrate, and NAD+ co-factor to activate the enzyme were mixed within a tube. After incubation at 37 °C for 3 h, the samples were run in 10% NuPAGE electrophoresis. Following western blotting, nitrocellulose membrane was incubated with anti-acetyl-lysine primary antibody.
PDH enzyme activity assays
A kit from Abcam was utilized to measure PDH activity according to manufacturer's instructions. Briefly, cultured cell (750 μg) or tissue extracts (100 μg) were loaded into the wells of a 96-well plate that was specific for PDH activity assay. The components of PDH enzyme were immunocaptured by the monoclonal antibodies coated on the wall of each well. The enzymatic activity of PDH was determined based on the production of NADH coupled to the reduction of a reporter dye, WST-1 to yellow color. Vmax and the absorbance of the reporter dye in each well were measured at 450 nm at room temperature using a kinetic program for 15 min (BioRad XMark Microplate spectrophotometer).
Measurement of reactive oxygen species (ROS) production
To determine if mimicking acetylation/deacetylation status of PDH can alter steady-state levels of oxidants originating from mitochondria in 293T cells, the cationic superoxide-sensitive dye MitoSOX™ Red (Invitrogen) was used. Cells were transfected and plated as described above. After 48 h, cells were trypsinized and washed with 5 mM pyruvate containing PBS once and then labeled with MitoSOX™ Red (2 μM, in 0.1% DMSO, 20 min) at 37 °C. Samples were analyzed using a Fortessa™ Flow Cytometer (Becton–Dickinson) (excitation 488 nm, emission 585 nm bandpass filter). The mean fluorescence intensity of 10,000 cells was analyzed in each sample and corrected for autofluorescence as compared to unlabeled cells. The MFI data were normalized to vector control 293T cell levels.
For quantitative analysis of oxidation products derived from MitoSOX™ Red, HPLC electrochemical analysis was performed [27-29]. Briefly, cells (≈1×106 cells) were preloaded with MitoSOX™ Red (2 μM/1 mL of media at 37 °C for 20 min). Cells were centrifuged, washed with PBS, counted and stored at −80 °C. On the day of HPLC analysis, samples were prepared as described by Zielonka et al. [30]. An ESA CoulArray system with a Phenomenex 250 mm × 4.6 mm Synergi 4 m Polar-RP 80A column was used to quantify oxidation products of MitoSOX™ Red, using 50 mM phosphate buffer, pH 2.6, mobile phase with gradients containing 10 – 60% acetonitrile. Standard curves were prepared from authentic standards of MitoSOX-2-OH-E+ and MitoSOX-E+ synthesized from MitoSOX™ Red using methods as outlined in [30]. Eight electrochemical channels were used for detection and quantitation set at 0, 200, 280, 365, 400, 450, 500, 600 mV; quantification of MitoSOX-2-OH-E+ was determined using the 200, 280 and 365 mV channels, and for MitoSOX-E+ the 280, 365, 400, 450 and 500 mV channels. HPLC data were adjusted for cell numbers and dilutions made during sample processing and analysis.
Lactate production
To measure extracellular lactate production, Cayman's Colorimetric Glycolysis cell-based assay kit was used according to the manufacturer's instructions (Cayman Chemical Company). Briefly, the principle of this kit is based on the oxidization of lactate by lactate dehydrogenase to generate pyruvate and NADH. Formed NADH interacts and reduces tetrazolium substrate to a colored formazan product. The intensity of color at 490 nm is proportional to the amount of lactate produced and therefore the rate of glycolysis. Cells were prepared as explained above. Supernatants were collected and centrifuged at 400 g for 5 min to remove cell debris. 10 μL supernatants were used for each reaction (sample, standards and controls).
Glucose uptake
To measure glucose utilization, “Glucose Uptake Colorimetric Assay Kit” from BioVision was utilized according to the manufacturer's instructions and as previously described [31]. 2-Deoxyglucose (2-DG) is taken into the cells by glucose transporters and phosphorylated to 2-DG-6-phosphate (2-DG6P). 2-DG6P is not further metabolized and accumulates in the cells. The amount of 2-DG6P in the cells is directly proportional to glucose uptake. Using this kit, 2-DG6P is oxidized to produce NADPH. Recycling amplification reactions involving NADPH, glutathione disulfide and glutathione lead to the formation of a colorful product from a substrate-5,5’-dithiobis-(2-nitrobenzoic acid) (DTNB). Absorbance was measured at 412 nm using a plate reader at 37 °C.
Measurement of proliferation rates and cell survival
Cells were grown in 10 cm petri dishes to reach about 90% confluence before irradiation exposures. Treated cells were harvested using trypsinization after 3 h, counted and plated in 12-and 24-well plates in triplicate for cell survival assays. For DCA exposure assays, cells were counted and plated into 24-well plates before DCA exposure. Cell proliferation and cell survival were determined using either Cell Counting Kit-8 from Dojindo Molecular Technologies, Inc. and/or Biorad automated cell counter as previously described [32,33].
Measurement of oxygen consumption rate
Cellular oxygen consumption rate (OCR) was determined using a Seahorse XF24 instrument (Seahorse Biosciences). For 293T cells, Seahorse plates were first coated with Cell-Tak Cell and Tissue Adhesive (BD Biosciences). 293T cells were seeded into the Seahorse plate at a density 250,000/well, and MCF7 cells were seeded at 25,000/well 16 h prior to running the Seahorse experiment. 75 μL of oligomycin A (an ATP synthase inhibitor) (final concentration, 2.5 μM), carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (a mitochondrial uncoupler) (final concentration, 10 μM), and antimycin/rotenone (complex III inhibitors) (final concentrations, 2 μM) were added into different ports of the Seahorse cartridge. Each experimental group was analyzed using 5 replicates in each analysis. O2 consumption rates (pmol/min) per 106 cells were measured and corrected for antimycin and rotenone. In addition, the rates of O2 utilization were calculated as amol cell−1 s−1, as previously described [34].
Statistical analysis
Unpaired Students's t test and one-way ANOVA with post-hoc analyses were performed via GraphPad Prism software.
RESULTS
SIRT3 deacetylates PDHA1 and increases the PDH activity in vitro
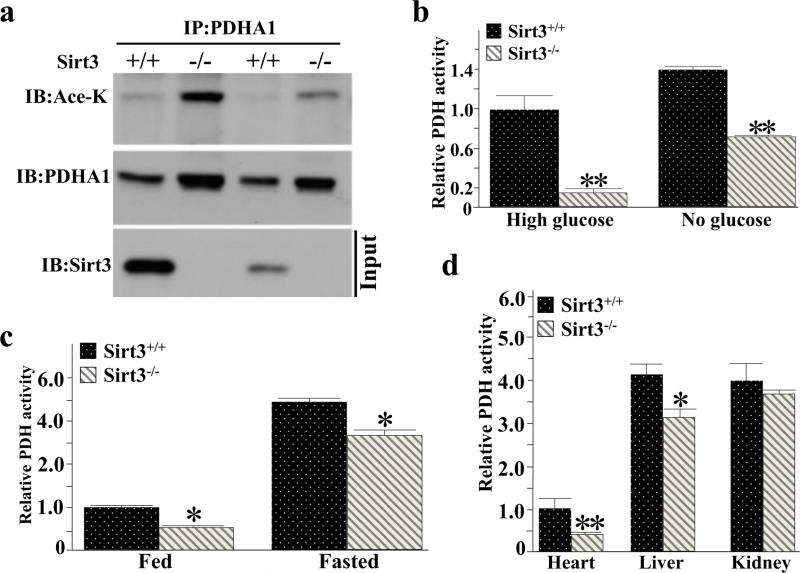
The genetic deletion of Sirt3 in mice results in hyper-acetylation of mitochondrial proteins [39] and cells lacking Sirt3 appear to generate ATP more from glycolytic pathways than oxidative phosphorylation [15,22]. To determine which downstream targets may account for this observation, a mass spectrometry analysis was done and the PDHA1 subunit of PDH was identified as a potential SIRT3 deacetylation target [40,41]. In addition, liver extracts from Sirt3+/+ and Sirt3−/− mice were immunoprecipitated (IPed) with anti-PDHA1 antibody, separated, and subsequently immunoblotted with an anti-pan acetyl-lysine showing that PDHA1 exhibited increased protein acetylation in mice lacking Sirt3 (Figure 1a). Consistent with this result, Sirt3−/− displayed a significant decrease in PDH activity in both high and no glucose, as determined using a colorimetric PDH activity kit from Abcam, as compared to wild-type MEF (Figure 1b). PDH activity was also decreased in the brain in the Sirt3−/− mice on an ad libitum diet and after fasting (Figure 1c), as compared to the control mice. Finally, PDH activities were also decreased in heart and liver murine tissues from the Sirt3−/− mice on an ad libitum diet (Figure 1d) while the protein levels of PDHA1 were very similar (Supplemental Figure S1).
Figure 1. SIRT3 deacetylates PDHA1 and increases the PDH activity in vitro.
(a) Sirt3+/+ and Sirt3−/− liver extracts (600 μg of protein) were IPed with anti-PDHA1 antibody, separated, and subsequently immunoblotted with an anti-pan acetyl-lysine and PDHA1 antibody.
(b) Sirt3+/+ and Sirt3−/− MEFs extracts were cultured in high or no glucose and extracts were used to determine PDH activity using a PDH activity Microplate assay kit (Abcam, Inc).
(c) The brains from the Sirt3+/+ and Sirt3−/− mice, on an ad libitum diet or after fasting (36 h), were harvested and PDH activities were determined as above.
(d) Sirt3+/+ and Sirt3−/− mouse heart, liver, and kidney were isolated and extracts were used to determine PDH activity as above.
PDH activities for all data panels are presented as relative changes compared to control cells or tissues. Error bars represent one standard deviation from the mean (*p<0.05 and **p<0.001).
Overexpression of SIRT3 increases PDH activity in vitro
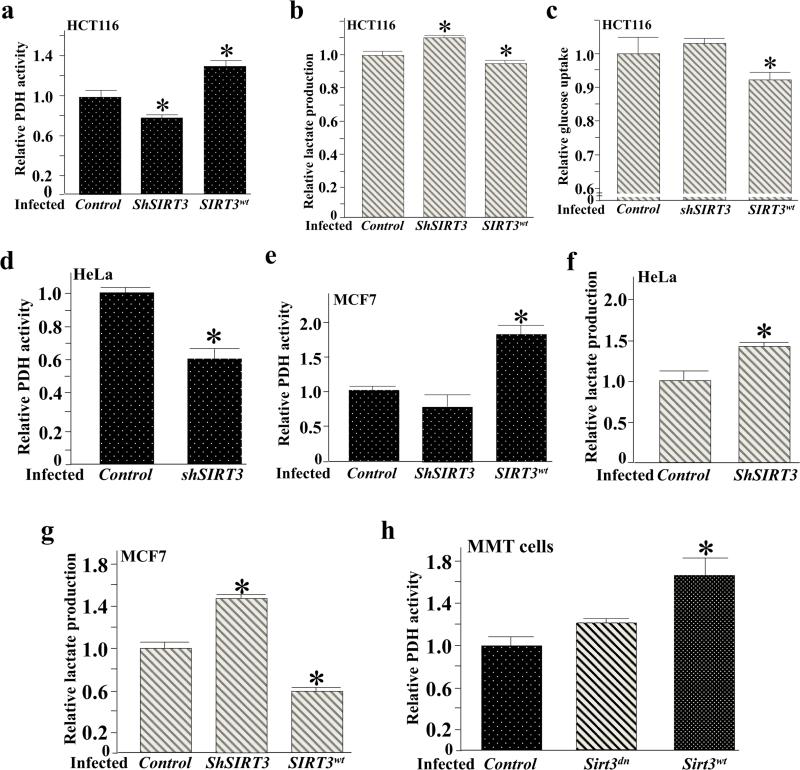
Since the Warburg effect is considered a common metabolic phenomenon in cancer cells, and SIRT3 gene deletion was reported in 20% of all human cancers, we decided to utilize several different cancer cell lines including breast, colon, cervical whose SIRT3 expressions genetically engineered to determine the roles of SIRT3 on PDH activity and glucose metabolism. To determine the role of SIRT3 on the activity of PDH, as well as on other cellular measurements of glycolysis, HCT116 (human colonic tumor cells) were infected with wild-type SIRT3, a control virus, as well as lenti-shSIRT3, to knockdown SIRT3. HCT116 cells infected with lenti-SIRT3WT exhibited an increase in PDH activity (Figure 2a) and a decrease in lactate production (Figure 2b), and glucose uptake (Figure 2c). In contrast, cells infected with lenti-shSIRT3 exhibited decreased PDH activity (Figure 2a), and increased lactate production (Figure 2b). In addition, HeLa (cervical cancer) and T47D (breast tumor) cells infected with lenti-shSIRT3 also exhibited a decrease in both PDH activity (Figures 2d and Supplemental Figure S2a) and an increase in lactate production (Figures 2f and Supplemental Figure S2b). MCF7 (breast tumor) cells infected with lenti-SIRT3WT exhibited an increase in PDH activity (Figure 2e) and a decrease in lactate production (Figure 2g). Finally, Sirt3−/− mouse mammary tumor cells (MMT) were infected with lenti-SIRT3WT displayed an increase in PDH activity as compared to cells infected with a SIRT3 deacetylation null (DN) lenti-SIRT3DN that exhibited less PDH activity (Figure 2h).
Figure 2. Overexpression of SIRT3 increases PDH activity in vitro.
(a-c) HCT116 cells were infected with Myc-Sirt3WT or shRNA and after 36 h cells were harvested and measured for (a) PDH activity, (b) lactate production, and (c) glucose uptake.
(d-e) HeLa and MCF7 cells were infected with either a control lentivirus or lenti-shSIRT3, or lenti-SIRT3WT and after 48 h, cells were harvested and measured for PDH activity.
(f-g) HeLa and MCF7 cells were infected with either a control lentivirus or lenti-shSIRT3, or lenti-SIRT3WT and after 48 h; cells were harvested and measured for lactate production.
(h) Sirt3−/− MMT cells were infected with either lenti-SIRT3DN or lenti-SIRT3WT and after 48 h cells were harvested and measured for PDH activity.
For all these experiments the change in PDH activity, lactate production, and glucose uptake are presented as relative changes compared to control cells or tissues. Error bars represent one standard deviation from the mean (*p<0.05 versus the control group).
SIRT3 interacts with and deacetylates PDHA1 subunit of PDC
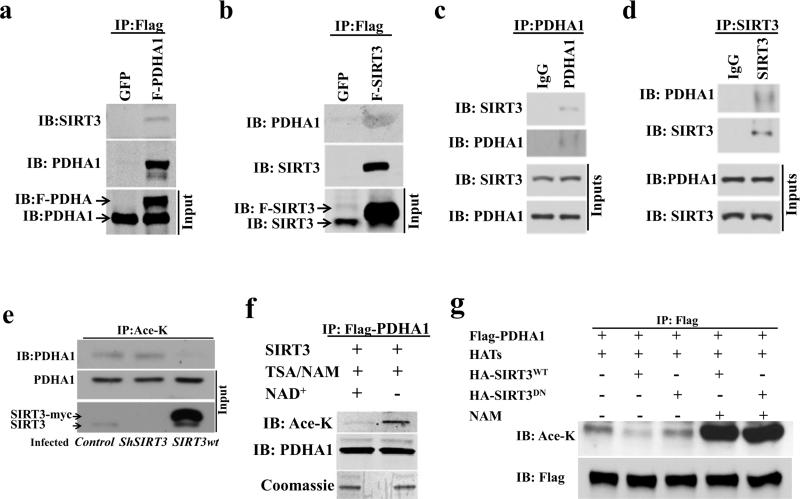
To determine if there is an interaction between PDHA1 and SIRT3, an overexpression system in 293T cells was used. 293T cells were transfected with Flag-PDHA1 (Figure 3a) or Flag-SIRT3 (Figure 3b) followed by immunoprecipitation (IP) with an anti-Flag antibody and samples were subsequently immunoblotted with either an anti-SIRT3 or anti-PDHA1 antibody. These results showed an interaction between SIRT3 and PDHA1 using exogenous proteins. This interaction was also confirmed for an endogenous interaction between PDHA1 and SIRT3 by reverse IP experiments using anti-PDHA1 or anti-SIRT3 antibodies (Figures 3c-d). Finally, immunofluorescence was used to determine that these two proteins co-localize in the mitochondria (Supplemental Figure S3a).
Figure 3. SIRT3 physically interacts and co-localizes with PDHA1.
(a-b) 293T cells were transfected with (a) Flag-PDHA1 or (b) Flag-SIRT3 and 48 h after transfection cell extracts were IPed with an anti-Flag antibody and subsequently immunoblotted with anti-SIRT3 and anti-PDHA1 antibodies. Input immunoblotted is shown as a control.
(c-d) 293T cells here harvested and endogenous (c) PDHA1 or (d) SIRT3 were IPed, separated, and immunoblotted with an anti-SIRT3 and anti-PDHA1 antibody.
(e) H1299 lung cancer cells were stained with anti-PDHA1 (red staining), anti-SIRT3 (green staining), or DAPI (purple staining) and subsequently merged. Scale bar, 10Pm.
(f) HCT116 cells were infected with Myc-Sirt3WT or shRNA and after 36 h extracts were IPed with and anti-pan acetyl-lysine antibody and immunoblotted with anti-PDHA1.
(g) Purified acetylated Flag-PDHA1 was mixed with recombinant SIRT3, without or with NAD+, and samples were immunoblotted with anti-pan acetyl-lysine and anti-PDHA1 antibodies.
(h) PDHA1 was co-transfected with acetyl-transferases (Tip60 and CBP) as well as HA SIRT3WT or a deacetylation null mutant (HA-SIRT3DN) gene and extracts were IPed with an anti-Flag antibody. Samples were immunoblotted with anti-pan acetyl-lysine and anti-Flag antibodies.
To determine if PDHA1 is a direct SIRT3 deacetylation target, HCT116 cells were infected with Myc-Sirt3WT or shSIRT3 and extracts were IPed with and anti-pan acetyl-lysine antibody and immunoblotted with anti-PDHA1. These results showed that enforced expression of SIRT3 decreased PDHA1 acetylation (Figure 3e). In addition, IPed purified acetylated Flag-PDHA1 was mixed with recombinant SIRT3, without or with NAD+, followed by immunoblotting with anti-pan acetyl-lysine and anti-PDHA1 antibodies. This in vitro deacetylation assay also showed that SIRT3 deacetylates PDHA1 (Figure 3f). Finally, Flag-PDHA1 was co-transfected with acetyl-transferases (Tip60 and CBP) as well as HA-SIRT3WT or a deacetylation null mutant (HA-SIRT3DN) gene and a tissue culture deacetylation assays also showed that SIRT3, but not the SIRT3 deacetylation mutant, deacetylates PDHA1 (Figure 3g).
SIRT3 deacetylates and activates PDH complex at lysine 321 of PDHA1 subunit
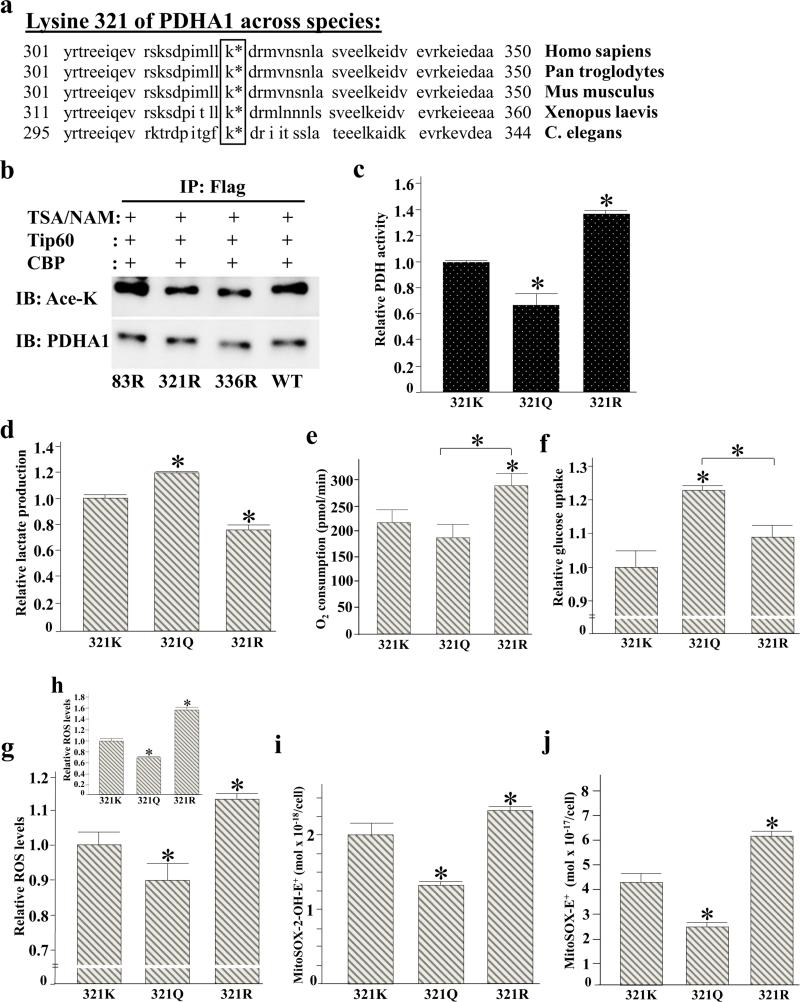
PDHA1 amino acid sequences across multiple divergent species were analyzed to determine if any lysines are evolutionary conserved which may provide a clue to potential reversible acetyl lysines that may be SIRT3 downstream deacetylation targets. Sequences between 300 and 350 are shown and K321 and K336 are conserved from human to C. elegans which might indicate their importance in the regulation and function of PDHA1 protein (Figure 4a). Previous mass spectrometry analyses investigating the acetylation of mitochondrial proteins using Sirt3+/+ and Sirt3−/− MEF cells, and hepatocytes revealed the acetylation of PDHA1 on multiple lysines [30]. To specifically focus on PDHA1 acetylation, mass spectrometry analysis was performed using an in vitro deacetylation assay with purified PDHA1 and SIRT3. Analysis of the identified acetylated peptides revealed that K83, K321, and K336 of PDHA1 can be acetylated.
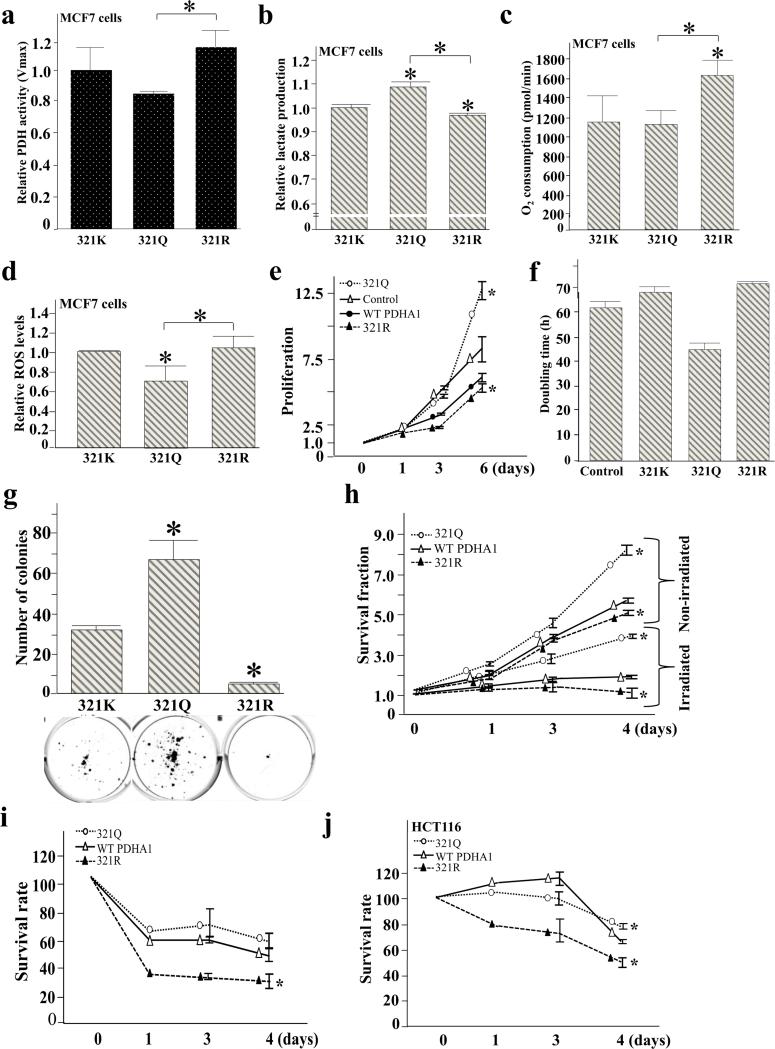
Figure 4. Acetylation of PDHA1 lysine 321 directs PDH enzymatic activity.
(a) Lysine 321 of PDHA1 is phylogenetically conserved in various species from human to the C. elegans. The amino acid sequence of PDHA1 between around 301 and 350 was compared among Homo sapiens (human), Pan troglodytes (chimpanzee), Mus musculus (house mouse), Xenopus laevis (African clawed frog), and Caenorhabditis elegans (roundworm). K321 in human PDHA1 sequence was conserved in all species investigated. In Xenopus and C. elegans, the sequence frame is slightly shifted. In Xenopus, K321 corresponds to K331; in C. elegans it corresponds to K315.
(b) 293T cells were transfected with Flag-PDHA1 as well as PDHA1 lysine mutants (Flag-PDHA1K83R, Flag-PDHA1K321R, and Flag-PDHA1K336R) with Tip60 and CBP and TSA / NAM, IPed with an anti-Flag antibody, and subsequently immunoblotted with an anti-pan acetyl-lysine and anti-PDHA1 antibody.
(c-i) 293T cells expressing shPDHA1 to knockdown endogenous PDHA1 were subsequently transfected with Flag-PDHA1K321K, Flag-PDHA1K321Q, or Flag-PDHA1K321R. Cell extracts were used to determine (c) PDH activity, (d) lactate production, (e) O2 consumption rates (pmol/min) per 106 cells were measured using the Seahorse Bioscience instrumentation and corrected for antimycin and rotenone. For 321K cells the basal OCR | 52 amol cell−1 s−1, (f) glucose uptake, and (g-h) relative oxidant production as determined by the fluorescence of MitoSOX E+ (g) relative oxidant production which was determined as in (g) was normalized by subtracting oxidant production from parallel samples grown MnSOD and mutant PDHA1 overexpressing cells (h). (i-j) For quantitative analysis of oxidation products derived from MitoSOX™ Red, HPLC electrochemical analysis was performed.
For all these experiments the change in PDH activity, lactate production, glucose uptake, and oxidant production are presented as relative changes compared to control cells or tissues. Error bars represent one standard deviation from the mean (*p<0.05 versus the control group).
It has previously been shown that substitution of a lysine with a glutamine mimics the acetylated lysine state, while substitution with an arginine mimics deacetylation [25,38,39]. Thus, mutating lysine 83, 321, or 336 to an arginine mimics a deacetylated lysine state, while substitution with a glutamine mimics an acetylated lysine state. To determine if K83, K321 and/or K336 are SIRT3 specific deacetylation targets, 293T cells were co-transfected with Flag-PDHA1, Flag-PDHA1K83R, Flag-PDHA1K321R, and Flag-PDHA1K336R in the presence of Tip60 and CBP and were treated with TSA and NAM. Then, cell extracts were IPed with an anti-Flag antibody and subsequently immunoblotted with an anti-pan acetyl-lysine and anti-PDHA1 antibody. The PDHA1 K321 and K336 arginine mutants, but not the K83 lysine mutant decreased the level of acetylation (Figure 4b), indicating that K321 and K336 are most possibly SIRT3 specific targets.
To determine if the acetylation status of lysine 321 directs PDH functional activity Flag-PDHA1K321, Flag-PDHA1K321Q, and Flag-PDHA1K321R were transfected into 293T cells constitutively expressing shPDHA1 that stably knocked down endogenous PDHA1. 293T cells expressing Flag-PDHA1K321R exhibited an increase in PDH activity (Figure 4c), decreased lactate production (Figure 4d), and increased O2 consumption (Figure 4e). In contrast, cells expressing Flag-PDHA1K321Q exhibited a decrease in PDH activity (Figure 4c), increased lactate production (Figure 4d), and increased glucose uptake (Figure 4f).
To determine if the acetylation status of PDH alters cellular ROS, cells expressing the genes above were used to measure ROS as determined by MitoSOX™ Red fluorescence. These experiments show that cells expressing Flag-PDHA1K321R exhibited increased cellular oxidant levels (Figure 4g and Supplemental Figure S4) in contrast to cells expression Flag-PDHA1K321Q. To confirm if increased MitoSOX™ Red fluorescence in Flag-PDHA1K321R expressing cells is due to increased oxidant levels, and not due to the increased uptake of the probe and/or oxidation byproducts, the experiments above (Figure 4g) were repeated and ROS was normalized using enforced expression of manganese superoxide dismutase (MnSOD) as well as the mutant PDHA1 gene. In these experiments the mitochondrial oxidant levels were normalize to ROS production without MnSOD expression. These results clearly show that cells expressing Flag-PDHA1K321R exhibited increased cellular oxidant levels (Figure 4h). Finally, these experiments were also repeated using HPLC with electrochemical detection (see methods section) and these results verified that the red fluorescence arose from the two-electron oxidation of MitoSOX™ Red to the ethidium analog, MitoSOX-2-OH-E+ and MitoSOX E+. As such, these results confirm those obtained using MitoSOX™ Red fluorescence technique (Figure 4i-j, Supplemental Figure S5).
Acetylation of PDHA1 K321 alters cancer cell proliferation
Consistent to 293T cells expressing mutant PDHA forms, we observed similar metabolic changes when we utilized PDHA1K321, Flag-PDHA1K321Q, and Flag-PDHA1K321R expressing MCF7 cells. Enforced expression of Flag-PDHA1K321R resulted in increased PDH activity relative to Flag-PDHA1K321Q expressing cells (Figure 5a). Expression of Flag-PDHA1K321R led to decreased lactate production (Figure 5b), and increased O2 consumption (Figure 5c), and the expression of Flag-PDHA1K321Q led to decreased ROS production in these cells (Figure 5d). To determine if the acetylation status of PDHA1 K321 alters the proliferation properties of PDHA1, lenti Flag-PDHA1K321, lenti Flag-PDHA1K321Q, and lenti Flag-PDHA1K321R were infected into MCF7 cells that constitutively express shPDHA1 and these engineered cell lines were used in several well-established assays that measure the proliferation and in vitro transformation [22,40-44]. These experiments showed that MCF7 cells with enforced expression of PDHA1K321Q, and expressing shPDHA1, exhibited increased proliferation, as determined by counting total cell numbers (Figure 5e), doubling time (Figure 5f), increased colony formation when plated at low density (Figure 5g), and cell survival when treated with 4 Gy and 8 Gy of ionizing radiation (Figure 5h and Supplemental Figure S3b), as compared to cells expressing PDHA1K321R or PDHA1K321.
Figure 5. Acetylation of PDHA1 K321 alters the in vitro transformative properties of PDH.
MCF7 cells expressing shPDHA1 to knockdown endogenous PDHA1 were subsequently transfected with Flag-PDHA1K321K, Flag-PDHA1K321Q, or Flag-PDHA1K321R.
(a) Relative PDH activity was measured.
(b) Relative lactate secreted into the media was measured.
(c) O2 consumption rates (pmol/min) per 106 cells were measured using the Seahorse instrument and corrected for antimycin and rotenone. For 321K cells the basal OCR ≈ 20amol cell−1 s−1 consistent with previous published OCR for MCF7 cells, 30 amol cell−1 s−1 [27].
(d) Cell extracts were labeled with MitoSOX™ Red. Oxidant production was determined using a flow cytometer as determined by the fluorescence of MitoSOX E+.
(e-f) MCF7 cells expressing shPDHA1 were infected with Flag-PDHA1K321K, Flag-PDHA1K321Q, or Flag-PDHA1K321R and the total number of cells was counted at days 1, 3, and 6 (e). Doubling times of these cells were determined (f).
(g) MCF7 cells expressing shPDHA1 were infected with Flag-PDHA1K321K, Flag-PDHA1K321Q, or Flag-PDHA1K321R and plated at low cell densities (250 cells per 60 mm plate). After 21 days, plates were stained with crystal violet, and the number of colonies for each group was quantified.
(h) The MCF7 cells described above were treated with and without 4 Gy of ionizing radiation and cell survival was determined using a colony formation assay.
(i) The MCF7 cells as mentioned above were treated with DCA (10 mM) and cell growth, as measured by total cell number, were determined at 1, 3, and 4 days and normalized to the untreated cells.
(j) HCT116 cells expressing shPDHA1 were infected with Flag-PDHA1K321K, Flag-PDHA1K321Q, or Flag-PDHA1K321R. Then, they were treated with 10 mM DCA and cell growth, as measured by total cell number, were determined at 1, 3, and 4 days, and normalized to the non-treated cells.
Error bars represent one standard deviation (*p<0.05 versus the control group).
Dichloroacetate (DCA) has been proposed to attenuate cancer cell proliferation by inhibiting upstream pyruvate kinase enzyme which consequently shifts metabolism from glycolysis to oxidative phosphorylation [39]. We tested if the expression of PDHA1K321R sensitizes MCF7 cells to anti-tumorigenic effects of DCA. MCF7 cells with enforced expression of PDHA1K321R displayed increased sensitivity to 10 mM DCA treatment, as compared to control cells, as determined by counting total cell numbers (Figure 5i). Likewise, HCT116 cells with enforced expression of PDHA1K321R also exhibited relatively higher sensitivity to 10 mM DCA treatment (Figure 5j). These experiments suggest that the acetylation status of K321 directs, at least in some part, the proliferation properties of PDHA1 consistent with the PDH activity results presented above.
DISCUSSION
Tumors exhibit metabolic reprogramming characterized by the preferential use of glucose, as first published by Otto Warburg in 1956 [13,14], and generally referred to as the “Warburg effect.” However, the mechanism(s) linking these processes remain largely elusive. Even though the Warburg effect is considered metabolic hallmark of most tumor cells, highly proliferative non-cancer cells also display high rates of glycolysis to maintain elevated cell proliferative capacity. Altered metabolism serves these highly proliferative cells not only providing biosynthetic building blocks such as amino acids, nucleic acids, and fatty acids for the generation of newly formed cells, but also assuring enough ATP through glycolysis to perform necessary cellular processes.
Cells lacking Sirt3 exhibit abnormally high levels of ROS as well as increased glucose consumption that has been suggested to promote, at least in some part, a tumor permissive phenotype both in vitro and in vivo [18,24,40]. In this regard, two independent studies by Finley et al. [21] and Bell et al. [44] have suggested a biochemical link between Sirt3 and HIF-1D and this regulatory node regulated glucose metabolism. In this regard, it was suggested that loss of SIRT3 increases HIF-1D target gene expression, glycolytic metabolism, and glucose-dependent cellular proliferation. In this study, we suggest an additional mechanism by which loss of SIRT3 and the aberrant hyper-acetylation of PDHA1 K321 might result in metabolic reprogramming (Figure 6) as well as the degree of tumor cell transformative properties. In MCF7 cells, deacetylation mimetic MCF7 cells showed significant increase in OCR and ROS production relative to acetylation mimetic cells. However, while 321R expressing MFC7 cells showed about 50% increase in OCR relative to control cells, the same cells didn't show significant increase in MitoSOX signal relative to control MCF7 cells. One of the reasons might be that control cells include wild type PDHA1 protein which may be activated or inactivated by deacetylation or acetylation naturally depending on different conditions at the time of measurement. In addition, due to fine tuning role of deacetylation on PDH activity, the metabolic changes were moderate, and to detect such small changes between mutant and control PDHA1 expressing cells with statistical significance were limited. According to our findings, the difference in colony formation ability between 321Q and 321R PDHA1 expressing MFC7 cells was very significant. Considering the fine tuning role of K321 deacetylation on PDH activity and moderate decrease in lactate production, we postulate that in addition to increasing PDH activity and ROS production, additional unidentified cellular changes in 321R expressing MCF7 cells might lead to a decrease in their colony formation ability in vitro. Metabolic changes, such as in PDH activity and ROS levels in 293T cells expressing 321K, 321Q and 321R genes were more pronounced than MCF7 cells probably because we had a chance to see immediate metabolic changes after transfection, before some other changes in the cells take place to mask or equilibrate the real change. In Figure 6, we show that deacetylation of PDH at K321 increases its activity which subsequently led to elevated ROS which had been shown by overexpression of mutant PDHA1 forms. However, SIRT3 has numerous substrates including MnSOD. Moreover, the amount of pyruvate and formation of various Krebs cycle intermediates might exhibit some antioxidant qualities. Therefore, combination of all of these factors determines the net ROS value in the cells.
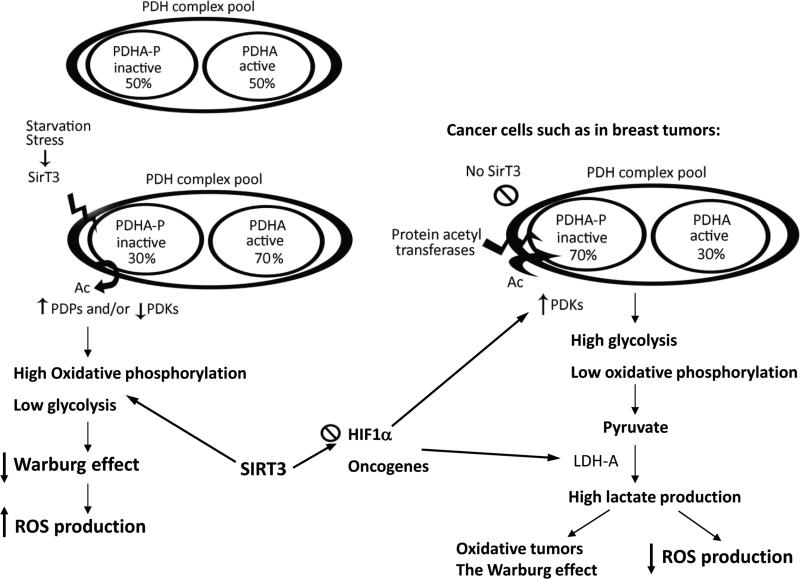
Figure 6. Summary: SIRT3 deacetylates PDHA1 at lysine 321 and decreases the Warburg effect.
PDH complexes in the mitochondria constitute a pool of active and inactive forms of the enzyme whose overall activity depends on the cell types and different physiological conditions. SIRT3 acts as a sensor for specific stress conditions such as starvation. When it gets activated, it deacetylates PDHA1 subunit of PDH. Deacetylation by SIRT3 increases the portion of active PDH. High PDH activity might increase the oxidative phosphorylation and ROS production, and decrease glycolysis. In many cancer cells, such as breast tumors, SIRT3 expression is significantly decreased. This results in hyper-acetylation of lysine residues (K321, K336, and possibly some others) and a decrease in PDH activity. Reduction in PDH activity causes higher glycolytic rates because pyruvate is not converted to acetyl-CoA, and this may ultimately increase the Warburg effect and decrease the production of oxidants in cancer cells.
When PDH activity was screened in Sirt3 wild-type and knockout tissues, acetylation/deacetylation of PDHA1 changed PDH activity to a relatively smaller degree than phosphorylation. This result seems reasonable since it has been proposed that the acetylome appears to play a fine-tuning role in the direction of downstream deacetylation targets and this fine-tuning of PDH activity may provide a more precise regulation in response to different physiological conditions and tissue cell types. For example, it has recently been shown using a diabetes skeletal muscle model that the acetylation of K336 can change the phosphorylation status of PDH [26]. However, this was not observed to change the catalytic activity of PDH enzyme in vitro. The authors suggested that other lysine(s) may more directly determine PDH activity under other conditions and our results suggest that PDH K321 may be one such reversible acetyl-lysine. Consistent to this idea, our results did not detect any increase in the activity of PDH complex when the PDHA1K336R lysine mutant was overexpressed in 293T cells. As such, it is proposed that the regulation of PDH activity directly, and PDC indirectly by the acetylome, may be complex and involve several different reversible acetyl-lysines that may be responding to different environmental nutrient availably conditions. While causative data to propose such an mechanism to direct mitochondrial enzymes is still lacking, it does appear that many different mitochondrial proteins contain more than one potential reversible acetyl-lysine [29,42] suggesting a complex interaction between the acetylome and mitochondrial energy generating protein complexes.
Many cancer cells re-wire and change the traditional cellular metabolism to stimulate mainly glycolysis even when the oxygen is present in the micro environment, thereby compensating their high demand for macromolecule synthesis [46]. In many tumors, SIRT3 expression is muted [15] and these tumors show features that are similar to the Warburg effect such as high lactate production and glucose consumption [22] and lower oxidant production because of higher rates of glycolysis and possibly pentose phosphate pathways and lower levels of electron transport chain [47]. Lower oxidant flux might be one of the reasons for the relative resistance of cancer cells expressing acetylated forms of PDHA1 (PDHA1K336Q) against ionizing radiation since ionizing radiation contributes to apoptosis of cancer cells partly by increasing the flux of potentially toxic oxidants. Additionally, DCA inhibits glycolysis by indirectly activating PDH, and is currently under investigation in clinical trials as a potential anticancer agent. Our data showed that cancer cells expressing deacetylated form of PDHA1 (PDHA1K336R) displayed higher sensitivity for anti-proliferative effects of DCA further suggesting the contribution of SIRT3 to the activation of PDH. The difference in metabolism between normal and cancer cells has a potential for cancer therapies. Mice lacking Sirt3 provide a unique in vivo murine model, mechanistically connecting aberrant acetylation, which is proposed to de-regulate with increasing age, the Warburg effect, and carcinogenesis.
Supplementary Material
Highlights.
SIRT3 interacts with PDHA1 and directs its enzymatic activity via changes in protein acetylation.
SIRT3 deacetylates PDHA1 lysine 321 and a PDHA1 mutant, mimicking a deacetylated lysine (PDHA1K321R) increases PDH activity.
The PDHA1 mutant, mimicking an acetylated lysine (PDHA1K321Q) decreases PDH activity. x PDHA1K321Q exhibited a more transformed in vitro cell phenotype as compared to PDHA1K321R.
ACKNOWLEDGMENTS
DG is supported by NCI-1R01CA152601-01, 1R01CA152799-01A1, 1R01CA168292-01A1, 1R01CA16383801A1, NIH R01 CA169046, NIH 5 P30 CA086862, and a Hirshberg Foundation for Pancreatic Cancer Research Seed Grant Award. We thank Dr. Navdeep Chandel and Sam Weinberg of Northwestern University for use of the Seahorse Bioscience instrumentation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Linn TC, Pettit FH, Reed LJ. Alpha-Keto Acid Dehydrogenase Complexes. X. Regulation of the Activity of the Pyruvate Dehydrogenase Complex from Beef Kidney Mitochondria by Phosphorylation and Dephosphorylation. Proc Natl Acad Sci U S A. 1969;62(1):234–241. doi: 10.1073/pnas.62.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters SJ. Regulation of PDH activity and isoform expression: diet and exercise. Biochem Soc Trans. 2003;31:1274–1280. doi: 10.1042/bst0311274. [DOI] [PubMed] [Google Scholar]
- 3.Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL. Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia. 2005;7:1–6. doi: 10.1593/neo.04373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pilegaard H, Birk JB, Sacchetti M, Mourtzakis M, Hardie DG, et al. PDH-E1alpha dephosphorylation and activation in human skeletal muscle during exercise: effect of intralipid infusion. Diabetes. 2006;55:3020–3027. doi: 10.2337/db06-0152. [DOI] [PubMed] [Google Scholar]
- 5.Knoechel TR, Tucker AD, Robinson CM, Phillips C, Taylor W, et al. Regulatory roles of the N-terminal domain based on crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands. Biochemistry. 2006;45:402–415. doi: 10.1021/bi051402s. [DOI] [PubMed] [Google Scholar]
- 6.McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, et al. Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem. 2008;283:22700–22708. doi: 10.1074/jbc.M801765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson BH. Lacticacidemia. Biochim Biophys Acta. 1993;1182:231–244. doi: 10.1016/0925-4439(93)90064-8. [DOI] [PubMed] [Google Scholar]
- 8.Roche TE, Cate RL. Evidence for lipoic acid mediated NADH and acetyl-CoA stimulation of liver and kidney pyruvate dehydrogenase kinase. Biochem Biophys Res Commun. 1976;72:1375–1383. doi: 10.1016/s0006-291x(76)80166-0. [DOI] [PubMed] [Google Scholar]
- 9.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, et al. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- 10.Stacpoole PW. The pyruvate dehydrogenase complex as a therapeutic target for age- related diseases. Aging Cell. 2012;11:371–377. doi: 10.1111/j.1474-9726.2012.00805.x. [DOI] [PubMed] [Google Scholar]
- 11.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 12.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 13.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 14.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim EA, Yang SJ, Choi SY, Lee WJ, Cho SW. Inhibition of glutamate dehydrogenase and insulin secretion by KHG26377 does not involve ADP-ribosylation by SIRT4 or deacetylation by SIRT3. BMB Rep. 2012;45:458–463. doi: 10.5483/bmbrep.2012.45.8.040. [DOI] [PubMed] [Google Scholar]
- 17.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, et al. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany NY) 2011;3:102–107. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, et al. Sirt3, Mitochondrial ROS, Ageing, and Carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, et al. SIRT3 Regulates Long-chain Acyl-CoA Dehydrogenase by Deacetylating Conserved Lysines Near the Active Site. J Biol Chem. 2013 doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Someya S, Yu W, Hallows WC, Xu J, Vann JM, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finley LW, Carracedo A, Lee J, Souza A, Egia A, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 Is a Mitochondrial Tumor Suppressor: A Scientific Tale That Connects Aberrant Cellular ROS, the Warburg Effect, and Carcinogenesis. Cancer Res. 2012;72:2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan F, Kerr H, Ross RA, Newton DJ, Belch JJ. Effects of poor glucose handling on arterial stiffness and left ventricular mass in normal children. Int Angiol. 2006;25:268–273. [PubMed] [Google Scholar]
- 24.Finley LW, Haigis MC. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med. 2012;18:516–523. doi: 10.1016/j.molmed.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62:3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, et al. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci U S A. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalyanaraman B, Dranka BP, Hardy M, Michalski R, Zielonka J. HPLC-based monitoring of products formed from hydroethidine-based fluorogenic probes--the ultimate approach for intra- and extracellular superoxide detection. Biochim Biophys Acta. 2014;1840:739–744. doi: 10.1016/j.bbagen.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielonka J, Hardy M, Kalyanaraman B. HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic Biol Med. 2010;46:329–338. doi: 10.1016/j.freeradbiomed.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto N, Ueda M, Sato T, Kawasaki K, Sawada K, Kawabata K, Ashida H. Measurement of glucose uptake in cultured cells. Curr Protoc Pharmacol. 2011 doi: 10.1002/0471141755.ph1214s55. Chapter 12:Unit 12.14.1-22. [DOI] [PubMed] [Google Scholar]
- 32.Buch K, Peters T, Nawroth T, Sänger M, Schmidberger H, et al. Determination of cell survival after irradiation via clonogenic assay versus multiple MTT Assay--a comparative study. Radiat Oncol. 2012;7:1. doi: 10.1186/1748-717X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi X, Yang P, Sun M, Yang Y, Li F. Decreased 1,25-Dihydroxyvitamin D3 level is involved in the pathogenesis of Vogt-Koyanagi-Harada (VKH) disease. Mol Vis. 2011;17:673–679. [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner BA, Venkataraman S, Buettner GR. The rate of oxygen utilization by cells. Free Radic Biol Med. 2011;51:700–712. doi: 10.1016/j.freeradbiomed.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombard DB, Zwaans BM. SIRT3: As Simple As It Seems. Gerontology. 2013 doi: 10.1159/000354382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 37.Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, et al. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One. 2012;7:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Gogol M, Carey M, Lee D, Seidel C, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 40.Gius D, Cao XM, Rauscher FJ, 3rd, Cohen DR, Curran T, et al. Transcriptional activation and repression by Fos are independent functions: the C terminus represses immediate-early gene expression via CArG elements. Mol Cell Biol. 1990;10:4243–4255. doi: 10.1128/mcb.10.8.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SH, Zhu Y, Ozden O, Kim HS, Jiang H, et al. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl Cancer Res. 2012;1:15–21. [PMC free article] [PubMed] [Google Scholar]
- 42.Gius DR, Ezhevsky SA, Becker-Hapak M, Nagahara H, Wei MC, et al. Transduced p16INK4a peptides inhibit hypophosphorylation of the retinoblastoma protein and cell cycle progression prior to activation of Cdk2 complexes in late G1. Cancer Res. 1999;59:2577–2580. [PubMed] [Google Scholar]
- 43.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, et al. A Mitochondria-K+ Channel Axis Is Suppressed in Cancer and Its Normalization Promotes Apoptosis and Inhibits Cancer Growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 44.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 46.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, et al. Tyrosine Phosphorylation of Mitochondrial Pyruvate Dehydrogenase Kinase 1 Is Important for Cancer Metabolism. Mol Cell. 2011;44:864–77. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.