Abstract
Self-assembled nano-micelles of amphiphilic polymers represent a novel anticancer drug delivery system. However, their full clinical utilization remains challenging because the quantitative structure-property relationship (QSPR) between the polymer structure and the efficacy of micelles as a drug carrier is poorly understood. Here, we developed a series of QSPR models to account for the drug loading capacity of polymeric micelles using the genetic function approximation (GFA) algorithm. These models were further evaluated by internal and external validation and a Y-randomization test in terms of stability and generalization, yielding an optimization model that is applicable to an expanded materials regime. As confirmed by experimental data, the relationship between microstructure and drug loading capacity can be well-simulated, suggesting that our models are readily applicable to the quantitative evaluation of the drug-loading capacity of polymeric micelles. Our work may offer a pathway to the design of formulation experiments.
Introduction
Cancer, as one of main diseases, will soon replace heart disease to become world's leading killer, seriously threatening human health [1]. Chemotherapy is an effective method for the treatment of cancer. However, the toxicity and side effects of anticancer drugs can be life threatening, and most of these drugs suffer from poor water solubility, have short half-lives, and can cause tumor cells to become drug resistant [2]. Extensive research is therefore being conducted on new formulations that might effectively improve the curative effect of anticancer drugs while minimizing or even eliminating their toxicity and side effects. One promising approach is to consider using a new drug delivery system to control the target delivery of drugs to the lesion [3,4]. Nano-micelles self-assembled from amphiphilic polymers represent a candidate system due to their many advantages, including small particle size, release behavior, improved drug solubility and the potential for both passive and active targeting ability [5–9]. Until now, many polymers have been extensively investigated for use in constructing micelles for drug delivery [10–16]. Linear block polymers were studied first due to their simple structures. Although some of these polymers reached the clinical evaluation stage, these polymers exhibit limited stability and burst release due to the dynamic nature of self-assembly [17–20]. Thus, high-grafting-density comb polymers and dendritic polymers have been developed to improve the stability and release performance of the drug-loaded micelle system [21–23]. According to the in vitro experiments, the rates of the drug release from the polymeric micelles were obviously different at different pH conditions. Specifically, the drug release rate at weakly acidic circumstance (around tumor site) was much higher than that at normal condition, indicating that the drug-loaded polymeric micelles exhibited pH-responsive. With the rapid development of controllable active polymerization, many methods, especially atom transfer radical polymerization (ATRP) and ring opening polymerization (ROP), have been used to control the polymeric composition, topological structure and to prepare functional polymers [9]. However, to obtain an appropriate drug delivery system, extensive experimentation is required; such experiments are not only financially costly but also are highly time-consuming. Thus far, studies on the structure-performance relationship of micelles have been confined to qualitative studies, including molecular simulation and mesoscopic simulation methods, which only describe the chemical morphology, microstructure and mesoscopic phase separation of polymers or colloids [24–26]. Very few studies of the quantitative relationships involved have been reported. Due to lack of information on the QSPR, it is difficult to fundamentally understand the nature of the interactions between the structure of polymer molecules and the drug-loading and drug-release performances. Herein, the QSPR is the major issue addressed.
The GFA algorithm offers a new approach to developing structure-property models [27–31]. QSPR models can be created automatically by combining the use of a genetic algorithm with statistical modeling tools. In this method, a GFA is used to solve function approximation. This algorithm is initially developed from two apparently disparate algorithms: Holland’s genetic algorithm and Friedman’s multivariate adaptive regression splines algorithm [32,33]. The GFA algorithm makes use of a population of many models and tests only the final, fully constructed model rather than generating a single model. The models are scored using Friedman's “lack of fit” (LOF) measure as the evaluation function [34,35]. This algorithm has been successfully applied to generate a variety of QSPR models.
QSPR studies are very useful for obtaining in-depth insights into structure-property relationships. In this article, the QSPR is studied via the GFA technique based on a series of four/six-arm star polymer structures and the data regarding the DOX-loading capacities of the micelles. The descriptors in the QSPR are also selected using a GFA algorithm. Both cross validations of multiple linear regression (MLR) and the leave-one-out (LOO) method were used to build and evaluate the QSPR models. The characteristics of these models, including fitting ability, predictive ability, stability and generalization ability, are evaluated by internal and external validation and the Y-randomization test. We also define the applicability domain of the optimization model. The results obtained provide important guidance for the design and synthesis of the desired polymers.
Materials and Methods
Experimental data and data splitting
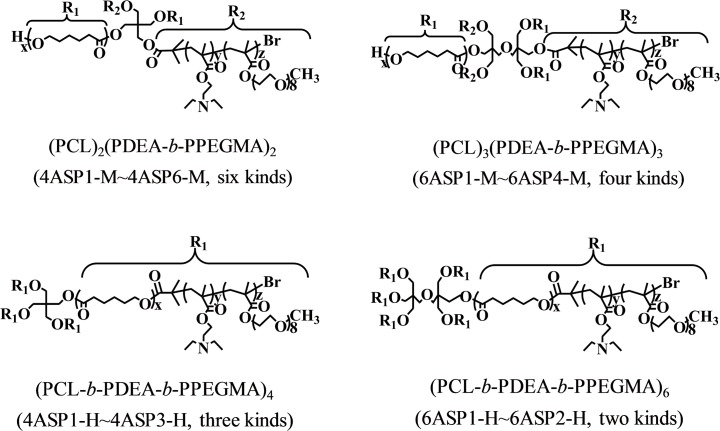
The research system used was star polymers, and the drug-loading capacities (LC, % w/w) of micelles self-assembled from these star polymers were used as the performance measure. In this study, 15 kinds of star polymers were used, including six kinds of four-miktoarm star polymers (PCL)2(PDEA-b-PPEGMA)2, four kinds of six-miktoarm star polymers (PCL)3(PDEA-b-PPEGMA)3, three kinds of four-homoarm star polymers (PCL-b-PDEA-b-PPEGMA)4, and two kinds of six-homoarm star polymers (PCL-b-PDEA-b-PPEGMA)6. Fifteen kinds of designed polymers were synthesized in our laboratory. Drug-loaded micelles were prepared using two DOX/polymer ratios (10/40 and 20/40 (mg/mg)), resulting in 30 data LC datasets (LC datasets were transformed using natural logarithms (ln) to better conform to the normal distribution), as shown in S1 Table [36–38]. All LC data were measured in aqueous solution at pH 7.4 and room temperature. All details are shown in Fig. 1 and S1 Table.
Fig 1. The structures of four- and six-armed polymers.
To build and validate a QSPR model of good general ability, the original dataset was divided into a training set and a test set using the Duplex method which was an effective way to select representative training and test sets [39]. Briefly, Euclidean distances were calculated between each pair of samples as follows: (1) The two samples that are furthest away from each other are selected for the training set. (2) From the remaining samples, the two samples that are furthest away from each other are included in the test set. (3) The remaining point that is furthest away from the two points in step (1) is included in training set, and the one furthest away from the two points in step (2) is included in the test set. Finally, the datasets were split into the training set (containing 22 data) and the test set (containing 8 data, but not less than five) [40]. See S1 Table for a detailed list.
Computational details regarding descriptors
Total fifty-two kinds of molecular descriptors (including physical and chemical descriptors, fragment counts, topological, spatial and energy descriptors, atomic volumes and surfaces, and atomistic descriptors which are suitable for describing the relationship between polymer molecular structure and micellar property) could been computed using the QSAR module of Materials Studio 5.0 (Accelrys Inc.) [41] and used as candidates, as shown in S2 Table. To obtain the lowest energy conformation of the polymer molecules, the geometric structures of all polymer molecules were constructed and optimized using the following three steps. First, the initial polymer structure was constructed in the Materials visualizer. Then, the polymer structures were minimized in the Discover Module by setting the optimization method to smart minimizer, the force field type to compass, the convergence level to ultra-fine, the non-bond methods to van der Waals and Coulomb interactions, and the summation method to atom-based. Finally, the structures were optimized using molecular dynamics simulation for the NVE and NVT ensembles in turn, and the temperature was set to 298K; the number of steps was set to 10,000. Then, values of 52 descriptors were calculated using the QSAR and Forcite Modules of Materials Studio 5.0 based on the polymer structure that was optimized as described above. These descriptor values were separated according to the quality ratios of the drug/polymer. Constant terms, near-constant values and pairwise-correlated descriptors (one of any two descriptors with a correlation coefficient greater than 0.99) were excluded in a pre-reduction step. The remaining 36 descriptors and their detailed values for the 22 polymers, showing in S3 and S4 Tables, were selected and calculated for the following research.
Model development
In this study, some methods were used to develop and evaluate a reliable model. GFA-MLR technique, derived from an analogy with the evolution of DNA [42], was used to generate a series of models. In GFA algorithm, an individual or model was represented as one-dimensional string of bits. It was a distinctive characteristic of GFA that it could create a population of models rather than a single model. GFA algorithm, selecting the basis functions genetically, developed better models than those made using stepwise regression methods.
And then, the models were estimated using the “lack of fit” (LOF), which was measured using a slight variation of the original Friedman formula [34,35], so that best model received the best fitness score. The LOF was determined by the following revised equation in Materials Studio 5.0:
| (1) |
where, SSE is the sum of squares of the errors, c is the number of terms in the model (other than the constant term), and d is a scaled smoothing parameter, which is used to evaluate the Friedman LOF statistic. Larger values of the smoothness parameter cause larger models to be penalized to a greater degree. p is the total number of descriptors contained in all model terms (ignoring the constant term), n is the total number of samples in the training set, and λ is a safety factor, which was assigned the value 0.99 to ensure that the denominator of the expression would never become zero.
The applicability domain for the best QSPR model should be defined [43]. In this work, we have applied the leverage approach to investigate the domain of applicability of polymers [44]. It is a great benefit of this approach that can draw the Williams plot (a plot of standardized predicted residuals versus leverage values) to observe the applicability domain of a QSPR model.
Model validation
The fitting ability, stability, reliability and predictive ability of the developed models were evaluated by some validation [43–46], including internal and external validation parameters.
Internal validation parameters
R 2: R 2 (the square of the correlation coefficient) describes the fraction of the total variation attributed to the model. The closer the value of R 2 is to 1.0, the better the regression equation explains the Y variable. R 2 is the most commonly used internal validation indicator and is expressed as follows:
| (2) |
where, are the experimental property, the predicted property and the mean experimental property of the samples in the training set, respectively.
R 2 cv: R 2 cv (the cross-validated correlation coefficient) is derived from cross validation and is the cross-validated equivalent of R 2. In general, the developed models were subjected to internal validation using the LOO-cross-validation method. R 2 cv is expressed as follows:
| (3) |
where, Y obs and have the same definitions as in Equation (2) above. is the LOO-predicted property of the samples in the training set. The closer this value is to 1.0, the better the predictive ability of the model [45].
RMSE: The root mean square error (RMSE) is dispersion degree of random error, presenting a more intuitive index of the fitting ability of the model [46], and is defined as follows:
| (4) |
where, i represents sample i and n is the total number of samples in the training set. The lower the value of RMSE is, the better the predictive ability of the model.
Y-randomization test: The Y-randomization test is a statistical method that is widely used to test the reliability and robustness of a model [47]. The purpose of the Y-randomization test is to detect and quantify chance correlations between the dependent variable and the descriptor. In this method, a QSPR model is recalculated for randomly reordered responses, and the descriptor matrix remained unchanged. The obtained models should exhibit significantly lower values of R 2 or R 2 cv than the original model because the relationship between the structure and the response has been broken. Y-randomization is performed through response scrambling with maximum iterations of 500; then, the mean values of R 2 or R 2 cv are calculated. If either R 2 or R 2 cv is higher than 0.5, the original model is suspected to be relevant, and the reliability of the QSPR model is doubtful [48].
External validation parameters
Internal validation is an essential step in QSPR model development. The desired internal validation results show that the model exhibits higher stability and prediction ability. However, no real prediction ability is shown for external samples. Therefore, the external predictive ability and extrapolation of the models should be evaluated.
R 2 pred: R 2 pred is termed the predictive R 2 of a development model and is an important parameter that is used to test the external predictive ability of a QSPR model [48]. R 2 pred is defined as follows:
| (5) |
where, are the experimental property, the predicted property and the mean of the experimental property of the samples in the test set, respectively.
and : and are developed to obtain a true predictive QSPR model based on the and parameters proposed by Roy et al [49]. , which has been found to be a better metric than the original , is the average value of and . is the absolute difference between and . If the value of is higher than 0.5, the value of should be lower than 0.2 [50]. In general, better models exhibit lower values. The equations for these parameters are shown in S5 Table.
Applicability domain
After internal and external validation, it cannot be claimed that this model will provide reliable results for any unknown sample, even though all evaluation indexes prove that the model is stable and reliable and exhibits good generalization and predictive ability. In fact, each model has its own applicability domain. If the predictive value of the sample lies within this applicability domain, the value is reliable. On the contrary, the model remains unreliable [50]. Thus, it is necessary to define the applicability domain of the model before it is applied to simulate unknown samples.
The leverage approach is generally used to define applicability domain of a model, and the leverage approach requires the assumption that the datasets used follow the normal distribution [44]. This approach can quantify the applied range of the model, and the results can be presented using the Williams plot, in which the leverage values (or Hat values) and cross-validated standardized residuals are used as the abscissa and ordinate, respectively. In this plot, two horizontal lines and one vertical line delimit a safety zone. The critical leverage h* (the vertical line) is generally fixed at 3(p + 1)/n (where n is the number of training compounds and p is the number of variables used in the model, respectively). The leverage (h i) of every sample is defined as follows:
| (6) |
where, x i is the descriptor row-vector of the query ith sample, X is the characteristic matrix of the training set, and m is the number of query samples. A higher leverage (h i > h*, outside the safety zone) indicates that the predicted response is unreliable because it goes beyond reasonable extrapolation. If the cross-validated standardized residual of the sample is lower or higher than three standard deviation units (represented as the two horizontal lines), the sample remains outside the reasonable range. Only values that fall within this area are considered reliable.
Results and Discussion
Determination of the optimal descriptor number
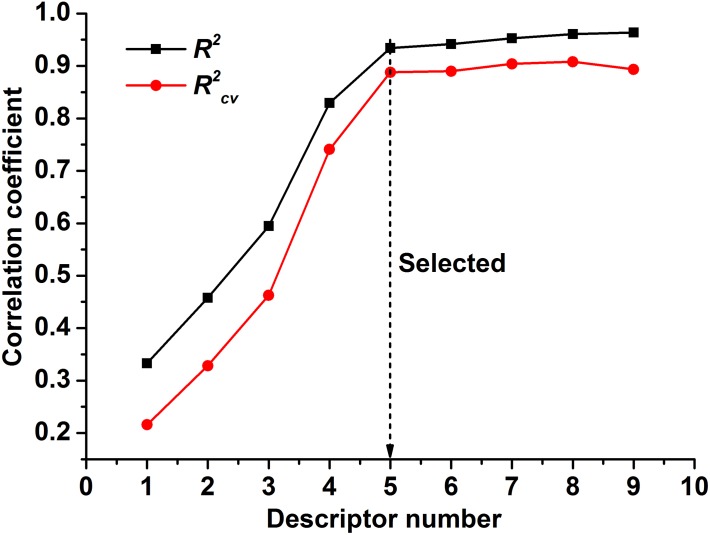
To select the descriptors that are most relevant to the ln(LC) of the polymeric micelles, 36 descriptors, which were calculated using Materials Studio, were used as inputs for the GFA algorithm. The optimal subset size was realized when further increases in the descriptor did not significantly improve model performance. Here, LOO-cross-validation was used to estimate the models. A plot of R 2 cv and R 2 against the number of selected descriptors is shown in Fig. 2 and indicates that the optimal model consists of five descriptors [39]. Moreover, R 2 cv is a key parameter regarding the predictive ability of the model. The closer the value of R 2 cv is to 1, the better prediction ability the model can deliver. For a good model, R 2 cv should be closer to R 2 (R 2 cv is usually lower). If R 2 cv is far smaller than R 2, then the possibility of data over-fitting in the regression model becomes significantly higher. When the number of descriptors is 5 (Fig. 2), R 2 cv fits R 2 most comfortably, indicating that the optimal subset size is five.
Fig 2. Dependence of the correlation coefficients R2cv and R 2 on the number of descriptors.
Model building
To develop the optimization model, we included 22 samples in the training set. The number of descriptors in the regression equation was 5, and Population and Generation were set to 1,000 and 5,000, respectively. The number of top equations returned was 10 (starting from the tenth model, the value of R 2 is lower than 0.9). Mutation probability was 0.1, and the smoothing parameter was 0.5. The models were scored based on Friedman’s LOF. The statistical parameters of the ten models are shown in Table 1.
Table 1. Statistical parameters of the ten GFA-MLR models.
| Model | R 2 | R 2 cv | RMSE(a) | F | R 2 pred | RMSE(b) | ||
|---|---|---|---|---|---|---|---|---|
| 1 | 0.934 | 0.888 | 0.0675 | 45.31 | 0.811 | 0.1121 | 0.7500 | 0.0406 |
| 2 | 0.930 | 0.876 | 0.0683 | 42.52 | 0.810 | 0.1127 | 0.7563 | 0.0276 |
| 3 | 0.924 | 0.864 | 0.0724 | 38.96 | 0.766 | 0.1250 | 0.7096 | 0.0518 |
| 4 | 0.924 | 0.864 | 0.0726 | 38.77 | 0.809 | 0.1128 | 0.7451 | 0.1064 |
| 5 | 0.919 | 0.849 | 0.0749 | 36.22 | 0.811 | 0.1122 | 0.7545 | 0.0291 |
| 6 | 0.918 | 0.851 | 0.0754 | 35.72 | 0.804 | 0.1142 | 0.7392 | 0.1269 |
| 7 | 0.913 | 0.838 | 0.0774 | 33.66 | 0.773 | 0.1230 | 0.7091 | 0.0025 |
| 8 | 0.913 | 0.837 | 0.0774 | 33.66 | 0.808 | 0.1132 | 0.7513 | 0.0404 |
| 9 | 0.909 | 0.829 | 0.0794 | 31.85 | 0.773 | 0.1230 | 0.7073 | 0.0102 |
| 10 | 0.883 | 0.779 | 0.0899 | 24.16 | 0.745 | 0.1302 | 0.6187 | 0.1872 |
RMSE (a): root mean square error of the training set; RMSE (b): root mean square error of the test set. F: The F test is a standard statistical test for the equality of the variances of two populations having normal distributions.
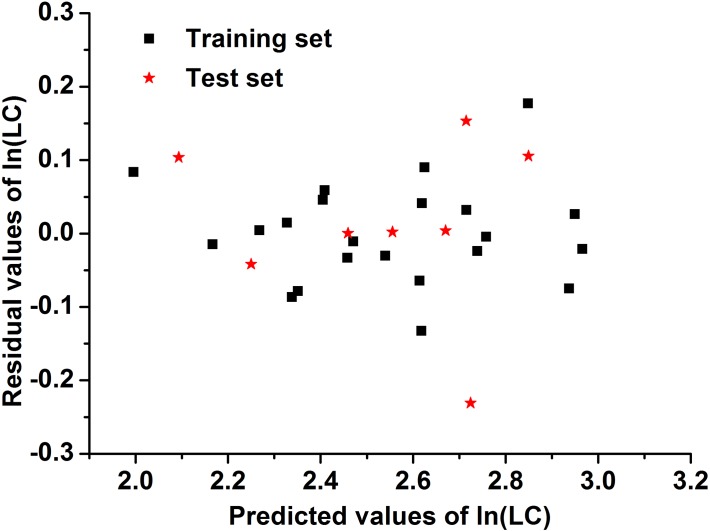
R 2, R 2 cv and F for Model 1 were 0.934, 0.888 and 45.31, respectively, and these values are the largest found among the ten models. Moreover, RMSE(a) was the smallest among the models, and the difference between R 2 and R 2 cv was the lowest. These results suggest that Model 1 exhibits the best fitting ability and the best internal predictive ability. To examine the stability of Model 1, the Y-randomization test was conducted, and a residual scatter diagram was plotted. After repeating the Y-randomization test more than 500 times, the mean values of R 2 and R 2 cv became 0.043 and 0.005, respectively, much lower than 0.5, indicating that Model 1 is more stable, and there is no “chance correlation” phenomenon occurring for Model 1. As seen in Fig. 3, the points are distributed irregularly and randomly, proving that Model 1 is more stable.
Fig 3. A scatter diagram of residuals for Model 1.
Regarding the external validation parameters for Model 1 (Table 1), R 2 pred is the highest (0.811) and RMSE(b) is the lowest (0.1121) among the models. The (0.75) of Model 1 is higher than 0.5, and the (0.04) is much lower than 0.2. These results show that Model 1 exhibits slightly higher external prediction ability than the other models.
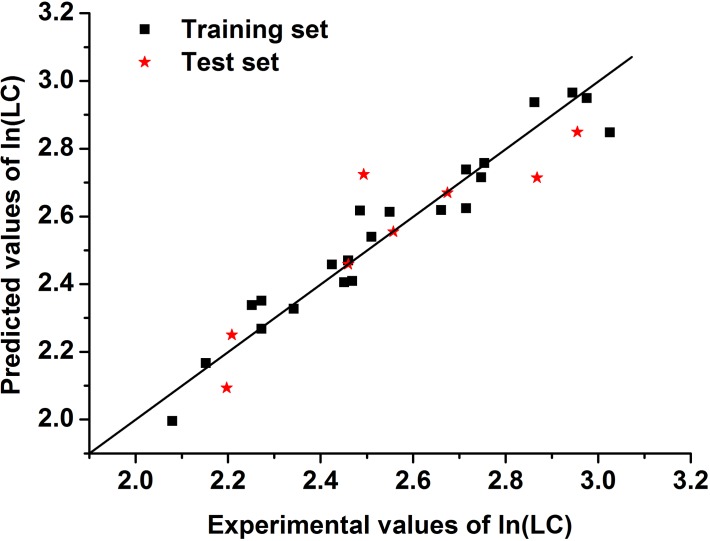
As seen in S6 Table, Model 1 shows the minimal residuals, possessing the optimal prediction ability. With regard to the optimization model, all of the predicted values of drug loading capacity of the samples in the training and test sets are close to their experimental values, as shown in S1 Table. Moreover, Fig. 4 shows that the sample points in the training and test sets are uniformly distributed along the line y = x, indicating that the residual errors of the predicted and experimental values are very low for Model 1. For example, the predicted value of No. 26 is 14.4, very similar to the experimental value 14.5, and the residual value (0.0040) is also much lower than 0.1.
Fig 4. The linear correlation diagram between the predicted values and experimental values of ln(LC) for the training and test sets used for Model 1.
S7 Table is the correlation matrix of the five descriptors in the optimization model. According to the previous research [51], all the correlation coefficients are less than 0.95, indicating the correlation analysis indicated that SSOV, SSA, EV TPE and IE are not highly correlated. Furthermore, if the multi-collinearity was present, in order to avoid its effects, probability (p) values of each coefficient are used to check whether multi-collinearity is affecting a correlation. It is generally accepted if the p-value is less than 0.05 [52]. Table 2 shows that all the p-values of five descriptors in Model 1 are very low (p ≤ 0.005), showing that multi-collinearity could not affect the correlation here.
Table 2. Different parameters of the GFA-MLR Model 1.
| Descriptors | Unstandardized coefficients | 95% Confidence interval of B | Standardized coefficient | p-value < | ||
|---|---|---|---|---|---|---|
| B | std. error | lower limit | upper limit | |||
| Intercept | 2.548 | 0.017 | 2.512 | 2.584 | — | 0.005 |
| SSOV | −1.953 | 0.143 | −2.256 | −1.650 | −7.601 | 0.005 |
| SSA | 1.101 | 0.096 | 0.898 | 1.305 | 3.966 | 0.005 |
| EV | 0.339 | 0.034 | 0.266 | 0.412 | 1.162 | 0.005 |
| TPE | 0.320 | 0.034 | 0.248 | 0.392 | 1.339 | 0.005 |
| IE | 0.525 | 0.053 | 0.413 | 0.636 | 2.138 | 0.005 |
Based on the above analysis, the 5-parameter version of Model 1 was selected as the optimization model, as shown in Table 2. The equation for the optimization model is as following:
| (7) |
SSOV: solvent surface occupied volume; SSA: solvent surface area; EV: ellipsoidal volume; TPE: total potential energy; IE: inversion energy. If the values of the five descriptors could be work out, the predicted LC could be given via the above equation. The values of five descriptors in training and test set are shown in S8 Table.
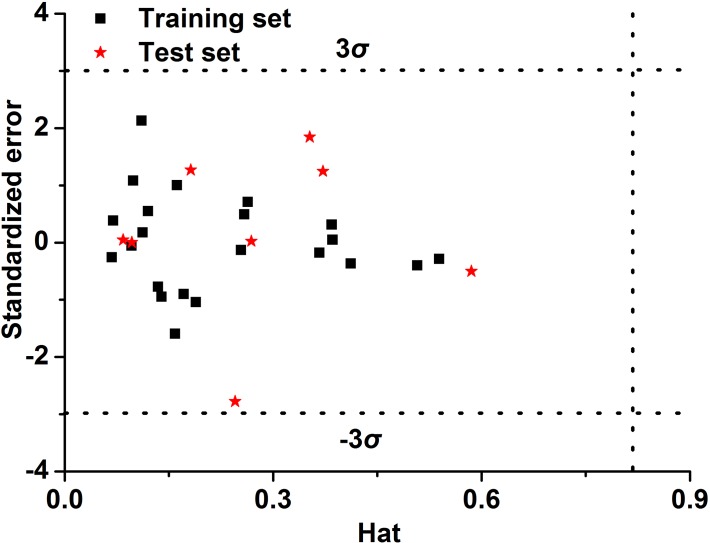
Applicability domain of the optimization model
Fig. 5 shows a Williams plot of the optimization model; this plot was developed using a leverage method to define the applicability domain of the model. All data points fall within the safety zone, except for one point, which is close to the bottom warning line but remains in the reasonable area. And the residual errors of the training and test sets are relatively smaller. Thus, the model was able to accurately predict the results.
Fig 5. Williams plot of the optimization model.
Descriptor interpretation and mechanism analysis
By interpreting the descriptors in the optimization model, it is possible to gain some insight into the factors that are likely to govern the ln(LC) of these polymeric micelles. The relative importance of the descriptors was determined based on their standardized regression coefficients. Table 2 shows that the most important descriptor is the SSOV, which is the solvent surface occupied volume of the polymers. This descriptor is mainly correlated with the hydrophilicity of a polymer molecule in aqueous solution. The SSA is a solvent surface area descriptor of the polymers and describes the hydrophobicity of polymer molecules in aqueous solution. EV is the ellipsoidal volume descriptor, which describes the volume of the ellipsoid of inertia, which is derived from the inertia tensor of the system. This descriptor is mainly correlated with the rigidity (indirectly reflecting the hydrophobicity) of polymer molecules in aqueous solution. IE is the inversion energy descriptor, which describes the energy required to transform a molecule from one spatial form (or invertomer) to another. This descriptor is a good indicator of the nature of the bonding between the atoms of a polymer.
Table 3 presents the relationship between the five descriptors and the three monomers. As shown in the table, positive correlations exist between PCL or PDEA and the five descriptors, whereas negative correlations exist between PPEGMA and the corresponding parameters. The correlation coefficients for the former group are slightly higher than those of monomers. An unknown synergistic effect might occur between the two hydrophobic blocks (PCL and PDEA).
Table 3. The correlation coefficient between the different segments and the five descriptors.
| Segment | SSOV | SSA | EV | TPE | IE |
|---|---|---|---|---|---|
| PCL | 0.55 | 0.31 | 0.60 | 0.54 | 0.72 |
| PDEA | 0.79 | 0.59 | 0.63 | 0.52 | 0.72 |
| PPEGMA | −0.21 | −0.03 | −0.51 | −0.01 | −0.33 |
| PCL+PDEA | 0.79 | 0.52 | 0.74 | 0.63 | 0.86 |
Table 4 shows the contributions of different segments to the descriptor values. The calculated values are based on the data presented in S9 Table using the PPEGMA monomer as a reference. For example, the contribution to SSOV caused by the DEAEMA monomer is calculated as follows: CDEA = (705.01/1583.56)/(187.28/533.45) = 1.27. With regard to the descriptor SSOV, different contributions resulted from the three monomers in the order CCL > CDEA > CPEGMA.
Table 4. The contributions of the tested monomers to the five descriptors.
| SSOV | SSA | EV | TPE | IE | |
|---|---|---|---|---|---|
| CCL | 1.36 | 1.52 | 0.29 | 0.85 | 10 |
| CDEA | 1.27 | 1.30 | 0.39 | 1.28 | 114 |
| CPEGMA | 1 | 1 | 1 | 1 | 1 |
As shown in Table 4, the hydrophobic segments (CL and DEA) in the polymer exhibited enhanced SSOV, resulting in a higher value of SSOV. A larger micellar core might be induced by segments that are more hydrophobic, thus providing a larger surface area for solvent and resulting in higher value of SSOV. Conversely, more hydrophilic segments (PEGMA) yield lower values of SSOV, possibly because the PEGMA segment distributed into the surface of the micelle. The same is true for SSA. The effect of the PEGMA segment on EV is greater than those of the CL and DEA segments. However, the relationship between hydrophilic segments and EV is negative, as shown in Table 3. The pH-sensitive DEA segment is the main factor for the descriptor TPE, and this segment exerts a positive affect. The DEA segment is also the largest contributor to the descriptor IE and provides a much greater contribution (up to 114-fold) than those of the CL (10-fold) and PEGMA (1-fold) segments. Equation (7) and S10 Table demonstrate that the descriptors SSOV, SSA and IE are the main factors in the optimization model. Because the contributions of the three descriptors are lower than that of EV, the coefficients of the three descriptors are much higher than that of EV; IE shows the most dramatic change among the five descriptors. The equation describing the model shows that the drug loading capacity can be enhanced by reducing SSOV and increasing the remaining four descriptors (SSA, EV, TPE and IE). According to the above analysis, although increases in CL and DEA enhance the values of SSOV (reducing LC) and the other four descriptors (increasing LC), the overall effect is to increase LC. An increase in PEGMA decreases the value of SSOV (increasing LC) but decreases the other descriptors, reducing the LC overall. In other words, to enhance the drug loading capacity of the micelles, the number of hydrophobic segments (DEA and CL) should be increased and the number of hydrophilic segments (PEGMA) should be decreased; this conclusion is consistent with the experimental observations.
Thus, the amounts of CL and DEA positively and significantly affect the drug loading capacity. The hydrophobic CL and DEA units are located in the core of the spherical micelle and provide sufficient space to accommodate the anticancer drug DOX in a synergistic manner. Decreasing the number of PEGMA units is also important for increasing the drug-loading capacity.
Conclusions
This work addresses the QSPR between a series of amphiphilic four/six-arm star polymer structures and their micelle drug-loading capacities. Our study developed and evaluated ten models based on the GFA, the LOF function, MLR and LOO-cross-validation methods. The optimal descriptor number of the model was determined to be five based on the statistical results of the LOO-cross-validated correlation coefficient (R 2 cv) and the square of the correlation coefficient (R 2). By analyzing the internal and external validation parameters of all models, the QSPR model was confirmed as optimal and to possess good fitting ability, good predictive ability and high stability. The influence of polymer structure on micelle drug-loading capacity was also analyzed. This study provides an effective approach for the design and synthesis of new star polymers with desired drug-loading capacities.
Supporting Information
(DOC)
(DOC)
(DOC)
(XLS)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by National Natural Science Foundation of China (No.21176090), Team Project of Natural Science Foundation of Guangdong Province, China (No.S2011030001366), Specialized Research Fund for the Doctoral Program of Higher Education of China (No.20130172110009), and Fundamental Research Funds for the Central Universities, China (No.2013ZP0010, 2013ZZ0059, 2014ZP0020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64: 9–29. 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 2. Tyrrell ZL, Shen Y, Radosz M. Fabrication of micellar nanoparticles for drug delivery through the self-assembly of block copolymers. Prog Polym Sci. 2010;35: 1128–1143. [Google Scholar]
- 3. Torchilin VP. Targeted polymeric micelles for delivery of poorly soluble drugs. Cell Mol Life Sci. 2004;61: 2549–2559. [DOI] [PubMed] [Google Scholar]
- 4. Feng ST, Li J, Luo Y, Bhargava SK, Bansal V. pH-Sensitive Nanomicelles for Controlled and Efficient Drug Delivery to Human Colorectal Carcinoma LoVo Cells. PLoS ONE. 2014;9: e100732 10.1371/journal.pone.0100732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siepmann J, Faisant N, Akiki J, Richard J, Benoit JP. Effect of the size of biodegradable microparticles on drug release: experiment and theory. J Control Release. 2004;96: 123–134. [DOI] [PubMed] [Google Scholar]
- 6. Zhang YH, Li XR, Zhou YX, Wang XN, Fan YT, Huang YQ, et al. Preparation and evaluation of poly (ethylene glycol)–poly (lactide) micelles as nanocarriers for oral delivery of cyclosporine A. Nanoscale Res Lett. 2010;5: 917–925. 10.1007/s11671-010-9583-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hira SK, Mishra AK, Ray B, Rytting E, Manna PP. Targeted Delivery of Doxorubicin-Loaded Poly (ε-caprolactone)-b-Poly (N-vinylpyrrolidone) Micelles Enhances Antitumor Effect in Lymphoma. PLoS ONE. 2014;9: e94309 10.1371/journal.pone.0094309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang CY, Kos P, Müller K, Schrimpf W, Troiber C, Lächelta U, et al. Native chemical ligation for conversion of sequence-defined oligomers into targeted pDNA and siRNA carriers. J Control Release. 2014;180: 42–50. 10.1016/j.jconrel.2014.02.015 [DOI] [PubMed] [Google Scholar]
- 9. Feng C, Lu G, Li Y, Huang X. Self-Assembly of Amphiphilic Homopolymers Bearing Ferrocene and Carboxyl Functionalities: Effect of Polymer Concentration, β-Cyclodextrin, and Length of Alkyl Linker. Langmuir. 2013;29: 10922–10931. 10.1021/la402335d [DOI] [PubMed] [Google Scholar]
- 10. Lavasanifar A, Samuel J, Kwon GS. Poly(ethylene oxide)-block-poly(L-amino acid) micelles for drug delivery. Adv Drug Deliv Rev. 2002;54: 169–190. [DOI] [PubMed] [Google Scholar]
- 11. Kim S, Kim JY, Huh KM, Acharya G, Park K. Hydrotropic polymer micelles containing acrylic acid moieties for oral delivery of paclitaxel. J Control Release. 2008;132: 222–229. 10.1016/j.jconrel.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim SW. Biomaterials to gene delivery. J Control Release. 2011;155: 116–118. 10.1016/j.jconrel.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiong XB, Falamarzian A, Garg SM, Lavasanifar A. Engineering of amphiphilic block copolymers for polymeric micellar drug and gene delivery. J Control Release. 2011;155: 248–261. 10.1016/j.jconrel.2011.04.028 [DOI] [PubMed] [Google Scholar]
- 14. Zhang CY, Yang YQ, Huang TX, Zhao B, Guo XD, Wang JF, et al. Self-assembled pH-responsive MPEG-b-(PLA-co-PAE) block copolymer micelles for anticancer drug delivery. Biomaterials. 2012;33: 6273–6283. 10.1016/j.biomaterials.2012.05.025 [DOI] [PubMed] [Google Scholar]
- 15. Yang YQ, Lin WJ, Zhao B, Wen XF, Guo XD, Zhang LJ, et al. Synthesis and physicochemical characterization of amphiphilic triblock copolymer brush containing pH-sensitive linkage for oral drug delivery. Langmuir. 2012;28: 8251–8259. 10.1021/la301099q [DOI] [PubMed] [Google Scholar]
- 16. Luo YL, Chen LL, Miao Y, Xu F. Novel AB4-type CTBN-b-mPEG PU micelle-like amphiphilic block copolymer micelles for prednisone drug release. Ind Eng Chem Res. 2013;52: 1571–1580. [Google Scholar]
- 17. Oerlemans C, Bult W, Bos M, Storm G, Nijsen JFW, Hennink WE. Polymeric micelles in anticancer therapy: targeting, imaging and triggered release. Pharm Res. 2010;27: 2569–2589. 10.1007/s11095-010-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bauhuber S, Liebl R, Tomasetti L, Rachel R, Goepferich A, Breunig M. A library of strictly linear poly (ethylene glycol)-poly (ethylene imine) diblock copolymers to perform structure–function relationship of non-viral gene carriers. J Control Release. 2012;162: 446–455. 10.1016/j.jconrel.2012.07.017 [DOI] [PubMed] [Google Scholar]
- 19. Kataoka K, Harada A, Nagasaki Y. Block copolymer micelles for drug delivery: Design, characterization and biological significance. Adv Drug Deliv Rev. 2012;64: 37–48. [DOI] [PubMed] [Google Scholar]
- 20. Xu Y, Thurber CM, Macosko CW, Lodge TP, Hillmyer MA. Poly (methyl methacrylate)-block-polyethylene-block-poly (methyl methacrylate) triblock copolymers as compatibilizers for polyethylene/poly (methyl methacrylate) Blends. Ind Eng Chem Res. 2014;53: 4718–4725. [Google Scholar]
- 21. Poon Z, Lee JA, Huang S, Prevost RJ, Hammond PT. Highly stable, ligand-clustered “patchy” micelle nanocarriers for systemic tumor targeting. Nanomedicine. 2011;7: 201–209. 10.1016/j.nano.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tatiya PD, Hedaoo RK, Mahulikar PP, Gite VV. Novel polyurea microcapsules using dendritic functional monomer: synthesis, characterization, and its use in self-healing and anticorrosive polyurethane coatings. Ind Eng Chem Res. 2013;52: 1562–1570. [Google Scholar]
- 23. Yang XL, Luo YL, Xu F, Chen YS. Thermosensitive mPEG-b-PA-g-PNIPAM comb block copolymer micelles: effect of hydrophilic chain length and camptothecin release behavior. Pharm Res. 2014;31: 291–304. 10.1007/s11095-013-1160-y [DOI] [PubMed] [Google Scholar]
- 24. Zheng LS, Yang YQ, Guo XD, Sun Y, Qian Y, Zhang LJ. Mesoscopic simulations on the aggregation behavior of pH-responsive polymeric micelles for drug delivery. J Colloid Interface Sci. 2011;363: 114–121. 10.1016/j.jcis.2011.07.040 [DOI] [PubMed] [Google Scholar]
- 25. Guo XD, Zhang LJ, Qian Y. Systematic multiscale method for studying the structure-performance relationship of drug-delivery systems. Ind Eng Chem Res. 2012;51: 4719–4730. [Google Scholar]
- 26. Luo Z, Jiang J. pH-sensitive drug loading/releasing in amphiphilic copolymer PAE–PEG: Integrating molecular dynamics and dissipative particle dynamics simulations. J Control Release. 2012;162: 185–193. 10.1016/j.jconrel.2012.06.027 [DOI] [PubMed] [Google Scholar]
- 27. Bahl J, Ramamurthi N, Gunturi SB. Novel algorithm to select basis functions in spline regression: applications in quantitative structure–activity relationship studies. J Chemom. 2012;26: 85–94. [Google Scholar]
- 28. Shahlaei M. Descriptor selection methods in quantitative structure–activity relationship studies: a review study. Chem Rev. 2013;113: 8093–8103. 10.1021/cr3004339 [DOI] [PubMed] [Google Scholar]
- 29. Mehra R, Nargotra A, Shah BA, Taneja SC, Vishwakarma RA, Koul S. Pro-apoptotic properties of parthenin analogs: a quantitative structure–activity relationship study. Med Chem Res. 2013;22: 2303–2311. [Google Scholar]
- 30. Bouton F, Durand M, Nardello-Rataj V, Borosy AP, Quellet C, Aubry JM. A QSPR Model for the Prediction of the “Fish-Tail” Temperature of CiE4/Water/Polar Hydrocarbon Oil Systems. Langmuir. 2010;26: 7962–7970. 10.1021/la904836m [DOI] [PubMed] [Google Scholar]
- 31. Fagerberg T, Zoete V, Viatte S, Baumgaertner P, Alves PM, Romero, et al. Prediction of cross-recognition of peptide-HLA A2 by melan-a-specific cytotoxic T lymphocytes using three-dimensional quantitative structure-activity relationships. PLoS ONE. 2013;8: e65590 10.1371/journal.pone.0065590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holland JH. Adaptation in natural and artificial systems: An introductory analysis with applications to biology, control, and artificial intelligence Ann Arbor: U Michigan Press; 1975. [Google Scholar]
- 33.Friedman J. Technical report no. 102. Laboratory for computational statistics. Stanford: U Stanford Press; 1990.
- 34. Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci. 1994;34: 854–866. [Google Scholar]
- 35. Friedman JH. Multivariate adaptive regression splines. Ann Stat. 1991;19: 1–67. [DOI] [PubMed] [Google Scholar]
- 36. Yang YQ, Zhao B, Li ZD, Lin WJ, Zhang CY, Guo XD, et al. pH-sensitive micelles self-assembled from multi-arm star triblock co-polymers poly (ε-caprolactone)-b-poly (2-(diethylamino) ethyl methacrylate)-b-poly (poly (ethylene glycol) methyl ether methacrylate) for controlled anticancer drug delivery. Acta Biomater. 2013;9: 7679–7690. 10.1016/j.actbio.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 37. Lin WJ, Nie SY, Zhong Q, Yang YQ, Cai CZ, Wang JF, et al. Amphiphilic miktoarm star copolymer (PCL)3-(PDEAEMA-b-PPEGMA)3 as pH-sensitive micelles in the delivery of anticancer drug. J Mater Chem B Mater Biol Med. 2014;2: 4008–4020. [DOI] [PubMed] [Google Scholar]
- 38. Lin WJ, Nie SY, Xiong D, Guo XD, Wang JF, Zhang LJ. pH-responsive micelles based on (PCL)2(PDEA-b-PPEGMA)2 miktoarm polymer: controlled synthesis, characterization, and application as anticancer drug carrier. Nanoscale Res Lett. 2014;9: 243–254. 10.1186/1556-276X-9-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lei BL, Xi LL, Li JZ, Liu HX, Yao XJ. Global, local and novel consensus quantitative structure-activity relationship studies of 4-(phenylaminomethylene) isoquinoline-1,3 (2H, 4H)-diones as potent inhibitors of the cyclin-dependent kinase 4. Anal Chim Acta. 2009;644:17–24. 10.1016/j.aca.2009.04.019 [DOI] [PubMed] [Google Scholar]
- 40. Alexander G, Alexander T. Beware of q2 . J Mol Graph Model. 2002;20: 269–276. [DOI] [PubMed] [Google Scholar]
- 41. Mhlanga P, Hassan WAW, Hamerton I, Howlin BJ. Using Combined Computational Techniques to Predict the Glass Transition Temperatures of Aromatic Polybenzoxazines. PLoS ONE. 2013;8: e53367 10.1371/journal.pone.0053367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci. 1994;34, 854–866. [Google Scholar]
- 43. Gharagheizi F, Eslamimanesh A, Ilani-Kashkouli P, Mohammadi AH, Richon D. QSPR molecular approach for representation/prediction of very large vapor pressure dataset. Chem Eng Sci. 2012;76: 99–107. [Google Scholar]
- 44. Atkinson AC. Plots, transformations, and regression: an introduction to graphical methods of diagnostic regression analysis Oxford: Clarendon Press; 1985. [Google Scholar]
- 45. Schüürmann G, Ebert RU, Chen J, Wang B, Kühne R. External Validation and Prediction Employing the Predictive Squared Correlation Coefficient—Test Set Activity Mean vs Training Set Activity Mean. J Chem Inf Model. 2008;48: 2140–2145. 10.1021/ci800253u [DOI] [PubMed] [Google Scholar]
- 46. Gramatica P. Principles of QSAR models validation: internal and external. QSAR Comb Sci. 2007;26: 694–701. [Google Scholar]
- 47. Roy K, Kabir H. QSPR with extended topochemical atom (ETA) indices: Modeling of critical micelle concentration of non-ionic surfactants. Chem Eng Sci. 2012;73: 86–98. [Google Scholar]
- 48. Gramatica P. Evaluation of different statistical approaches for the validation of quantitative structure–activity relationships ECVAM, Ispra: 2004;177. [Google Scholar]
- 49. Roy K, Kabir H. QSPR with extended topochemical atom (ETA) indices: Exploring effects of hydrophobicity, branching and electronic parameters on logCMC values of anionic surfactants. Chem Eng Sci. 2013;87: 141–151. [Google Scholar]
- 50. Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H. Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model. 2012;52: 396–408. 10.1021/ci200520g [DOI] [PubMed] [Google Scholar]
- 51. Babel MS, Gupta AD, Pradhan P. A multivariate econometric approach for domestic water demand modeling: an application to Kathmandu, Nepal. Water Resour Manag. 2007;21: 573–589. [Google Scholar]
- 52. Dearden JC, Cronin MTD, Kaiser KLE. How not to develop a quantitative structure–activity or structure–property relationship (QSAR/QSPR). SAR QSAR Environ Res. 2009;20: 241–266. 10.1080/10629360902949567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(XLS)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.