Abstract
Higher rates of alcohol use and other drug-dependence have been observed in some Native American populations relative to other ethnic groups in the U.S. Previous studies have shown that alcohol dehydrogenase (ADH) genes and aldehyde dehydrogenase (ALDH) genes may affect the risk of development of alcohol dependence, and that polymorphisms within these genes may differentially affect risk for the disorder depending on the ethnic group evaluated. We evaluated variations in the ADH and ALDH genes in a large study investigating risk factors for substance use in a Native American population. We assessed ancestry admixture and tested for associations between alcohol-related phenotypes in the genomic regions around the ADH1-7 and ALDH2 and ALDH1A1 genes. Seventy-two (72) ADH variants showed significant evidence of association with a severity level of alcohol drinking-related dependence symptoms phenotype. These significant variants spanned across the entire 7 ADH gene cluster regions. Two significant associations, one in ADH and one in ALDH2, were observed with alcohol dependence diagnosis. Seventeen (17) variants showed significant association with the largest number of alcohol drinks ingested during any 24-hour period. Variants in or near ADH7 were significantly negatively associated with alcohol-related phenotypes, suggesting a potential protective effect of this gene. In addition, our results suggested that a higher degree of Native American ancestry is associated with higher frequencies of potential risk variants and lower frequencies of potential protective variants for alcohol dependence phenotypes.
Keywords: Alcoholism, Low coverage sequencing, Admixture, Alcohol metabolizing enzymes
1 Introduction
Higher rates of alcohol use and other substance dependence have been observed in some Native American populations relative to other ethnic groups in the U.S. (Compton et al., 2007). Although there is variation between tribes, the U.S. Indian Health Service has cited substance dependence as one of the most urgent health problems facing Native Americans (IHS, 1997). The causes of higher rates of alcohol and other substance dependence in the Native Americans are thought to have both environmental and genetic components (Ehlers and Gizer, 2013).
One persisting theory concerning Native American drinking hypothesizes that Native Americans metabolize alcohol differently than other ancestral groups, resulting in physiological consequences that includes a “loss of control” following alcohol consumption and subsequently problem drinking (Leland, 1976). Empirical studies of alcohol drinking in the laboratory, in Native Americans, provide little support for such theories (Garcia-Andrade et al., 1997). However, investigations of potential differences in alcohol metabolism are a logical avenue of research that may be capable of explaining some of the variance in ethnic differences in response to alcohol and in the development of alcohol dependence.
Ethanol is primarily metabolized in the liver and the upper digestive track by alcohol dehydrogenase (ADH) that converts ethanol to acetaldehyde, and aldehyde dehydrogenase (ALDH) that subsequently converts acetaldehyde to acetate. Previous studies have indeed shown that the ADH genes and the ALDH genes may affect the risk of development of alcohol dependence (Bosron et al., 1993; Chen et al., 2009; Edenberg, 2007). ADH and ALDH exist in multiple isozymes that differ in their kinetic properties. Polymorphisms within the genes that encode for the isozymes vary in their allele frequencies between ethnic groups, and thus may differentially affect risk depending on the ethnic group evaluated. For instance, the ALDH2*2 allele, which is partially responsible for the alcohol-induced flushing reaction, is common in East Asian but rare in other ethnic group including NA (Shen et al., 1997; Wall et al., 1992; Rex et al., 1985).
Variants in ADH1B and ADH1C may also influence drinking behavior. For example, the ADH1B*2 (rs1229984) and ADH1B*3 (rs2066702) alleles each result in an amino acid change producing a more efficient enzyme that allows for the more rapid accumulation of acetaldehyde (Carr et al., 1989; Hurley et al., 1990). As a result, both polymorphisms exhibit a protective relation with alcohol dependence and related phenotypes (e.g., ADH1B*2 - MacGregor et al., 2009; Thomasson et al., 1991, 1994; and ADH1B*3 - Edenberg et al., 2006; Ehlers et al., 2007; McCarthy et al., 2010; Wall et al., 2003). Notably, both associations were recently confirmed in a large scale GWAS meta-analysis of alcohol dependence (Gelernter et al., 2014). Candidate gene studies have also reported significant associations between alcohol dependence and other ADH genes, most prominently ADH1C (Higuchi et al., 1995; Kuo et al., 2008) and ADH4 (Edenberg et al., 2006; MacGregor et al., 2009). Rare variants in the ADH gene cluster have also been found that are significantly associated with alcohol dependence in European-American, European-Australian and African-American populations (Zuo et al., 2013).
Allele distributions in Navajo and Sioux are similar to Euro-American but not to Japanese and African-Americans (Bosron et al., 1988; Rex et al., 1985). Ehlers et al. (2012) has found that variants in ADH1B may be protective against alcoholism in Native American and Mexican American populations. Gizer et al. (2011) reported a significant association between an ADH4 variant and alcohol dependence in the same Native American sample, and Mulligan et al. (2003) reported associations between ADH1C variants and alcohol dependence in an independent sample of Native Americans. Importantly, however, these findings cannot explain the high prevalence of alcoholism in Native American populations, given that none of the associated variants are private to Native American ancestral groups.
One obstacle to the discovery of genes associated with complex phenotypes, such as alcohol dependence, has been the inability to conduct comprehensive genome wide sequencing (GWS) of population samples and to develop statistical models that incorporate such variables as family relatedness and ethnic admixture. Recent advances in GWS techniques and analytical methods have made possible the identification of both rare and common variants in family studies of novel populations enriched for substance dependence phenotypes, such as Native Americans. The present report is part of a larger study exploring risk factors for substance dependence in an American Indian community in the west (Ehlers et al., 2004a, 2008; Gilder et al., 2004, 2006, 2007, 2008). DNA obtained from this community sample has recently been sequenced using low coverage whole genome sequencing (WGS) (Bizon et al., 2014). Utilizing these data, the aims of the present study were twofold. The first aim was to investigate the relations between alcohol dependence and genetic variants in the genes involved in alcohol metabolism within the Native American sample population. The seven alcohol dehydrogenase genes (ADH7-ADH1C-ADH1B-ADH1A-ADH6-ADH4-ADH5) are in a single cluster region on chromosome 4q21-24 with each gene coding for a unique isozyme. The two aldehyde dehydrogenase genes involved in alcohol metabolism are ALDH1A, located on chromosome 9q21.13, and ALDH2, located on chromosome 12q24.2 (Ehlers and Gizer, 2013).
The second aim was to leverage the significant admixture and family relatedness to further explore the relations between these genes and alcohol dependence. To accomplish this aim we assessed ancestry admixtures and used a variance component approach (Schork, 1992) to test for associations between alcohol-related phenotypes in the genomic regions around the ADH and ALDH genes. Additionally, we examined the ancestry background in the genomic regions of interest and their influence on the phenotypes.
2 Materials and Methods
2.1 Participants and phenotypes
The protocol of the study was approved by the Scripps Institutional Internal Review Board and Indian Health Council, a tribal review group overseeing health issues for the reservations where recruitments took place. Written informed consent was obtained from each participant after study procedures had been fully explained. Participants were compensated for their time spent in the study.
Seven hundred and eight (708) Native Americans, from extended pedigrees, participated in the study. The characteristics of this population have been previously described (Ehlers et al., 2011). Participants who had at least one-sixteenth self-reported American Indian heritage were targeted and recruited for the study as previously described (Ehlers et al., 2004a). All subjects were assessed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) in order to collect demographic information and to make DSM-IV diagnoses (American Psychiatric Association Task Force on DSM-IV, 1994). The SSAGA is a polydiagnostic psychiatric interview that has undergone both reliability and validity testing and been successfully used in Native American populations (Bucholz et al., 1994; Hesselbrock et al., 1999; Wall et al., 2003). Six hundred and ninety seven sequenced samples (n = 697) were eventually used in the association studies. The average age of the analyzed samples was 31.3 years old (range 18-82, std=13.2 yrs), with 42.8% being male. The average years of education was 11.57 (std = 1.56). 42.4% of participants (n = 295) had at least 50% self-reported Native American heritage as indicated from their federal Indian blood quantum.
Tested phenotypes included: (i) alcohol dependence diagnosis defined by DSM-IV, (ii) the number of DSM-IV alcohol dependence symptoms related to drinking ranging from 0 to 4, e.g. 1) drank more than intended/more days in a row or when promised self wouldn't for three or more times; 2) drunk when didn't want to three or more times; 3) during drinking or recovering from the effects of drinking had little time for anything else; and 4) given up or greatly reduced important activities to drink (Ehlers et al., 2004b), (iii) the number of DSM-IV alcohol withdrawal symptoms ranging from 0 to 4 (Ehlers et al., 2004b), e.g. 1) the shakes (trembling of the hands), unable to sleep, anxiety or depression, sweating, rapid heart rate, nausea or vomiting, feeling physically weak, headache, auditory or visual hallucinations; 2) seizures; and 3) delirium tremens (DTs), and (iv) the largest number of alcohol drinks (max drinks) ever consumed in a 24-hour period. These phenotypes were chosen as previous analyses had demonstrated that they had a significant genetic component in this population (Ehlers et al., 2004b; Ehlers and Gizer, 2013). Among participants, 46% (n=320) had an alcohol dependence diagnosis; 60% (n=420) reported experiencing at least one alcohol dependence drinking symptom; 20% (n=142) reported experiencing at least one withdrawal symptom. The distribution of the reported largest number of drinks ever consumed in a 24-hour period is particularly skewed with a long right tail: median = 20 drinks, range = [0.5,166], skewness = 2.08 and Pearson's measure of kurtosis = 8.55.
2.2 Sequencing
Blood derived DNA was sequenced using Illumina low-coverage whole genome sequencing (LCWGS), as well as genotyped using an Affymetrix Exome1A chip. The pair-end sequencing was performed on HiSeq2000 sequencers (Illumina). About 80% of the samples have coverage between 3X and 12X, approximately evenly distributed. Reads from whole genome sequencing were aligned using BMA, and realigned near indels with GATK. Variants were called using both GATK Unified Genotyper following the best-practices for low-coverage samples (DePristo et al., 2011) and the LD-aware variant caller Thunder (Li et al., 2011). Imputation was done using the program Thunder. Qualities of variant calling were assessed through a comparison between the sequencing results to genotypes generated on the exome array for the same set of subjects (Bizon et al., 2014).
2.3 Candidate gene regions
We tested for associations between alcohol dependence, alcohol dependence drinking symptoms, alcohol withdrawal symptoms, and the maximum number of alcohol drinks consumed in any 24-hour period in the genomic regions around the ADH and ALDH genes, including 10k basepairs upstream and downstream from each of the genetic regions. Given the sample size (n = 697), variants of less than 0.01 minor allele frequency (MAF) were excluded. The ADH gene cluster region (ADH7-ADH1C-ADH1B-ADH1A-ADH6-ADH4-ADH5) is on chromosome 4: 99,982,130–100,366,894. The region had 3302 SNPs called, of which 1679 had MAF ≥ 0.01. The ALDH1A1 gene is on chromosome 9: 75,505,578–75,705,358. It had 1466 SNPs, of which 397 had MAF ≥ 0.01. The ALDH2 gene is on chromosome 12 : 112,194,346–112,257,789. It had 328 SNPs, of which only 83 had MAF ≥ 0.01.
2.4 Ancestry estimates
To assess ancestry and admixture proportions in the samples, we used a supervised clustering approach implemented in the ADMIXTURE software (Alexander et al., 2009) and clustered subject data into four clusters corresponding to four major continental populations: African, East Asian, European, and Native American. These populations were defined by the individuals contained in a reference panel containing genotype information at about 300k strand-unambiguous SNPs. The admixture estimates were further refined through a noise reduction approach via bootstrapping (Libiger and Schork, 2012).
In addition to global ancestries estimated for each individual with SNPs spanning entire genomes, we also estimated local ancestry admixture. A sliding window of 200 consecutive SNPs in the reference panel moved across each chromosome at the step of 50 SNPs. Local ancestry and admixture proportions were estimated using SNPs within each window for all samples. Local ancestries for each candidate gene region were taken to be the ancestry estimates for the window on which the region is centered. Each candidate region being investigated fell within a single such window. We decided on a window of a fixed-width with respect to the number of ancestry-informative markers rather than using only the markers within a candidate gene region as too few ancestry-informative markers are insufficient to yield meaningful admixture estimates.
Figure 1 shows the distributions of the global Native American ancestries estimated using the whole genome, and the local Native American ancestries using SNPs around candidate regions. The correlation coefficients between estimated local Native American ancestry and global Native American ancestry of all samples were 0.55, 0.49 and 0.59 for regions around ADH cluster (Figure 1B), ALDH1A1, and ALDH2, respectively. That the local and global ancestry estimates were positively correlated is expected; that the correlation was moderate yet not strong justifies the inclusion of local ancestry estimates in the analysis in addition to the global ancestry estimates.
Figure 1.
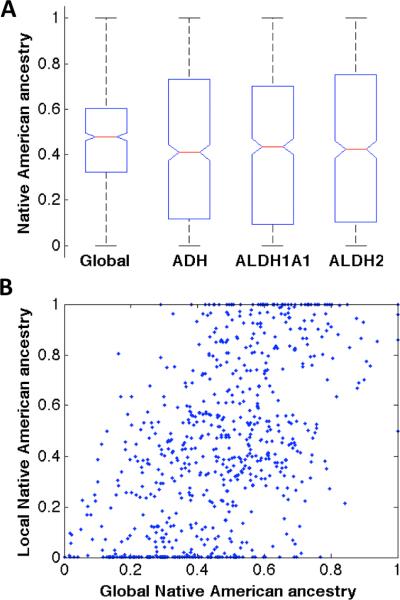
Global and local Native American ancestry estimates. (A) Native American (NA) ancestry distribution in the samples. Global ancestry was estimated using variants distributed across the whole genome; local ancestry was estimated using variants in and around each of the candidate regions (ADH, ALDH1A1, ALDH2). (B) Local NA ancestry of the ADH region plotted against global NA ancestry of all samples, each point representing an individual (Pearson's r = 0.55).
2.5 Association analysis
Many of the samples had a mixed ethnic background as indicated by self-report and as shown by admixture analysis from genotypes (Figure 1A). The samples were composed of 161 families of variable sizes. It has been established that both population substructure (Helgason et al., 2005) and genetic relatedness (Weir et al., 2006) may cause inflations in test statistics and potentially spurious associations. To control for admixture and familial relatedness, a variance component approach as implemented in EMMAX (Kang et al., 2010) was used in the association tests, with global and local ancestry admixture estimates as described in section 2.4 further incorporated as covariates. The local ancestry estimates were moderately correlated with the global ancestry estimates for the genomic regions concerned. Additional covariates included gender, age, and age-squared. To obtain correct distributions of the test statistic from EMMAX under a null hypothesis, we used a permutation procedure that randomly shuffled the phenotype values of the samples tested. One million permutations were performed. False discovery rates (FDR) controlled by Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995) were used to set significant p values from the test statistics of the association test. We report the FDR-adjusted p values (Yekutieli and Benjamini, 1999).
We further examined the ancestral backgrounds of the genomic regions under investigation, and stratified the samples by their degree of NA ancestry and phenotype values to determine whether a variant that is significantly associated with one or more alcohol-related phenotypes has a larger impact on those with higher NA heritage.
3 Results
Seventy-two (72) variants showed significant evidence of association with the alcohol dependence drinking symptoms phenotype (Table I: ph=S). These significant variants spanned the entire seven ADH gene cluster. The majority of them however resided in ADH4-ADH6-ADH1AADH1B, in both genic and intergenic regions. Interestingly, most of these variants reside in non-coding regions. All but one of the intergenic variants are in strong linkage disequilibrium (LD) with at least one of the ADH genic variants (r2 > 0.8). Three of these variants are novel and not found in dbSNP (National Center for Biotechnology Information, 2013). The alternative allele frequencies of the sixty-nine (69) variants that were positively associated with the phenotype were significantly higher in our samples than those in the 1000 Genome Project (1000 Genomes Project Consortium et al., 2012), while the three negatively associated variants had slightly lower allele frequencies than those in the 1000 Genome Project (data not shown).
Table I.
Variants significantly associated with alcohol dependence phenotypes.
| Position | R | A | dbSNP id | Gene | Location | p FDR | ph | MAF | Hh | Hl | Lh | Ll |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100017594 | G | A | rs145341314 | ADH5/4 | Intergenic | 0.0026 | S | 0.323 | 0.700 | 0.573 | 0.094 | 0.069 |
| 100036065 | T | C | - | ADH5/4 | Intergenic | 0.0026 | S | 0.294 | 0.667 | 0.565 | 0.063 | 0.055 |
| 100037140 | T | C | rs187939013 | ADH5/4 | Intergenic | 0.0410 | S | 0.336 | 0.667 | 0.573 | 0.094 | 0.123 |
| 100051161 | C | A | rs7692974 | ADH4 | Intron7/6 | 0.0234 | S | 0.334 | 0.700 | 0.565 | 0.094 | 0.123 |
| 100057074 | G | A | - | ADH4 | Intron6/5 | 0.0029 | S | 0.028 | 0.100 | 0.015 | 0.000 | 0.000 |
| 100058770 | C | A | rs10026860 | ADH4 | Intron5/4 | 0.0246 | S | 0.335 | 0.700 | 0.565 | 0.094 | 0.123 |
| 100061531 | C | T | rs6840413 | ADH4 | Intron4/3 | 0.0234 | S | 0.334 | 0.700 | 0.565 | 0.094 | 0.123 |
| 100062338 | C | T | rs35882815 | ADH4 | Intron4/3 | 0.0226 | S | 0.335 | 0.700 | 0.565 | 0.094 | 0.123 |
| 100066287 | C | T | rs4148884 | ADH4/6 | Intergenic | 0.0175 | S | 0.326 | 0.700 | 0.565 | 0.063 | 0.103 |
| 100067337 | A | G | rs2226896 | ADH4/6 | Intergenic | 0.0026 | S | 0.306 | 0.700 | 0.551 | 0.063 | 0.048 |
| 100068799 | T | A | - | ADH4/6 | Intergenic | 0.0082 | S | 0.278 | 0.667 | 0.507 | 0.063 | 0.041 |
| 100069458 | A | G | rs10022174 | ADH4/6 | Intergenic | 0.0125 | S | 0.326 | 0.700 | 0.558 | 0.063 | 0.103 |
| 100070816 | A | G | rs12649136 | ADH4/6 | Intergenic | 0.0026 | S | 0.306 | 0.700 | 0.551 | 0.063 | 0.048 |
| 100072567 | C | T | rs2156731 | ADH4/6 | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100075175 | C | T | rs57058570 | ADH4/6 | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100076962 | T | C | rs72679827 | ADH4/6 | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100085463 | C | T | rs61681818 | ADH4/6 | Intergenic | 0.0026 | S | 0.311 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100088567 | T | C | rs12499210 | ADH4/6 | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100094364 | C | T | rs36139400 | ADH4/6 | Intergenic | 0.0026 | S | 0.311 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100102320 | C | G | rs12504491 | ADH4/6 | Intergenic | 0.0026 | S | 0.311 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100104829 | C | T | rs55926943 | ADH4/6 | Intergenic | 0.0026 | S | 0.310 | 0.700 | 0.565 | 0.063 | 0.048 |
| 100108104 | C | T | rs58964688 | ADH4/6 | Intergenic | 0.0029 | S | 0.311 | 0.700 | 0.565 | 0.063 | 0.062 |
| 100115380 | C | T | rs138293424 | ADH4/6 | Intergenic | 0.0026 | S | 0.311 | 0.700 | 0.558 | 0.063 | 0.055 |
| 100116074 | G | A | rs12502056 | ADH4/6 | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.551 | 0.063 | 0.048 |
| 100120992 | A | T | rs12499710 | ADH4/6 | Intergenic | 0.0026 | S | 0.310 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100125868 | G | A | rs12507078 | ADH6 | Intron8 | 0.0026 | S | 0.309 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100138736 | A | C | rs7692358 | ADH6 | Intron1 | 0.0088 | S | 0.316 | 0.700 | 0.565 | 0.063 | 0.075 |
| 100139366 | C | T | rs4699734 | ADH6 | Intron1 | 0.0026 | S | 0.308 | 0.700 | 0.558 | 0.063 | 0.055 |
| 100145441 | G | A | rs7375388 | ADH6/1A | Intergenic | 0.0026 | S | 0.318 | 0.683 | 0.565 | 0.094 | 0.055 |
| 100145719 | G | A | rs55641355/rs7375429 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.551 | 0.063 | 0.055 |
| 100147453 | G | A | rs58584512 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100148946 | C | T | rs143770077 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100151262 | T | G | rs7669335 | ADH6/1A | Intergenic | 0.0030 | S | 0.311 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100152009 | T | G | rs148671455 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100152864* | G | A | rs6813954 | ADH6/1A | Intergenic | 0.0458 | M | 0.370 | 0.011 | 0.065 | 0.641 | 0.796 |
| 100157010 | A | G | rs58660705 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100157132 | G | C | rs60468511 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.558 | 0.063 | 0.055 |
| 100157669 | C | T | rs7654607 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100157771 | G | T | rs60345995 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.551 | 0.063 | 0.055 |
| 100161162 | T | C | rs12501204 | ADH6/1A | Intergenic | 0.0026 | S | 0.310 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100164269 | T | C | rs149215710 | ADH6/1A | Intergenic | 0.0036 | S | 0.310 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100168980 | C | G | rs4699381 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100169797 | G | C | rs72679867 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100171239 | C | T | rs2010612 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100173158 | C | T | rs12504443 | ADH6/1A | Intergenic | 0.0026 | S | 0.308 | 0.700 | 0.558 | 0.063 | 0.048 |
| 100181911 | G | A | rs113289860 | ADH6/1A | Intergenic | 0.0026 | S | 0.310 | 0.700 | 0.551 | 0.063 | 0.055 |
| 100181934 | T | C | rs72679874 | ADH6/1A | Intergenic | 0.0026 | S | 0.309 | 0.700 | 0.551 | 0.063 | 0.055 |
| 100183973 | A | G | rs7663369 | ADH6/1A | Intergenic | 0.0026 | S | 0.313 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100185431 | G | A | rs12506882 | ADH6/1A | Intergenic | 0.0026 | S | 0.312 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100186063 | C | T | rs58827689 | ADH6/1A | Intergenic | 0.0026 | S | 0.312 | 0.700 | 0.565 | 0.063 | 0.055 |
| 100188396 | T | C | rs17028765 | ADH6/1A | Intergenic | 0.0329 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100195815 | G | T | rs12512110 | ADH6/1A | Intergenic | 0.0326 | S | 0.295 | 0.667 | 0.536 | 0.063 | 0.055 |
| 100200153 | C | T | rs28364333 | ADH1A | Intron8 | 0.0329 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100202705 | G | T | rs12504321 | ADH1A | Intron6 | 0.0270 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100203037 | C | T | rs12504365 | ADH1A | Intron6/3UTR | 0.0428 | S | 0.297 | 0.667 | 0.544 | 0.063 | 0.062 |
| 100203156 | A | G | rs12512053 | ADH1A | Intron6/3UTR | 0.0439 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100203176 | T | C | rs12508502 | ADH1A | Intron6/3UTR | 0.0439 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100203447 | A | C | rs2276332 | ADH1A | Intron6/Exon2 | 0.0432 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100206912 | T | C | rs28364304 | ADH1A | Intron3 | 0.0326 | S | 0.296 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100211399 | T | C | rs3805325 | ADH1A | Intron1 | 0.0269 | S | 0.297 | 0.667 | 0.544 | 0.063 | 0.055 |
| 100215587 | C | T | rs72679893 | ADH1A | Intron1 | 0.0134 | S | 0.302 | 0.667 | 0.536 | 0.094 | 0.055 |
| 100219582 | G | A | rs12505816 | ADH1A | Intron1 | 0.0141 | S | 0.300 | 0.667 | 0.529 | 0.094 | 0.055 |
| 100223957 | A | C | rs111797757 | ADH1A | Intron1 | 0.0134 | S | 0.301 | 0.667 | 0.536 | 0.094 | 0.055 |
| 100225336 | G | A | rs56948765 | ADH1A | Intron1 | 0.0192 | S | 0.303 | 0.667 | 0.544 | 0.094 | 0.055 |
| 100226777 | A | C | rs73832758 | ADH1A | Intron1 | 0.0193 | S | 0.302 | 0.667 | 0.536 | 0.094 | 0.055 |
| 100227288 | G | C | rs28914794 | ADH1A | Intron1 | 0.0192 | S | 0.302 | 0.667 | 0.544 | 0.094 | 0.055 |
| 100228945 | T | C | rs17033 | ADH1B | 3UTR | 0.0192 | S | 0.299 | 0.667 | 0.529 | 0.094 | 0.055 |
| 100233126 | C | T | rs28914775 | ADH1B | Intron6/7 | 0.0193 | S | 0.301 | 0.667 | 0.536 | 0.094 | 0.055 |
| 100233892 | C | T | rs28914770 | ADH1B | Intron6/7 | 0.0175 | S | 0.302 | 0.667 | 0.544 | 0.094 | 0.055 |
| 100253409 | C | T | rs1497372 | ADH1C | Downstream | 0.0185 | D | 0.230 | 0.335 | 0.214 | 0.254 | 0.104 |
| 0.0038 | S | 0.230 | 0.450 | 0.203 | 0.250 | 0.116 | ||||||
| 100294612 | A | C | rs1846039 | ADH1C | Upstream | 0.0491 | M | 0.459 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100296248 | C | T | rs10030511 | ADH1C | Upstream | 0.0491 | M | 0.459 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100296684 | T | C | rs35208813 | ADH1C | Upstream | 0.0491 | M | 0.460 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100296748 | T | C | rs35796885 | ADH1C | Upstream | 0.0491 | M | 0.460 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100299450 | G | A | rs9997820 | ADH1C | Upstream | 0.0491 | M | 0.459 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100299641 | C | A | rs1908962 | ADH1C | Upstream | 0.0458 | M | 0.490 | 0.565 | 0.548 | 0.531 | 0.352 |
| 100299978 | T | C | rs1837932 | ADH1C | Upstream | 0.0491 | M | 0.459 | 0.587 | 0.565 | 0.609 | 0.432 |
| 100304921* | C | T | rs2654841 | ADH7 | Downstream | 0.0491 | M | 0.459 | 0.413 | 0.436 | 0.391 | 0.568 |
| 100306839* | G | A | rs2654842 | ADH7 | Downstream | 0.0475 | M | 0.462 | 0.413 | 0.436 | 0.391 | 0.568 |
| 100310907* | C | T | rs284782 | ADH7 | Downstream | 0.0458 | M | 0.431 | 0.413 | 0.419 | 0.328 | 0.500 |
| 100312545* | G | T | rs2654843 | ADH7 | Downstream | 0.0458 | M | 0.464 | 0.413 | 0.436 | 0.391 | 0.568 |
| 100313611* | G | A | rs284783 | ADH7 | Downstream | 0.0458 | M | 0.432 | 0.413 | 0.419 | 0.328 | 0.500 |
| 100320597* | A | G | rs2851015 | ADH7 | Downstream | 0.0458 | M | 0.463 | 0.413 | 0.436 | 0.391 | 0.580 |
| 100322445* | T | C | rs991316 | ADH7 | Downstream | 0.0458 | M | 0.370 | 0.250 | 0.307 | 0.375 | 0.568 |
| 100322497* | C | G | rs1442491 | ADH7 | Downstream | 0.0458 | M | 0.370 | 0.250 | 0.307 | 0.375 | 0.568 |
| 100322498* | C | A | rs991315 | ADH7 | Downstream | 0.0458 | M | 0.370 | 0.250 | 0.307 | 0.375 | 0.568 |
| 100326449* | T | C | rs72681947 | ADH7 | Downstream | 0.0326 | S | 0.094 | 0.000 | 0.015 | 0.125 | 0.171 |
| 100327104* | T | A | rs72681948 | ADH7 | Downstream | 0.0326 | S | 0.094 | 0.000 | 0.015 | 0.125 | 0.171 |
| 100334449* | T | A | rs17588403 | ADH7 | Intron8 | 0.0270 | S | 0.095 | 0.000 | 0.022 | 0.125 | 0.171 |
| 112241720+ | G | A | rs190914158 | ALDH2 | Exon11/12 | 0.0090 | D | 0.023 | 0.079 | 0.012 | 0.007 | 0.000 |
R: reference allele. A: alternative allele. pFDR: FDR-adjusted p-value.
ph: phenotype (S: dependence drinking symptoms; D: dependence diagnosis; M: max-drink).
MAF: alternative allele frequency.
Hh, Hl,Lh,Ll: alternative allele frequency of subset samples (H/L: high/low NA heritage, h/l: high/low values of respective phenotype).
Negative association.
Variant is on chromosome 12; all other are on chromosome 4.
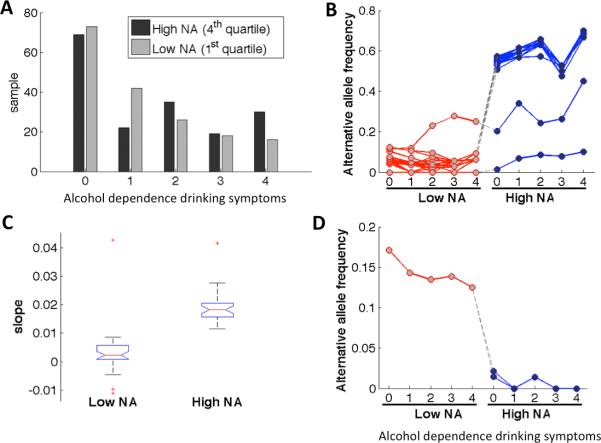
To further understand the relation between the significant variants and the Native American ancestry and their potential impacts on the phenotype, we stratified the samples by their degrees of local NA ancestry and their levels of severity, i.e., the number, of alcohol dependence drinking symptoms. Figure 2A shows a histogram of samples in the first and the fourth quartiles of NA ancestries around the ADH regions. The first quartile by degree of estimated local NA ancestry is considered as having a low degree (≤ 12%) of NA heritage, while the fourth quartile a high degree (≥ 73%). The number of alcohol dependence drinking symptoms ranged from 0 to 4. For each of the significant variants, we obtained its alternative allele frequencies in each of the sample subsets: samples of low or high NA heritages at various levels of dependence drinking symptoms (Figure 2B). Alternative allele frequencies in samples of high (H) or low (L) NA heritages and having 4 (h) or 0 (l) dependence drinking symptoms are also listed in the last four columns (Hh, Hl, Lh, Ll) of Table I. Note that for variants positively associated with the dependence drinking phenotype, the samples of high NA heritage had higher alternative allele frequencies than those of low NA heritage, suggesting that these variants were more prevalent in the Native Americans. Further, in the samples of high NA heritage, those having the largest number of dependence drinking symptoms (Hh) had higher allele frequencies than those having no drinking symptoms (Hl). In contrast, the three ADH7 variants that were negatively associated with this phenotype seemed more prevalent in the samples of low NA heritage and became rare in those of high NA heritage (Figure 2D). For each variant, we also regressed allele frequency on the dependence drinking variable for samples of low or high NA heritages respectively. Boxplots of the slopes of the linear regression lines are shown in Figure 2C for the 69 variants that were positively associated with the phenotype. The slopes for samples of high NA heritage were all positive and significantly higher than those of low NA heritage (p-value < 10−4), suggesting that they may represent risk factors for dependence drinking symptoms in the NA population, particularly of those with higher NA heritage.
Figure 2.
Samples stratified by degree of local NA ancestry and the severity level of alcohol dependence drinking symptoms. (A) Alcohol dependence drinking symptoms of samples with low and high NA heritages. (B/D) Alternative allele frequencies of 69/3 significant variants positively/negatively associated with dependence drinking in each sample subset. Each line represents a variant. (C) Linear regression of allele frequencies of significant variants (positively associated with dependence drinking) in samples of low and high NA heritages onto the number of dependence drinking symptoms.
One variant in ADH1C and one variant in ALDH2 were found to be significantly positively associated with DSM-IV alcohol dependence (Table I: ph=D). The last four columns of Table I list the alternative allele frequencies of these two variants in the subsets of samples having high NA heritage and being diagnosed with or without alcohol dependence (Hh, Hl), or having low NA heritage and with or without alcohol dependence (Lh, Ll). Of note, SNP rs1497372 (4:100253409) of ADH1C appeared to be significantly associated with both the severity level of dependence drinking symptoms (S) and alcohol dependence (D) phenotypes. The other significant variant, rs190914158 in ALDH2, had a low allele count in the sampled population (see last row in Table I). It appeared even rarer in the 1000 Genomes Project with a minor allele frequency of 0.0009, corresponding to two alleles out of 1092 total genomes. In contrast, we found that thirty (30) subjects in our 697 analyzed samples were heterozygous and one homozygous for this variant. Figure 3A shows the distribution of alcohol dependence in samples with high or low local NA ancestry. In the samples of low NA, there was only one heterozygous subject on variant rs190914158, who was alcohol dependent. In the samples of high NA heritage, there were the one homozygous who was alcohol dependent, and 15 heterozygous samples, 13 of whom were alcohol dependent, and two not dependent.
Figure 3.
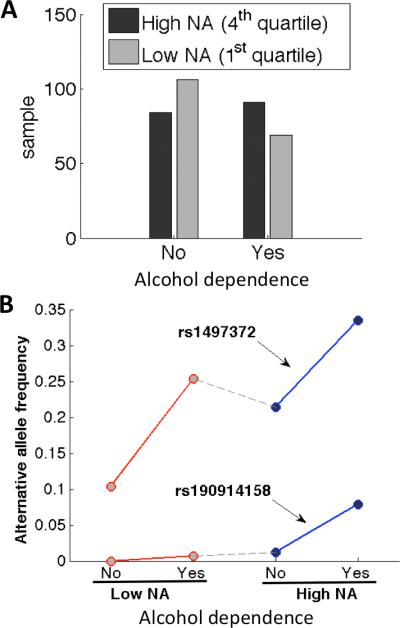
Samples stratified by degree of local NA ancestry and the alcohol dependence diagnosis. (A) Alcohol dependence of samples with low and high NA heritages. (B) Alternative allele frequencies of 2 significant variants positively associated with alcohol dependence in each sample subset.
Seventeen (17) variants showed significant association with the largest number of alcoholic drinks consumed in a 24-hour period (Table I: ph=M), of which ten (10) variants were negatively associated with the phenotype. Most of the associated variants were in the intergenic region between ADH1C and ADH7, and nine of the ten negatively associated variants were downstream of ADH7. The majority of variants were in weak LD with variants in either ADH1C or ADH1B with r2's between 0.15 and 0.20, with the exception of rs6813954 which was in LD with variants in ADH1A (r2=0.41). To examine the potential contribution of ancestry to the phenotype, we again stratified the samples by their degrees of local Native American ancestry and their phenotype magnitudes. Because the maximum number of drinks consumed in a 24-hour period was not categorical, we binned the samples into five roughly equal-sized groups. For each bin, the samples were further split by the quartiles of the degree of local NA ancestry. Figure 4A shows a histogram of samples in the first (low degree) and the fourth (high degree) quartiles of NA ancestry of the binned samples. The last four columns of Table I list the alternative allele frequencies of these variants in the subset samples having high (H) or low (L) NA heritage and in the two extreme (l: ≤ 10 or h: ≥ 40) bins. Figure 4B shows the allele frequencies of the variants that were negatively associated with the max drink phenotype in each of the sample subsets. Figure 4C shows the allele counts of the samples in the two extreme bins, maximum drink ≤ 10 and ≥ 40, for one of the negatively associated variant rs6813954 located between ADH6 and ADH1A. The allele counts in each of the categories were strikingly different. Normalized allele frequencies of the samples with high degree of NA heritage were 0.0109 for the subset of samples having no fewer than 40 max drinks or 0.0645 for the subset of samples having no more than 10 max drinks respectively, and 0.6406 and 0.7955 respectively for the samples of low degree NA. The allele frequency of this variant is 0.6648 in the 1000 Genomes Project. The ADH7 variants exhibited similar but less striking differences (Figure 4B).
Figure 4.
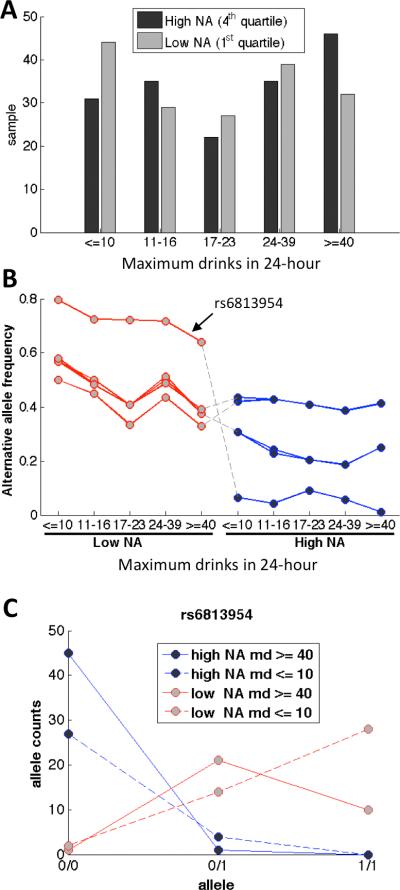
Samples stratified by degree of local NA ancestry and the maximum drinks (md) ever consumed in a 24-hour period. (A) Maximum drinks in 24-hour of samples with low and high NA heritages. (B) Alternative allele frequencies of 10 significant variants negatively associated with max drink phenotype in each sample subset. (C) Allele counts of a potentially protective variant in each sample subset (md = maximum drinks).
4 Discussions
The primary aim of the present study was to investigate the relations between alcohol dependence and genetic variants in the seven alcohol dehydrogenase genes (ADH7-ADH1C-ADH1B-ADH1A-ADH6-ADH4-ADH5) and two aldehyde dehydrogenase genes (ALDH1A and ALDH2) responsible for encoding the primary enzymes responsible for alcohol metabolism. In addition to DSM-IV-defined alcohol dependence, we conducted association analyses with a count of dependence drinking symptoms phenotype, a count of alcohol withdrawal symptoms, and a lifetime estimate of the largest number of alcoholic drinks consumed in a 24-hour period.
Ninety (90) variants showed significant evidence of association with one of the phenotypes (72 ADH variants for alcohol dependence drinking symptoms; 17 for the largest number of alcoholic drinks consumed in a 24-hour period; rs1497372 of ADH1C and rs190914158 of ALDH2 for alcohol dependence). Many of the significant variants are in non-coding regions, suggesting that they might be potentially involved in regulatory functions. No variant was significantly associated with alcohol withdrawal symptoms. Of note, most of the loci showed associations related to increased risk for the measured alcohol use phenotypes, though a subset of 13 loci showed protective effects against the development of these phenotypes. One of these variants, rs6813954, is located between ADH6 and ADH1A with the remaining 12 located in or near ADH7, suggesting a potential protective effect of variants within this gene. Of the seven ADH genes, ADH7 codes for the enzyme with the highest maximal activity for ethanol, and it is expressed in stomach rather than liver. Previous studies have suggested that ADH7 plays a protective role against alcoholism (although the protect variant reported was an intronic SNP), potentially through an epistatic interaction with another variant in the region, and that its effect cannot simply be explained by LD with other nearby ADH gene variants (Han et al., 2005; Osier et al., 2004).
Several genome-wide association studies and subsequent replication studies have also supported the involvement of the ADH gene cluster in the development of alcohol use phenotypes (e.g., Frank et al., 2012; Gelernter et al., 2013; Park et al., 2013). The present study provides further support for this association; however, there has been a lack of consistency across studies in terms of the specific genes within the ADH cluster and the specific SNPs within those genes that show the strongest evidence for association. A likely explanation for this lack of consistency is the extent of LD in the region spanning multiple ADH genes, thus making it difficult to identify the causal variants.
Given that an aim of the present study was to examine genetic risk factors for alcohol use phenotypes that might be specific to Native American populations, we identified three variants that had not been reported in dbSNP and may be novel to the Native American population under study. These variants were all located in intronic regions and thus, their potential impact on ADH gene function and expression is difficult to determine. The variant in ALDH2, rs190914158, that was associated with alcohol dependence showed a 25-fold increase in prevalence of its minor allele in our NA samples relative to the populations sampled in the 1000 Genomes Project. The variant is a synonymous SNP that is not predicted to be damaging by Poly-Phen2 or SIFT, making it difficult to draw strong conclusions regarding its role in the development of alcohol use phenotypes.
Although the present report did not identify variants specific to the Native American population that could be considered causal, the findings of the present study did suggest that there appear to be elevated frequencies of alleles associated with increased risk for alcohol use phenotypes and reduced frequencies of alleles associated with protection against alcohol use phenotypes. Specifically, we observed that a higher degree of NA ancestry was associated with higher level of alcohol dependence drinking, and further, that variants associated with alcohol dependence drinking symptoms were more prevalent among samples showing higher degrees of NA ancestry at the ADH gene cluster compared to samples of lower degrees of NA ancestry in this region. Importantly, this finding was significant even after controlling for genome-wide measures of ancestry, suggesting that genetic liability for alcohol dependence phenotypes is increased in this region relative to other ancestral groups.
In addition to informing molecular genetic studies of alcohol use studies in Native Americans, these results have broader practical implications for sequencing studies being conducted in admixed populations. The NA community samples under investigation in this report presented a unique analytic challenge because of underlying population substructure and complex, extended pedigree structures. To address this, we used a mixed model approach as implemented in EMMAX given that mixed models are thought to be a practical and comprehensive approach that simultaneously addresses confounding effects resulting from population stratification, family structure and cryptic relatedness (Price et al., 2010). We supplemented this approach, which considers these factors at the genome-wide level, with a unique set of analyses that included local ancestry information to evaluate whether differences in origin of a specific chromosomal region could be associated with alcohol use phenotypes. As additional admixed populations are studied using sequencing technologies, we believe this approach may provide further insights into the genetic architecture of complex disease across different ancestral groups.
Despite the potential impact of these findings, some limitations should be noted. First, many SNPs within the candidate gene region showed high levels of LD with one another. As a result, treating each test as independent when correcting for multiple comparisons, as was done in the present report using the Benjamini–Hochberg FDR, may have been overly conservative. While this approach provided strong protections against Type I errors, we believe that a less conservative approach that estimates the number of independent tests as a function of LD and provides a critical p-value based on this calculation as suggested by (Nyholt, 2004) and (Li et al., 2012) may have yielded more statistical power for the present analyses. Second, we examined only a couple of genomic regions in the present report. Given that alcohol dependence is a complex disorder and its genetic component is likely resulting from polygenic effects (Whitfield et al., 1998), we intend to extend our investigations to the complete genome in future studies, including investigations into both common and relatively rare variants.
In summary, the present study suggests that low-coverage WGS combined with ancestry and admixture analyses can identify significant variants associated with alcohol dependence phenotypes in the regions of the major alcohol metabolizing enzymes. Our results taken together suggest that a higher degree of Native American ancestry is associated with higher frequencies of potential risk variants and lower frequencies of potential protective variants for alcohol dependence phenotypes in these gene regions. Incorporating information concerning a persons ADH gene profile may be useful information to provide to members of this population in prevention programs that are targeting high-risk drinking behaviors.
Acknowledgments
Funding for this study was provided by grants from the National Institutes of Health (NIH); from the National Institute on Alcoholism and Alcohol Abuse (NIAAA) and the National Center on Minority Health and Health Disparities (NCMHD) 5R37 AA010201 (CLE) and the National Institute of Drug Abuse (NIDA) DA030976 (CLE, IRG, KCW, NJS). NIAAA, NCMHD and NIDA had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication. NJS, QP and OL are supported in part by NIH grants R21 DA033813, U19 AG023122, R01 DA030976, R01 MH094483, R01 HL089655, R01 AG035020, R01 MH093500 and R01 MH100351. IRG and KCW are supported in part by NIH grant DA030976.
Footnotes
The authors declare no conflict of interest.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A Brooks LD, DePristo MA Durbin RM, Handsaker RE Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association Task Force on DSM-IV . Diagnostic and Statistical Manual of Mental Disorder (DSM-IV) American Psychiatric Association; Washington, DC.: 1994. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statis. Soc. B. 1995;57(1):289–300. [Google Scholar]
- Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, Mieczkowski PA, Sailsbery JK, Wang X, Ehlers CL, Wilhelmsen KC. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15(1):85. doi: 10.1186/1471-2164-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosron WF, Rex DK, Harden CA, Li TK, Akerson RD. Alcohol and aldehyde dehydrogenase isoenzymes in Sioux North American Indians. Alcohol Clin Exp Res. 1988;12(3):454–5. doi: 10.1111/j.1530-0277.1988.tb00225.x. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Ehrig T, Li TK. Genetic factors in alcohol metabolism and alcoholism. Semin Liver Dis. 1993;13(2):126–35. doi: 10.1055/s-2007-1007344. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–58. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Carr LG, Xu Y, Ho WH, Edenberg HJ. Nucleotide sequence of the ADH2(3) gene encoding the human alcohol dehydrogenase beta 3 subunit. Alcohol Clin Exp Res. 1989;13:594–596. doi: 10.1111/j.1530-0277.1989.tb00383.x. [DOI] [PubMed] [Google Scholar]
- Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact. 2009;178(1-3):2–7. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Compton WM, Thomas YF, Stinson FS, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV drug abuse and dependence in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2007;64(5):566–76. doi: 10.1001/archpsyc.64.5.566. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry. 2013;170(2):154–64. doi: 10.1176/appi.ajp.2012.12010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Montane-Jaime K, Moore S, Shafe S, Joseph R, Carr LG. Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin Exp Res. 2007;31:216–220. doi: 10.1111/j.1530-0277.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG. Association of aldh1 promoter polymorphisms with alcohol-related phenotypes in southwest California Indians. Alcohol Clin Exp Res. 2004a;28(10):1481–6. doi: 10.1097/01.alc.0000141821.06062.20. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 mission Indians. Am J Psychiatry. 2004b;161(7):1204–10. doi: 10.1176/appi.ajp.161.7.1204. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Phillips E. P3 components of the event-related potential and marijuana dependence in southwest California Indians. Addict Biol. 2008;13(1):130–42. doi: 10.1111/j.1369-1600.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Gilder DA, Wilhelmsen KC. Linkage analyses of stimulant dependence, craving, and heavy use in American Indians. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(7):772–80. doi: 10.1002/ajmg.b.31218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Liang T, Gizer IR. ADH and ALDH polymorphisms and alcohol dependence in Mexican and Native Americans. The American Journal of Drug and Alcohol Abuse. 2012;38(5):389–394. doi: 10.3109/00952990.2012.694526. PMID: 22931071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. The firewater myth and response to alcohol in mission Indians. Am J Psychiatry. 1997;154(7):983–8. doi: 10.1176/ajp.154.7.983. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, et al. Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Molecular psychiatry. 2014;19:41. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilder DA, Wall TL, Ehlers CL. Comorbidity of select anxiety and affective disorders with alcohol dependence in southwest California Indians. Alcohol Clin Exp Res. 2004;28(12):1805–13. doi: 10.1097/01.alc.0000148116.27875.b0. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Dixon M, Corey L, Phillips E, Ehlers CL. Co-morbidity of select anxiety, affective, and psychotic disorders with cannabis dependence in southwest California Indians. J Addict Dis. 2006;25(4):67–79. doi: 10.1300/J069v25n04_07. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Corey L, Ehlers CL. Factors associated with remission from cannabis dependence in southwest California Indians. J Addict Dis. 2007;26(4):23–30. doi: 10.1300/J069v26n04_04. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Lau P, Corey L, Ehlers CL. Factors associated with remission from alcohol dependence in an American Indian community group. Am J Psychiatry. 2008;165(9):1172–8. doi: 10.1176/appi.ajp.2008.07081308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Edenberg HJ, Gilder DA, Wilhelmsen KC, Ehlers CL. Association of Alcohol Dehydrogenase Genes with Alcohol-Related Phenotypes in a Native American Community Sample. Alcoholism: Clinical and Experimental Research. 2011;35(11):2008–2018. doi: 10.1111/j.1530-0277.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Oota H, Osier MV, Pakstis AJ, Speed WC, Odunsi A, Okonofua F, Kajuna SLB, Karoma NJ, Kungulilo S, Grigorenko E, Zhukova OV, Bonne-Tamir B, Lu RB, Parnas J, Schulz LO, Kidd JR, Kidd KK. Considerable haplotype diversity within the 23kb encompassing the adh7 gene. Alcohol Clin Exp Res. 2005;29(12):2091–100. doi: 10.1097/01.alc.0000191769.92667.04. [DOI] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the scan. Addiction. 1999;94(9):1361–70. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152:1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ, Bosron WF. Expression and kinetic characterization of variants of human beta 1 beta 1 alcohol dehydrogenase containing substitutions at amino acid 47. J Biol Chem. 1990;265:16366–16372. [PubMed] [Google Scholar]
- Indian Health Service . U S Indian Health Service, Division of Program Statistics: Trends in Indian Health. U S Department of Health and Human Services, Public Health Service, Indian Health Service; Rockville, MD: 1997. [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH Genes With Alcohol Dependence in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland J. Firewater myths: North American Indian drinking and alcohol addiction. Publications Division, Rutgers Center of Alcohol Studies; New Brunswick, NJ.: 1976. [Google Scholar]
- Li MX, Yeung JMY, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131(5):747–56. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21(6):940–51. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libiger O, Schork NJ. A method for inferring an individual's genetic ancestry and degree of admixture associated with six major continental populations. Front Genet. 2012;3:322. doi: 10.3389/fgene.2012.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DM, Pedersen SL, Lobos EA, Todd RD, Wall TL. ADH1B*3 and response to alcohol in African-Americans. Alcohol Clin Exp Res. 2010;34:1274–1281. doi: 10.1111/j.1530-0277.2010.01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC. Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet. 2003;113:325–336. doi: 10.1007/s00439-003-0971-z. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information . Database of Single Nucleotide Polymorphisms (dbSNP) dbSNP Build ID; 2013. p. 139. [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ. Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res. 1994;18:640–643. doi: 10.1111/j.1530-0277.1994.tb00923.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry. 2003;160:41–46. doi: 10.1176/appi.ajp.160.1.41. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Nightingale BN, O'Brien ME, Heath AC, Birley AJ, Martin NG. Molecular biology of alcohol dependence, a complex polygenic disorder. Clin Chem Lab Med. 1998;36(8):633–6. doi: 10.1515/CCLM.1998.111. [DOI] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. Journal of Statistical Planning and Inference. 1999;82(1999):171–196. [Google Scholar]
- Zuo L, Zhang H, Malison RT, Li CSR, Zhang XY, Wang F, Lu L, Lu L, Wang X, Krystal JH, Zhang F, Deng HW, Luo X. Rare ADH variant constellations are specific for alcohol dependence. Alcohol and Alcoholism. 2013;48(1):9–14. doi: 10.1093/alcalc/ags104. [DOI] [PMC free article] [PubMed] [Google Scholar]