Abstract
Insect Odorant Receptors (ORs) comprise an enormous protein family that translates environmental chemical signals into neuronal electrical activity. These heptahelical receptors are proposed to function as ligand-gated ion channels and/or to act metabotropically as G protein-coupled receptors (GPCRs). Resolving their signalling mechanism has been hampered by the lack of tertiary structural information and primary sequence similarity to other proteins. We use amino acid evolutionary covariation across these ORs to define restraints on structural proximity of residue pairs, which permit de novo generation of three-dimensional models. The validity of our analysis is supported by the location of functionally important residues in highly constrained regions of the protein. Importantly, insect OR models exhibit a distinct transmembrane domain packing arrangement to that of canonical GPCRs, establishing the structural unrelatedness of these receptor families. The evolutionary couplings and models predict odour binding and ion conduction domains, and provide a template for rationale structure-activity dissection.
Introduction
In almost all of the many millions of insect species, repertoires of several dozen to several hundred highly divergent Odorant Receptors (ORs) are responsible for detecting a myriad of volatile chemical signals in the environment1. By transducing binding of specific odours into sensory neuron activity, insect ORs play critical roles in mediating olfactory behaviours such as mosquito host-seeking2, moth mate identification3 and drosophilid pathogen avoidance4.
Surprisingly, the insect OR signalling mechanism has been challenging to define5, 6. Like G protein-coupled receptors (GPCRs), insect ORs contain seven putative transmembrane helices (TMHs)7, 8. This observation prompted a long-standing supposition that the insect receptors – similar to their mammalian counterparts – act metabotropically8-12. However, insect ORs have the opposite membrane orientation to GPCRs and function as heteromeric complexes (of unknown stoichiometry) of an odour ligand-specific “tuning” OR and a co-receptor, ORCO (formerly called OR83b)13-16. Moreover, functional analyses in heterologous cells provided evidence that OR/ORCO complexes act as odour-gated cation channels11, 17-19. However, the lack of similarity of insect ORs to known ion channel classes has made it difficult to dissect the molecular basis of this ionotropic function20.
An important way to help resolve the mechanistic basis of insect OR signalling would be to obtain three-dimensional structural information of these receptors. Unfortunately, recombinant expression of these polytopic transmembrane proteins is technically very challenging21, which has precluded X-ray crystal structure determination. Furthermore, because insect ORs have no significant similarity to sequences of known three-dimensional structure, homology modelling22, 23 is not possible. A complementary approach to predict both protein structure and functionally-important sites is to use information contained within sequence variation amongst members of large protein families24. Interactions between pairs of amino acids that are important for protein structure and function impose evolutionary restraints on the sets of mutations acceptable at interacting sites25. Reciprocally, identification of such evolutionary couplings (ECs) within primary sequences of isostructural proteins can provide information on distance restraints between amino acid pairs and thereby insight into higher-order structure and functional domains25. Identified coevolving residues can also, depending upon the analysis method25, reflect distally coupled residues important for allosteric communication within proteins26, 27. Here, we applied this approach to identify ECs across the large insect OR family, which we use to build de novo the first three-dimensional models of these proteins and identify functionally important sites.
Results
We used an extended version of the EVfold-transmembrane method28 (Fig. 1a) to compute evolutionary couplings for selected insect ORs, starting with multiple sequence alignments containing a set of 5907 known and newly-annotated receptors (Supplementary Table 1 and Supplementary Data 1-4). From these alignments we extracted patterns of amino acid coevolution to produce a “contact map” of evolutionarily constrained pairs of residues along the OR sequence (Fig. 1b, Supplementary Fig. 1, and Supplementary Data 5-8). These evolutionary couplings (ECs) were used in two ways: first, to probe functional sites, since evolutionary co-conservation reveals selection pressures beyond single position conservation (Supplementary Data 9-10); second, to produce three-dimensional models by combining ECs with secondary structure predictions to fold the OR polypeptide (Fig. 1c and Supplementary Data 11-14).
Figure 1. Evolutionary couplings in insect olfactory receptors.
(a) Left: The structural proximity of residues in a protein leaves a visible record of amino acid covariation in the protein family sequence alignment due to the evolutionary pressure to maintain favourable interactions. By analysing these patterns with a statistical model of sequence coevolution, Evolutionary Couplings (ECs) between residue pairs can be inferred and used to predict three-dimensional (3D) contacts. Right: Schematic of the EVfold-transmembrane prediction workflow (see Methods). A multiple alignment of insect OR protein sequences is used to identify ECs between pairs of coevolving amino acids with EVfold-PLM. After discarding pairs that are inconsistent with predicted membrane topology and secondary structure, the remaining highest-ranking EC pairs are used as distance restraints on an extended polypeptide to fold a set of 3D OR models.
(b) Contact map representation of the top 200 predicted ECs for D. melanogaster OR85b. The axes represent the indices along the OR85b primary sequence, along which predicted transmembrane helical segments (TMHs) are annotated as blue bars and predicted helical secondary structure as grey bars. Black dots represent ECs between pairs of residues; the representation is mirror-symmetric along the diagonal. The lines of ECs parallel and anti-parallel to the diagonal of the contact map are characteristic of the helix packing arrangements observed in alpha-helical transmembrane proteins28; the dashed orange lines highlights one of these between TMH1 and TMH2. Three high-density regions of ECs within the N-terminal tail, extracellular loop 2 and intracellular loop 3 are highlighted by red dashed circles (see also Fig. 3a).
(c) Top-ranked predicted 3D structural models for OR85b (left, model 140_12) and ORCO (right, model 310_2; the ORCO-specific intracellular loop 2 insertion is not modelled) viewed from within the membrane bilayer (side view) and from the extracellular face (top view). Models are colour-coded from N-terminus (blue) to C-terminus (red). As expected for members of the same protein family, the 3D models agree in their overall helical packing arrangement; the observed differences may be caused by actual structural differences of different subfamilies in the OR protein family sequence space and/or by inaccuracies in sequence alignment, statistical inference and 3D modelling.
We used D. melanogaster OR85b (a tuning OR that responds to 2-heptanone and several esters29, 30) and ORCO as anchors for these analyses to obtain models for both a ligand-specific OR and the co-receptor. ORs and ORCO have the same transmembrane topology16, 17, 31 and can – with the exception of an ORCO-specific ~70 amino acid insertion in intracellular loop 2 (IL2) – be confidently aligned over their whole length (Supplementary Data 2). We therefore expect OR85b and ORCO to share the same overall three-dimensional (3D) fold and our predicted ECs and structural models to be similar. However, differences in alignments arising from distinct anchor sequences that cover different parts of the sequence space of the protein family may capture subfamily-specific restraints25.
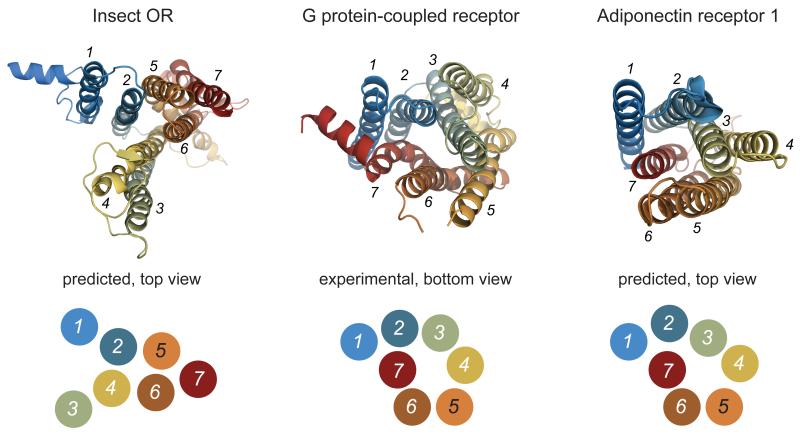
Examination of the predicted contact maps for OR85b and ORCO revealed a number of lines of ECs parallel or perpendicular to the diagonal within predicted TMHs (Fig. 1b, orange dashed box. and Supplementary Fig. 1), which are characteristic of parallel and anti-parallel helix arrangements in alpha-helical membrane proteins28. This observation provides one validation of our approach as the EC analysis is given no prior knowledge that insect ORs are transmembrane proteins. Importantly, the predicted helical contacts bear no resemblance to those of GPCRs or to those of the Adiponectin Receptor 1, a seven TMH protein that has the same membrane orientation as insect ORs32, but which appear to have a convergent helical arrangement to that of GPCRs (Fig. 2)28. These observations indicate that insect ORs adopt a seven TMH packing arrangement that is distinct from GPCR structures. Thus, the shared heptahelical secondary structure of these receptor families is, in contrast to initial assumptions8, 9, likely to be coincidental and not necessarily indicative of a shared signalling mechanism.
Figure 2. An Evolutionary Coupling-based insect OR model has a heptahelical packing arrangement distinct from G-protein coupled receptors.
Top: Comparison of the helical packing arrangements of insect ORs, a G protein-coupled receptor (GPCR) and the Adiponectin receptor 1 (AR1); the structures are colour-coded from N-terminus (blue) to C-terminus (red). The OR is the top-ranked model of OR85b (see Fig. 1c); the GPCR is the crystal structure of the β2-adrenergic receptor (PDB 2RH155); the AR1 structure is the top-ranked model described previously28. As the N-termini of ORs and AR1s are located in the cytosol, but the N-terminus of GPCRs is located extracellularly (or lumenally), the GPCR structure is shown in the opposite orientation so that the packing arrangements are visually comparable. Structures were rotated so that the positioning of TMH1 and TMH2 agrees, as far as possible, between the different molecules. Bottom: corresponding simplified two-dimensional representations of the helical contacts highlighting the difference between the helical packing arrangement of insect ORs when compared to GPCRs and AR1s.
In analyses with both OR85b and ORCO, three regions of the sequences contain patches of residues with multiple, strong ECs (N-terminal tail, extracellular loop 2 (EL2) and intracellular loop 3 (IL3)/TMH7 (Fig. 1b and Supplementary Fig. 1, red dashed circles, and Fig. 3a). These EC clusters suggest these regions are under strong evolutionary constraint and are therefore predictive of functional importance28. Remarkably, although molecular insights into insect ORs are still in their infancy – and dispersed across several different “model” receptors – experimental validation of the importance of all three regions is available. A single amino acid mutation in the N-terminus of the D. melanogaster pheromone receptor OR67d (C23W) abolishes receptor function in vivo33; mapping of this residue onto the contact map by identifying the homologous position in OR85b (F11) and ORCO (A23) revealed it to lie within the N-terminal cluster of ECs (Fig. 1b, Fig. 3a, Supplementary Fig. 1 and Supplementary Table 2), suggesting that this segment fulfils an important role in all ORs; we test this prediction below. The second patch of EC-enriched residues, in EL2, contains a number of predicted beta-turns separated by conserved proline residues; mutation of these, and adjacent residues, in several different ORs strongly or completely disrupts function34. Although our analysis is unable to predict the precise conformation of this loop, the presence of strongly constrained residues in this region is consistent with the detrimental effect of their mutation. Lastly, IL3 forms at least part of the molecular interface for assembly of ORCO and ligand-specific ORs into heteromeric complexes16, which may explain the very high frequency of strong ECs within this region (Fig. 3a). We are currently unable to distinguish ECs that mediate intramolecular versus intersubunit interactions in these receptors. Nevertheless, a systematic mutational analysis of cysteine residues in D. melanogaster ORCO revealed a particularly important contribution of those cysteines in IL3 to forming functional homomeric and/or heteromeric ion channels35 (Supplementary Table 2). In summary, although these observations do not validate the ECs and structural models per se, the striking coincidence of known functionally-important sites with highly coupled residues gives basic confidence in our results.
Figure 3. Highly coupled OR residues coincide with experimentally characterised functional regions.
(a) Positions with above-average EC strength (top 25% of sites, blue spheres) on the top-ranked model of OR85b (140_12; Supplementary Data 13) (membrane-integral side view). Strongly coupled residues cluster around three regions of the model: (i) N-terminus, (ii) extracellular loop 2, (iii) intracellular loop 3 and along the span of TMH7. Many of the strongly-coupled TMH7 residues do not feature in the contact map (Fig. 1b), because they were excluded as structural contacts with our standard transmembrane clash filter.
(b) Experimentally characterised residues in different OR and ORCO proteins (spheres coloured by different shades of blue according to functional categorisation; Supplementary Table 2) mapped onto the 3D model of OR85b (140_12), based on a sequence alignment of OR and ORCO sequences (Supplementary Data 2). Amongst many residues whose mutation have general deleterious effects on ion channel function, only a few residues influence ion selectivity (which are strong candidates for pore-lining residues), including two sites in ORCO (double asterisks marking residues at the extracellular end of TMH6 and TMH7) and two in a tuning OR (B. mori OR1) (single asterisks marking residues at the intracellular end of TMH5 and TMH6) (see text and Supplemental Table 2 for details).
To validate the predictive power of our ECs in identifying important residues in insect ORs, we focussed on the N-terminal cytoplasmic region of ORCO, whose role (if any) is unknown. We generated a version of this receptor (ORCO6Mut) bearing mutations in six of the top-ranked constrained residues in the sequence of D. melanogaster ORCO (A23, M24, F30, M31, H32, N33) (Supplementary Data 10); several of these residues feature as pairs in the top ranked ORCO ECs (i.e. M24-F30, M24-M31, M24-H32; Supplementary Data 6). We also generated an ORCO mutant bearing a small deletion spanning these residues (ORCOΔ23-33). Wildtype and mutant ORCO proteins were expressed in Xenopus oocytes for functional analysis by two-electrode voltage clamp recording of ligand-evoked current responses20. We first co-expressed ORCO with OR47a or OR85b, whose most potent agonists are pentyl acetate and 2-heptanone, respectively18, 30, 36. Wild type ORCO co-expressed with these receptors can reconstitute dose-dependent odour-evoked currents (Fig. 4a-b). By contrast, ORCO6Mut and ORCOΔ23-33 have highly diminished or abolished response to these odours, respectively (Fig. 4a-b). In a second set of experiments, we tested current responses of wildtype and mutant ORCOs to VUAA1, an artificial small-molecule agonist that can activate this co-receptor when expressed alone (without a tuning OR partner)37. Wildtype ORCO produced robust currents upon VUAA1 presentation, but neither mutant protein responded to this ligand (Fig. 4c). Taken together, our computational and experimental data support a previously unappreciated function for the ORCO N-terminus; that the tuning receptor OR67d is also sensitive to a mutation in this region33, suggests that this sequence may have a conserved role across the OR family. Determining the precise contribution of the N-terminus to receptor function – for example, in folding, trafficking and/or complex assembly – will nevertheless require further experimentation beyond the scope of this study.
Figure 4. Functional analysis of the ORCO N-terminus.
(a-c) Left: Representative whole cell current traces to the indicated stimuli with two-electrode voltage-clamp in Xenopus oocytes injected with cRNAs for the indicated combinations of wildtype or mutant D. melanogaster ORs. ORCO6Mut contains amino acid substitutions in the six top-ranked N-terminal residues (A23S, M24A, F30A, M31A, H32A, N33A); ORCOΔ23-33 bears a deletion of this region (A23-N33). Ligand solutions were applied for 3 s (arrowheads). Right: Quantification of current amplitudes (mean ± SEM; n = 5 oocytes for wildtype ORCO and ORCO6Mut; n = 4 for ORCOΔ23-33).
Finally, we asked whether our analysis could provide insight into the proposed function of insect ORs as ligand-gated ion channels. Four studies have experimentally-defined mutations that affect (directly or indirectly) ligand recognition properties of different receptors30, 38-40 (Supplementary Table 2). Notably, when mapped onto the OR85b model, these cluster in the external half of the TM2-4 region (Fig. 3b), suggesting that this region comprises a part of a ligand-binding pocket. The proposition that the relatively long EL2 loop might form a “lid” that could cover/uncover this pocket34 is inline with the location of these residues.
Although ORs do not bear strong similarity to known ion channels, previous characterisation of a sequence suggested to bear some resemblance to a K+ channel selectivity filter in D. melanogaster ORCO TMH6 (T393VVGYLG399) demonstrated a role in controlling K+ permeability11 (Supplementary Table 2). Independently, a systematic screen of residues in Bombyx mori ORCO showed mutation of Y464 (in TMH7) – and the equivalent Y478 in D. melanogaster ORCO – affected K+ selectivity20. Strikingly, this residue lies directly opposite the TVVGYLG sequence in our model, suggesting that this region forms a key part of the pore (Fig. 3b, double asterisks). Channel properties depend upon both ORCO and the tuning OR, indicating that both subunits contribute to formation of the ion conducting passage18, 20, 41, 42. We therefore mapped the position of additional residues that have direct or indirect effects upon channel function20 (Supplementary Table 2). The majority of these are in TMH5-7 region (Fig. 3b), consistent with this region forming a central part of the ion conducting channel; notably, the only two of these residues that have effects on K+ ion selectivity (D299 and E356 in B. mori OR120) – as opposed to more general deleterious effects on ion channel function – are also located directly opposite each other at the cystosolic end of TMH5 and TMH6 (Fig. 3b, single asterisks). Future determination of the stoichiometry of the heteromeric complex and identification of inter-subunit ECs may permit modelling of its quaternary structure to visualise how this C-terminal region forms the ion-conducting channel. The function of these receptors in odour-evoked signalling clearly depends upon other regions too, such as the N-terminus and EL2.
Discussion
Our analysis of ECs in insect ORs has allowed us to build the first three-dimensional models of this receptor family. These proteins appear to define a novel fold, as searches with DALI43 did not reveal highly significant similarity of our models to experimentally-determined membrane protein structures (data not shown). It is, however, important to keep in mind that our current models are coarse-grained and the accuracy of the folded 3D structures is limited by multiple factors, including: (i) the availability of OR sequences, (ii) accurate secondary structure predictions, and (iii) the confounding of monomer folding by the presence of multimer contacts, conformational plasticity and functional couplings, which are not easily disambiguated. As additional receptor sequences become available and as the EVfold method itself is further developed to fold multimers, future OR models – which we plan to update on the EVfold website (http://evfold.org) – will be more accurate. It will be of particular interest to determine whether ORCO and ligand-specific ORs have distinct structural features. For ORCO-specific predictions, many hundreds of additional ORCO sequences from other insects are required. Similarly, increased sequence availability and improvement of alignments will aid analysis of the divergent tuning ORs. Nevertheless, the current models are both consistent with available and new experimental data and offer a number of predictions – notably, through revelation of highly constrained residues (Supplementary Data 9-10) – to guide and visualise structure-function analyses of these unusual sensory receptors. Moreover, these predictions may be the first step to assist in the design of novel pharmacological reagents to manipulate OR function and thereby control olfactory behaviours of pest insects.
Methods
Annotation of OR sequences
New OR sequences from nine additional drosophilid genomes (Supplementary Table 1 and Supplementary Data 1) were identified by TBLASTN44 using D. melanogaster OR protein sequences as queries.
Multiple sequence alignment
All identified OR sequences were compiled into a FASTA database (Supplementary Data 1) and aligned iteratively with either D. melanogaster OR85b or D. melanogaster ORCO as reference sequences using jackhmmer with default parameters45. In the ORCO-anchored alignment, homologues that split into two separate aligned domains (due to the presence of the ORCO-specific insertion in IL2) were re-merged using custom Python scripts. In the OR85b alignment, the inclusion E-value (1E-40) was manually chosen to maximise EC contrast over residual background couplings. In both cases, alignment columns with more than 50% gaps were excluded from evolutionary coupling analysis as described28.
Evolutionary couplings inference from sequence variation
Evolutionary couplings were calculated with an updated version of our protocol (EVfold-PLM) based on a pseudolikelihood maximisation statistical inference procedure46-48 instead of the original mean-field approximation24. Sequences in the alignment were downweighted at a 90% identity threshold to reduce the influence of sampling bias on statistical inference. Webserver and code for evolutionary couplings calculations are available on evfold.org.
De novo folding from protein sequences
The folding protocol is described in more detail elsewhere24, 28. In brief, evolutionary couplings were filtered using a combination of predicted transmembrane topology and predicted secondary structure. Transmembrane topology was predicted using PolyPhobius49 and validated by consensus prediction using TOPCONS50 and MEMSAT-SVM51. Secondary structure was predicted using PSIPRED52 and low confidence predictions of helix or beta-strand in ORCO (reliability index < 3) were set to coil. Increasing numbers of top-ranked evolutionary couplings were then used to fold D. melanogaster OR85b and ORCO from an extended polypeptide with CNS distance geometry and simulated annealing53. Local secondary structure was constrained based on predicted topology and secondary structure. Due to the lack of alignment coverage on the ORCO-specific insertion in IL2 (residues 247-329), this region was not modelled to avoid interference with the folding of the membrane-integral domain.
Clustering and ranking of predicted models
All generated protein models were clustered and ranked using the default quality assessment protocol of EVfold-transmembrane24, 28. The ranking procedure uses a score composed of the agreement of each model with predicted lipid exposure, predicted secondary structure, and evolutionary couplings. Models with structural knots are excluded from consideration as top-ranked models. Additional structure-based clustering ensures that high-scoring outlier structures can be excluded from consideration as confident predictions.
Model visualisation and annotation
Protein models were visualised and annotated using PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC. To annotate functionally analysed residues of different ORs (Supplementary Table 2) on the common model, the equivalent positions in OR85b and ORCO were determined using the jackhmmer multiple sequence alignment anchored around OR85b (Supplementary Data 2).
Functional analysis of ORs in Xenopus oocytes
Mutant orco plasmid constructs were generated by standard cloning procedures; primer sequences are as follows: ORCO6Mut (forward: 5′-GCGCGAATTCGCCACCACCATGACAACCTCGATG-3′, reverse: 5′-CAGGCCGGAGTACTTCGCCGACCGGATGTTGGGCAT-3′, and forward: 5′-AAGTACTCCGGCCTGGCCGCGGCCGCCTTCACGGGCGGCAGT-3′, reverse: 5′-GCGCCTCGAGTTACTTGAGCTGCAC-3′) ORCOΔ23-33 (forward: 5′-TTCACGGGCGGCAGTGCCTTC-3′, reverse: 5′-CCGGATGTTGGGCATCAGGTC-3′). cRNAs were synthesised from linearised modified pSPUTK vector54 containing wildtype or mutant versions of D. melanogaster ORCO, OR47a or OR85b. Stage V to VII oocytes were treated with 2 mg/ml of collagenase B (Roche Diagnostics, Tokyo, Japan) in Ca2+-free saline solution (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES, pH 7.5) for 1-2 h at 18°C. Oocytes were microinjected either with 6.25 ng cRNA encoding a ligand tuning OR (OR47a or OR85b) and 6.25 ng cRNA encoding ORCO (Fig. 4a-b), or 12.5 ng of ORCO cRNA (Fig. 4c). Injected oocytes were incubated for 3-4 days at 18°C in Barth’s solution supplemented with 84.7 mg/l of gentamycin. Whole-cell currents were recorded using the two-electrode voltage-clamp technique as previously described3. Intracellular glass electrodes were filled with 3 M KCl. Signals were amplified with an OC-725C amplifier (Warner Instruments, Hamden, CT, USA), low-pass filtered at 50 Hz and digitised at 1 kHz. Ligand solution was delivered via a silicon tube connected to a computer-driven solenoid. Prior to experiments, pentyl acetate (CAS: 628-63-7; Wako, Osaka, Japan) and 2-heptanone (CAS: 110-43-0; TCI, Tokyo, Japan) were directly diluted into the bath solution (115 mM NaCl, 2.5 mM KCl, 1.8 mM BaCl2, and 10 mM HEPES, pH 7.2). VUAA1 (CAS: 525582-84-7; Vitas-M Laboratory, Moscow, Russian Federation) was prepared in dimethylsulphoxide as 20 mM, 66.7 mM or 200 mM stock solutions, which were then diluted into the bath solution at 0.15% to give the final desired concentration.
Supplementary Material
Acknowledgements
We are grateful to our colleagues for supplying OR protein sequence files, and thank Vanessa Ruta and members of the Benton lab for discussions and comments on the manuscript. T.A.H. and D.S.M. are supported by the National Institutes of Health (NIH R01 award GM106303). Research in K.T.’s laboratory is supported by the ERATO Touhara Chemosensory Signal Project from JST, Japan. Research in R.B.’s laboratory is supported by the University of Lausanne, European Research Council Starting Independent Researcher and Consolidator Grants (205202 and 615094) and the Swiss National Science Foundation (31003A_140869).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139:45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degennaro M, et al. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 4.Stensmyr MC, et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Benton R. Chemical sensing in Drosophila. Curr. Opin. Neurobiol. 2008;18:357–363. doi: 10.1016/j.conb.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa T, Vosshall LB. Controversy and consensus: noncanonical signaling mechanisms in the insect olfactory system. Curr. Opin. Neurobiol. 2009;19:284–292. doi: 10.1016/j.conb.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 8.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 9.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai T, et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. USA. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wicher D, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 12.Stengl M, Funk NW. The role of the coreceptor Orco in insect olfactory transduction. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2013;199:897–909. doi: 10.1007/s00359-013-0837-3. [DOI] [PubMed] [Google Scholar]
- 13.Lundin C, et al. Membrane topology of the Drosophila OR83b odorant receptor. FEBS Lett. 2007;581:5601–5604. doi: 10.1016/j.febslet.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- 15.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLOS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smart R, et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 19.Silbering AF, Benton R. Ionotropic and metabotropic mechanisms in chemoreception: ‘chance or design’? EMBO Rep. 2010;11:173–179. doi: 10.1038/embor.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa T, Pellegrino M, Sato K, Vosshall LB, Touhara K. Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLOS ONE. 2012;7:e32372. doi: 10.1371/journal.pone.0032372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carraher C, Nazmi AR, Newcomb RD, Kralicek AV. Recombinant expression, detergent solubilisation and purification of insect odorant receptor subunits. Protein Expr. Purif. 2013 doi: 10.1016/j.pep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLOS ONE. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks DS, Hopf TA, Sander C. Protein structure prediction from sequence variation. Nat. Biotechnol. 2012;30:1072–1080. doi: 10.1038/nbt.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 27.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 28.Hopf TA, Colwell LJ, Sheridan R, Rost B, Sander C, Marks DS. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Nichols AS, Luetje CW. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85B contributes to ligand-receptor interactions. J. Biol. Chem. 2010;285:11854–11862. doi: 10.1074/jbc.M109.058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsitoura P, et al. Expression and membrane topology of Anopheles gambiae odorant receptors in lepidopteran insect cells. PLOS ONE. 2010;5:e15428. doi: 10.1371/journal.pone.0015428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamauchi T, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 33.Jin X, Ha TS, Smith DP. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu P, Leal WS. Probing insect odorant receptors with their cognate ligands: insights into structural features. Biochem. Biophys. Res. Commun. 2013;435:477–482. doi: 10.1016/j.bbrc.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner RM, et al. Mutational analysis of cysteine residues of the insect odorant co-receptor (Orco) from Drosophila melanogaster reveals differential effects on agonist- and odorant/tuning receptor-dependent activation. J. Biol. Chem. 2014;289:31837–31845. doi: 10.1074/jbc.M114.603993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 37.Jones PL, Pask GM, Rinker DC, Zwiebel LJ. Functional agonism of insect odorant receptor ion channels. Proc. Natl. Acad. Sci. USA. 2011;108:8821–8825. doi: 10.1073/pnas.1102425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature. 2011;478:511–514. doi: 10.1038/nature10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leary GP, et al. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. USA. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes DT, Wang G, Zwiebel LJ, Luetje CW. A Determinant of Odorant Specificity Is Located at the Extracellular Loop 2-Transmembrane Domain 4 Interface of an Anopheles gambiae Odorant Receptor Subunit. Chem. Senses. 2014;39:761–769. doi: 10.1093/chemse/bju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pask GM, Jones PL, Rutzler M, Rinker DC, Zwiebel LJ. Heteromeric Anopheline odorant receptors exhibit distinct channel properties. PLOS ONE. 2011;6:e28774. doi: 10.1371/journal.pone.0028774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nichols AS, Chen S, Luetje CW. Subunit Contributions to Insect Olfactory Receptor Function: Channel Block and Odorant Recognition. Chem. Senses. 2011 doi: 10.1093/chemse/bjr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eddy SR. Accelerated Profile HMM Searches. PLOS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ekeberg M, Lovkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 47.Balakrishnan S, Kamisetty H, Carbonell JG, Lee SI, Langmead CJ. Learning generative models for protein fold families. Proteins. 2011;79:1061–1078. doi: 10.1002/prot.22934. [DOI] [PubMed] [Google Scholar]
- 48.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. USA. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kall L, Krogh A, Sonnhammer EL. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics. 2005;21(Suppl 1):i251–257. doi: 10.1093/bioinformatics/bti1014. [DOI] [PubMed] [Google Scholar]
- 50.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37:W465–468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinf. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 54.Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem. Biophys. Res. Commun. 2003;305:964–969. doi: 10.1016/s0006-291x(03)00863-5. [DOI] [PubMed] [Google Scholar]
- 55.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.