Abstract
Objectives
Structural aberration in chromosomes characterizes almost all human solid cancers and analysis of those alterations may reveal the history of chromosomal instability. However, the clinical significance of massive chromosomal abnormality in ovarian high-grade serous carcinoma (HGSC) remains elusive. In this study, we addressed this issue by analyzing the genomic profiles in 455 ovarian HGSCs available from The Cancer Genome Atlas (TCGA).
Methods
DNA copy number, mRNA expression, and clinical information were downloaded from the TCGA data portal. A chromosomal disruption index (CDI) was developed to summarize the extent of copy number aberrations across the entire genome. A Cox regression model was applied to identify factors associated with poor prognosis. Genes whose expression was associated with CDI were identified by a 2-stage multivariate linear regression and were used to find enriched pathways by Ingenuity Pathway Analysis.
Results
Multivariate survival analysis showed that a higher CDI was significantly associated with a worse overall survival in patients. Interestingly, the pattern of DNA copy number alterations across all the chromosomes was similar between tumors with high and low CDI, suggesting they did not arise from different mechanisms. We also observed that expression of several genes was highly correlated with the CDI, even after adjusting for local copy number variation. We found that molecular pathways involving DNA damage response and mitosis were significantly enriched in these CDI-correlated genes.
Conclusion
Our results provide a new insight into the role of chromosomal rearrangement in the development of HGSC and the promise of applying CDI in risk-stratifying HGSC patients, perhaps for different clinical managements. The genes whose expression is correlated with CDI are worthy of further study to elucidate the mechanism of chromosomal instability in HGSC.
Keywords: Chromosomal instability, Ovarian cancer, High-grade serous carcinoma
Introduction
Structural aberration in chromosomes is a cardinal feature inherent to almost all solid tumors [1,2]. Those changes are the results of an underlying chromosomal instability leading to chromosomal missegregation and reiterative cycles of DNA strand breaks and rejoining. It has been thought that such chromosomal instability generates a genetically diversified repertoire of tumor cells. Those tumor clones harboring amplification of oncogenes and deletion of tumor suppressors fuel the Darwinian selection and subsequent clonal evolution and expansion to establish clinically detected tumors [3]. Thus, analysis of structural alterations in cancer genome may reveal the history of prior chromosomal instability and help identify genomic signatures that are associated with pathogenesis and treatment outcome.
Ovarian high-grade serous carcinoma (HGSC) represents the most common and aggressive type of ovarian neoplasms [4]. HGSC most likely arises from fallopian tube epithelium as a precursor non-invasive lesion known as “serous tubal intraepithelial carcinoma” which may disseminate to ovary and peritoneal tissues [5]. As a result, the proposal of tubal origin of HGSC calls into question if early stage diseases of HGSC have ever existed, featuring HGSC as a unique human neoplastic disease from this perspective of tumor progression. Previous genome-wide studies and recently The Cancer Genome Atlas (TCGA) have demonstrated an exaggerated level of chromosomal alterations including DNA copy number gain and loss [6,7]. However, recurrent somatic mutations are uncommon except in TP53 of which mutations occurs in essentially all HGSCs [7]. In this study, we used ovarian HGSC as a disease model to determine if the overall levels of chromosomal disruption as reflected by “segments” of DNA copy number changes correlate with clinical outcome and molecular features by analyzing data from 455 HGSCs available from the TCGA dataset.
Materials and methods
DNA copy number, mRNA expression and clinical information were downloaded from the TCGA (https://tcga-data.nci.nih.gov/tcga/). Specifically, segmented data from the Agilent 1×1 M copy number was used to calculate the chromosomal disruption index (CDI) while gene-level summaries of copy number were used in linear models to identify and characterize associations between variables. We also obtained gene-level estimates of mRNA expression summarized from custom Agilent G4502A 07 expression arrays.
To determine the overall level of chromosomal disruption in individual HGSCs, we developed the CDI, which was defined as the total number of discrete segments of copy number variation (CNV) with an estimated fold change of more than 2 (4 copies) or less than 1/2, (1 copy). While varying the threshold leads to slightly different results, the indices for various thresholds were highly correlated and qualitatively very similar (Fig. S1).
Cox regression was used to evaluate associations between copy number or gene expression and clinical outcomes including overall survival and progression free survival. A multivariate model which included several known prognostic factors such as patient age, tumor stage, and residual disease volume was applied to identify genes for which copy number and/or expression had an independent prognostic value. A 2-stage multivariate linear regression, including the same clinical covariates, was used to identify genes whose expression was associated with CDI, after adjusting for the cis effects of copy number changes on gene expression. Spearman correlation was used to measure association among amplicons, and between amplicons, gene expression and number of non-synonymously mutated genes with statistical significance determined in terms of false discovery rate, by comparison to a permutation distribution. GenometriCorr [8], an R package recently developed to assess the spatial correlation of two sets of genomic intervals, was employed to compare different sets of amplicons in terms of their genomic locations.
To determine if specific transcription factors might be recruited to facilitate chromosomal breakage in HGSC, we identified hotspots of chromosomal breakage and correlated those with the locations of 480 transcription factor target gene sets having at least 5 target genes. We downloaded target sets for transcription factors from TRANSFAC using the Automated Sequence Annotation Pipelines (ASAP) [9]. The list used in these analyses was obtained and frozen on December 8, 2010. To identify hotspots of chromosomal breakage, we plotted the start and end points of each significantly, copy-number altered segment of DNA (fold change of more than 2 (4 copies) or less than 1/2, (1 copy)). A smooth, spline-based estimate of the distribution of breakpoints within each chromosome was calculated and thresholded at 5% of the peak value to isolate hotspots. Within the selected regions, peaks were identified by calculating the first and second derivatives of the spline approximation. The locations of peaks were compared to locations of transcription factor targets using GenometriCorr.
To identify the pathways associated with high CDI, canonical pathway analyses were generated through the use of IPA (Ingenuity® Systems, www.ingenuity.com), which identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Genes whose expression showed significant positive correlation with CDI (p<.001) and were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. Fisher's exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone. Benjamini–Hochberg method was applied to adjust the p-value for multiple comparisons.
Results
Chromosome disruption is an independent predictor for overall survival of high-grade ovarian serous carcinoma
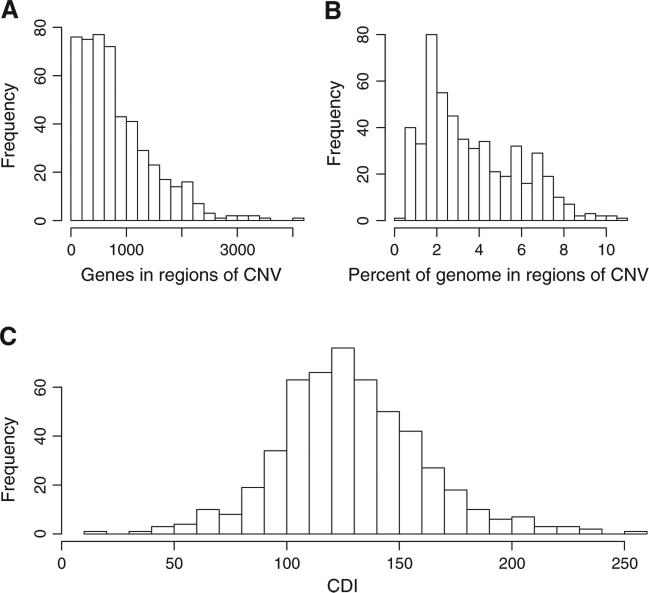
Several interesting findings were observed by global analysis of the ovarian HGSC genome using the TCGA data set. First, examining the DNA copy number data revealed an exaggerated, yet highly variable, number of discrete DNA segments showing copy number variations (CNVs) including copy number gain or loss, suggestive of a history of accumulated chromosomal disruptions during tumor evolution. Using stringent criteria (>2-fold) for calling copy number changes, we observed that a HGSC, on average, contained 100–150 distinct CNVs covering 1.9%–5.3% of the cancer genome and harboring 350–1150 genes. For those extreme cases, more than 10% of the genome and nearly 5000 genes were affected (Fig. 1A and B). Although the degree of chromosomal disruption is highly variable across tumors, it is striking that the distribution of CDI among HGSCs followed a bell-shaped curve with a single mode (Fig. 1C). It is not possible to identify a separate, distinct population of super-disruptors at the upper end of the CDI distribution or of chromosomally stable disease at the lower, thereby suggesting a single disease process of HGSC based on CDI.
Fig. 1.
Extent of copy number alterations in HGSC samples. (A) The number of genes falling into altered regions in each sample. (B) The proportion of the genome implicated in copy number alterations by sample. (C) The distribution of chromosomal disruption index (CDI) by sample, calculated as the number of copy number altered segments of DNA.
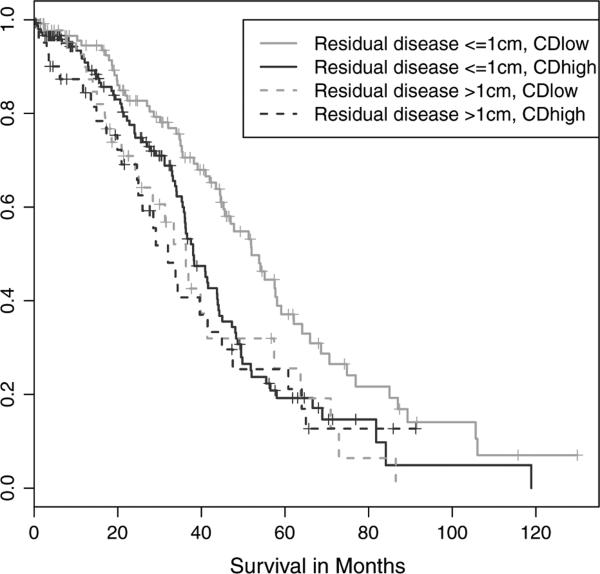
To assess if tumors with different CDI would have different clinical outcome, we modeled the relationship between chromosome disruption and overall survival in a multivariate Cox regression model with patient age, tumor stage, and residual tumor volume included as covariates. Stage proved to have little prognostic value in this dataset—as seen in the summarized patient characteristics (Table 1), almost all samples were stage IIIC; but age, residual tumor volume and CDI were all significantly correlated with patients' overall survival. Our analysis convincingly confirmed the hypothesis, with those HGSCs exhibiting high CDI having a significantly shorter overall survival than those with lower CDI (p=0.0021). The main trends were illustrated in a Kaplan–Meier plot (Fig. 2), showing that the CDI and the residual tumor size were independent predictors of the overall survival. Sub-optimal surgical debulking (residual tumor of more than 1 cm) was associated with worse outcome regardless of CDI but the degree of chromosomal disruption significantly stratified risk in the optimally debulked group.
Table 1.
Summary of clinical characteristics.
| Race (455/455) | 83.1% White, 10.1% Asian, 4.4% Black, 2.4% other |
|---|---|
| Age (430/455) | 30.5-87.5 years, median = 58.7 years, mean = 59.8 years |
| Stage (428/455) | 16.4% stage IV, 73.6% stage IIIc, 5.4% stage IIIa-b, 4.7% stage 2 |
| Residual disease (356/455) | 75.8% achieved optimal surgical debulking (<1 cm residual tumor) |
The analyses presented here were based on HGSC 455 tumor samples from the NCI/NHGRI Cancer Genome Atlas Project. The distributions of age, race, stage and residual disease were summarized here. In cases of missing data, distributions were calculated for annotated samples, and the annotated sample size provided.
Fig. 2.
Overall survival by CDI and residual tumor. A Kaplan–Meier analysis of TCGA samples showed that among patients with residual tumor volumes less than 1 cm (solid lines), those with CDI less than the median (green line) had distinctly better outcomes than those with CDI greater than the median (blue line).
Normal cell contamination, especially from lymphocytes, may bias DNA copy number analysis [10] by decreasing the signal-to-noise ratio of SNP array experiments [11], thereby lowering the CDI calculated. In addition, it has been reported that an increase of intratumoral lymphocytes is associated with better prognosis in some cancers [12–17]. Therefore, a lymphocytic infiltration is a possible confounding factor in the assessment of the association between CDI and prognosis. We addressed this issue by using PTPRC (CD45), CD4, and CD8 expression as surrogate markers for leukocyte, CD4+ lymphocyte, and CD8+ lymphocyte contamination, respectively. As suspected, CDI is negatively correlated with PTPRC expression (Spearman Cor.=−0.30, p=3.663e−11) and CD4 expression (Spearman Cor.=−0.23, p=4.356e−7), though not significantly correlated with CD8 expression (Cor.=−0.06, p=0.2099). Furthermore, both PTPRC and CD4 were marginally associated with better prognosis in a univariate Cox regression model (p=0.061 and 0.047, respectively). However, in multivariate Cox models including age, residual tumor, and CDI, both PTPRC and CD4 were not identified as an independent prognostic factor (p=0.36 and 0.17, respectively), while CDI remained a significant predictor of overall survival (p=0.0025 and 0.0013, respectively). This result suggested that leukocyte or lymphocyte contamination did not account for the significant association between CDI and overall survival.
Since inactivation in the BRCA DNA repair pathway is an early event in the pathogenesis of HGSC [7,18] and could potentially control the genome integrity of ovarian carcinoma, we determined if the “BRCAness” due to inactivating germline, somatic mutations or promoter hypermethylation of BRCA1 and BRCA2 was associated with CDI. The mutation status of BRCA1 and BRCA2 was available for 316 of the samples in our analysis dataset, including a total of 98 HGSCs with “BRCAness” (64 mutations and 34 promoter methylation). On average, BRCA-altered samples had 11.66 fewer copy number segment alterations per tumor than BRCA-WT samples (119.94 vs 131.60) with a t-statistic of −3.28 and a p-value of 0.0012. Interestingly, loss of BRCA1/2 by mutation or methylation was strongly associated with an increased number of non-synonymous somatic mutations per sample. On average, 60.95 mutations were detected in samples with BRCAness vs. 41.45 in BRCA-WT samples, p=5.276e−07. Both loss of BRCA and increased number of mutated genes per sample were individually associated with better survival, (BRCA loss: coef=−0.439, p=0.02; mutated gene count: coef=−0.008, p=0.026). However, in a multivariate model that included both BRCA status and the number of non-synonymously mutated genes per sample in addition to CDI, a high CDI remained significantly correlated to greater risk of death (p=0.032) while neither BRCA alteration nor the number of non-synonymously mutated genes were identified as significant, independent prognostic factors (p=0.18 and 0.12, respectively).
Common hotspots of chromosome breakage were shared by high-grade serous carcinoma with different chromosome disruption index
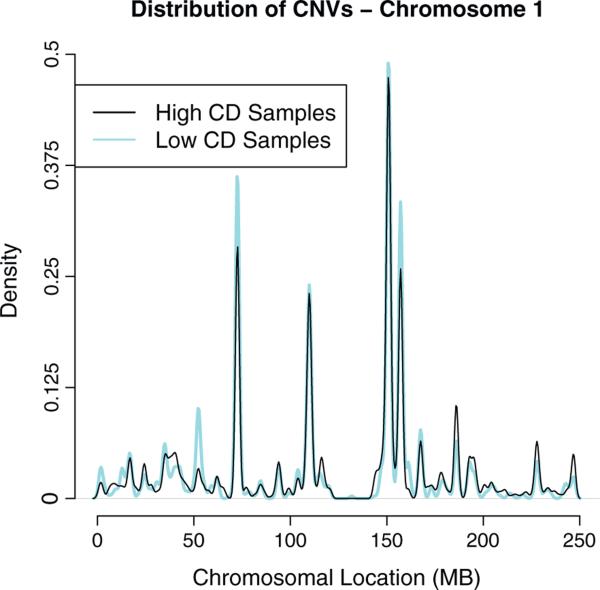
There are at least two possibilities that account for the significant association between CDI and overall survival. First, it is possible that HGSCs with high CDI represent a molecularly distinct group of diseases that develops with a different pathogenesis from those with low CDI. Alternatively, the high levels of chromosomal disruption may be characteristic of HGSC in general, so that the samples with the highest CDI are merely the extremes of a continuum of HGSC developing from the same molecular etiology. Under this model, the CDI might be viewed as a time stamp, with the most advanced tumors having had the opportunity to accumulate more clonal changes. In this study, we found two independent lines of evidence in favor of the single molecular etiology hypothesis. First, the distribution of CDI among HGSCs followed a bell-shaped curve with a single mode (Fig. 1C). If there were distinct disease subgroups, we could expect to observe multiple modes in their distribution. Second, we found that the CNVs occur at similar genomic locations regardless of their CDI. In this analysis, we segregated the HGSCs into equally sized high-, intermediate- and low-CDI groups, and then compared the genomic locations of the CNVs observed in the high-CDI group to those seen in low-CDI using the GenometriCorr measure. We demonstrated that the genomic locations of CNVs in both groups faithfully overlapped each other (Fig. S2). Co-localization of CNV distributions in a representative chromosome 1 was shown in Fig. 3.
Fig. 3.
Distribution of copy number events across chromosome 1. The CNV events identified along chromosome 1 concentrate in a number of hotspots. Interestingly, there was a nearly perfect co-location of CNVs obtained from HGSCs with high (black) and low CDI (gray).
The presence of CNV hotspots prompted us to seek the possible common cause of DNA breakage in HGSCs. Haffner et al. associated double strand break in prostate cancer with androgen receptor binding sites, determining that androgen signaling activity recruits TOP2B to androgen receptor target sites, mediating double strand break at those sites [19]. We sought to determine whether a similar mechanism might operate in ovarian cancer, particularly suspecting that estrogen receptor could play such a role in ovarian cancer [20]. Accordingly, we localized hotspots of chromosomal breakage across the genome, and tested for proximity to the targets of a number of transcription factors (see Materials and methods for details). In total, we considered 480 transcription factors each having at least 5 target genes (Table S1). The strongest associations were to SP2, PMX2B, TEF5, ZFP148, RARB, and PAX6 isoform1, though none of the transcription factor target sets was statistically significantly associated with the breakpoints, even before multiple-test correction.
Chromosomal disruption correlated with expression of genes involved in mitotic checkpoints and DNA damage response
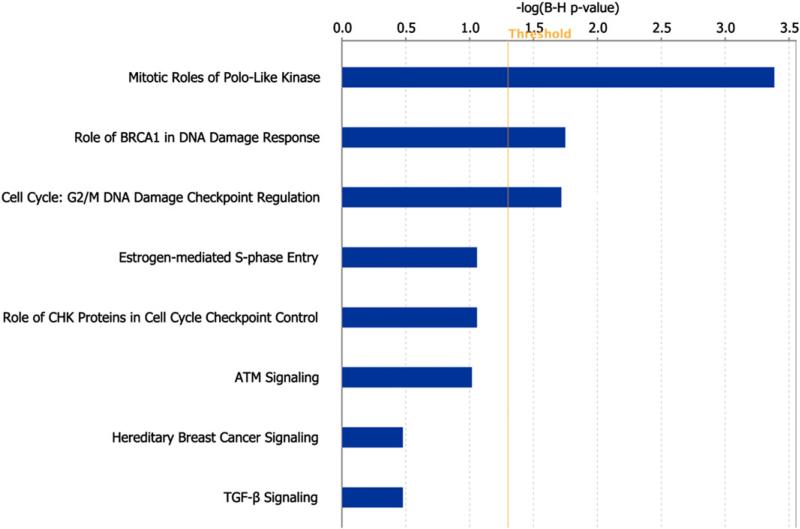
Finally, we tried to identify the molecular pathways associated with chromosomal disruption. We performed a 2-stage linear regression to determine the association between CDI and expression of genes, after adjusting for their copy numbers (Table S2). Genes whose expression was significantly positively correlated with CDI (p<0.001) were identi-fied and used to find enriched pathways through the Ingenuity Pathway Analysis (IPA). Using Benjamini–Hochberg adjusted p-value<0.05 as the significance level, we identified three canonical pathways, including polo-like kinase, BRCA1-mediated DNA damage response and G2/M DNA damage checkpoint, that may contribute to generate high levels of chromosome disruption (Fig. 4). Genes involved in these three pathways were summarized in Table S3.
Fig. 4.
Ingenuity Pathway Analysis of genes whose expression correlated with CDI. Polo-like kinase, BRCA1-mediated DNA damage response and G2/M DNA damage checkpoint pathways were statistically significantly enriched with genes whose expression correlated positively with CDI.
Discussion
Chromosomal instability is an acquired phenotype that characterizes almost all human solid tumors. While it is challenging to directly measure such instability in human tumors as samples obtained from at least two time points are generally required for comparison, most studies infer chromosomal aberration status by capturing the genome-wide “snapshot” of accumulated chromosomal disruption based on ploidy assessment and karyotype [21]. Using those methods, investigators have reported that an increase in both numerical and structural chromosomal changes is associated with worse clinical outcome and drug resistance in several cancer types [22–26]. In order to enhance the resolution in evaluating the result of chromosomal instability, we developed a chromosome disruption index (CDI) which is defined by the discrete subchromosomal segments showing DNA copy number gain or loss as a result of reiterated chromosomal breakage and rearrangement. Using this tool to analyze the TCGA ovarian carcinoma database, we found that ovarian high-grade serous carcinoma (HGSC), a highly lethal tumor, was characterized by widespread chromosomal disruption in its genome. More interestingly, we found that a higher degree of chromosomal disruption (i.e., increased CDI) was independently associated with shorter overall survival, a result suggesting that CDI was a marker for poor clinical response in patients who received the platinum-based chemotherapy. To our best knowledge, this is the first report demonstrating the biological significance of chromosomal disruption in ovarian cancer and our findings should have several biological and translational implications.
Though variable degrees of chromosome disruption were observed in each HGSC sample, we demonstrated that among HGSCs with different degrees of chromosomal disruption, there existed common chromosomal regions susceptible to disruption; the presence of these hotspots suggested an underlying mechanism that caused chromosome breakage at common locations. One such possibility is transcription-induced double strand break: transcription factors such as androgen receptor and estrogen receptor can mediate target-specific double strand break via topoisomerase [19,27]. We tested this hypothesis by assessing the co-localization between the common breakpoints and the target sites of several transcription factors, including estrogen receptor. We found, however, that all transcription factor target gene sets did not geographically correlate with the hotspots of chromosome disruption, suggesting that transcription-induced double strand break could not merely explain the presence of breakpoint hotspots in HGSCs. Alternatively, the breakage hotspots may correspond to common fragile sites, which are genomic regions susceptible to double strand breaks upon oncogene-induced replication stress [28,29]. This may lead to chromo-some breakage and rearrangement at fragile sites, which in turn may contribute to cancer development [30,31]. Since the distribution of fragile sites can be cell-type specific [28], it would be interesting to compare the distribution of fragile sites between HGSC and its putative precursor lesion, serous tubal intraepithelial carcinoma.
In this study, the expression levels of leukocyte- and lymphocyte-associated markers (CD45, CD4 and CD8) did not appear as a strong outcome predictor as the number of tumor-infiltrating lymphocytes shown in previous reports using immunohistochemistry [12–17]. One explanation is that the current study analyzed whole tumor tissues using array platforms, therefore it is not able to distinguish tumor-infiltrating lymphocytes from intravascular lymphocytes. Furthermore, the current study using array approach is not able to estimate the number of tumor-associated lymphocytes given that the expression levels of lymphocyte markers were relative to the overall transcription levels of the tumor and there was a variation in expression of lymphocyte markers for tumor-associated lymphocytes.
Analyzing the set of genes whose expression was highly correlated with high CDI, we identified polo-like kinase pathway as the top pathway significantly associated with a high degree of chromosome disruption. Polo-like kinase, especially PLK1, is one of the key members orchestrating the complex process involving cell division, and specifically it plays crucial roles in entrance of mitosis, centrosome maturation, bipolar spindle formation, chromosome segregation, cytokinesis, and exit of mitosis [32,33]. Deregulation of PLK1 expression, which has been reported to correlate with worse clinical outcome in a variety of cancers [34], may contribute to chromosomal instability, thereby facilitating oncogenic transformation [35]. Stratifying cancers with CDI may help select patients for therapeutics targeting PLK1, which have been under development [36].
The CDI developed in this study may represent one of the first genome-wide markers to classify ovarian cancer patients into different outcome groups. Such stratification may have an impact on clinical management of ovarian cancer patients. For example, the patients who are in the worse survival group may benefit from aggressive therapeutic regimens and being considered to enter emerging clinical trials for new treatment options. Moreover, the analysis platform, i.e., SNP array, has been widely accessible and the method has been relatively established and standardized. The cost for the assay has been precipitously reduced in recent years with a price range that could be similar to routine HER2 fluorescence in situ hybridization test. Therefore, it warrants future studies using different cohorts to validate the clinical utility of CDI in HGSC patients.
In summary, we applied a new computational method to measure the overall chromosomal disruption in HGSC genome with a basis on calculating the total number of amplified and deleted chromosomal segments. Our results provide biological significance of chromosomal disruption in the development of HGSC and, at the same time, generate future research directions worth exploring. For example, the pathways and genes associated with high CDI should be further studied to elucidate the mechanism of chromosomal instability in HGSC. Moreover, our data also indicate the promise of applying CDI in risk-stratifying HGSC patients, possibly for different clinical managements.
Supplementary Material
HIGHLIGHTS.
▶ Chromosome disruption index (CDI) was an independent predictor for overall survival of high-grade serous carcinoma.
▶ Hotspots of chromosome breakage were found across different high-grade serous carcinoma.
▶ Pathways related to mitosis and DNA damage repair were enriched with high CDI-associated genes.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2012.11.031.
Conflict of interest statement
There is no duality of interest for any authors.
References
- 1.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462(2–3):247–53. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Cho KR, Shih I-M. Ovarian cancer. Annu Rev Pathol Mech Dis. 2009;4(1):287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn E, Kurman RJ, Vang R, Sehdev AS, Han G, Soslow R, et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma—evidence supporting the clonal relationship of the two lesions. J Pathol. 2011;226(3):421–6. doi: 10.1002/path.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo KT, Guan B, Feng Y, Mao TL, Chen X, Jinawath N, et al. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69(9):4036–42. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favorov A, Mularoni L, Cope LM, Medvedeva Y, Mironov AA, Makeev VJ, et al. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol. 2012;8(5):e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrew Kossenkov, Manion Frank J, Eugene Korotkov, Moloshok Thomas D, Ochs Michael F. ASAP: automated sequence annotation pipeline for web-based updating of sequence information with a local dynamic database. Bioinformatics. 2003;19(5):675–6. doi: 10.1093/bioinformatics/btg056. [DOI] [PubMed] [Google Scholar]
- 10.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quanti-fication of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li A, Liu Z, Lezon-Geyda K, Sarkar S, Lannin D, Schulz V, et al. GPHMM: an integrated hidden Markov model for identification of copy number alteration and loss of heterozygosity in complex tumor samples using whole genome SNP arrays. Nucleic Acids Res. 2011;39(12):4928–41. doi: 10.1093/nar/gkr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svennevig JL, Lunde OC, Holter J, Bjørgsvik D. Lymphoid infiltration and prognosis in colorectal carcinoma. Br J Cancer. 1984;49(3):375. doi: 10.1038/bjc.1984.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ménard S, Tomasic G, Casalini P, Balsari A, Pilotti S, Cascinelli N, et al. Lymphoid infiltration as a prognostic variable for early-onset breast carcinomas. Clin Cancer Res. 1997;3(5):817–9.. [PubMed] [Google Scholar]
- 14.Roxburgh CSD, Salmond JM, Horgan PG, Oien KA, McMillan DC. Tumour inflammatory infiltrate predicts survival following curative resection for node-negative colorectal cancer. Eur J Cancer. 2009;45(12):2138–45. doi: 10.1016/j.ejca.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102(51):18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104(9):3360–5. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke B, Tinker AV, Lee C-H, Subramanian S, van de Rijn M, Turbin D, et al. Intraepithelial T cells and prognosis in ovarian carcinoma: novel associations with stage, tumor type, and BRCA1 loss. Mod Pathol. 2008;22(3):393–402. doi: 10.1038/modpathol.2008.191. [DOI] [PubMed] [Google Scholar]
- 18.Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 19.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–75. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju B-G, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisom-erase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312(5781):1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 21.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122(4):1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57(7):941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 23.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38(9):1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 24.Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71(5):1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106(21):8671–6. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17(24):7704–11. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haffner MC, De Marzo AM, Meeker AK, Nelson WG, Yegnasubramanian S. Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target? Clin Cancer Res. 2011;17(12):3858–64. doi: 10.1158/1078-0432.CCR-10-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28(1):22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 30.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27(23):3256–64. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 31.Dillon LW, Burrow AA, Wang Y-H. DNA instability at chromosomal fragile sites in cancer. Curr Genom. 2010;11(5):326–37. doi: 10.2174/138920210791616699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barr FA, Silljé HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5(6):429–40. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 33.Donaldson MM, Tavares AAM, Hagan IM, Nigg EA, Glover DM. The mitotic roles of Polo-like kinase. J Cell Sci. 2001;114(13):2357–8. doi: 10.1242/jcs.114.13.2357. [DOI] [PubMed] [Google Scholar]
- 34.Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24(2):287–91. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 35.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24(2):267–76. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 36.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6(4):321–30. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.