Summary
Neutrophils serve critical roles in inflammatory responses to infection and injury, and mechanisms governing their activity represent attractive targets for controlling inflammation. The commensal microbiota is known to regulate the activity of neutrophils and other leucocytes in the intestine, but the systemic impact of the microbiota on neutrophils remains unknown. Here we utilized in vivo imaging in gnotobiotic zebrafish to reveal diverse effects of microbiota colonization on systemic neutrophil development and function. The presence of a microbiota resulted in increased neutrophil number and myeloperoxidase expression, and altered neutrophil localization and migratory behaviours. These effects of the microbiota on neutrophil homeostasis were accompanied by an increased recruitment of neutrophils to injury. Genetic analysis identified the microbiota-induced acute phase protein serum amyloid A (Saa) as a host factor mediating microbial stimulation of tissue-specific neutrophil migratory behaviours. In vitro studies revealed that zebrafish cells respond to Saa exposure by activating NF-κB, and that Saa-dependent neutrophil migration requires NF-κB-dependent gene expression. These results implicate the commensal microbiota as an important environmental factor regulating diverse aspects of systemic neutrophil development and function, and reveal a critical role for a Saa-NF-κB signalling axis in mediating neutrophil migratory responses.
Introduction
Leucocytes such as neutrophils and macrophages are key mediators and effectors of inflammatory stimuli and represent attractive therapeutic targets for controlling acute and chronic inflammation. The complex community of microorganisms residing within the intestine (gut microbiota) has been identified as an important environmental factor regulating leucocyte function within the intestinal compartment (Abt and Artis, 2009). However, the presence of microbiota appears to also have profound systemic effects on leucocytes. Peripheral neutrophils collected from germ-free (GF) rodents display reduced phagocytosis, microbicidal activity, and production of superoxide anion and nitric oxide compared with ex-GF animals colonized with normal microbiota (conventionalized or CONVD) (Ohkubo et al., 1990; 1999; Clarke et al., 2010). Similarly, macrophages collected from peritoneal exudate in GF animals display reduced superoxide anion production and microbicidal activity, and impaired chemotaxis compared with CONVD controls (Jungi and McGregor, 1978; Morland and Midtvedt, 1984; Czuprynski and Brown, 1985; Mitsuyama et al., 1986; Oliveira et al., 2005). The importance of the microbiota on systemic inflammation is further underscored by reports that multiple rodent models of spondyloarthritis do not develop disease when raised under GF conditions (Taurog et al., 1994; Rehakova et al., 2000), but disease can be initiated upon gut colonization with specific bacteria (Rath et al., 1996; Sinkorova et al., 2008).
Although recent research has yielded an abundance of new information about the impact of gut microbiota on intestinal leucocyte biology and immunity (Abt and Artis, 2009), gut microbiota effects on systemic leucocyte biology remain relatively unresolved. Our current information of the systemic effects of microbiota on neutrophils is largely derived from ex vivo experiments conducted on neutrophils collected from peripheral blood or bone marrow from GF and CONVD mammals (Ohkubo et al., 1990; 1999; Clarke et al., 2010). However, studies generated from ex vivo neutrophils may not be representative of the systemic population and do not fully recapitulate the native physiologic context of live tissues. Finally, mammals are not amenable to the high-resolution in vivo microscopy required to comprehensively define the systemic impact of microbiota on neutrophils. As a result, the specific aspects of systemic neutrophil activity affected by microbiota are not fully understood.
The zebrafish has several features that make it an attractive model to study the roles of commensal microbiota on systemic neutrophil biology. First, zebrafish are optically transparent from fertilization through early adulthood, permitting high-resolution imaging of host–microbe interactions in the intact physiologic context of a living vertebrate (Rawls et al., 2007; Kanther et al., 2011). Second, the zebrafish has innate and adaptive immune systems that share extensive homology with those of humans and other mammals (Kanther and Rawls, 2010). Likewise, the digestive tracts of zebrafish and mammals are similar, including an intestine, pancreas, liver and gall bladder (Ng et al., 2005; Wallace et al., 2005). Third, we have developed methods for rearing zebrafish under GF conditions and colonizing GF zebrafish with members of the commensal microbiota (Pham et al., 2008).
Previous analyses of gnotobiotic zebrafish and mice have revealed that the presence of a microbiota causes extensive alterations in diverse aspects of host immunity and physiology. Reciprocally, host-mediated mucosal factors such as antimicrobial proteins, IgA, mucins, and inflammation alter microbial community composition and function (reviewed in Abt and Artis, 2009; Kanther and Rawls, 2010; Hooper et al., 2012; Tremaroli and Backhed, 2012). This complex interplay between host and microbial factors is central to the maintenance of homeostasis. However the host signalling pathways that mediate microbial cues to regulate systemic leucocyte responses remain unresolved.
Serum amyloid A (Saa) is a circulating HDL-associated apolipoprotein and acute phase protein. The Saa gene family (3 in humans, 4 in mice, 1 in zebrafish) is conserved across vertebrates (Fig. S1), suggesting important biological roles. Saa genes are expressed by multiple tissues including liver, intestinal epithelium (Eckhardt et al., 2010), and macrophages (Meek et al., 1992) and are markedly induced by diverse inflammatory stimuli (Uhlar and Whitehead, 1999) including gut microbiota (Hooper et al., 2001; Rawls et al., 2006; Ivanov et al., 2009; Kanther et al., 2011). Serum Saa protein level is a salient biomarker for inflammatory disorders including IBD (Okahara et al., 2005; Noble et al., 2008), necrotizing enterocolitis, sepsis (Ng et al., 2010), and chronic obstructive pulmonary disease (Bozinovski et al., 2008). The precise roles of Saa in inflammation remain elusive because both pro- and anti-inflammatory actions have been reported. Reported pro-inflammatory roles for Saa include stimulation of extracellular matrix (ECM)-degrading enzymes such as MMP9 (Lee et al., 2005), recruitment of neutrophils and monocytes (Badolato et al., 1994; Su et al., 1999; Connolly et al., 2010), suppression of neutrophil apoptosis (Christenson et al., 2008), stimulation of granulocytosis (He et al., 2009), opsonization of Gram-negative bacteria (Shah et al., 2006), Nlrp3 inflammasome activation (Ather et al., 2011; Niemi et al., 2011), and stimulation of pro-inflammatory cytokines such as IL1β (Patel et al., 1998; Lee et al., 2005; Cheng et al., 2008; Niemi et al., 2011). In contrast, numerous reports cite anti-inflammatory effects of Saa on neutrophils, including inhibition of MPO production (Renckens et al., 2006), oxidative burst (Linke et al., 1991; Gatt et al., 1998), and migration (Gatt et al., 1998), and induction of IL10 expression (Shah et al., 2006; Cheng et al., 2008; De Santo et al., 2010). These diverse effects may be due to Saa's ability to stimulate signalling events through multiple transmembrane receptors, including formyl peptide receptor 2 (Fpr2/Fprl1) (Su et al., 1999), receptor for advanced glycosylation end-products (RAGE) (Cai et al., 2007), scavenger receptor class B type I (Scarb1/CLA-1) (Baranova et al., 2005), and Toll-like receptor 2 (Tlr2) (He et al., 2009). The signal transduction pathways that act downstream of Saa to regulate gene expression include extracellular signal-regulated kinases 1 and 2 (ERK1/2), p38, c-Jun N-terminal kinase (JNK), Akt and NF-κB (Baranova et al., 2005; Jijon et al., 2005; Cheng et al., 2008). However, the relationship between these pathways and the distinct immune cellular responses evoked by Saa remain unclear. Importantly, in vivo genetic analysis of Saa has been complicated by the fact that the human and mouse genomes encode 3 and 4 paralogous Saa genes respectively (Fig. S1). We previously showed that colonization with a normal microbiota in zebrafish results in NF-κB-dependent induction of saa expression in the distal intestine, liver and swim bladder (Kanther et al., 2011). However, the in vivo roles of Saa in systemic neutrophil biology, as well as neutrophil requirements for NF-κB in these responses, remain unclear. In this study, we took advantage of the fact that the zebrafish genome encodes only a single Saa gene to define the requirement for Saa in microbiota-induced neutrophil responses. Our results reveal novel roles for the microbiota on systemic neutrophil biology including increased number and migratory behaviour and suggest that Saa-dependent neutrophil migration requires NF-κB signalling.
Results
Microbiota promotes increased neutrophil number and pro-inflammatory gene expression
To investigate the impact of the commensal microbiota on zebrafish myeloid lineages, we queried results from a microarray-based functional genomic comparison of gene expression in whole zebrafish at 6 days post fertilization (dpf) that had been raised GF or conventionalized since 3dpf (CONVD). Functional categorization of the resulting list of microbiota-regulated transcripts revealed enrichment for genes involved in leucocyte development and function (Kanther et al., 2011). CONVD zebrafish displayed relative increases in transcript levels for 17 genes known to be specifically expressed by myeloid leucocytes, including the zebrafish homologue of mammalian myeloperoxidase mpx (also called mpo; Table 1) (Rawls et al., 2004; 2006; Kanther et al., 2011). Our previous whole-mount in situ hybridization analysis of mpx mRNA in GF and CONVD zebrafish suggested that this increase in mpx transcript level could be due to increased neutrophil number or increased mpx mRNA levels in individual neutrophils (Kanther et al., 2011). To test if these transcript differences were associated with alterations in neutrophil number, we used transgenic Tg(mpx:GFP) zebrafish that robustly express GFP specifically in neutrophils (Renshaw et al., 2006). Stereomicroscopic evaluation of GFP(+) neutrophil number and localization in whole 6dpf GF and CONVD zebrafish revealed a qualitative increase in neutrophil number throughout the animal (Fig. 1A). Flow cytometry of GFP(+) neutrophils from dissociated 6dpf Tg(mpx:GFP) fish confirmed a significant increase in total steady-state number of neutrophils per animal in CONVD compared with GF animals (Fig. 1B). Quantitative RT-PCR in flow-sorted neutrophils from GF and CONVD larvae revealed significant increases in mpx mRNA in sorted neutrophils from CONVD animals (Fig. 1C). Colonization with a commensal microbiota therefore results in significant increases in neutrophil mpx expression together with increases in steady-state neutrophil number.
Table 1.
Elevated transcript levels for myeloid lineage genes in CONVD compared with GF zebrafish larvae.
| Gene name | FCa | Referenceb |
|---|---|---|
| myeloid-specific peroxidase (mpx) | 8.0 | Bennett et al., 2001; Lieschke et al., 2001 |
| matrix metalloproteinase 9 (mmp9) | 7.2 | Yoong et al., 2007 |
| microfibrillar-associated protein 4 (mfap4) | 6.4 | Zakrzewska et al., 2010 |
| matrix metalloproteinase 13a (mmp13a) | 5.1 | Qian et al., 2005; Yoong et al., 2007 |
| neutrophil cytosolic factor 1 (ncf1) | 3.0 | Qian et al., 2005 |
| lymphocyte cytosolic plastin 1 (lcp1) | 2.2 | Bennett et al., 2001 |
| CCAAT/enhancer binding protein, beta (cebpb) | 2.1 | Thisse et al., 2001 |
| coronin, actin binding protein, 1A (coro1a) | 2.0 | Song et al., 2004 |
Transcript fold change (FC) in 6dpf CONVD compared with GF zebrafish larvae from Kanther et al., (2011).
Reference establishing myeloid lineage expression for the respective zebrafish gene.
Fig. 1.
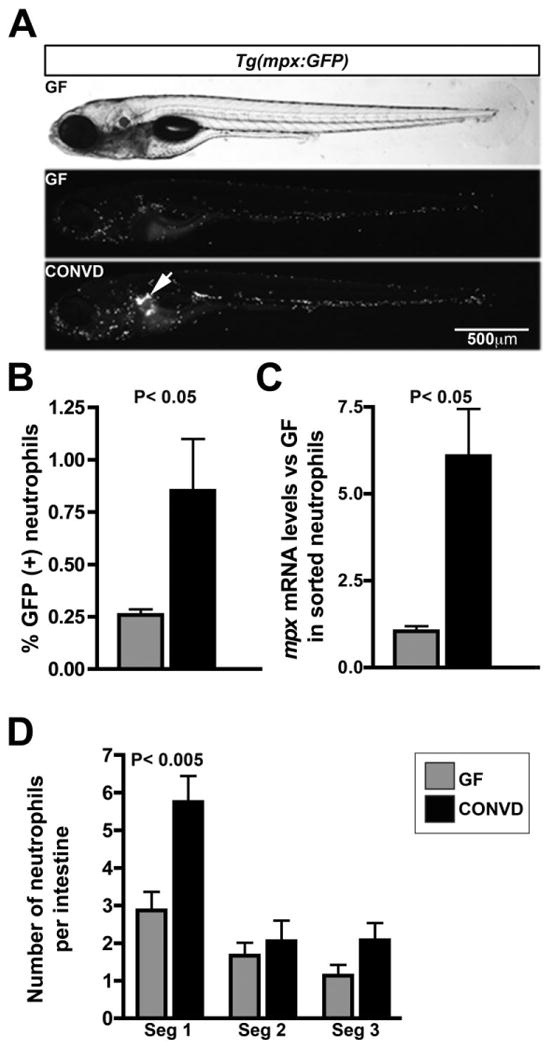
Microbiota regulates neutrophil localization, number and stimulates inflammatory biomarkers. A. Live 6dpf Tg(mpx:GFP) zebrafish show GFP(+) neutrophil localization as a function of microbial status. Note the increased neutrophil localization in the kidney (white arrow) in CONVD animals. B. Flow cytometry analysis reveals that the percent frequency of GFP(+) neutrophils in dissociated 6dpf Tg(mpx:GFP) zebrafish is higher in CONVD compared with GF animals. C. qRT-PCR for mpx mRNA levels in sorted neutrophils. D. Quantification of mean total number of GFP(+) neutrophils associated with dissected intestines by segment. Data are representative of 8–10 guts per microbial condition from two biological replicates. Significant Student's t-test P-values are shown.
Microbiota regulates tissue distribution and migration of neutrophils
We imaged whole 6dpf GF and CONVD Tg(mpx:GFP) zebrafish to evaluate the effect of microbiota on neutrophil localization and migration. Consistant with our previous mpx RNA whole-mount in situ hybridization results (Kanther et al., 2011), CONVD zebrafish displayed increased GFP expression in the kidney, a site of definitive haematopoiesis (Fig. 1A). Since the intestine harbours microbial communities that are markedly denser than that of the surrounding water, we analysed the frequency and distribution of GFP(+) neutrophils associated with intestines dissected from Tg(mpx:GFP) GF and CONVD larvae. We observed increased numbers of GFP(+) neutrophils in the intestines of CONVD Tg(mpx:GFP) zebrafish compared with GF controls, most significantly in the proximal region (segment 1) of the intestine (Fig. 1D). To determine if these changes in localization were associated with altered neutrophil migratory behaviours, we used confocal microscopy to quantify migration of individual neutrophils in live 6dpf GF and CONVD Tg(mpx:GFP) fish. Compared with GF controls, neutrophils in CONVD animals displayed significantly elevated migration velocity and decreased meandering (i.e. increased directional migration) compared with GF controls (Fig. 2, Movies S1 and S2). These results indicate that the microbiota regulates systemic neutrophil localization and migratory activity.
Fig. 2.
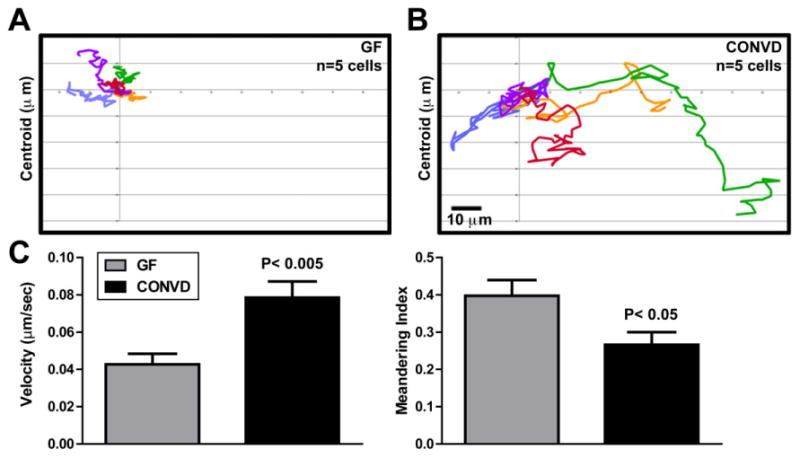
Microbiota induces systemic neutrophil migration. Neutrophil tracking analysis of the CHT from 15-min movies of live 6dpf GF (A) and CONVD (B) Tg(mpx:GFP) zebrafish reveals increased migratory activity in CONVD. C. Quantification of neutrophil migration velocity and meandering index in 6dpf GF and CONVD zebrafish (calculated from 29 neutrophils per condition). Significant Student's t-test P-values are shown. See also Movies S1 and S2.
Microbiota promotes neutrophil recruitment to extra-intestinal injury
To determine if the observed effects of microbiota on neutrophil number, localization, and migration have functional consequences, we used a well-established injury model in which a portion of the tail fin in larval zebrafish is resected and the recruitment of leucocytes to the wound is quantified over time (Renshaw et al., 2006; Yoo and Huttenlocher, 2011). Although early (1 h) GFP(+) neutrophil recruitment to the wound was slightly higher in GF animals compared with CONVD controls, later evaluation at 3, 6 and 15 h after injury revealed significantly more neutrophils recruited to the wound in CONVD animals (Fig. 3). These results confirm that colonization with a microbiota augments the host's capacity for recruiting neutrophils to extra-intestinal injury.
Fig. 3.
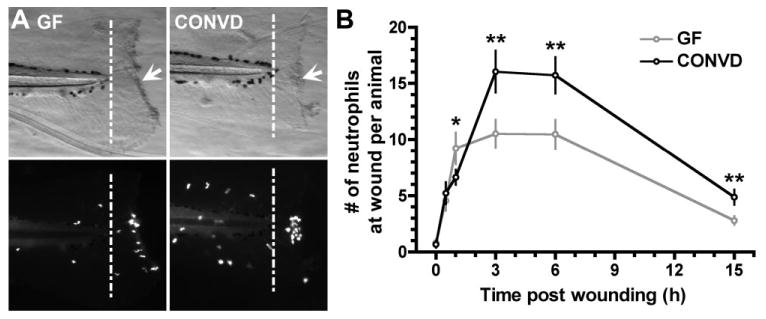
Microbiota promotes neutrophil recruitment to tail wounds in GF and CONVD Tg(mpx:GFP) zebrafish. A. Brightfield and GFP fluorescence images of 6dpf GF and CONVD zebrafish tails 3 h post injury. B. Mean numbers of neutrophils recruited to wound site (posterior to the end of the notochord marked by white dashed line) at time points post injury as indicated. Data represents 8–10 fish per condition per time point. Significant Student's t-test P-values are shown: *P < 0.05 and **P < 0.005 vs GF.
Saa is required for increases in neutrophil migration and suppression of inflammatory biomarkers following colonization with a microbiota
We next sought to define the role of zebrafish saa on neutrophil responses to the microbiota. Injection of zebrafish embryos with morpholino antisense oligonucleotides (MO) targeting saa resulted in partial knockdown of saa transcript through 6dpf (Fig. S2A). By comparing 6dpf GF and CONVD Tg(mpx:GFP) zebrafish injected with a MO targeting saa (saa-MO) or a standard negative control MO (ctrl-MO), we found that saa knockdown did not qualitatively alter the effects of the microbiota on neutrophil tissue distribution (Fig. 4A). Using a computational approach to quantify GFP(+) neutrophil number in whole live zebrafish (Ellett and Lieschke, 2012), we observed that the microbiota-induced increases in neutrophil number (Fig. 4B) and mpx mRNA levels (Fig. 4C) were also not affected by saa knockdown. In contrast, in vivo imaging of 6dpf GF and CONVD Tg(mpx:GFP) fish revealed striking tissue-specific saa-dependent alterations in microbiota-induced neutrophil migration behaviour (Movies S3–5 and S6). In ctrl-MO animals, neutrophils associated with the CHT, intestine, and fin displayed significantly increased migration velocity in the presence of a microbiota. In saa-MO animals, the microbiota caused a similar increase in fin neutrophils but failed to increase neutrophil migration velocity in the CHT and intestine (Fig. 4F). The presence of a microbiota in ctrl-MO animals caused a significant reduction in the meandering index of neutrophils associated with the CHT but not those in the intestine or fin. In saa-MO animals, the effect of the microbiota on CHT neutrophil meandering was significantly attenuated (Fig. 4G). Strikingly, neutrophils associated with the intestine in saa-MO animals displayed a microbiota-dependent decrease in meandering that was not observed in ctrl-MO controls (Fig. 4G). These results suggest multiple novel in vivo roles for Saa in regulating tissue-specific neutrophil migratory behaviours. These alterations in microbiota-induced neutrophil migration in saa-MO fish were accompanied with significant increases in microbiota-dependent induction of inflammatory biomarkers ncf1 and il1b (Fig. 4D and E). These results reveal a potential anti-inflammatory role for saa in suppressing inflammatory gene expression and complex tissue-specific roles in neutrophil migration responses to commensal microbiota.
Fig. 4.
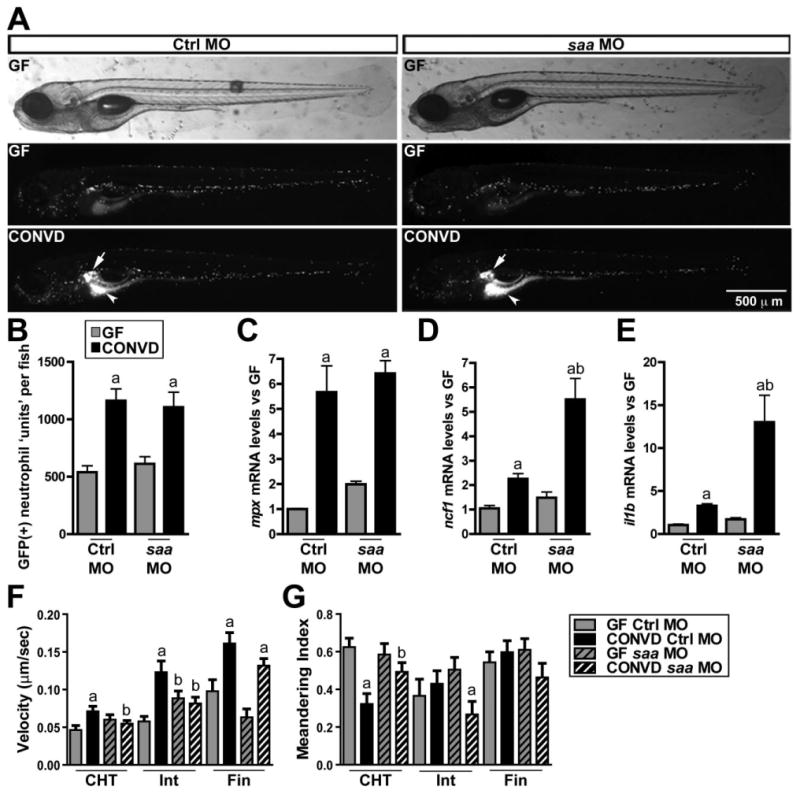
saa mediates systemic neutrophil migration in response to the microbiota. Comparisons of 6dpf GF and CONVD Tg(mpx:GFP) zebrafish injected with either standard control (Ctrl MO) or a morpholino targeting saa (saa MO). A. Images of whole live 6dpf Tg(mpx:GFP) zebrafish show GFP(+) neutrophil localization including increased concentration of neutrophils in the kidney (white arrow) and intestine (white arrow head) in CONVD Ctrl MO and saa MO zebrafish. B. Neutrophil units quantified by GFP densitometry from whole animal images similar to those shown in A. C. qRT-PCR for mpx mRNA in sorted neutrophils. qRT-PCR for ncf1 (D) and il1b (E) mRNA in whole 6dpf zebrafish. Quantification of neutrophil velocity (F) and meandering index (G) in the CHT, intestine (Int), and fin (calculated from 7–29 neutrophils per tissue per condition). Student's t-test P-values are indicated: a, P < 0.05 compared with GF condition in same genotype; b, P < 0.05 compared with Ctrl MO in same microbial condition. See also Movies S3–S6.
Saa-dependent induction of neutrophil migration requires NF-κB activity
Saa has been shown to promote neutrophil migration in mammals (He et al., 2009; Connolly et al., 2010) and we have shown that SAA activates NF-κB signalling in mammalian cells (Jijon et al., 2005). Because the NF-κB transcription factor has been linked to neutrophil migration (Penzo et al., 2010), we hypothesized that increased Saa levels in response to microbiota might activate NF-κB and NF-κB-dependent immune cell migration. To test this, we turned to cell culture where the cell autonomous roles of SAA and NF-κB could be readily evaluated. Culture methods for purified zebrafish neutrophils have not been established, therefore we first tested the effects of SAA on the PAC2 zebrafish fibroblast cell line. Western blot analysis showed that SAA induced phosphorylation of the NF-κB protein inhibitor IκBα, a key process for canonical NF-κB activity (Fig. 5A). Immunofluorescence analysis showed that SAA promoted nuclear translocation of the NF-κB transcriptional subunit RelA/p65 (data not shown). To confirm that SAA functionally impacts NF-κB signalling, we investigated transcriptional activity using pikbaa:Luc gene reporter system (Kanther et al., 2011). Luciferase activity increased ∼3-fold in pikbaa:Luc-transfected PAC2 cells following SAA stimulation, a level similar to LPS stimulation (Fig. 5B). Importantly, increased NF-κB activity was associated with SAA-induced accumulation of NF-κB target genes mmp9 and ikbaa mRNA (Fig. 5C and D). These findings demonstrate that SAA induces NF-κB signalling and expression of NF-κB target genes in zebrafish cells. We next sought to directly test the functional impact of NF-κB signalling in Saa-induced neutrophil migration using mouse peritoneal neutrophils. Using a transwell migration assay, we observed that peritoneal neutrophil motility in response to SAA increased by ∼2-fold (Fig. 6A and B). Neutrophil migration was reduced by 82% when the NF-κB inhibitor Bay 11–7082 (BAY) was co-incubated with SAA (Fig. 6A and B). Treatment with the protein synthesis inhibitor cycloheximide (CHX) strongly attenuated SAA-induced neutrophil transmigration (73%), suggesting that NF-κB-mediated gene expression is necessary for neutrophil movement (Fig. 6A and B). These findings implicate microbiota-induced SAA expression as an important host response regulating neutrophil behaviour.
Fig. 5.
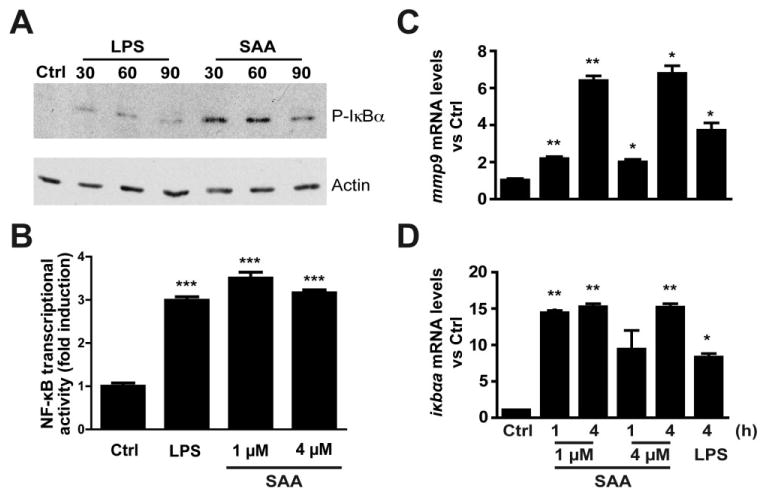
SAA stimulation of a zebrafish cell line results in activation of the canonical NF-κB pathway and induces expression of NF-κB target genes. A. Western blot of zebrafish PAC-2 cells shows rapid phosphorylation of IκBα proteins after LPS (10 μg ml−1) or SAA (4 μM) stimulation. B. Zebrafish PAC-2 cells transfected with ikbaa-luciferase gene reporter (pikbaa:Luc) show increased luciferase activity upon stimulation with LPS (10 μg ml−1) or SAA (1, 4 μM). C and D. qRT-PCR using primers for mmp9 and ikbaa, predicted NF-κB target genes, demonstrate induction upon stimulation of PAC-2 cells with LPS (10 μg ml−1) or SAA (1, 4 μM) normalized to 18S ribosomal RNA [rRNA]). Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. 6.
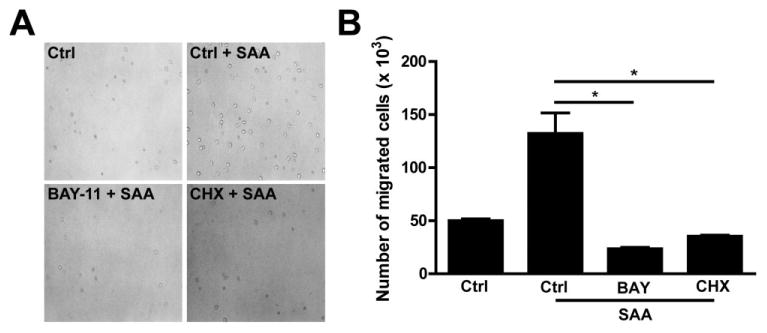
SAA promotes neutrophil migration and requires NF-κB and protein synthesis. A. Peritoneal isolated murine neutrophils were pretreated for 1 h with BAY 11-7082 (BAY, 25 μM) or cycloheximide (CHX, 50 μg ml−1) and then plated into the top well of a Transwell system. The cells' migration in response to SAA (2.08 μM, 25 μg ml−1) in the bottom well was enumerated after 2.5 h. Representative light images of neutrophils migrated into bottom wells. Magnification 200×. B. Quantitative measurements of migrated neutrophils. Data are expressed as mean ± SEM. *P < 0.05. Results are representative of two independent experiments.
Discussion
The majority of microbes on the human body reside in the intestine, where they are known to contribute significantly to intestinal physiology and mucosal immunity. There is, however, increasing evidence that the influence of the microbiota extends beyond the confines of the intestine to other tissues and their pathologies (McFall-Ngai et al., 2013). Therefore, the identification of the cellular and molecular mechanisms by which the microbiota shapes the systemic physiology of animal hosts is an important research objective. An improved understanding of the microbiota's impact on systemic leucocyte function is particularly needed due to the implication of the microbiota in the aetiology of inflammatory diseases in intestinal and extra-intestinal compartments. Previous analysis of the microbiota's impact on neutrophil biology in mammals has been limited to ex vivo comparisons of neutrophils collected from peripheral blood or bone marrow (Ferencik et al., 1985; Ohkubo et al., 1990; 1999; Clarke et al., 2010). Here we have utilized the transparency of the zebrafish to provide the first comprehensive view of the microbiota's systemic impact on in vivo neutrophil function. Our results reveal diverse consequences of microbiota colonization on neutrophil homeostasis and behaviour, as well as recruitment of neutrophils to injury. We also show a mechanistic role for a SAA-NF-κB signalling axis in microbiota-dependent neutrophil migration. These findings underscore the potential of the microbiota to influence the systemic physiology of animal hosts and provide an important new conceptual framework for understanding the microbiota's roles in inflammatory diseases.
Our observations of increased systemic neutrophil number in CONVD compared with GF zebrafish larvae reveal a novel role for the microbiota in defining the steady state neutrophil population. Systemic neutrophil number might be influenced by differences in microbiota composition or husbandry practices, because a recent comparison of starved GF and conventionally raised zebrafish larvae in a different zebrafish facility did not reveal differences in neutrophil number (Galindo-Villegas et al., 2012). Inflammatory stimuli can regulate the hematopoietic compartment in zebrafish, as injection with LPS induces myelopoiesis (Liongue et al., 2009) and bacterial infection or tail wounding induces mobilization of neutrophils from the CHT (Yoo and Huttenlocher, 2011; Deng et al., 2013). Our study using commensal microbial colonization adds a novel aspect to bacteria-host interaction by showing that microbial cues regulate myelopoietic programs.
In vivo imaging of GFP(+) neutrophils in gnotobiotic zebrafish revealed significant influences of microbiota on neutrophil localization and migratory behaviour. We detected a quantitative increase in neutrophil localization in the intestines of CONVD versus GF zebrafish, with the most marked increases in the proximal intestine. This is consistent with a previous study by Bates and colleagues that reported an increase in MPO(+) cell number in the distal intestine of CONVD versus GF zebrafish (Bates et al., 2007). These microbiota-associated increases in intestinal neutrophil number may be due to tissue-specific alterations in neutrophil recruitment or retention, or may simply reflect the observed overall increase in systemic neutrophil number. Sites of haematopoiesis are dynamic during zebrafish development, and occur in the CHT and kidney at the larval stages under study here (for review see Kanther and Rawls, 2010). We observed salient qualitative increases in GFP(+) neutrophil localization to the kidney region in CONVD zebrafish. The significance of this microbiota-induced neutrophil localization remains unknown, and could be indicative of altered granulopoiesis in the kidney hematopoietic tissue or linked to microbiota-induced NF-κB activation in the adjacent swim bladder (Kanther et al., 2011). Time-lapse in vivo microscopy revealed that microbiota-induced alterations in neutrophil localization were accompanied by significantly increased neutrophil migration velocity in all evaluated tissues and decreased neutrophil meandering in the CHT region. These observed tissue-specific influences of the microbiota on neutrophil behaviour underscore the utility of in vivo imaging in the zebrafish for defining the regional impact of microbial colonization status.
These systemic impacts of the microbiota on neutrophil development are predicted to have diverse functional consequences on host immunity and inflammation. In support, we found that the presence of a microbiota is associated with significant alterations in inflammatory gene expression in neutrophils. Our previous genomic comparison of whole GF and CONVD zebrafish larvae revealed that transcripts encoding multiple myeloid markers including neutrophil-specific mpx were increased in the presence of the microbiota (Kanther et al., 2011). Moreover, we previously identified individual bacterial species sufficient to induce mpx in gnotobiotic zebrafish (Rawls et al., 2004; 2006; Kanther et al., 2011) and showed that this response requires functional bacterial flagella (Rawls et al., 2007). Here we found that mpx mRNA levels were increased in sorted neutrophils from CONVD compared with GF zebrafish larvae. This is consistent with previous analysis of MPO levels in neutrophils from gnotobiotic mammals (Ferencik et al., 1985), suggesting an evolutionarily conserved role for microbiota in controlling neutrophil gene expression programs. The breadth and impact of microbiota-induced alterations in neutrophil transcription remain to be defined. However, a recent genetic analysis in zebrafish revealed that mpx functions to downregulate H2O2 gradients established at sites of injury and thereby contributes to the resolution of inflammation (Pase et al., 2012). This data suggests that increased mpx expression in neutrophils in the presence of the microbiota might serve as an anti-inflammatory response to commensal microbial colonization. Consistent with a previous study (Galindo-Villegas et al., 2012), we also found that the presence of a microbiota significantly increased the number of neutrophils recruited to a fin injury. This could be due to increased recruitment or retention of neutrophils in the wound in the presence of a microbiota, or could reflect the observed overall increase in systemic neutrophil number in those animals. Although GF neutrophils express less mpx transcript, other microbiota-induced responses must cause this wound recruitment phenotype because mpx-deficient neutrophils develop normally and migrate normally to a fin wound (Pase et al., 2012). Notably, GF fish recruited more neutrophils exclusively at an early time point (1 h) after injury, suggesting neutrophils might be slower to arrive but accumulate in greater numbers in the wound of colonized animals. Together, these results establish that the microbiota regulates neutrophil function as well as development, and suggest that identification of the underlying molecular mechanisms could provide potential therapeutic targets for controlling inflammation.
Our results identify a novel role for Saa in regulating neutrophil migratory behaviour in response to microbiota colonization. SAA proteins are known to be produced by multiple vertebrate tissues and cell types in response to various stimuli including gut microbiota. Indeed, we showed that zebrafish saa transcript levels in distal intestine, liver, and swim bladder are elevated upon colonization with a microbiota via myd88-dependent and NF-κB-dependent mechanisms (Kanther et al., 2011). However the functional consequence of commensal microbiota-induced SAA on neutrophil activity remained unknown. The existence of a single Saa orthologue in zebrafish allowed us to test the requirement of saa function on neutrophil responses to the microbiota using MO knockdown. Microbiota-induced alterations in neutrophil number, localization and mpx levels were unaffected by saa knockdown, suggesting that these host responses do not require saa. In contrast, neutrophil migratory responses to the microbiota were strongly affected by saa knockdown. Microbiota-induced increases in neutrophil velocity were attenuated in the CHT and intestines of saa-MO fish but not in the fin. This reveals novel tissue-specific roles for saa in neutrophil migration, and suggests that neutrophils located in these tissue compartments might have different sensitivities or accessibility to Saa protein. The ability of the microbiota to induce a lower meandering index in the CHT was also attenuated following saa knockdown, suggesting that this host response also requires Saa. Knockdown of saa additionally caused an unexpected reduction in meandering index in the intestine, consistent with an increased inflammatory tone in that tissue. In support of this notion, saa knockdown was associated with increased expression of inflammatory biomarkers il1b and ncf1, reminiscent of increased susceptibility to DSS colitis in mice deficient for Saa1 and Saa2 (Eckhardt et al., 2010). Importantly, MO injection resulted in only a partial knockdown of wild-type saa transcript. Therefore saa may have additional functions that could be revealed by stronger loss of function approaches.
Saa induces neutrophil migration in mammals as well as zebrafish. Saa is known to activate NF-κB signalling in mammalian cells, and NF-κB has also been linked to mammalian neutrophil migration. To test whether Saa induces neutrophil migration by activating NF-κB, we turned to cell culture platforms where the cell autonomous effects of Saa can be readily evaluated. We find that zebrafish fibroblasts, like mammalian cells, respond to Saa exposure by inducing the NF-κB signalling pathway and downstream transcriptional targets. Transwell migration assays using murine peritoneal neutrophils revealed that Saa-dependent induction of neutrophil migration requires NF-κB-dependent gene expression. We previously found that the ability of the microbiota to induce zebrafish saa required NF-κB (Kanther et al., 2011), indicating that the NF-κB pathway is involved at multiple steps in this process. Together, our data support a model in which the presence of a microbiota results in NF-κB-dependent induction of saa in multiple tissues, which leads to systemic NF-κB-dependent increases in neutrophil migration. In parallel, the microbiota causes altered neutrophil number, localization, and mpx expression using saa-independent mechanisms. Since the NF-κB pathway regulates gene transcription in multiple tissues and cell types (Kanther et al., 2011), in vivo analysis of the cell-autonomous roles of NF-κB in neutrophil behaviour will require new approaches for controlling the NF-κB pathway specifically in the neutrophil lineage. Although this study focused exclusively on neutrophils, we anticipate that the microbiota might have similar effects on other leucocyte lineages. Additional studies are needed to determine the similarities and differences between leucocyte responses to colonization by commensal microbiota and infection with pathogenic microbes. Of particular interest are the mechanisms by which gut microbes might regulate aspects of haematopoiesis and mobilization of immune cells to distinct target tissues such as the gut. An improved understanding of how commensal and pathogenic microbes control systemic neutrophil function could lead to the development of new probiotic, antibiotic and pharmacologic approaches for controlling neutrophil activity and inflammation to reduce incidence and severity of human IBD and other inflammatory diseases.
Experimental procedures
Animal husbandry
All experiments using zebrafish and mice were performed using protocols approved by the Animal Studies Committee of the University of North Carolina at Chapel Hill. Conventionally raised wild-type (TL strain) and Tg(BACmpx:GFP)i114 [hereafter referred to as Tg(mpx:GFP)] (Renshaw et al., 2006) zebrafish were maintained as described (Flynn et al., 2009; Kanther et al., 2011). Production using in vitro fertilization methods, colonization, maintenance and sterility testing of GF zebrafish was performed as described (Pham et al., 2008). GF and CONVD animals were reared at an average density of 1.3 animals per ml in sterile vented tissue culture flasks (Cellstar) housed in an air incubator at 28.5°C on a 14 h light cycle. Wild-type 8- to 12-wk-old C57BL/6 mice were maintained under specific pathogen free conditions.
In vivo imaging
For time-lapse imaging, zebrafish were anesthetized in 4× Tricaine (MS-222; Sigma-Aldrich), and then mounted in 1% low melting point agarose containing 1× Tricaine on glass bottom dishes (MatTek Corporation). Solidified agarose containing fish was then covered in sterile GZM containing 2× Tricaine. Time-lapse movies were captured using a Zeiss 510 Meta Laser Scanning Confocal Microscope at a rate of 1 frame every 15 s for 5 or 15 min. For live whole animal imaging, zebrafish were anesthetized as described above, mounted in 3% methylcellulose, and imaged using a Leica M205 FA stereomicroscope.
Tail wounding assay
GF and CONVD 6dpf Tg(mpx:GFP) zebrafish were anesthetized as described above. Fish were mounted in 3% methylcellulose on a 35 mm Petri dish by placing only the anterior part of the fish into the methylcellulose. The posterior part of the fish was covered with sterile GZM. Fish were then observed under a LeicaS6E StereoZoom stereomicroscope with a Leica L2 cold light illuminator. Tail amputations were performed posterior to the end of the notochord using a scalpel. GF and CONVD animals were revived in sterile GZM containing antibiotics (AB-GZM) (Pham et al., 2008), and kept at 28.5°C. Fish were collected at time points indicated and euthanized in 8× Tricaine. Larvae were fixed in 4% paraformaldehyde overnight at 4°C, and washed 3 times for 10 min and 3 times for 1 h in PBS + 0.2% Tween. Fish were then mounted in 3% methylcellulose and imaged using a Leica M205 FA stereomicroscope. Numbers of GFP(+) neutrophils posterior to the notochord were quantified in 8–10 fish for each condition.
Morpholino injections and validation
Zebrafish embryos at the 1–2 cell stage were injected with morpholinos (GeneTools LLC) targeting saa (saa.i2e3, 0.9 pmol per embryo; GTCCTTTGCACTTCAAAAATAGAGT), or standard control MO (0.9 pmol per embryo; Gene Tools LLC) using a Drummond Nanoject II microinjector. Efficacy of splice-blocking MOs was measured by RT-PCR. cDNA was prepared from pools of whole larvae at 6dpf as described (5–15 larvae per pool) (Rawls et al., 2007), and 10 ng of cDNA was used as a template in PCR reactions using gene-specific primers (forward: CTTGCTGTGCTGGTGATGTT; reverse: AGTCTTCTGGGGGT CATCTTC). We resolved PCR amplicons on 2% agarose gels to detect morphant transcripts (Fig. S2).
Flow cytometry analysis
Tg(mpx:GFP) zebrafish were reared under GF and CONVD conditions. GF and CONVD 6dpf larvae were pooled and killed (50 animals per condition per experiment). Excess media was removed and animals were finely chopped using sterile razor blades in a 10 cm Petri dish, diluted in 1.5 ml 5% fetal bovine serum in Hanks' balanced salt solution (FBS/HBSS), and then transferred to sterile Eppendorf tubes. Cells were pelleted at 1000 g for 5 min at 4°C, and cell pellets were washed with 1 ml FBS/HBSS and pelleted again at 1000 g for 5 min at 4°C. Cell pellets were then treated with 500 μl 10 mg ml−1 collagenase/ dispase liberase (Roche) solution in FBSS/HBSS for 35 min at room temperature with vigorous shaking. Digestion was stopped using 500 μl stopping solution [100 μl 0.5 M EDTA (pH 8.0) in 9.9 ml FBS/HBSS]. Cells were pelleted at 1000 g for 5 min at 4°C. The cell pellets were resuspended in 500 μl FBS/HBSS and passed through 40 μm mesh filter (BD Falcon), and the mesh was washed twice with 250 μl FBSS/HBSS. Cells were then brought to final volume of 1 ml of FBS/HBSS prior to sorting using a MoFlo sorting flow cytometer (Beckman Coulter). Conventionally raised non-transgenic and transgenic controls (pools of 50 fish per genotype) were prepared as above and used to define fluorescence and cell size gates. Single cell suspensions from GF and CONVD zebrafish were sorted for GFP fluorescence using Summit software. For RNA analysis GFP(+) neutrophils were collected in 1 ml cold FBSS/HBSS and stored at 4°C. These cells were then pelleted at 1000 g for 5 min at 4°C. The supernatant was then removed and replaced with 1 ml Trizol (Invitrogen). Cells were then prepared as described below for quantitative RT-PCR analysis.
Cell tracking analysis
Tracking analysis of EGFP-expressing neutrophils was performed in Volocity (Improvision, Perkin Elmer). The locations of individual cells were tracked in each frame of time-lapse images (5 or 15 min) captured as described above using Volocity's ‘track objects manually’ tool. Tracking was performed as described in the Volocity User Guide. 3–5 time-lapse movies per experimental condition were analysed. For tissue-specific analyses, tracked cells were categorized as either caudal hematopoietic tissue (CHT), intestine, or fin cells based on their location for the duration of the movie.
Zebrafish cell culture and stimulation
PAC-2 zebrafish embryonic fibroblast cells were grown at 28°C in Leibowitz L-15 medium supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals) and penicillin G (50 U ml−1; Life Technologies) in 0.5% CO2. Cell lines were used between passages 15 and 35. Cells were grown to near confluency (90%) in 6-well plates (Costar), and were stimulated with LPS (10 μg ml−1; from Escherichia coli 0111:B4; Sigma) or SAA (1, 4 μM; PeproTech) for the specified amount of time in media containing 1% FBS. Immunofluorescence assays for RelA/p65 were performed as described (Kanther et al., 2011).
Western immunoblot analysis
PAC-2 cells were stimulated with LPS (10 μg ml−1) or SAA (4 μM) at specified time points. Cells were harvested and lysed in 1× Laemmli buffer, and the protein concentration was measured using a Bio-Rad quantification assay (Bio-Rad Laboratories). Western blot for IκBα (S32; Cell Signaling) and actin (MP Biomedicals) was performed as described previously (Kanther et al., 2011).
Transfection and luciferase activity assays
For transfections, PAC-2 cells were seeded into 12-well tissue culture plates (∼5 × 105 cells per well) and grown in 1 ml medium with 1% FBS at 28°C to −70% confluency. Transfections with 0.2 μg ml−1 of the previously described pikbaa:Luc were performed using Lipofectamine 2000 (Invitrogen) as described by the manufacturer (Kanther et al., 2011). After 24 h transfection, fresh medium was supplied and cells were stimulated with LPS (10 μg ml−1) or SAA (4 μM) for 24 h. Cells were then lysed and luciferase activity was determined using an LMax luminometer microplate reader (Molecular Devices, Sunnyvale, CA, USA). Results were normalized for extract protein concentrations measured with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA).
Quantitative RT-PCR analysis
To isolate RNA from whole zebrafish larvae, 6dpf larvae (5–30 larvae per group) were anesthetized in 4× Tricaine (Sigma-Aldrich), placed in 1 ml Trizol (Invitrogen) taking care to remove any excess media, and then repeatedly passed through a 25-gauge needle (BD Syringe) until homogenized. Samples were vortexed for 30 s, incubated at room temperature for 5 min, supplemented with 0.2 ml chloroform per sample, vortexed for 30 sec, and incubated at room temperature for 2 min. The samples were then centrifuged at 12 000 g for 15 minutes at 4°C, and then the colourless upper phase containing the RNA was transferred to a new RNAse-free tube. An equal volume of 70% RNAse-free ethanol was added to each tube containing the upper phase, and then RNA was isolated using PureLink™ RNA Mini Kit (Ambion) following the manufacturer's specifications. To isolate RNA from primary sorted zebrafish neutrophils or cultured zebrafish PAC2 fibroblasts, we used Trizol (Invitrogen) following manufacturer's specifications. Total RNA was used in reverse transcription and quantitative PCR assays using gene-specific primers for 18S (forward: CACTTGTCCCTCTAAGAAGTTGCA; reverse: GGTT GATTCCGATAACGAACGA), mpx (forward: TCCAAAGCTATG TGGGATGTGA; reverse: GTCGTCCGGCAAAACTGAA), ncf1 (forward: TTCATCTCGCCGTCAGACTCGTTT; reverse: TGTAC ACATAGTGCTGGCTGGGAA), il1b (forward: TGGACTTCG CAGCACAAAATG; reverse: GTTCACTTCACGCTCTTGGATG), ikbaa (forward: GCCGTGCAGATCATCAAAC; reverse: CCGC TGTAGTTAGGGAAGGT), and mmp9 (forward: CATCACTG AAATCCAGAAGGAGCTT; reverse: GTTCACCATTGCCTGA GATCTTC) as described (Kanther et al., 2011).
Neutrophil isolation and migration assay
Mice were injected intraperitoneally with 2.5 ml of 3% Fluid Thioglycollate Medium (Difco Laboratories) previously auto-claved for 15 min under 15 psi. Mice were euthanized with CO2 intoxication and neutrophils in the peritoneal cavity were retrieved by lavage with 10 ml of ice-cold HBSS supplemented with 1.5 mM ethylene diamine-tetraacetic acid (EDTA). Neutrophils were resuspended in 0.5% FBS RPMI 1640 medium and pre-treated for 1 h with the NF-κB inhibitor BAY-11–7082 (25 μM; Calbiochem), or cycloheximide (50 μg ml−1; Sigma). Cells were plated at ∼2 × 106 per insert in 6-well Transwells (Corning) with 3 μm pore in the presence of SAA (25 μg ml−1) and incubated at 37°C and 5% CO2 for 2.5 h. Neutrophils were imaged and counted as previously described (Sun et al., 2012).
Phylogenetic analysis
Protein sequences were exported from GenBank into the Cipres Science Gateway v3.2 (Miller et al., 2010) and aligned using Muscle. Phylogenetic trees were inferred using maximum likelihood in RAxML HPC2 7.3.1 on XSEDE using a GAMMA model and a BLOSUM62 protein substitution matrix. The best-scoring ML tree was identified and prepared using Dendroscope v1.4. Rapid bootstrap resampling (1000 replicates) was used to test the robustness of inferred topologies. Multiple sequence alignments were prepared using Boxshade.
Statistical methods
Statistical analysis was performed using unpaired two-tailed Student's t-test, or one-way analysis of variance (anova) followed by Tukey's post-test. Values were calculated using GraphPad Prism software, and P < 0.05 was considered significant.
Supplementary Material
Supporting information: Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Phylogenetic analysis of Saa protein sequences. A. Maximum likelihood phylogenetic tree of human (Homo sapiens, Hs) SAA1 (NP_000322), SAA2 (NP_110381) and SAA4 (NP_006503); mouse (Mus musculus, Mm) Saa1 (NP_033143), Saa2 (NP_035444), Saa3 (NP_035445) and Saa4 (NP_035446); chicken (Gallus gallus, Gg) Saa (ADF56353); rainbow trout (Oncorhynchus mykiss, Om) Saa (NP_001117908); Tetraodon (Tetraodon nigroviridis, Tn) Saa (CAF99678); amphioxus (Branchiostoma belcheri, Bb) Saa (BAB97379); and zebrafish (Danio rerio, Dr) Saa (NP_001005599) and SaaL1 (NP_956429). Zebrafish SaaL1 is included as an outgroup. Bootstrap support (≥ 50%) is shown as results from 1000 bootstrap replicates. B. Multiple sequence alignment of Saa proteins. Identical residues are highlighted in black, and similar residues are highlighted in grey.
Movie S1. Neutrophil migration in GF zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish imaged at a rate of one frame every 15 s for 15 min reveals decreased neutrophil migration compared with CONVD controls (compared with Movie S2).
Movie S2. Neutrophil migration in CONVD zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish imaged at a rate of one frame every 15 s for 15 min reveals increased neutrophil migration compared with GF controls (compared with Movie S1).
Movie S3. Neutrophil migration in GF Ctrl-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish injected with standard control MO (Ctrl MO) were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S4–S6).
Movie S4. Neutrophil migration in CONVD Ctrl-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish injected with standard control MO (Ctrl MO) were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3, S5 and S6).
Movie S5. Neutrophil migration in GF saa-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish injected with saa MO were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3, S4 and S6).
Movie S6. Neutrophil migration in CONVD saa-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish injected with saa MO were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3–S5).
Fig. S2. PCR validation of saa MO knockdown. Injection of 1-cell stage embryos with 9 pmol saa.i2e3 morpholino (saa MO) results in ∼150 bp reduction in a subset of saa transcripts in both GF and CONVD larvae at 6dpf. This is consistent with saa.i2e3 MO causing exclusion of exon 3 (154 bp) from saa transcripts. These saa MO-induced splicing defects were not observed following injection of a standard control morpholino (Ctrl MO). Data from two biological replicate pools (#1 and #2; 5–15 larvae per pool) are shown. Note that the total saa transcript levels are increased in CONVD compared with GF controls, but that the magnitude of saa transcript increase varies between replicates.
Acknowledgments
The authors are grateful to Gray Camp and Joan Kalnitsky (Microbiology and Immunology Flow Cytometry Core Facility) for assistance with flow cytometry, and Michael Chua and Neal Kramarcy (Michael Hooker Microscopy Facility) for assistance with microscopy. This work was supported by grants from the NIH (DK073695, DK081426 and DK094779 to J.F.R., DK047700 and DK073338 to C.J.), the NIDDK Center for Gastrointestinal Biology and Disease (DK034987), and a Pew Scholars in the Biomedical Sciences award to J.F.R.
References
- Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr Opin Gastroenterol. 2009;25:496–502. doi: 10.1097/MOG.0b013e328331b6b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ather JL, Ckless K, Martin R, Foley KL, Suratt BT, Boyson JE, et al. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J Immunol. 2011;187:64–73. doi: 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badolato R, Wang JM, Murphy WJ, Lloyd AR, Michiel DF, Bausserman LL, et al. Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J Exp Med. 1994;180:203–209. doi: 10.1084/jem.180.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova IN, Vishnyakova TG, Bocharov AV, Kurlander R, Chen Z, Kimelman ML, et al. Serum amyloid A binding to CLA-1 (CD36 and LIMPII analogous-1) mediates serum amyloid A protein-induced activation of ERK1/2 and p38 mitogen-activated protein kinases. J Biol Chem. 2005;280:8031–8040. doi: 10.1074/jbc.M405009200. [DOI] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopoly-saccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- Bozinovski S, Hutchinson A, Thompson M, Macgregor L, Black J, Giannakis E, et al. Serum amyloid a is a biomarker of acute exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177:269–278. doi: 10.1164/rccm.200705-678OC. [DOI] [PubMed] [Google Scholar]
- Cai H, Song C, Endoh I, Goyette J, Jessup W, Freedman SB, et al. Serum amyloid A induces monocyte tissue factor. J Immunol. 2007;178:1852–1860. doi: 10.4049/jimmunol.178.3.1852. [DOI] [PubMed] [Google Scholar]
- Cheng N, He R, Tian J, Ye PP, Ye RD. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol. 2008;181:22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson K, Bjorkman L, Tangemo C, Bylund J. Serum amyloid A inhibits apoptosis of human neutrophils via a P2X7-sensitive pathway independent of formyl peptide receptor-like 1. J Leukoc Biol. 2008;83:139–148. doi: 10.1189/jlb.0507276. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M, Marrelli A, Blades M, McCormick J, Maderna P, Godson C, et al. Acute serum amyloid A induces migration, angiogenesis, and inflammation in synovial cells in vitro and in a human rheumatoid arthritis/SCID mouse chimera model. J Immunol. 2010;184:6427–6437. doi: 10.4049/jimmunol.0902941. [DOI] [PubMed] [Google Scholar]
- Czuprynski CJ, Brown JF. Phagocytes from flora-defined and germfree athymic nude mice do not demonstrate enhanced antibacterial activity. Infect Immun. 1985;50:425–430. doi: 10.1128/iai.50.2.425-430.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santo C, Arscott R, Booth S, Karydis I, Jones M, Asher R, et al. Invariant NKT cells modulate the suppressive activity of IL-10-secreting neutrophils differentiated with serum amyloid A. Nat Immunol. 2010;11:1039–1046. doi: 10.1038/ni.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt ER, Witta J, Zhong J, Arsenescu R, Arsenescu V, Wang Y, et al. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol. 2010;10:133. doi: 10.1186/1471-230X-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Lieschke GJ. Computational quantification of fluorescent leukocyte numbers in zebrafish embryos. Methods Enzymol. 2012;506:425–435. doi: 10.1016/B978-0-12-391856-7.00046-9. [DOI] [PubMed] [Google Scholar]
- Ferencik M, Bergendi L, Mandel L, Kovaru F, Stefanovic J. Lysosomal enzyme activities in polymorphonuclear leukocytes, macrophages, serum, and spleen of conventional, germ-free, and antigen-free Minnesota miniature swine. Folia Microbiol (Praha) 1985;30:65–75. doi: 10.1007/BF02922500. [DOI] [PubMed] [Google Scholar]
- Flynn EJ, 3rd, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J Lipid Res. 2009;50:1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Villegas J, Garcia-Moreno D, de Oliveira S, Meseguer J, Mulero V. Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. Proc Natl Acad Sci USA. 2012;109:E2605–E2614. doi: 10.1073/pnas.1209920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt ME, Urieli-Shoval S, Preciado-Patt L, Fridkin M, Calco S, Azar Y, Matzner Y. Effect of serum amyloid A on selected in vitro functions of isolated human neutrophils. J Lab Clin Med. 1998;132:414–420. doi: 10.1016/s0022-2143(98)90112-3. [DOI] [PubMed] [Google Scholar]
- He RL, Zhou J, Hanson CZ, Chen J, Cheng N, Ye RD. Serum amyloid A induces G-CSF expression and neutrophilia via Toll-like receptor 2. Blood. 2009;113:429–437. doi: 10.1182/blood-2008-03-139923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijon HB, Walker J, Hoentjen F, Diaz H, Ewaschuk J, Jobin C, Madsen KL. Adenosine is a negative regulator of NF-kappaB and MAPK signaling in human intestinal epithelial cells. Cell Immunol. 2005;237:86–95. doi: 10.1016/j.cellimm.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Jungi TW, McGregor DD. Impaired chemotactic responsiveness of macrophages from gnotobiotic rats. Infect Immun. 1978;19:553–561. doi: 10.1128/iai.19.2.553-561.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Sun X, Muhlbauer M, Mackey LC, Flynn EJ, 3rd, Bagnat M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-kappaB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Kim MK, Park KS, Bae YH, Yun J, Park JI, et al. Serum amyloid A stimulates matrix-metalloproteinase-9 upregulation via formyl peptide receptor like-1-mediated signaling in human monocytic cells. Biochem Biophys Res Commun. 2005;330:989–998. doi: 10.1016/j.bbrc.2005.03.069. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- Linke RP, Bock V, Valet G, Rothe G. Inhibition of the oxidative burst response of N-formyl peptide-stimulated neutrophils by serum amyloid-A protein. Biochem Biophys Res Commun. 1991;176:1100–1105. doi: 10.1016/0006-291x(91)90397-p. [DOI] [PubMed] [Google Scholar]
- Liongue C, Hall CJ, O'Connell BA, Crosier P, Ward AC. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood. 2009;113:2535–2546. doi: 10.1182/blood-2008-07-171967. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek RL, Eriksen N, Benditt EP. Murine serum amyloid A3 is a high density apolipoprotein and is secreted by macrophages. PNAS. 1992;89:7949–7952. doi: 10.1073/pnas.89.17.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE) 2010;2010:1–8. [Google Scholar]
- Mitsuyama M, Ohara R, Amako K, Nomoto K, Yokokura T, Nomoto K. Ontogeny of macrophage function to release superoxide anion in conventional and germfree mice. Infect Immun. 1986;52:236–239. doi: 10.1128/iai.52.1.236-239.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morland B, Midtvedt T. Phagocytosis, peritoneal influx, and enzyme activities in peritoneal macrophages from germfree, conventional, and ex-germfree mice. Infect Immun. 1984;44:750–752. doi: 10.1128/iai.44.3.750-752.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, et al. Formation of the digestive system in zebrafish: III. Intestinal epithelium morphogenesis. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Ng PC, Ang IL, Chiu RW, Li K, Lam HS, Wong RP, et al. Host-response biomarkers for diagnosis of late-onset septicemia and necrotizing enterocolitis in preterm infants. J Clin Invest. 2010;120:2989–3000. doi: 10.1172/JCI40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi K, Teirila L, Lappalainen J, Rajamaki K, Baumann MH, Oorni K, et al. Serum amyloid A activates the NLRP3 inflammasome via P2×7 receptor and a cathepsin B-sensitive pathway. J Immunol. 2011;186:6119–6128. doi: 10.4049/jimmunol.1002843. [DOI] [PubMed] [Google Scholar]
- Noble CL, Abbas AR, Cornelius J, Lees CW, Ho GT, Toy K, et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut. 2008;57:1398–1405. doi: 10.1136/gut.2008.148395. [DOI] [PubMed] [Google Scholar]
- Ohkubo T, Tsuda M, Tamura M, Yamamura M. Impaired superoxide production in peripheral blood neutrophils of germ-free rats. Scand J Immunol. 1990;32:727–729. doi: 10.1111/j.1365-3083.1990.tb03216.x. [DOI] [PubMed] [Google Scholar]
- Ohkubo T, Tsuda M, Suzuki S, El Borai N, Yamamura M. Peripheral blood neutrophils of germ-free rats modified by in vivo granulocyte-colony-stimulating factor and exposure to natural environment. Scand J Immunol. 1999;49:73–77. doi: 10.1046/j.1365-3083.1999.00456.x. [DOI] [PubMed] [Google Scholar]
- Okahara S, Arimura Y, Yabana T, Kobayashi K, Gotoh A, Motoya S, et al. Inflammatory gene signature in ulcerative colitis with cDNA macroarray analysis. Aliment Pharmacol Ther. 2005;21:1091–1097. doi: 10.1111/j.1365-2036.2005.02443.x. [DOI] [PubMed] [Google Scholar]
- Oliveira MR, Tafuri WL, Afonso LC, Oliveira MA, Nicoli JR, Vieira EC, et al. Germ-free mice produce high levels of interferon-gamma in response to infection with Leishmania major but fail to heal lesions. Parasitology. 2005;131:477–488. doi: 10.1017/S0031182005008073. [DOI] [PubMed] [Google Scholar]
- Pase L, Layton JE, Wittmann C, Ellett F, Nowell CJ, Reyes-Aldasoro CC, et al. Neutrophil-delivered Myeloperoxidase dampens the Hydrogen Peroxide burst after tissue wounding in Zebrafish. Curr Biol. 2012;22:1818–1824. doi: 10.1016/j.cub.2012.07.060. [DOI] [PubMed] [Google Scholar]
- Patel H, Fellowes R, Coade S, Woo P. Human serum amyloid A has cytokine-like properties. Scand J Immunol. 1998;48:410–418. doi: 10.1046/j.1365-3083.1998.00394.x. [DOI] [PubMed] [Google Scholar]
- Penzo M, Molteni R, Suda T, Samaniego S, Raucci A, Habiel DM, et al. Inhibitor of NF-kappa B kinases alpha and beta are both essential for high mobility group box 1-mediated chemotaxis [corrected] J Immunol. 2010;184:4497–4509. doi: 10.4049/jimmunol.0903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F, Zhen F, Ong C, Jin SW, Meng Soo H, Stainier DY, et al. Microarray analysis of zebrafish cloche mutant using amplified cDNA and identification of potential downstream target genes. Dev Dyn. 2005;233:1163–1172. doi: 10.1002/dvdy.20444. [DOI] [PubMed] [Google Scholar]
- Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Balish E, et al. Normal luminal bacteria, especially bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta(2) microglobulin transgenic rats. J Clin Invest. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI. In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci USA. 2007;104:7622–7627. doi: 10.1073/pnas.0702386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehakova Z, Capkova J, Stepankova R, Sinkora J, Louzecka A, Ivanyi P, Weinreich S. Germ-free mice do not develop ankylosing enthesopathy, a spontaneous joint disease. Hum Immunol. 2000;61:555–558. doi: 10.1016/s0198-8859(00)00122-1. [DOI] [PubMed] [Google Scholar]
- Renckens R, Roelofs JJ, Knapp S, de Vos AF, Florquin S, van der Poll T. The acute-phase response and serum amyloid A inhibit the inflammatory response to Acinetobacter baumannii Pneumonia. J Infect Dis. 2006;193:187–195. doi: 10.1086/498876. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Shah C, Hari-Dass R, Raynes JG. Serum amyloid A is an innate immune opsonin for Gram-negative bacteria. Blood. 2006;108:1751–1757. doi: 10.1182/blood-2005-11-011932. [DOI] [PubMed] [Google Scholar]
- Sinkorova Z, Capkova J, Niederlova J, Stepankova R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum Immunol. 2008;69:845–850. doi: 10.1016/j.humimm.2008.08.296. [DOI] [PubMed] [Google Scholar]
- Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci USA. 2004;101:16240–16245. doi: 10.1073/pnas.0407241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SB, Gong W, Gao JL, Shen W, Murphy PM, Oppenheim JJ, Wang JM. A seven-transmembrane, G protein-coupled receptor, FPRL1, mediates the chemotactic activity of serum amyloid A for human phagocytic cells. J Exp Med. 1999;189:395–402. doi: 10.1084/jem.189.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Threadgill D, Jobin C. Campylobacter jejuni induces colitis through activation of mammalian target of rapamycin signaling. Gastroenterology. 2012;142:86–95. doi: 10.1053/j.gastro.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, et al. Expression of the zebrafish genome during embryogenesis. 2001 [WWW document]. URL http://zfin.org.
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Huttenlocher A. Spatiotemporal photolabeling of neutrophil trafficking during inflammation in live zebrafish. J Leukoc Biol. 2011;89:661–667. doi: 10.1189/jlb.1010567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoong S, O'Connell B, Soanes A, Crowhurst MO, Lieschke GJ, Ward AC. Characterization of the zebrafish matrix met alloproteinase 9 gene and its developmental expression pattern. Gene Expr Patterns. 2007;7:39–46. doi: 10.1016/j.modgep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:e1–e11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information: Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Fig. S1. Phylogenetic analysis of Saa protein sequences. A. Maximum likelihood phylogenetic tree of human (Homo sapiens, Hs) SAA1 (NP_000322), SAA2 (NP_110381) and SAA4 (NP_006503); mouse (Mus musculus, Mm) Saa1 (NP_033143), Saa2 (NP_035444), Saa3 (NP_035445) and Saa4 (NP_035446); chicken (Gallus gallus, Gg) Saa (ADF56353); rainbow trout (Oncorhynchus mykiss, Om) Saa (NP_001117908); Tetraodon (Tetraodon nigroviridis, Tn) Saa (CAF99678); amphioxus (Branchiostoma belcheri, Bb) Saa (BAB97379); and zebrafish (Danio rerio, Dr) Saa (NP_001005599) and SaaL1 (NP_956429). Zebrafish SaaL1 is included as an outgroup. Bootstrap support (≥ 50%) is shown as results from 1000 bootstrap replicates. B. Multiple sequence alignment of Saa proteins. Identical residues are highlighted in black, and similar residues are highlighted in grey.
Movie S1. Neutrophil migration in GF zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish imaged at a rate of one frame every 15 s for 15 min reveals decreased neutrophil migration compared with CONVD controls (compared with Movie S2).
Movie S2. Neutrophil migration in CONVD zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish imaged at a rate of one frame every 15 s for 15 min reveals increased neutrophil migration compared with GF controls (compared with Movie S1).
Movie S3. Neutrophil migration in GF Ctrl-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish injected with standard control MO (Ctrl MO) were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S4–S6).
Movie S4. Neutrophil migration in CONVD Ctrl-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish injected with standard control MO (Ctrl MO) were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3, S5 and S6).
Movie S5. Neutrophil migration in GF saa-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) GF zebrafish injected with saa MO were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3, S4 and S6).
Movie S6. Neutrophil migration in CONVD saa-MO zebrafish larvae. Live 6dpf Tg(mpx:GFP) CONVD zebrafish injected with saa MO were imaged at a rate of one frame every 15 s for 5 min (compared with Movies S3–S5).
Fig. S2. PCR validation of saa MO knockdown. Injection of 1-cell stage embryos with 9 pmol saa.i2e3 morpholino (saa MO) results in ∼150 bp reduction in a subset of saa transcripts in both GF and CONVD larvae at 6dpf. This is consistent with saa.i2e3 MO causing exclusion of exon 3 (154 bp) from saa transcripts. These saa MO-induced splicing defects were not observed following injection of a standard control morpholino (Ctrl MO). Data from two biological replicate pools (#1 and #2; 5–15 larvae per pool) are shown. Note that the total saa transcript levels are increased in CONVD compared with GF controls, but that the magnitude of saa transcript increase varies between replicates.


