Abstract
Actin filament nucleation and branching by Arp2/3 complex is activated by nucleation-promoting factors (NPFs), whose C-terminal WCA region contains binding sites for actin (W) and Arp2/3 complex (CA). It is debated whether one or two NPFs are required for activation. Here, we present conclusive evidence in support of the two-NPF model and show that actin plays a crucial role in the interactions of two mammalian NPFs, N-WASP and WAVE2, with Arp2/3 complex. Competition between actin-WCA and glia maturation factor (GMF) for binding to Arp2/3 complex suggests that during activation the first actin monomer binds at the barbed end of Arp2. Based on distance constrains obtained by time-resolved fluorescence resonance energy transfer, we define the relative position of the two actin-WCAs on Arp2/3 complex and propose an atomic model of the 11-subunit transitional complex.
The proteins that catalyze the formation of new actin filaments in cells, known as actin filament nucleators, play essential roles in the control of cytoskeleton remodeling in processes such as cell motility, vesicle trafficking and endocytosis1, 2, 3. Filament nucleators overcome the rate-limiting step for actin polymerization, i.e. the formation of actin dimers and trimers4, thus determining the time and location for polymerization. Among actin filament nucleators, Arp2/3 complex is unique in its ability to mediate both nucleation and branching of actin filaments5, 6. Arp2/3 complex consists of seven proteins, including the actin-related proteins 2 and 3 (Arp2 and Arp3), which function as an actin-like dimer during nucleation7, 8. The remaining five subunits of the complex, ArpC1-5, mediate regulatory interactions with various cofactors and branching interactions with the mother filament8, and form a scaffold for stabilization of the Arp2-Arp3 heterodimer7. By itself, Arp2/3 complex is inactive7, 9, 10. It is activated by multiple inputs, including ATP11, the mother filament12, and proteins called nucleation-promoting factors (NPFs)9, 13. Classical NPFs, such as WASP, N-WASP, WAVE and WHAMM, are generally unrelated, but they all contain a C-terminal WCA (WH2, Central, Acidic) region featuring binding sites for actin (W)14 and Arp2/3 complex (CA)12, 15. The WCA region of NPFs is sufficient to catalyze the nucleation and branching reaction9, whereas their other domains are typically implicated in regulation and localization16, 17. WCA brings together ATP-actin and ATP-Arp2/3 complex and induces a conformational change in the complex that promotes side binding to the mother filament18 and formation of a branch (daughter) filament that grows from the barbed end of the Arps at a 70° angle relative to the mother filament8, 10.
Different methods have been used to map the interactions of WCA with Arp2/3 complex, including chemical crosslinking19, 20, 21, 22, NMR19, 20, direct interaction of purified components23, SAXS24, transfers of photoactivatable labels25, and x-ray crystallography26. Collectively, these results implicate subunits Arp2, Arp3, ArpC1 and ArpC3 in interactions with WCA. Some of these subunits are more than 50Å apart in the structure of the inactive complex7, which given the short length of the WCA polypeptide is difficult to reconcile with a single WCA binding site. Two groups have now reported that Arp2/3 complex binds two NPFs25, 26, which leads to more efficient activation of the complex and is also consistent with the observation that in cells NPFs are frequently clustered on membranes or bound to dimeric partners27. However, these reports were recently disputed by another study that found that Arp2/3 complex bound a single NPF in the presence or the absence of actin28.
Here, we test the two competing models of activation, placing special emphasis on the role of actin in the interaction of WCA with Arp2/3 complex. We conclude that the WCAs of two different NPFs, N-WASP and WAVE2, bind with 2:1 stoichiometry to Arp2/3 complex, both in isolation and when bound to actin. Based on competition experiments with glia maturation factor (GMF) and distance measurements by time-resolved fluorescent energy transfer (TR-FRET), we further show that the first actin subunit binds at the barbed end of Arp2 and propose a detailed atomic model of the transitional complex formed by two actin-WCAs and Arp2/3 complex.
Results
Preparation of stable actin-WCA complexes
In cells, where the concentration of actin monomers is high (> 100 μM)29, NPFs are likely to be pre-bound to actin, which interacts with relatively high affinity (Kd < 1.0 μM) with the W domains of NPFs12, 14, 30, 31. Because the actin subunits bound to NPFs are thought to contact the barbed end of Arp2, Arp3 or both, it is likely that NPFs interact very differently with Arp2/3 complex depending on whether or not they are pre-bound to actin. However, testing this hypothesis has proven difficult, due to spontaneous polymerization of actin, which interferes with biochemical and structural studies of Arp2/3 complex requiring high sample concentrations under physiological salt conditions. While latrunculin (A or B) has been used in some studies to prevent actin polymerization25, 32, this toxin binds in the nucleotide-binding cleft at the pointed end of the actin monomer and is thus expected to interfere with proper interaction with the Arps, as suggested by a recent study28 and analogous to its effect on actin-actin interactions33. To circumvent these problems, we devised two different strategies, namely crosslinking of actin to WCA and substitution of the N-terminal region of the W domain in WCA by gelsolin segment 1 (GS1) (Fig. 1). Both approaches avoid the need for latrunculin, producing highly stable actin-WCA complexes that capture the intermediate polymerization nucleus formed by Arp2/3 complex, WCA and actin. Additionally, both approaches inhibit Arp2/3 complex nucleation, simplifying the analysis.
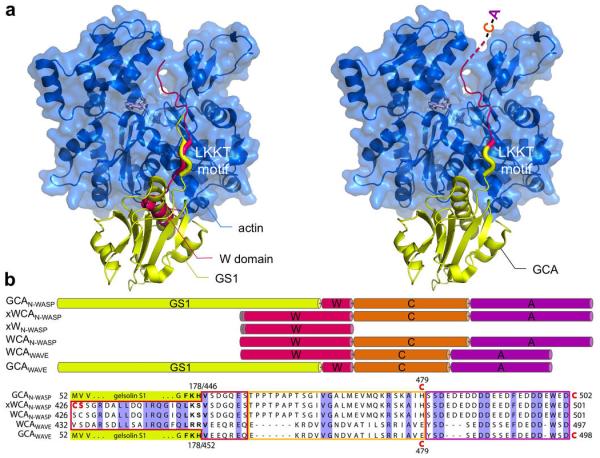
Figure 1.
Design of WCA constructs. (a) Superimposition of the structures of actin (blue) bound to GS135 (yellow) and W14 (red), and design of GCA constructs in which the N-terminal portion of W is replaced by GS1. The motif ‘LKKT’, present in both GS1 and the W domain, was used as the fusion site. (b) Schematic representation and sequence of WCA constructs of N-WASP and WAVE2 (W, red; C, orange; A, magenta; GS1, yellow). Residues that were mutated are highlighted in red. N-WASP residues 426CS427 were mutated to 426SC427 in constructs xWN-WASP and xWCAN-WASP for direct, unconstrained crosslinking to actin Cys-374. Single cysteine variants of constructs GCAN-WASP (H479C or C502 added at the C-terminus) and GCAWAVE (E479C or C498 added at the C-terminus) were made for labeling with fluorophores. Residues in the LKKT motif are highlighted in bold.
Crosslinking, which we reported previously24, is based on the structures of W domains bound to actin14, and takes advantage of the proximity of actin’s reactive Cys-374 to the N-terminal helix of the W domain, in which we introduced a cysteine residue for direct disulfide bond formation (Fig. 1b). We determined the crystal structure of a W domain crosslinked to actin in this manner and found it to be nearly identical to those of non-crosslinked W-actin complexes34. Here, we made two constructs for crosslinking of the WCA and W regions of N-WASP to actin, xWCAN-WASP and xWN-WASP (Fig. 1). Crosslinking, however, has drawbacks; it precludes the use of additional cysteine substitutions for labeling of WCA with fluorophores (used here) and impedes the use of reducing agents necessary to preserve the integrity of Arp2/3 complex over longer periods of time.
Our second approach resolves these shortcomings by taking advantage of the presence of a ‘LKKT’ sequence motif in the linker between gelsolin domains 1 and 2, which is structurally equivalent to that of the W domain14, 35 (Fig. 1a). Accordingly, we substituted the N-terminal portion of the W domain in the WCAs of N-WASP and WAVE2 with GS1. The resulting constructs are referred to here as GCAN-WASP and GCAWAVE (Fig. 1 and Supplementary Fig. S1). In addition, cysteine mutants of these constructs were made for labeling with fluorophores (Fig. 1b).
Previously, we found that crosslinking of WCA to actin strongly inhibited the nucleation activity of Arp2/3 complex (20 nM) in the presence of 2 μM actin (6% pyrene labeled)24. Under identical conditions, we found here that 200 nM GCAN-WASP has an even stronger inhibitory effect on polymerization (Supplementary Fig. S2). NPFs dissociate from branch junctions immediately after nucleation36. GS1, which binds with picomolar affinity to actin37 and crosslinked WCA do not dissociate (see below), and probably inhibit polymerization by interfering with actin subunit addition at the barbed end of the first two actins. The fact that inhibition is greater for GCAN-WASP than xWCA is consistent with the larger size of GS1 compared to the W domain; i.e. the smaller W domain obstructs, but does not fully block barbed end monomer addition.
Two N-WASP WCAs bind to Arp2/3 complex ± actin
There are differences between the two studies that reported interaction of Arp2/3 complex with two WCAs25, 26. Using isothermal titration calorimetry (ITC), one of the studies established that yeast Arp2/3 complex bound two CAs of yeast Wsp1p, but both yeast and mammalian Arp2/3 complex bound only one CA of mammalian N-WASP26. This study also concluded that actin-WCA bound with weaker affinity to Arp2/3 complex than CA alone. In contrast, the other study found that the stoichiometry of the mammalian complex was 2:1 (WCA-Arp2/3 complex), both in the presence or the absence of actin-latrunculin B25. And both studies were challenged by a work that found that Arp2/3 complex binds a single WCA with or without actin28. Here, we tested the two competing models of activation, addressing specifically the role of actin in the interaction of mammalian Arp2/3 complex and NPFs. First we analyzed the binding of WCAN-WASP alone to Arp2/3 complex by ITC, which resulted in an exothermic reaction (Fig. 2a) with similar profile to that of yeast Arp2/3 complex with Wsp1p CA26. While the reaction was consistent with two WCA binding sites (inflection point observed near 1:1 molar ratio), stoichiometry and affinity are interconnected during fitting of ITC data to a binding isotherm. Therefore, different fitting models were tested for each ITC experiment reported here (see examples in Supplementary Fig S3). In this case, the best fit corresponded to a two-independent-site binding model (Figs. 2a and 3 and Supplementary Table S1). The affinity of site-1 was found to be ~9-fold higher than that of site-2 (Kd of 20 vs. 182 nM), and both affinities were an order of magnitude higher than reported for the yeast complex26.
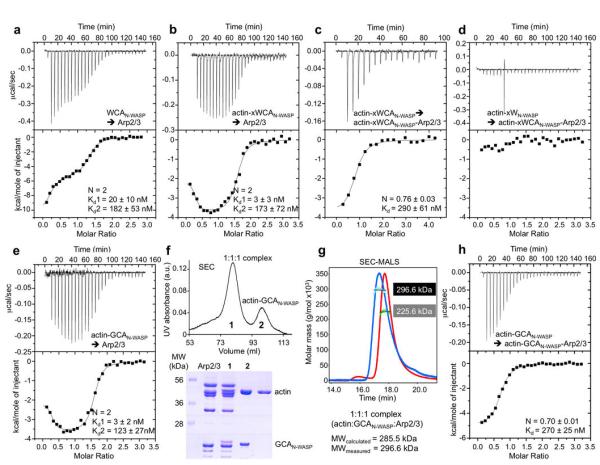
Figure 2.
Two N-WASP WCAs bind to Arp2/3 complex with or without actin. (a) ITC titration of 157 μM WCAN-WASP into 10 μM Arp2/3 complex. The best fit of the data corresponded to a two-site binding isotherm (Supplementary Table S1). The reported errors correspond to the standard deviation of the fit. (b) Titration of 189 μM actin-xWCAN-WASP into 12.5 μM Arp2/3 complex. (c) Titration of 136 μM actin-xWCAN-WASP into 4 μM 1:1:1 actin-xWCAN-WASP-Arp2/3 complex purified by gel filtration after the experiment shown in part b (Supplementary Fig. S4). (d) Titration of 86 μM actin-xWN-WASP into 5 μM purified 1:1:1 actin-xWCAN-WASP-Arp2/3 complex. (e) Titration of 147 μM actin-GCAN-WASP into 9.9 μM Arp2/3 complex. (f) Purification by gel filtration of the saturated complex resulting from the experiment shown in part e. Analysis by SDS-PAGE reveals that peak-1 consists of Arp2/3 complex with bound actin-GCAN-WASP and peak-2 consists of actin-GCAN-WASP dissociating from the weak-affinity site-2. (g) SEC-MALS analysis of peak-1 reveals a 1:1:1 actin-GCAN-WASP-Arp2/3 complex with mass 296.6 kDa (calculated mass 285.5 kDa). Arp2/3 complex alone is shown as a control (measured mass 225.6 kDa, calculated mass 223.6 kDa). (h) Titration of 7 μM purified 1:1:1 actin-GCAN-WASP-Arp2/3 complex into 106 μM actin-GCAN-WASP.
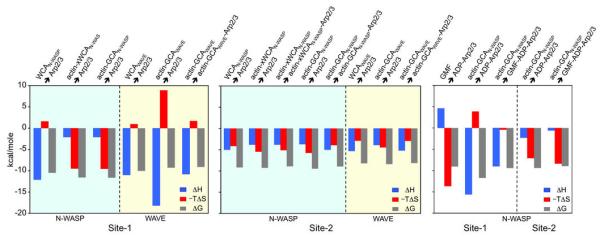
Figure 3.
Schematic representation of the thermodynamic parameters of Arp2/3 complex interactions with WCA species. Experiments are color-coded: cyan, titrations of WCA variants of N-WASP; yellow, titrations of WCA variants of WAVE2 and white, titrations of GMF and GCAN-WASP into ADP-Arp2/3 complex. The data is separated into binding sites 1 and 2.
The titration of crosslinked actin-xWCAN-WASP into Arp2/3 complex also fitted best to a two-independent-site model (Fig. 2b). In the presence of actin, the affinity of site-2 remained mostly unchanged, while that of site-1 appeared to increase somewhat (Kd of 3 vs. 20 nM). However, these high affinity values fall near the accuracy limits of our ITC instrument (Kd ≤ 10 nM), and thus a precise comparison is not warranted. It is more informative to compare the thermodynamic parameters of the titrations (Fig. 3 and Supplementary Table S1). For site-2, we did not observed significant differences in thermodynamic parameters ± actin. In contrast, there was a major change in site-1, from an enthalpy-driven reaction in the absence of actin to an entropy-driven reaction in presence of actin. These results suggest that at least for N-WASP actin has no appreciable contribution to the interaction at site-2, but it contributes substantially to the binding energy at site-1.
To experimentally validate the 2:1 model used for fitting of the ITC data, the saturated actin-xWCAN-WASP-Arp2/3 complex was purified by gel filtration (Supplementary Fig. S4a), which resulted in separation of two species. Characterization of these species by SDS-PAGE and multi-angle light scattering (MALS) revealed that the first peak consisted of a stable 1:1:1 complex (actin-xWCAN-WASP-Arp2/3 complex) and the second peak corresponded to actin-xWCAN-WASP dissociating from the weaker-affinity site-2 (Supplementary Fig. S4b). The 1:1:1 complex corresponds to the one studied by us previously by SAXS24, and clearly shows that in that study one actin-xWCAN-WASP was lost during gel filtration. Re-titration of actin-xWCAN-WASP into the purified 1:1:1 complex resulted in binding to a single site, restoring a 2:2:1 actin-xWCAN-WASP-Arp2/3 complex (Fig. 2c). The affinity, thermodynamic parameters and profile of the titration clearly show that this binding corresponds to site-2 of the initial titration (Figs. 2b,c and 3 and Supplementary Table S1). Importantly, crosslinked actin-xWN-WASP (lacking CA) did not bind to the purified 1:1:1 complex (Fig. 2d), further suggesting that actin does not contribute much to the interaction at site-2, and that CA interactions with Arp2/3 complex account for most of the binding energy at this site.
Next, we titrated actin-GCAN-WASP into Arp2/3 complex. The titration and thermodynamic parameters of the reaction were strikingly similar to those observed with actin-xWCAN-WASP (Figs. 2b,e and 3). GCAN-WASP and xWCAN-WASP differ only in the way they interact with actin (through GS1 or crosslinked W domain), and the striking similarity of these two titrations appears to cross-validate both methods of stabilization of WCA complexes with actin. In both cases, the presence of actin led to a dramatic change in the binding isotherm for site-1 (molar ratios ≤ 1). After confirming that dissociation of Arp2/3 complex subunits did not occur during titration (Supplementary Fig. S5a), we repeated this experiment twice, using lower protein concentrations, to better resolve the transition at site-1. Independent of protein concentration, all the titrations fitted to two-binding-site isotherms, with similar binding affinities and thermodynamic parameters (Supplementary Figs. S5).
As above, the saturated 2:2:1 actin-GCAN-WASP-Arp2/3 complex was purified by gel filtration and analyzed by SDS-PAGE and MALS, which again revealed two species: a 1:1:1 complex and actin-GCAN-WASP dissociating from site-2 (Fig. 2f,g). The 1:1:1 complex was re-titrated with actin-GCAN-WASP, which bound to a single site, restoring the saturated 2:2:1 complex (Fig. 2h). Here again, the affinity of the interaction and the thermodynamic parameters and profile of the titration clearly show that this binding corresponds to site-2 of the initial titration (Figs. 2e,h and 3).
Two WAVE2 WCAs bind to Arp2/3 complex ± actin
To test whether these results could be generalized to other NPFs, we performed similar experiments with WAVE2. First, we titrated the WCA of WAVE2 into Arp2/3 complex, which also resulted in two binding sites (Fig. 4a). However, the affinities of sites 1 and 2 were respectively 2- and 6-fold lower than for N-WASP. The titration of actin-GCAWAVE also fitted best to a two-independent-site model (Fig. 4b and Supplementary Fig. 3b), but again their affinities were significantly lower than for actin-GCAN-WASP (Supplementary Table S1). Analysis by MALS of the saturated complex resulting from the ITC titration revealed a reaction boundary (Fig. 4c), consistent with complex dissociation. Indeed, when this complex was purified by gel filtration, two peaks were obtained, which by SDS-PAGE corresponded to dissociating actin-GCAWAVE and Arp2/3 complex with a sub-stoichiometric amount of actin-GCAWAVE bound (Fig. 4d). Thus, in contrast to N-WASP (Fig. 2f,g), actin-GCAWAVE dissociates during gel filtration not only from site-2, but also partially from site-1, consistent with the lower affinity of this NPF. This was confirmed by re-titrating the purified complex with actin-GCAWAVE, which resulted in two binding sites (Fig. 4e).
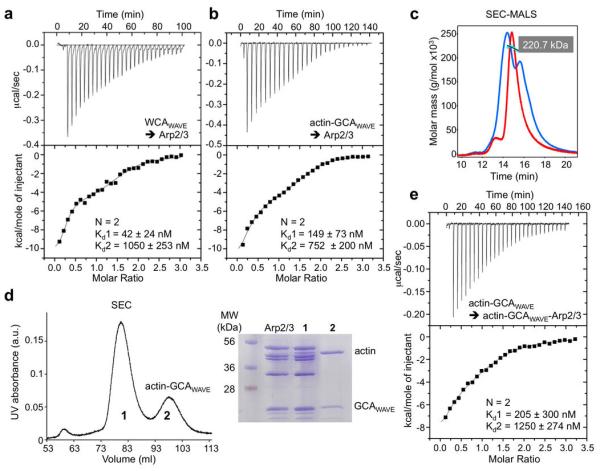
Figure 4.
wo WAVE2 WCAs bind to Arp2/3 complex with or without actin. (a) Titration of 127 μM WCAWAVE into 8.9 μM Arp2/3 complex. The best fit of the data corresponded to a two-site binding isotherm (Supplementary Table S1). The reported errors correspond to the standard deviation of the fit. (b) Titration of 167 μM actin-GCAWAVE into 11 μM Arp/3 complex. (c) Analysis by MALS of the saturated complex after the experiment shown in part b reveals a reaction boundary (blue trace) with two peaks on each side of the Arp2/3 complex control (red trace). The masses of these peaks could not be accurately measured. (d) Purification by gel filtration of the saturated complex from the experiment shown in b. Analysis by SDS-PAGE revealed that peak-1 contained Arp2/3 complex and a sub-stoichiometric amount of actin-GCAWAVE, whereas peak-2 contained actin-GCAWAVE dissociating from Arp2/3 complex. Note that in this gel, GCAWAVE overlaps with subunit ArpC3. (e) The titration of 74.4 μM actin-GCAWAVE into peak-1 of the gel filtration (at ~4.9 μM) displayed a similar profile as the titration into Arp2/3 complex alone (part b) and was best fitted to a two-site binding model.
These results reveal differences and similarities between N-WASP and WAVE. WAVE2’s WCA is 10-aa shorter than that of N-WASP (Fig. 1b), with deletions proceeding the C and A regions that might constrain binding to Arp2/3 complex and reduce the number of acidic amino acids in the A region. These differences may explain why WAVE2’s WCA had lower affinity for Arp2/3 complex in the presence or the absence of actin, and contrary to N-WASP its affinity for site-1 decreased slightly in the presence of actin (Figs. 2 and 4). Also different from N-WASP, the interaction of WAVE2’s WCA with site-1 was enthalpy-driven in the absence or the presence of actin, whereas that of N-WASP became entropy-driven in the presence of actin (Fig. 3). Yet, both NPFs bound with 2:1 stoichiometry to Arp2/3 complex in the presence or the absence of actin. Moreover, when bound to actin, the interactions of both NPFs with site-1 underwent large changes in thermodynamic parameters, whereas their interactions with site-2 were mostly unchanged by actin (Fig. 3).
The first actin subunit binds at the barbed end of Arp2
Different studies have distinctly proposed that the first actin subunit binds at the barbed end of Arp224, 25, 32 or Arp320, 26, 38, but with the exception of the SAXS analysis of the 1:1:1 actin-xWCA-Arp2/3 complex, which placed the first actin at the barbed end of Arp224, there is no direct evidence in support of either model. To address this question, we used competition between actin-GCAN-WASP and GMF for binding to Arp2/3 complex. The ADF/cofilin-family member GMF interacts with Arp2/3 complex and promotes debranching39. We recently found that GMF interacts very weakly with ATP-Arp2/3 complex but forms a relatively tight 1:1 complex with ADP-Arp2/3 complex40. A recent structure of this complex showed that GMF binds at the barbed end of Arp241. We reasoned that if the first actin subunit binds at the barbed end of Arp2, it should compete with GMF binding.
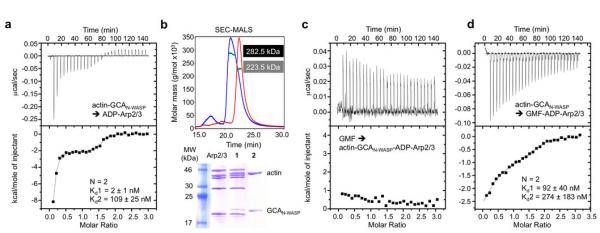
To test this idea, we first established that actin-GCAN-WASP could bind ADP-Arp2/3 complex (Fig. 5a). At saturation, the stoichiometry of the interaction was 2:2:1 actin-GCAN-WASP-Arp2/3 complex, and the affinities of both sites were almost identical to those observed in the ATP state. In contrast, we observed a dramatic change in the character of the interaction at site-1, from entropy-driven binding in the ATP state to enthalpy-driven binding in the ADP state, similar to the interaction in the absence of actin (Fig. 3 and Supplementary Table S1). Because the only difference between these two experiments is the nucleotide state of Arp2/3 complex (the nucleotide state on actin does not change through simple dialysis, Methods), this finding is consistent with a previous study that revealed a nucleotide-dependent conformational change in Arp2/3 complex11. Purification by gel filtration of the 2:2:1 complex formed in the ADP state produced two well-separated species, identified by SDS-PAGE and MALS as a 1:1:1 actin-GCAN-WASP-Arp2/3 complex (measured mass 282.5 kDa) and actin-GCAN-WASP dissociating from the lower-affinity site-2 (Fig. 5b). As before40, GMF bound ADP-Arp2/3 complex with a Kd ~ 0.2 μM (Supplementary Fig. S6). However, GMF did not bind to the purified 1:1:1 complex (Fig. 5c), suggesting direct competition with actin-GCAN-WASP for binding to Arp2.
Figure 5.
ompetition between GMF and GCAN-WASP for binding to ADP-Arp2/3 complex. (a) Titration of 112 μM actin-GCAN-WASP into 8.0 μM ADP-Arp/3 complex. The best fit of the data was to a two-site binding isotherm (Supplementary Table S1). The reported errors correspond to the standard deviation of the fit. (b) Analysis by MALS and SDS-PAGE of the two species resulting from gel filtration (not shown) of the saturated complex after the experiment shown in part a. Peak-1 consisted of a 1:1:1 actin-GCAN-WASP-Arp2/3 complex in the ADP state (measured mass 282.5, calculated mass 285.5 kDa) and peak-2 contains actin-GCAN-WASP dissociating from site-2. Arp2/3 complex alone is shown as a control. (c) Titration of 90 μM GMF into 6.3 μM purified 1:1:1 actin-GCAN-WASP-Arp2/3 complex (part b, peak-1). (d) Titration of 107 μM actin-GCAN-WASP into 7.7 μM saturated GMF-Arp/3 complex (from the experiment shown in Supplementary Fig. S6).
To check that the lack of binding of GMF was not due to long-range allosteric effects, we inverted the order of the titration; actin-GCAN-WASP was titrated into the 1:1 complex of GMF with ADP-Arp2/3 complex (Fig. 5d). This resulted in a ~ 46-fold drop in the affinity of actin-GCAN-WASP for site-1 and large changes in thermodynamic parameters (Fig. 3 and Supplementary Table S1), whereas the affinity of site-2 decreased only 2.5-fold and the thermodynamic parameters were mostly unchanged. The affinity of actin-GCAN-WASP for site-1 in the ADP state is ~ two orders of magnitude higher than that of GMF (Kd 2 vs. 200 nM, Fig. 5a and Supplementary Fig. S6), such that it was expected that actin-GCAN-WASP would displace GMF from the complex. Yet, the large changes in thermodynamic parameters and affinity for site-1 strongly suggest that GMF, which binds at the barbed end of Arp241, competes with the binding of actin-GCAN-WASP to this site, and thus the first actin subunit most likely binds at the barbed end of Arp2.
Steady-state FRET confirms two actin-WCA binding sites and reveals a conformational change
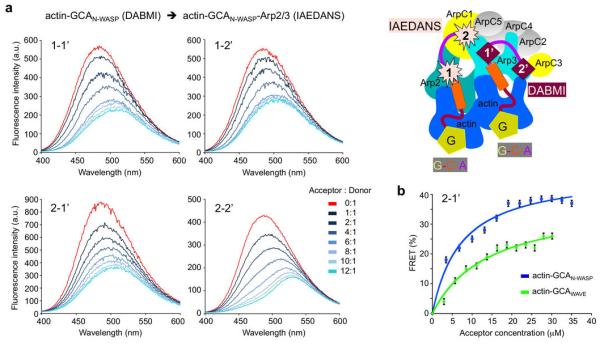
To independently test the two-site binding model, we used steady-state FRET, for which cysteine residues were introduced by mutagenesis at the end of the C region or added C-terminally to the A region in constructs GCAN-WASP and GCAWAVE (Fig. 1b). We refer to these two labeling sites as positions 1 for the donor (or 1’ for the acceptor) and 2 (or 2’), respectively. The single mutant constructs were labeled with either the donor probe IAEDANS or the acceptor probe DABMI, a fluorescence pair sensitive to energy transfer in the range of distances ~30-65 Å. For each experiment, an actin-GCAN-WASP-Arp2/3 complex (or actin-GCAWAVE-Arp2/3 complex) labeled with the donor IAEDANS at position 1 or 2 was pre-purified as described above (Figs. 2f and 4d). These complexes were then titrated with actin-GCAN-WASP (or actin-GCAWAVE) labeled at position 1’ or 2’ with the acceptor DABMI. Fluorescence quenching only occurred in the presence of DABMI-labeled GCA, and was observed for all the combinations of probe positions for both N-WASP (Fig. 6) and WAVE2 (Supplementary Fig. 7). Additionally, a significant shift toward longer wavelengths (red shift) was observed in all the titrations. A red shift of IAEDANS signal corresponds to a more solvent-exposed environment of the probe42, which is consistent with a conformational change taking place in Arp2/3 complex upon binding of actin-WCA to site-2. While the occurrence of a red shift precludes an accurate quantitative analysis, tentative affinity values for site-2 were obtained by fitting the titrations with the smallest red shift, corresponding to probe positions 2 and 1’ (Fig. 6b, Supplementary Fig. 7c). The resulting affinities appeared underestimated compared to the more reliable values obtained by ITC (Kd values 7 μM vs. ~200 nM for actin-GCAN-WASP and 14 μM vs. ~1 μM for actin-GCAWAVE). While these affinities are likely distorted by the red shift and the presence of the probes, this analysis confirms the presence of a second binding site for actin-WCA.
Figure 6.
teady state FRET analysis of actin-GCA binding to Arp2/3 complex. (a) Fluorescence quenching during titration of acceptor-labeled actin-GCAN-WASP into donor-labeled 1:1:1 actin-GCAN-WASP-Arp2/3 complex. Probe sites and titration ratios are indicated. A cartoon representation illustrates the relative positions of the donor (IAEDANS) and acceptor (DABMI) probes in these experiments. (b) FRET as a function of acceptor concentration for the 2-1’ titrations (with smallest red shift) of actin-GCAN-WASP (blue) and actin-GCAWAVE (green, experiment shown in Supplementary Fig. S7). Data points represent mean ± s.e.m. Data fitting resulted in Kd values of 7 and 14 μM for actin-GCAN-WASP and actin-GCAWAVE, respectively.
Time-resolved FRET defines the relative position of two WCAs on Arp2/3 complex
The observation of energy transfer for all the probe positions opened the way for TR-FRET experiments that could reveal the relative distance between probes in the complex. For these experiments, we used actin-GCAN-WASP-Arp2/3 complex that can be purified with 1:1:1 stoichiometry (i.e. site-1 is fully occupied). As above, a pre-purified 1:1:1 complex containing the donor IAEDANS at position 1 or 2 was mixed with actin-GCAN-WASP containing the acceptor DABMI at one of the two labeling positions. All four possible inter-probe distances (1-1’, 1-2’, 2-1’ and 2-2’) were determined by fitting the fluorescence decays using a double exponential function (Supplementary Fig. S8 and Methods). All the distances determined were tightly clustered in the range 53.8 to 60.4 Å (Table 1).
Table 1.
Distances from time-resolved FRET and model
| Distance | τD (ns) | τDA (ns) | R0 (Å) | RDA(Å) | RModel(Å) |
|---|---|---|---|---|---|
| 1-1’ | 14.22 | 11.13 | 44.85 | 55.53 | 56.0 |
| 1-2’ | 14.22 | 12.18 | 44.85 | 60.41 | 56.5 |
| 2-1’ | 14.52 | 11.39 | 45.01 | 55.82 | 59.7 |
| 2-2’ | 14.58 | 10.84 | 45.04 | 53.78 | 55.1 |
| 1-2 | 15.70 | 8.36 | 45.59 | 47.45 | 46.7 |
Another distance constraint was obtained using a double cysteine mutant of GCAN-WASP labeled at Cys-479 and Cys-502 with the donor and acceptor probes (Fig. 1b). This distance could be only measure for site-1, using the purified 1:1:1 complex, since the saturation state of the weaker affinity site-2 cannot be precisely determined, and any unbound actin-GCAN-WASP would contribute to the fluorescence. Double labeling of GCAN-WASP was carried out by under-labeling with IAEDANS, followed by labeling of all the unreacted cysteines with DABMI (Methods). This labeling strategy produces a mixture of donor-donor, acceptor-acceptor and donor-acceptor species. This mixture was analyzed by reverse phase HPLC, which together with a recording of DABMI absorbance at 502 nm, detection of IAEDANS fluorescence in peak fractions, and deconvolution of the 280 nm absorbance profile, resulted in a 32% estimate of the donor-acceptor fraction (Supplementary Fig. S9a,b). To determine the distance between probes, we fitted the donor-acceptor decay using a linear combination of two double-exponential functions (Supplementary Fig. S9c and Methods), fixing the molar fraction of the donor-acceptor decay to 32%. This analysis resulted in a distance estimate of 47.5 Å between the two probes at site-1.
The ensemble of FRET distances obtained here provides key constraints for modeling of two WCAs on the surface of Arp2/3 complex.
Discussion
Despite nearly 20 years of intensive investigation since the discovery of Arp2/3 complex43, only recently was it found that optimal activation requires two NPFs25, 26, 27. This fact was difficult to uncover, first because in isolation most NPFs appeared to be monomeric, but also because formation of the short-lived transitional complex is a multi-step and multi-factorial process that requires NPFs, ATP, actin monomers and culminates with binding to the mother filament44. The two-NPF model accounted for the fact that multiple binding partners of NPFs are dimeric, and that clustering at membranes would allow for two NPFs to work synergistically to activate Arp2/3 complex27. Strikingly, reinterpretation of most of the published data is also consistent with the two-NPF model25. This includes studies showing that subunits Arp2, Arp3, ArpC1 and ArpC3, which are distantly separated in the structure7, can all be crosslinked to NPFs19, 20, 21, 22. Photoactivatable biotin coupled to cysteine residues along the CA polypeptide also transfers onto these four subunits25. Spin labels at the C-termini of the C and A regions causes NMR line broadening of ArpC3 resonances, suggesting that these labels fall within ~25Å of ArpC320. Recombinant ArpC1 binds directly to WCA, and this interaction depends on the presence of the conserved tryptophan at the C-terminus of the A region23. SAXS analysis of a 1:1:1 actin-xWCA-Arp2/3 complex places the first actin subunit at the barbed end of Arp224. X-ray crystallography shows three residues of the A region of bovine N-WASP, 502EWE504, bound to Arp326. Collectively, these studies implicate subunits Arp2, Arp3, ArpC1 and ArpC3 in interactions with NPFs, which can only be explained by the two-NPF model. This model also provides an explanation for how cortactin NTA and N-WASP WCA can co-bind Arp2/3 complex21.
Despite these considerations, the two-NPF model was immediately challenged by a study that found that Arp2/3 complex interacts with a single NPF, both in the absence or the presence of actin28. The ongoing debate, and the fact that our own SAXS analysis of the 1:1:1 complex24 was used as supporting evidence in the latter study, motivated us to analyze the two competing models of Arp2/3 complex activation using different approaches. The results presented here provide strong evidence in support of the two-NPF model, and extend beyond this debate to also provide a likely pathway of the activation reaction and a testable atomic model of the transitional complex. The main findings of this work are: 1) The WCA regions of two mammalian NPFs, N-WASP and WAVE2, both bind with 2:1 stoichiometry to mammalian Arp2/3 complex, 2) In the presence of actin, the stoichiometry of the interaction is also 2:2:1, 3) Actin has little effect on the affinity of site-2, but changes the affinity and thermodynamic parameters of site-1 in an NPF-dependent manner, 4) The 2:2:1 stoichiometry is further demonstrated by purification of a half-saturated 1:1:1 complex that can bind a second actin-NPF, 5) Actin-GCAN-WASP binds Arp2/3 complex in the ADP state, interacting with two sites and with similar affinities as in the ATP state, but with markedly different thermodynamic parameters at site-1, consistent with a nucleotide-dependent conformational change11. The similar affinity of actin-WCA for ADP- and ATP-Arp2/3 complex is also consistent with the finding that ATP hydrolysis is not required for NPF release36. 6) GMF, which binds at the barbed end of Arp241, competes with the binding of actin-GCAN-WASP to site-1, suggesting that the first actin subunit is delivered at the barbed end of Arp2, 7) Fluorescence energy transfer between donor-acceptor probes on two actin-GCAs independently confirms the 2:2:1 stoichiometry of the interaction, 8) Steady-state FRET also reveals a major red shift upon saturation of site-2, indicative of a conformational change, and 9) We obtained a set of five inter-probe distances that combined with published data allow us to build an atomic model of the transitional complex.
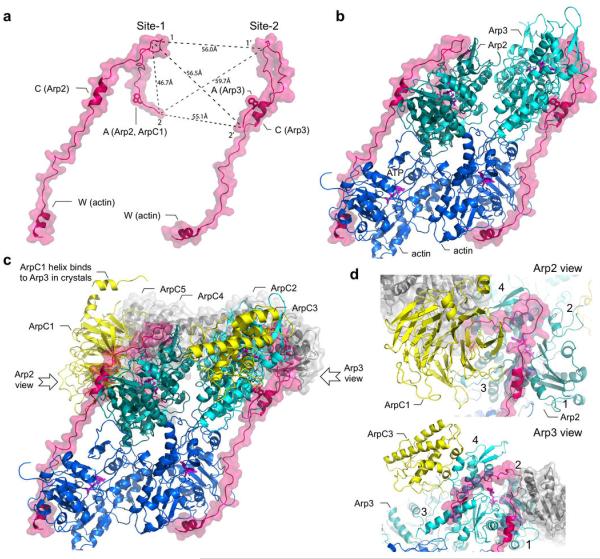
To build this model (Fig. 7 and Supplementary Movie S1), we started from the structure of inactive Arp2/3 complex containing three amino acids of the A region bound to Arp326. Subunit Arp2, which is partially disordered in this and other structures of Arp2/3 complex7, 45, was taken from the structure of the complex with GMF in which it was visualized41. In the inactive conformation, Arp2 obstructs binding to the barbed end of Arp3, which is a strong argument in favor of a movement of Arp2 as a first step in the activation process24, 25. For this, we superimposed the highest-resolution model available of the actin filament46 onto subunit Arp3, which defines the expected position of Arp2 upon activation, as well as those of the first two actin subunits at the barbed ends of Arp2 and Arp3. Note, that the only conformational change required is a rotation of Arp2, which does not generate steric clashes24 (Supplementary Movie S1). The structure of W bound to actin14 was then superimposed onto the actin subunits at the barbed ends of Arp2 and Arp3. In the structure of Arp2/3 complex, a helix of subunit ArpC1 (residues 297-305) of a symmetry-related complex makes a crystal packing contact with Arp3, which is thought to mimic the binding of the C region to Arp326. Consistent with this proposal, this helix, which is conspicuously detached from the rest of ArpC1 (Fig. 7), is proposed to mediate similar contacts between subdomain 1 and 3 of an actin subunit in the mother filament7, 8. Moreover, like the helix in the C-region47, the ArpC1 helix has an amphiphilic character, and its hydrophobic side faces Arp3, analogous to the interactions of several actin-binding proteins with actin48, 49. We thus built a model of CA bound to Arp3 (site-2), by superimposing the helix of the C region onto the ArpC1 helix, and finding a likely path for the acidic linker between this helix and 498EWE500 (mouse N-WASP numbering) along a positive surface on Arp3. The helix of the C-region was assumed to bind to Arp2 (site-1) in a way analogous to its interaction with Arp3, with the remaining, non-helical portion of the C-region extending toward the pointed end of Arp2. To this point, our model is precisely in agreement with that proposed by Padrick et al.25, and overlaps with two other models regarding the interactions of CA with Arp224 and Arp326. We also observe a strong correlation between experimentally measured FRET distances for probe pairs at positions 1-1’ and 1-2’ and the corresponding distances between Cα atoms in the model (Table 1 and Fig. 7).
Figure 7.
Model of the transitional complex formed by Arp2/3 complex, two actin subunits and two WCAs. (a) Two WCAs in their proposed Arp2/3 complex-bound conformation, showing the distances between Cα atoms of residues labeled in FRET experiments. (b) Proposed pseudo-symmetric interaction of two WCAs with Arp2-actin and Arp3-actin arranged in a filament-like conformation. (c) Illustration of the 11-subunit transitional complex. Subunits ArpC1 and ArpC3, which biochemical data place near the binding site of WCAs, are colored yellow. Subunits ArpC2, ArpC4 and ArpC5, thought not to interact with NPFs, are colored gray. (d) Close-up views of sites-1 (Arp2-ArpC1) and site-2 (Arp3-ArpC3). Subdomains 1-4 of Arp2 and Arp3 are indicated. See also Supplementary Movie S1.
No detailed model has yet been proposed for the A region at site-1. However, it has been abundantly documented that this region contacts both Arp2 and ArpC119, 20, 21, 22, 23, 25. Therefore, our model must explain this data, while also accounting for the FRET distances. Many models were tested while trying to satisfy these constraints. Models that placed the A region on the exposed surface of ArpC1 produced distances > 90 Å for the probe at position 2, i.e. much longer than measured by FRET. On the contrary, models in which the A region was directed toward Arp3 resulted in distances of < 30 Å for this probe. These models were rejected, since these distances would not be measurable using our FRET pair. Other models in which the A region was directed toward subunits ArpC2, ArpC4 and ArpC5 were not only inconsistent with the biochemical data, but invariably also failed to satisfy the ensemble of the FRET distances. To our surprise, the only model that satisfied all the criteria was based on the concept of pseudo-symmetry in the way CA interacts with Arp2 and Arp3 (Table 1, Fig. 7 and Supplementary Movie S1). In this model, the A region threads through a channel at the interface between Arp2 and ArpC1. Importantly, when CA first encounters Arp2 and ArpC1 in the inactive complex, these two subunits are separated and the binding surface is fully exposed on both sides. In other words, this model predicts that the A region adds affinity between Arp2 and ArpC1 by creating a new binding interface between these two subunits. As a final step, the stereochemistry of the resulting 11-subunit transitional complex (Arp2/3 complex, 2 actin subunits and 2 WCAs) was regularized using the program Phenix50.
While the model satisfies the FRET constraints and accounts for published data, it is important to understand its limitations. First, it is based on the assumption that during activation the overall structure of the complex remains mostly unchanged, except for the position of Arp2 and smaller changes at the interface between actin-WCA and subunits Arp2, ArpC1 and Arp3. It is likely that other changes occur, particularly at the interface with the mother filament. The model is also based on the assumption that Arp2, Arp3 and the actin subunits bound at their barbed ends adopt a filament-like conformation. However, deviations from this structure are likely, particularly that the filament model is only defined to limited (non-atomic) resolution46. Finally, while we have aimed to satisfy FRET distances, it is important to note that these distances contain measurement and fitting errors and that the position of the fluorophores is not precisely determined.
Nevertheless, the proposed atomic model accounts for the overwhelming majority of the published data19, 20, 21, 22, 23, 25,26,27,24 and will be available for independent testing. Because, at both sites the CA polypeptide meanders near the pointed ends of Arp2 and Arp3, this model also accounts for EM data showing extra density at the pointed ends of the Arps when WCA is bound to Arp2/3 complex51. This is also the surface that contacts the mother filament in the branch8, suggesting that steric hindrance of CA with the mother filament and W with subunits adding to the branch may together trigger NPF dissociation. As proposed above, steric hindrance with subunits adding to the branch may explain how the GCA design traps the transitional complex by blocking Arp2/3 complex-dependent nucleation (Supplementary Fig. S2), which can now be illustrated by superimposing GS1-actin onto the first two actin subunits of the transitional complex (Supplementary Fig. S10). This superimposition also illustrates that GS1 does not obstruct interactions of the first two actin subunits with Arp2/3 complex or with each other.
In summary, based on the ensemble of the results presented here we propose the following sequence of events. Two actin-NPFs bind to Arp2 and Arp3 in a pseudo-symmetric manner (Fig. 7 and Supplementary Movie 1). Competition with GMF (Fig. 5) demonstrates that actin-NPF binds first to Arp2 and ArpC1 (site-1), adding an actin subunit at the barbed of Arp2. This produces a partial conformational change that frees the barbed end of Arp3, allowing for the binding of a second actin-NPF to Arp3. Steady-state FRET (Fig. 6 and Supplementary Fig. S7) shows that this second binding event is also accompanied by a conformational change, which is anticipated to complete the transition of Arp2 and Arp3 into a filament-like conformation and promote binding of Arp2/3 complex to the mother filament.
Methods
Proteins
Arp2/3 complex was purified from bovine brain24. Actin was purified from rabbit skeletal muscle52. The cDNA encoding for mouse N-WASP (UniProt Q91YD9) was obtained from ATCC (clone 3169027). Construct WCAN-WASP was amplified by PCR and cloned between the NdeI and EcoRI sites of vector pTYB12 (New England BioLabs), comprising a chitin-binding domain for affinity purification and an intein domain for self-cleavage after purification (primers listed in Supplementary Table S2). Mutations S426C and C427S in construct xWCAN-WASP were introduced with the forward primer during cloning. Construct xWN-WASP, carrying mutations S426C and C427S, was synthesized. The cDNA for expression of mouse WCAWAVE (UniProt Q8BH43) was synthesized (Genewiz) and cloned as above. The cDNA encoding for human gelsolin (UniProt P06396) was purchased from ATCC (clone MGC-39262), and segment 1 (GS1) was cloned as above. The hybrid constructs GCAN-WASP and GCAWAVE were obtained by introducing silent mutations in the GS1 and CA genes that introduced NpsI restriction sites for hybridization in the reverse and forward primers, respectively. The ligation products were then cloned as above. Point mutations H479C in GCAN-WASP and E479C in GCAWAVE were introduced using the QuickChange mutagenesis kit (Qiagen). C-terminal cysteine residues were introduced into constructs GCAN-WASP (Cys-502) and GCAWAVE (Cys-498) using the reverse primers during cloning. All the proteins were expressed in BL21(DE3) cells (Invitrogen), grown in Terrific Broth medium at 37°C until the OD600 reached a value of 1.5-2. Expression was induced with 0.5 mM isopropylthio- β-D-galactoside and carried out for 16 hours at 20°C. Cells were harvested by centrifugation, re-suspended in 20 mM HEPES pH 7.5, 500 mM NaCl, 1 mM EDTA and 100 μM PMSF and lysed using a microfluidizer (Microfluidics). All the proteins were first purified on a chitin affinity column, followed by additional purification as follows: WCA constructs were purified by HPLC using a reverse phase C18 column and a 0-90% gradient of CH3CN in 0.1% TFA and GCA constructs were purified through an ion exchange MonoQ column (Pharmacia) in 20 mM HEPES pH 7.5 and a 50-1000 mM NaCl gradient. Human GMFγ (UniProt O60234) was expressed and purified as reported40. Protein concentration was determined spectrophotometrically, using calculated extinction coefficients.
Labeling with fluorophores
Cysteine mutants of GCA constructs were reduced with 20 mM DTT for 30 min at room temperature. DTT was removed on a PD10 column (GE Healthcare) in 20 mM HEPES pH 7.5, 100 mM NaCl, 10 mM EDTA. Single-labeling was performed overnight, at 4°C in the dark, using a 20-fold molar excess of 1,5-IAEDANS or DABMI (Molecular Probes) in the presence of 0.1 mM TCEP. The reaction was stopped by addition of 10 mM DTT. Unreacted probe was removed on a Sephadex G-50 column (GE Healthcare). Labeling was confirmed by mass spectrometry on a Voyager DE-PRO (Applied Biosystems).
Double-labeled GCAN-WASP was prepared by labeling first with a 10-fold molar excess of 1,5-IAEDANS (less reactive probe) for 3 h at room temperature. Unreacted probe was removed as above. Partially labeled GCAN-WASP was then incubated with a 20-fold molar excess of DABMI overnight at 4°C in the dark. After removal of unreacted probe, double-labeled GCAN-WASP was analyzed by HPLC on a reverse-phase C18 analytical column (Waters) using a 0-90% CH3CN gradient and 0.1% TFA. The multi-component spectrum was de-convoluted using the program Origin (Supplementary Fig. S9).
Preparation of complexes of WCA variants with actin and Arp2/3 complex
The cross-linked complexes actin-xWCAN-WASP and actin-xWN-WASP were prepared as reported24, 34. To prepare actin-GCA complexes, actin was mixed with a 1.5 molar excess of GCAN-WASP or GCAWAVE, followed by purification through an SD200HL 26/600 in Arp buffer (20 mM HEPES pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.2 mM ATP). The same protocol was used to purify 1:1:1 complexes of labeled actin-GCA bound to Arp2/3 complex.
Isothermal Titration Calorimetry (ITC)
ITC measurements were carried out on a VP-ITC apparatus (MicroCal). Samples where dialyzed for two days against Arp buffer (with either 0.2 mM ATP or ADP). Titrations were done at 25°C (ATP state) or 20°C (ADP state), the latter temperature being optimal for analysis of endothermic binding of GMFγ40. Titrations consisted of 10 μl injections, lasting for 10 s and spaced 300 s apart. The concentration (specified in figures) of the titrant was ~ 14-fold higher than that of the binding partner in the 1.44 ml cell. The heat of binding was corrected for the small exothermic heat of injection, determined by injecting ligand into buffer. Data were analyzed using the program Origin (OriginLab Corporation).
In competition experiments with GMF, actin-GCAN-WASP and Arp2/3 complex were dialyzed side-by-side for two days against Arp buffer supplemented with 0.2 mM ADP, resulting in ADP incorporation into Arp2/3 complex (purified Arp2/3 complex has no nucleotide bound45), but not actin. Indeed, actin bound to GS1 does not exchange nor hydrolyze the bound ATP, unless hexokinase and glucose are added to the dialysis buffer, which we verified through determination of the crystal structure (not shown).
Size-exclusion chromatography–multi-angle light scattering (SEC-MALS)
Samples (100 μl at 1–2 mg ml−1) were fractionated on a TSK-gel Super SW2000 column (Tosoh Bioscience) coupled to an Agilent 1100 HPLC system (Agilent Technologies). The molecular species separated by the column were analyzed through a DAWN HELEOS MALS detector and an Optilab rEX refractive index detector and their masses calculated with program Astra (Wyatt Technology).
Steady-state fluorescent energy transfer (FRET)
Steady-state fluorescence emission spectra ( λem = 400–600 nm) were recorded on a Cary Eclipse fluorometer (Varian) with excitation at λex = 337 nm. DABMI-labeled actin-GCA complexes at 300 μM were titrated into 300 μl of 3 μM pre-purified, IAEDANS-labeled actin-GCA-Arp2/3 complex and incubated for 10 min prior to measurements. Experiments were performed in Arp buffer (with 0.2 mM ATP) at 15°C. Final spectra were corrected for dilution. Data were fitted using the equation F(x) = Fmax x / (Kd + x) with the program KaleidaGraph (Synergy Software).
Time-resolved FRET (TR-FRET)
Fluorescence decays were recorded using a TimeMaster T-4 stroboscope lifetime fluorometer (Photon Technology International) with excitation at λex = 337 nm and emission at λem = 497 nm. Donor fluorescence decays, FD(t), were fitted with the function FD,fit(t), consisting of the convolution of a multi-exponential decay, FD(t), with the instrument response function (IRF) determined from light scattering by water,
| [1] |
where τDi is the donor-only fluorescence lifetime. In all cases, FD(t) was best fitted to two exponentials, n = 2 in Eq. 1 (Supplementary Fig. S8). The observed fluorescence decays of labeled actin-GCA bound to Arp2/3 complex FD+A(t) can be assumed to be a linear combination of fluorescence decays from uncoupled donors, FD(t), and donors coupled to acceptors, FDA(t). The acceptor is assumed to increase the fluorescence decay rate only through energy transfer. In this case, FD+A(t) is fitted by FD+A,fit(t):
| [2] |
where τDAi is the fluorescence lifetime of the donor in the presence of the acceptor. All the donors were assumed to be coupled to acceptors in intermolecular FRET samples, and therefore the mole fraction of donor only, XD, was set to 0, and the best fit was obtained with n = 2. In the case of intramolecular FRET samples, XD was varied in the fit defined by Eq. 2, and n was set to 4 (Supplementary Fig. S9). Distances were calculated from the average donor 〈τD〉 and donor-acceptor 〈τDA〉 lifetimes, defined as according to Eq. 3,
| [3] |
where RDA is the calculated separation between the donor and acceptor dipoles and R0 is the Förster distance defined by,
| [4] |
where QD is the quantum yield, κ2 is the orientation factor (assumed to be 2/3), n is the refractive index of protein in aqueous solution (1.4), and J is the overlap integral between the donor emission and acceptor absorption spectra. The quantum yield was determined at λex = 350 nm by comparison with a solution of quinine sulfate (1-5 μM in 0.1 M H2SO4), with absolute quantum yield of 0.753. Fluorescence decays for intermolecular FRET were analyzed using the program FeliX32 (PTI), and decays for intramolecular FRET were analyzed with the program FargoFit54.
Supplementary Material
Acknowledgements
Supported by National Institutes of Health grant R01 GM073791 to RD. DJK was supported by grant PF-13-033-01-DMC from the American Cancer Society. We thank Yale E. Goldman for help with TR-FRET experiments.
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Critical reviews in biochemistry and molecular biology. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 6.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 8.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 11.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 13.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 14.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20:217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends in cell biology. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Smith BA, Daugherty-Clarke K, Goode BL, Gelles J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1285–1290. doi: 10.1073/pnas.1211164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreishman-Deitrick M, et al. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–15256. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- 21.Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 22.Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 23.Pan F, Egile C, Lipkin T, Li R. ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J Biol Chem. 2004;279:54629–54636. doi: 10.1074/jbc.M402357200. [DOI] [PubMed] [Google Scholar]
- 24.Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E472–479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proceedings of the National Academy of Sciences of the United States of America. 1082011:E463–471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaucher JF, Mauge C, Didry D, Guichard B, Renault L, Carlier MF. Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex. The Journal of biological chemistry. 2012;287:34646–34659. doi: 10.1074/jbc.M112.394361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, Small JV. F- and G-actin concentrations in lamellipodia of moving cells. PloS one. 2009;4:e4810. doi: 10.1371/journal.pone.0004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 31.Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structural Basis for the Actin-Binding Function of Missing-in-Metastasis. Structure. 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small Molecules CK-666 and CK-869 Inhibit Actin-Related Protein 2/3 Complex by Blocking an Activating Conformational Change. Chemistry & biology. 2013 doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 34.Rebowski G, Namgoong S, Boczkowska M, Leavis PC, Navaza J, Dominguez R. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J Mol Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irobi E, et al. Structural basis of actin sequestration by thymosin-beta4: implications for WH2 proteins. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nature cell biology. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 37.Way M, Pope B, Gooch J, Hawkins M, Weeds AG. Identification of a region in segment 1 of gelsolin critical for actin binding. The EMBO journal. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Gandhi M, et al. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Current biology : CB. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boczkowska M, Rebowski G, Dominguez R. Glia maturation factor (GMF) interacts with Arp2/3 complex in a nucleotide state-dependent manner. The Journal of biological chemistry. 2013;288:25683–25688. doi: 10.1074/jbc.C113.493338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan Q, Nolen BJ. Structural basis for regulation of Arp2/3 complex by GMF. Nature Structural & Molecular Biology. 2013;20:1062–1068. doi: 10.1038/nsmb.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prochniewicz E, Thomas DD. Differences in structural dynamics of muscle and yeast actin accompany differences in functional interactions with myosin. Biochemistry. 1999;38:14860–14867. doi: 10.1021/bi991343g. [DOI] [PubMed] [Google Scholar]
- 43.Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgs HN, Pollard TD. Regulation of actin polymerization by Arp2/3 complex and WASp/Scar proteins. The Journal of biological chemistry. 1999;274:32531–32534. doi: 10.1074/jbc.274.46.32531. [DOI] [PubMed] [Google Scholar]
- 45.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 47.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 48.Dominguez R, Holmes KC. Actin structure and function. Annual review of biophysics. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu XP, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. The EMBO journal. 2012;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. Pt B. [DOI] [PubMed] [Google Scholar]
- 53.Scott TG, Spencer RD, Leonard NJ, Weber G. Emission Properties of Nadh Studies of Fluorescence Lifetimes and Quantum Efficiencies of Nadh, Acpyadh, and Simplified Synthetic Models. Journal of the American Chemical Society. 1970;92:687. &. [Google Scholar]
- 54.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.