Abstract
Background
Chromosomal rearrangements are a major driving force in shaping genome during evolution. Previous studies show that translocated genes could undergo elevated rates of evolution and recombination frequencies around these genes can be altered. Based on the recently released genome sequences of Triticum urartu, Aegilops tauschii, Brachypodium distachyon and bread wheat, an analysis of interchromosomal translocations in the hexaploid wheat genotype ‘Chinese Spring’ (‘CS’) was conducted based on chromosome shotgun sequences from individual chromosome arms of this genotype.
Results
A total of 720 genes representing putative interchromosomal rearrangements was identified. They were distributed across the 42 chromosome arms. About 59% of these translocated genes were those involved in the well-characterized translocations involving chromosomes 4A, 5A and 7B. The other 41% of the genes represent a large numbers of putative interchromosomal rearrangements which have not yet been described. The number of the putative translocation events in the D subgenome was about half of those presented in either the A or B subgenomes, which agreed well with that the times of interaction between the A and B subgenomes almost doubled that between either of them and the D subgenome.
Conclusions
The possible existence of a large number of interchromosomal rearrangements detected in this study provide further evidence that caution should be taken when using synteny in ordering sequence contigs or in cloning genes in hexaploid wheat. The identification of these putative translocations in ‘CS’ also provide a base for a systematic evaluation of their presence or absence in the full spectrum of bread wheat and its close relatives, which could have significant implications in a wide array of fields ranging from studies of systematics and evolution to practical breeding.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-015-0313-5) contains supplementary material, which is available to authorized users.
Keywords: Interchromosomal rearrangements, Wheat genome, Translocation, Comparative genomics, Chinese Spring
Background
Chromosomal translocations are frequently associated with genomic disorders in human [1], animals [2] and microbes [3,4]. They are also a major driving force in shaping genome during evolution [1-5] and translocated genes could undergo elevated rates of evolution [2,6]. Results from previous studies also indicate that chromosomal translocations can alter levels of recombination and the chances of getting the desired recombinants will be diminished if the targeted gene is located near or at a translocation breakpoint [7].
The interchromosomal translocations involving chromosomes 4A, 5A and 7B in hexaploid wheat have been well documented [8-10]. The rearrangement between chromosomes 4A and 5A also exists in the diploid A genome donor of Triticum urartu [11] as well as in the diploid wheat T. monococcum [9], indicating that the 4/5 translocation predates the polyploidization event forming tetraploid wheat. Similarly, the translocation between chromosome 7B and the rearranged chromosome 4A appears to also exist in the tetraploid wheat [9], indicating that this translocation had occurred before the second polyploidization event which formed hexaploid wheat.
Several additional interchromosomal rearrangements have been described. One of these is a reciprocal translocation between chromosomes 5B and 7B which was very prevalent in West European wheats in the 1960s and 1970s [12] and is likely widespread in modern varieties [7]. Another example is the interchromosomal translocation between chromosomes 5B and 6B present in landraces of tetraploid wheat from Ethiopia [13]. It is of interest to note that both of these interchromosomal translocations seem to be present only in genotypes from specific geographical regions. Although it is not clear whether any of them is associated with modified morphological characteristics, their highly localized geographical distributions suggest that they could be associated with adaptability.
Previous studies on chromosomal translocation in wheat have been based on either cytology [14,15] or molecular markers [8,9]. These techniques have only limited resolutions which allow only the detection of rearrangements involving large chromosome segments. Recent progress in genome sequencing and single-copy FISH [16] offers the potential to drastically enhance the power of detecting chromosomal rearrangements in polyploidy wheats and their relatives. Based on the recently released genome sequences of T. urartu [17], Aegilops tauschii [18], B. distachyon [19] and bread wheat [20], genes bordering each of the main translocation and inversion breakpoints on chromosomes 4A, 5A and 7B of the modern bread wheat genome were determined [21]. These genetic resources have also been exploited to assess interchromosomal rearrangements in the bread wheat genome and results obtained are reported here.
Methods
Data collection and analysis
Non-redundant orthologous gene sequences among CDSs (coding sequences) of B. distachyon, T. urartu, Ae. tauschii and wheat deletion bin-mapped ESTs (expressed-sequence tags) were identified in a recent study [22]. These sequences were analysed against the ‘CS’ shotgun sequences using the BLAST+ blastn algorithm with an E-value threshold of 10−5 (this value was applied in all subsequent BLAST analyses). The arm locations of genes on chromosome 3B were also identified previously [22]. For each of the non-redundant genes, the three best hits across the entire ‘CS’ genome were extracted. A gene was deemed to represent a putative interchromosomal rearrangement if any two of these three best hits were on different chromosomes from a given homoeologous group but the other one was on a chromosome belonging to a different homoeologous group [8,23]. For example, if the best three hits for a given gene were on chromosome arms 1AL, 1BL, and 2DS, the gene was considered to represent a putative translocation from 1DL to 2DS. For these genes, an additional 7 hits were then considered and the chromosomal locations were visually inspected again. Additional hits with different chromosomal locations from the first 3 hits were kept only. Only single-copy genes (the best 3 hits locating on the three chromosome arms belonging to a single homoeologous group) and those with simple duplications (3 of the best hits representing a single-copy genes and the other 3 hits representing a single homoeologous group, e.g. Bradi2g55820.1: 1AL, 1BL, 3BL, 2AL, 2BL, 2DL) were used in this study. For easy description, they were all classified as single-copy genes in this study. Genetic map locations of wheat contigs were obtained from http://www.wheatgenome.org/. [20]. A translocation was arbitrarily defined as the presence of at least two neighbouring genes with a maximum distance of 1.0 cM on at least one of the three wheat subgenomes.
Configuration of chromosome 4A
The arm ratio of chromosome 4A was reversed due to translocations between chromosomes of 4A, 5A and 7B [8-10,24,25]. As suggested previously [22], we used ‘original 4AS’and ‘original 4AL’ to refer to the arms of the ancestral version of this chromosome and ‘modern 4AS’ and ‘modern 4AL’ to refer to the modern arm configuration of this chromosome.
Validation of gene locations by PCR (polymerase chain reaction) amplification
Chromosome locations of a small number of genes identified from the above analysis were arbitrarily selected and validated using the euploid and nullisomic-tetrasomic and ditelosomic lines of ‘CS’. Annealing temperatures used ranges from 62 to 70°C depending on the primers (Additional file 1: Table S1). PCR products were separated on 1.5% agarose gels. If PCR products with similar sizes were generated from each of the wheat lines assessed, they were digested with a suitable restriction enzyme selected based on sequence alignments among the three subgenomes and then separated on gels.
Results
A total of 720 single-copy genes representing interchromosomal rearrangements were detected. These genes were located on each of the 42 chromosome arms (Table 1, Figure 1). Map locations for about 64% of these genes on at least one of the three subgenomes are known (Additional file 2: Table S2). Seven of these genes were further assessed by PCR amplification against the euploid and nullisomic-tetrasomic lines of ‘CS’. Primers designed for two of these genes (AEGTA13263, TRIUR3_25897) failed to amplify. PCR products were successfully obtained for the other five genes and chromosome locations deduced from the chromosome shotgun sequences from the International Wheat Genome Sequencing Consortium (IWGSC) were confirmed for all of them (Figure 2).
Table 1.
Locations of the 720 single-copy genes representing interchromosomal rearrangements on each of the 42 chromosome arms*
| Chr. arm | 1AS | 1AL | 1BS | 1BL | 1DS | 1DL | 2AS | 2AL | 2BS | 2BL | 2DS | 2DL | 3AS | 3AL | 3BS | 3BL | 3DS | 3DL | 4AS | 4AL | 4BS | 4BL | 4DS | 4DL | 5AS | 5AL | 5BS | 5BL | 5DS | 5DL | 6AS | 6AL | 6BS | 6BL | 6DS | 6DL | 7AS | 7AL | 7BS | 7BL | 7DS | 7DL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1AS | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 1AL | 1 | 2 | 1 | 1 | 5 | |||||||||||||||||||||||||||||||||||||
| 1BS | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 1BL | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||
| 1DS | 1 | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| 1DL | 3 | 11 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||||
| 2AS | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 2AL | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 2BS | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 2BL | 1 | 1 | 1 | 2 | ||||||||||||||||||||||||||||||||||||||
| 2DS | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||
| 2DL | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | |||||||||||||||||||||||||||||||
| 3AS | 1 | 1 | 1 | 1 | 2 | |||||||||||||||||||||||||||||||||||||
| 3AL | 1 | 1 | 3 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||||||||||||
| 3BS | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||||||||||||||
| 3BL | 1 | |||||||||||||||||||||||||||||||||||||||||
| 3DS | 3 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||
| 3DL | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||
| 4AS | 1 | |||||||||||||||||||||||||||||||||||||||||
| 4AL | 1 | 1 | 1 | 71 | 1 | |||||||||||||||||||||||||||||||||||||
| 4BS | 1 | |||||||||||||||||||||||||||||||||||||||||
| 4BL | 2 | 1 | 2 | 3 | 1 | |||||||||||||||||||||||||||||||||||||
| 4DS | ||||||||||||||||||||||||||||||||||||||||||
| 4DL | 1 | |||||||||||||||||||||||||||||||||||||||||
| 5AS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | ||||||||||||||||||||||||||||||||
| 5AL | 1 | 3 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 4 | 1 | 1 | 167 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 2 | 9 | 2 | |||||||||||||||||
| 5BS | 1 | 2 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||
| 5BL | ||||||||||||||||||||||||||||||||||||||||||
| 5DS | ||||||||||||||||||||||||||||||||||||||||||
| 5DL | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| 6AS | 1 | |||||||||||||||||||||||||||||||||||||||||
| 6AL | 1 | 3 | ||||||||||||||||||||||||||||||||||||||||
| 6BS | 1 | 1 | 1 | 3 | 1 | 1 | 2 | 2 | 1 | 3 | ||||||||||||||||||||||||||||||||
| 6BL | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 3 | ||||||||||||||||||||||||||||||||
| 6DS | 1 | 3 | 3 | 1 | 1 | 1 | 4 | 2 | ||||||||||||||||||||||||||||||||||
| 6DL | ||||||||||||||||||||||||||||||||||||||||||
| 7AS | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||
| 7AL | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||
| 7BS | 1 | 1 | 1 | 2 | 1 | 2 | 177 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||
| 7BL | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||
| 7DS | 1 | |||||||||||||||||||||||||||||||||||||||||
| 7DL |
*Genes were transferred from chromosome arms in column to those in the row.
Figure 1.
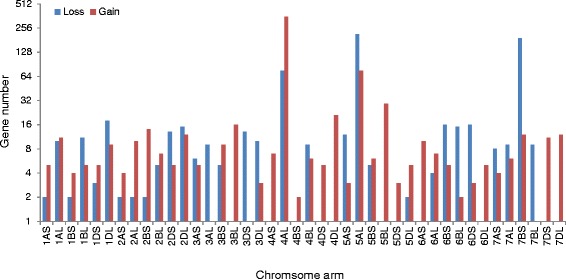
Distribution of the 720 single-copy genes and genes with simple duplication representing interchromosomal rearrangements among the 42 chromosome arms of bread wheat.
Figure 2.
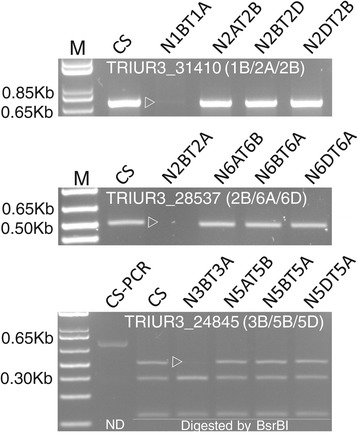
PCR profiles of the hexaploid wheat genotype ‘Chinese Spring’ (‘CS’) nullisomic-tetrasomic (NT) lines with primers for 3 of the genes involved in putative interchromosomal rearrangements. Locations of the genes detected from the chromosome shotgun sequences are given in brackets. 1Kb plus DNA ladder was used as the size marker (M). ND: non-digested control of the PCR product from ‘CS’. Fragments missing from the aneuploid lines of ‘CS’ are marked with open triangles.
Based on the presence of at least two neighbouring genes with a map distance of no more than 1.0 cM in at least one of the three subgenomes, 21 groups of genes representing interchromosomal rearrangement were detected. The 42 events of interchromosomal rearrangement represented by these 21 groups of genes were located on 18 of the 21 wheat chromosomes: 17 in the A subgenome, 17 in the B subgenome and the other 8 in the D subgenome (Figure 3). These 42 translocation events involved a total of 443 genes. Map locations were known for 333 of them (or 75%) in the A subgenome, 213 (48%) in the B subgenome, and 303 (68%) in the D subgenome (Additional file 2: Table S2).
Figure 3.
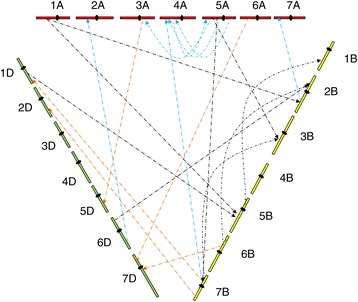
Distribution of the 42 interchromosomal translocation events among chromosomes of the three bread wheat subgenomes.
As expected, some of these translocations represent chromosome segments highly conserved between Brachypodium and wheat. One of the examples is represented by the two genes indicating a translocation from 1AL to 2BL (with homoeologous arm locations on 1BL, 1DL and 2BL, respectively). They are neighbouring genes in Brachypodium (Bradi3g20170 and Bradi3g20190). They also have the same map locations on both 1BL (83.82 cM) and 2BL (82.72 cM), the only two chromosome arms on which their locations are available. Another example is the segment translocated from 1DL to 5BL (with homoeologous locations on 1AL, 1BL and 5BL, respectively) represented by Bradi2g14170 and Bradi2g14240 (Additional file 2: Table S2). However, some of the other groups representing interchromosomal translocations consist of genes with similar locations in wheat but apparently having originated from different Brachypodium chromosomes. One of these groups is the one representing a translocation from 1AL to 5BL (with chromosome arm locations on 1BL, 1DL and 5BL, respectively), which consists of genes from two separate Brachypodium chromosomes (Additional file 2: Table S2).
Not unexpectedly, the three dominating groups of genes representing interchromosomal translocations were those representing the well-known translocations involving chromosomes 4A, 5A and 7B. They consisted of 177, 167, 71 and 9 genes, respectively (Table 1), and in total accounted for about 59% of the 720 genes detected in this study. Apart from them, the group with the largest number of genes was the one representing a translocation from 1DL to 5BL (with homoeologous arm locations on 1AL, 1BL and 5BL, respectively). This group consists of 11 genes, which was even larger than those representing the 5AL segment on the modern 7BS (with chromosome arm locations on 5BL, 5DL and 7BS, respectively) (9 genes) resulted from the 4A/5A/7B translocations (Additional file 2: Table S2). Judged by their map locations, the 11 genes transferred from 1DL to the modern 5BL likely represent more than one translocation event. The terminal locations of Bradi2g14170, Bradi2g14240 and Bradi2g26795 infer a translocation from the terminal end of 1DL to the terminal end of the modern 5BL. However, the map locations of several others genes (including Bradi2g26980, Bradi3g28810, TRIUR3_00282, and TRIUR3-27958) indicate the existence of a second translocation involving an interstitially located segment from 1DL to 5BL (Additional file 2: Table S2). Each of the other groups representing interchromosomal translocations contained 4 or less of the single-copy genes used in this study. For those with known map locations, genes in each of the groups were all closely linked, indicating that each of these gene groups likely represented a single interchromosomal translocation event.
Discussion
Taken advantage of the recently released chromosome arm-based sequences of the bread wheat genome, we conducted a systematic assessment of interchromosomal translocation in this important crop species. Even with the strict criteria used in this study, genes indicating the presence of interchromosomal rearrangement were detected on each of the 42 chromosome arms. About 59% of these genes were due to the well-known translocations involving chromosomes 4A, 5A and 7B. The other 41% of the translocated genes represent a large number of putative translocations scattered across chromosomes belonging to each of the three wheat subgenomes. Together with those putative intrachromosomal rearrangements reported earlier [22], the presence of such a large number of rearranged genes provides further evidence that caution should be taken when exploiting synteny in studying any of the three wheat genomes. As chromosomal translocations can alter levels of recombination and the chances of getting desired recombinants will be diminished if the gene conditioning the resistance is located near or at the translocation breakpoints [7], efficient breeding needs a clear understanding of how widespread these rearrangements exist in different wheat types and their close relatives.
For improving the likelihood that only genuine interchromosomal rearrangements were detected, only single-copy genes and those with simple duplications were selected for this study. The number of genes representing putative rearrangements could be significantly increased if the selection criteria were relaxed. For example, when the additional hits were tolerated, the number of genes representing the translocation from 6BS to 7DL would be increased from the currently 2 to 16. These additional genes with known map locations share a single location on either of the two non-translocated chromosome arms, 6AS or 6DS (Table 2), indicating that they likely represent a single chromosome segment on each of these two chromosomes. Thus it is not unreasonable to speculate that these genes could also represent a single segment on the third member of the homoeologous chromosome arm, 6BS, before the genes in concern were translocated to chromosome arm 7DL.
Table 2.
Genes representing a translocation from 6BS to 7DL
| Gene name | Locations* | |||||||
|---|---|---|---|---|---|---|---|---|
| Details of the best 3 hits | Best 3 hits | Additional hits | ||||||
| Contig_ID | Linkage (cM) | Contig_ID | Linkage (cM) | Contig_ID | Linkage# (cM) | |||
| Bradi1g35550.1 | 6AS_4384393 | NA | 6DS_2091402 | 81.58 | 7DL_3388333 | NA | 6AS,6DS,7DL | |
| Bradi3g06916.1 | 6AS_3107342 | NA | 6DS_2095270 | 81.58 | 7DL_3331018 | 111.08 | 6AS,6DS,7DL | |
| Bradi1g35592.1 | 6AS_4428500 | 60.96 | 6DS_2122226 | 81.58 | 7DL_3332022 | NA | 6AS,6DS,7DL | 2DL,5DL |
| Bradi3g05560.1 | 6AS_4392917 | 60.96 | 6DS_2080416 | 81.58 | 7DL_3328281 | 142.97 | 6AS,6DS,7DL | 1BS,3DL,4AL,7DS |
| Bradi3g05810.1 | 6AS_4403468 | 60.96 | 6DS_2098202 | 81.58 | 7DL_3393819 | 152.76 | 6AS,6DS,7DL | 4AL,4DS,7AL |
| Bradi3g05960.1 | 6AS_4357746 | 60.96 | 6DS_2125465 | 81.58 | 7DL_3358080 | 160.68 | 6AS,6DS,7DL | 5BL,5DL,7AL |
| Bradi3g07970.1 | 6AS_4351841 | 60.96 | 6DS_2058543 | 81.58 | 7DL_3349115 | 111.08 | 6AS,6DS,7DL | 4AL,4BS,4DS,5BL,7BL |
| TRIUR3_05619 | 6AS_4354230 | 60.96 | 6DS_2082494 | 81.58 | 7DL_3392566 | 191.24 | 6AS,6DS,7DL | 1AL,1BL,1DL,7DS |
| TRIUR3_15544 | 6AS_4357746 | 60.96 | 6DS_2125465 | 81.58 | 7DL_3358080 | 160.68 | 6AS,6DS,7DL | 5BL,5DL,7AL |
| AEGTA43678 | 6AS_4359749 | NA | 6DS_2056711 | 81.58 | 7DL_3390442 | NA | 6AS,6DS,7DL | 6DL,7DS |
| AEGTA43755 | 6AS_4365865 | NA | 6DS_2086887 | 81.58 | 7DL_3342428 | 120.28 | 6AS,6DS,7DL | 3B,4AS,4DL,7BS,7DS |
| Bradi3g06670.1 | 6AS_4384393 | NA | 6DS_2091402 | 81.58 | 7DL_3388333 | NA | 6AS,6DS,7DL | 5BL,5BS,5DL,5DS,6AL,6DL,7BS |
| Bradi3g16020.1 | 6AS_148467 | NA | 6DS_2077560 | 81.58 | 7DL_3361916 | NA | 6AS,6DS,7DL | 4DL,5AL,5BL,7AL |
| Contig36859 | 6AS_4324849 | NA | 6DS_1084274 | 81.58 | 7DL_3331672 | NA | 6AS,6DS,7DL | 1AL,1BL,2AL,2BL,5BL,5DL,6AL |
| Contig99362 | 6AS_148467 | NA | 6DS_2077560 | 81.58 | 7DL_3361916 | NA | 6AS,6DS,7DL | 4DL,5AL,5BL,5DL |
| TRIUR3_17134 | 6AS_3958480 | NA | 6DS_2065488 | 81.58 | 7DL_3396087 | 160.28 | 6AS,6DS,7DL | 1AL,1AS,1BS,2AL,2AS,7BL |
*4AS and 4AL refer to the short and long arms, respectively, of the modern chromosome 4A.
*NA indicates map locations not available.
The genes selected for this study were likely to be significantly under-estimated for another three reasons: Firstly, genes which detected sequences on one or two of the three wheat subgenomes were not considered. As the genome sequences used in this study is known to be incomplete [20], additional genes meeting the selection criteria used will likely become available with the improved genome coverage. Secondly, single chromosome arms are the smallest unit resolved for most of the genes used in this study. Although more than one segment on a modern chromosome arm could having been translocated from different parts of the wheat genome [21], the limited numbers of single-copy genes used in this study and the lack of map locations for many of them (Additional file 2: Table S2) made it difficult to reliably identify multiple rearrangement events within a single chromosome arm. For the same reasons, reliable identification of genes immediately flanking the breakpoints to precisely characterize the majority of the putative rearrangements may have to wait. Thirdly, the lack of map locations for many of the selected genes (Additional file 2: Table S2) also hampered the selection of neighbouring genes used for declaring the presence of interchromosomal rearrangements. Thus, additional interchromosomal rearrangements are likely to be detected when map locations for more genes become available. Clearly, although synteny has been extensively and successfully used in many applications [26-29], the possible existence of such a high percentage of translocated genes suggest that caution need to be taken when applying this approach at the gene level.
Previous studies indicate that the D subgenome of wheat has several unique features. For example, molecular marker analysis has showed that the D subgenome is much less polymorphic than either the A or B subgenome [30,31]. Chromosome shotgun sequences of ‘CS’ showed that, among the three bread wheat subgenomes, the class I elements of transposons (retroelements) were the least abundant but the class II elements (DNA transposons) were the most abundant in the D subgenome [20]. The significantly smaller number of the putative interchromosomal translocation events detected in the D subgenome in this study seems to be another addition to the unique features of this subgenome. However, considering the A and B subgenomes joined together much earlier than the D subgenome in the two polyploidization events leading to the formation of the hexaploid wheat [20,32], the interchromosomal rearrangements seem to have occurred at a similar frequency among them.
Similar to those in other plant species, current systematics of wheat and its close relatives is based mainly on morphological characteristics. It is known that the principal differences between some of the species, such as those among the various hexaploid forms of wheat, are due to different alleles of one or two single genes [33]. Considering that translocations are often associated with significant disorders in various species including mammals, birds and bacteria [1-4], it is not unanticipated that changes due to these chromosomal rearrangements could be more drastic than those due to single genes in wheat as well. The identification of the putative interchromosomal translocations in this study and those intrachromosomal rearrangements in earlier studies [22,25,34,35] in the hexaploid wheat genotype ‘CS’ paved the way for a systematic assessment of their presence or absence across the full spectrum of bread wheat and its close relatives, which would not only allow the classification of bread wheat and its relatives on a more scientific basis but also facilitate the exploitation of genes from the wild relatives in wheat breeding programs.
Conclusions
A total of 720 genes representing putative interchromosomal rearrangements in the bread wheat genotype ‘CS’ was detected in this study and the number of the genes was likely significantly underestimated. These genes were distributed on each of the 42 chromosome arms and they suggested the presence of a large number of putative interchromosomal rearrangements in this genotype. Together with those intrachromosomal rearrangements reported in earlier studies, these results provide further evidence showing that extensive structural differences likely exist among the three subgenomes of ‘CS’. An effort is urgently required to clarify which of these rearrangements were induced during the production of the aneuploids used for generating the shotgun sequences and which were specific to the genotype ‘CS’. A clear understanding of the presence or absence of these chromosomal rearrangements in the full spectrum of bread wheat and its close relatives could dramatically improve our capacity in wheat genome research as well as in new variety breeding.
Acknowledgements
The work reported in this publication was supported by CSIRO (Commonwealth Scientific and Industrial Organisation, Australia). JM is grateful to the Sichuan Agricultural University and the China Scholarship Council for funding his visit to CSIRO Agriculture Flagship. The authors are grateful to Drs Evans Lagudah (CSIRO) and Peng Zhang (Plant Breeding Institute, The University of Sydney) for providing the aneuploid stocks of ‘Chinese Spring’. The authors are also very grateful to Prof Katrien Devos (University of Georgia, USA) for constructive discussion during the preparation of this publication, and to Dr Karen Aitken (CSIRO) for her critical reading of the manuscript.
Abbreviations
- CDS
Coding sequence
- CS
Chinese Spring
- IWGSC
The international wheat genome sequencing consortium
- EST
Expressed-sequence tag
- PCR
Polymerase chain reaction
Additional files
Primer sequences used for validating chromosomal locations of selected genes.
Details of the 720 Single-copy genes representing possible interchromosomal rearrangements in wheat.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
This study was designed by CL, JM, and JS. Data collection, analysis and experiments were conducted by JM, JS, and ZZ. YW, Y-LZ, GY, and JD contributed to the design of the study and the preparation of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jian Ma, Email: plantgbmj@hotmail.com.
Jiri Stiller, Email: Jiri.stiller@csiro.au.
Zhi Zheng, Email: zhengzhi1103@aliyun.com.
Yuming Wei, Email: ymwei@sicau.edu.cn.
You-Liang Zheng, Email: ylzheng@sicau.edu.cn.
Guijun Yan, Email: guijun.yan@uwa.edu.au.
Jaroslav Doležel, Email: dolezel@ueb.cas.cz.
Chunji Liu, Email: Chunji.liu@csiro.au.
References
- 1.Brown JD, O’Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- 2.Burt DW, Bruley C, Dunn IC, Jones CT, Ramage A, Law AS, et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402(6760):411–2. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- 3.Colson I, Delneri D, Oliver SG. Effects of reciprocal chromosomal translocations on the fitness of Saccharomyces cerevisiae. EMBO Rep. 2004;5(4):392–8. doi: 10.1038/sj.embor.7400123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrow JD, Cooper VS. Evolutionary effects of translocations in bacterial genomes. Genome Biol Evol. 2012;4(12):1256–62. doi: 10.1093/gbe/evs099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankoff D, Nadeau JH. Chromosome rearrangements in evolution: From gene order to genome sequence and back. Proc Natl Acad Sci U S A. 2003;100(20):11188–9. doi: 10.1073/pnas.2035002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao W, Golding GB. Does gene translocation accelerate the evolution of laterally transferred genes? Genetics. 2009;182(4):1365–75. doi: 10.1534/genetics.109.104216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Law C, Worland A. The control of adult‐plant resistance to yellow rust by the translocated chromosome 5BS‐7BS of bread wheat. Plant Breed. 1997;116(1):59–63. doi: 10.1111/j.1439-0523.1997.tb00975.x. [DOI] [Google Scholar]
- 8.Liu C, Atkinson M, Chinoy C, Devos K, Gale M. Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor Appl Genet. 1992;83(3):305–12. doi: 10.1007/BF00224276. [DOI] [PubMed] [Google Scholar]
- 9.Devos K, Dubcovsky J, Dvořák J, Chinoy C, Gale M. Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet. 1995;91(2):282–8. doi: 10.1007/BF00220890. [DOI] [PubMed] [Google Scholar]
- 10.Nelson JC, Sorrells ME, Van-Deynze A, Lu YH, Atkinson M, Bernard M, et al. Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5, and 7. Genetics. 1995;141(2):721. doi: 10.1093/genetics/141.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King I, Purdie K, Liu C, Reader S, Pittaway T, Orford S, et al. Detection of interchromosomal translocations within the Triticeae by RFLP analysis. Genome. 1994;37(5):882–7. doi: 10.1139/g94-125. [DOI] [PubMed] [Google Scholar]
- 12.Riley R, Coucoli H, Chapman V. Chromosomal interchanges and the phylogeny of wheat. Heredity. 1967;22:233–48. doi: 10.1038/hdy.1967.29. [DOI] [Google Scholar]
- 13.Belay G, Merker A. Cytogenetic analysis of a spontaneous 5B/6B translocation in tetraploid wheat landraces from Ethiopia, and implications for breeding. Plant Breed. 1998;117(6):537–42. doi: 10.1111/j.1439-0523.1998.tb02203.x. [DOI] [Google Scholar]
- 14.Naranjo T, Roca A, Goicoechea P, Giraldez R. Arm homoeology of wheat and rye chromosomes. Genome. 1987;29(6):873–82. doi: 10.1139/g87-149. [DOI] [Google Scholar]
- 15.Valárik M, Bartoš J, Kovářová P, Kubaláková M, De Jong J, Doležel J. High‐resolution FISH on super‐stretched flow‐sorted plant chromosomes. Plant J. 2004;37(6):940–50. doi: 10.1111/j.1365-313X.2003.02010.x. [DOI] [PubMed] [Google Scholar]
- 16.Danilova TV, Friebe B, Gill BS. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet. 2014;127(3):715–30. doi: 10.1007/s00122-013-2253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling H-Q, Zhao S, Liu D, Wang J, Sun H, Zhang C, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- 18.Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–5. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- 19.Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, et al. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7282):763–8. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 20.International-Wheat-Genome-Sequencing-Consortium A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Stiller J, Berkman PJ, Wei Y, Rogers J, Feuillet C, et al. Sequence-based analysis of translocations and inversions in bread wheat (Triticum aestivum L.) PLoS One. 2013;8(11):e79329. doi: 10.1371/journal.pone.0079329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Stiller J, Wei Y, Zheng Y-L, Devos KM, Doležel J, et al. Extensive pericentric rearrangements in the bread wheat (Triticum aestivum L.) genotype ‘Chinese Spring’ revealed from chromosome shotgun sequence data. Genome Biol Evol. 2014;6(11):3039–48. doi: 10.1093/gbe/evu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi L, Friebe B, Gill BS. Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome. 2006;49(12):1628–39. doi: 10.1139/g06-123. [DOI] [PubMed] [Google Scholar]
- 24.Mickelson-Young L, Endo T, Gill B. A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor Appl Genet. 1995;90(7):1007–11. doi: 10.1007/BF00222914. [DOI] [PubMed] [Google Scholar]
- 25.Miftahudin. Ross K, Ma XF, Mahmoud A, Layton J, Milla MAR, et al. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics. 2004;168(2):651–63. doi: 10.1534/genetics.104.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, et al. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics. 2004;168(4):2169–85. doi: 10.1534/genetics.104.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvão VC, Nordström KJ, Lanz C, Sulz P, Mathieu J, Posé D, et al. Synteny‐based mapping‐by‐sequencing enabled by targeted enrichment. Plant J. 2012;71(3):517–26. doi: 10.1111/j.1365-313X.2012.04993.x. [DOI] [PubMed] [Google Scholar]
- 28.Ghedin E, Bringaud F, Peterson J, Myler P, Berriman M, Ivens A, et al. Gene synteny and evolution of genome architecture in trypanosomatids. Mol Biochem Parasitol. 2004;134(2):183–91. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Grant D, Cregan P, Shoemaker RC. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc Natl Acad Sci U S A. 2000;97(8):4168–73. doi: 10.1073/pnas.070430597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen AM, Barker GL, Berry ST, Coghill JA, Gwilliam R, Kirby S, et al. Transcript‐specific, single‐nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.) Plant Biotechnol J. 2011;9(9):1086–99. doi: 10.1111/j.1467-7652.2011.00628.x. [DOI] [PubMed] [Google Scholar]
- 31.Cadalen T, Boeuf C, Bernard S, Bernard M. An intervarietal molecular marker map in Triticum aestivum L. Em. Thell. and comparison with a map from a wide cross. Theor Appl Genet. 1997;94(3–4):367–77. doi: 10.1007/s001220050425. [DOI] [Google Scholar]
- 32.Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345(6194):1250092. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- 33.Miller T. Wheat breeding: its scientific basis. London: Chapman and Hall; 1987. Systematics and evolution; pp. 1–30. [Google Scholar]
- 34.Conley E, Nduati V, Gonzalez-Hernandez J, Mesfin A, Trudeau-Spanjers M, Chao S, et al. A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics. 2004;168(2):625–37. doi: 10.1534/genetics.104.034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linkiewicz A, Qi L, Gill B, Ratnasiri A, Echalier B, Chao S, et al. A 2500-locus bin map of wheat homoeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics. 2004;168(2):665–76. doi: 10.1534/genetics.104.034835. [DOI] [PMC free article] [PubMed] [Google Scholar]


