Abstract
Background
Autophagy is a cellular lysosomal degradation mechanism has been implicated in chronic liver diseases and hepatocellular carcinoma (HCC). Association of autophagy defect with the development of human HCC has been shown in transgenic mouse model.
Aim
We performed this study to verify whether a defect in autophagy would play a role in human hepatocellular carcinoma (HCC).
Methods
Archival tissue sections of 20 patients with HCC with or without hepatitis C virus (HCV) infection were studied. All slides were immunostained using monoclonal antibodies to p62 and glypican-3 with appropriate positive and negative controls. The expression of p62 and glycican-3 in the HCC and the surrounding non-tumor were semiquantitated. The cytoplasmic staining was graded as negative, weak or strong.
Results
Positive p62 staining was found in 20 out of 20 (100%) HCCs and negative staining was observed in 20 out of 20 non-tumor areas and cirrhotic nodules. Positive glypican-3 staining was found in 70% of HCCs and negative staining was seen in all non-tumor areas. An autophagy defect leading to increased expression of p62 and glypican-3 was also seen in the HCC cell line (Huh-7.5), but not in the primary human hepatocytes. Activation of cellular autophagy in Huh-7.5 cells efficiently cleared p62 and glypican-3 expression and inhibition of autophagy induced the expression of p62 and glypican-3.
Conclusions
This study shows that p62 is increased in HCC compared to the surrounding non-tumorous liver tissue suggesting that human HCCs are autophagy defective. We provide further evidence that glypican-3 expression in HCC may be also related to defective autophagy. Our study indicates that p62 immunostain may represent a novel marker for HCC.
Keywords: Hepatocellular Carcinoma, Autophagy, Glypican-3, p62, Immunostaining
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the incidence of HCC is increasing in the Western world (El-Serag et al., 2007; Befeler et al. 2002; Thorgeirsson et al., 2002). In the majority of cases, HCC develops as a result of chronic inflammation and cirrhosis secondary to hepatitis B and hepatitis C viral infection (HBV, HCV) and non-viral etiologies including non-alcoholic and alcoholic fatty liver diseases (Rustgi et al., 1987). HCCs detected at a very early stage are treatable, but HCCs diagnosed at later stages are difficult to treat and have worse prognosis. Therefore early diagnosis and development of newer targeted therapy are urgently needed to improve HCC patient survival.
The serum alpha-fetoprotein (AFP) level has been used as a marker for diagnosis and early detection of HCC (Johnson et al., 2001). However, it is not specific for HCC since elevated AFP levels have also been detected in a considerable number of patients with chronic liver disease and liver cirrhosis (Collier et al., 1998; Sherman M., 2001). Recently, a number of studies have demonstrated that glypican-3 is another reliable tumor marker for hepatocellular carcinomas (Jakubovic et al., 2007; Shirakawa et al., 2009; Kandil et al., 2007; Mounajjed et al., 2013). Glypican-3 immunostaining shows strong membranous and cytoplasmic staining of HCC and the expression was undetectable in normal and cirrhotic livers. Glypican-3 is a membrane-bound proteoglycan localized on the cell membrane of hepatocellular carcinoma (Filmus et al., 2008). Glypican-3 has been considered a better marker compared to AFP in the diagnosis of early HCC (Capurro et al., 2003; Hippo et al., 2004; Wang et al., 2006; Man et al., 2005; Yamauch et al., 2005). The mechanism as to why glypican-3 is expressed at a high level only in the tumor and not in the surrounding non-tumor liver is unknown. This could be due to the incomplete understanding of the complex molecular mechanisms linking to the multifactorial etiology involved in hepatocarcinogenesis. Recent studies have suggested that glypican-3 expression in HCC is regulated by the expression of Sulfatase-2, c-myc and microRNAs (Li et al., 2012; Lai et al., 2008; Maurel et al., 2013). Since these reports have not been consistent, additional mechanisms of glypican-3 regulation in HCC need to be explored. A search for a highly reliable tissue marker for the detection of early HCC is needed.
Autophagy is an evolutionary conserved lysosomal degradation process occurring in chronic liver diseases including viral hepatitis, alcoholic liver disease and fatty liver disease (Eskelinen et a., 2009). Cellular autophagy, which is maintained during chronic liver injury, may play an important role in the sustainment of chronic liver disease and cancer development (Rautou et al., 2010; Kotsafti et al., 2012; Raitou et al., 2011). In this regard, there is evidence to suggest that mice with deletion of autophagy related genes, i.e. ATG 5 and ATG 7 develop liver adenoma (Chen et al., 2009; Takamura et al., 2011; Cui et al., 2013). The involvement of a highly conserved cellular autophagy process in human hepatocarcinogenesis is unknown and needs to be explored.
This study was performed to verify our hypothesis that defective autophagy may play a role in the development of hepatocellular carcinoma in humans. Cellular autophagy is a multistep process that begins with initiation step, followed by nucleation step, elongation step and finally maturation step [28]. It is thought that several cellular compartments including the endoplasmic reticulum (ER), the Golgi apparatus and plasma membrane participate in the autophagy process. Cellular autophagy process is controlled by complex interactions of many cellular proteins, but how these protein-protein interactions when altered lead to autophagy defect is not well understood. To confirm that HCC cells are autophagy defective, we examined the accumulation of p62, which has been used as a marker for autophagy deficiency (Chen et al., 2009). This protein is used as an index for autophagy flux measurement since p62 is degraded during the autophagy process (Puissant et al., 2012; Kirkin et al., 2009). A monoclonal antibody to p62 was used to determine autophagy defect in HCC samples. The expression of p62, an autophagy defect marker, was compared with glypican-3, another reliable marker for human hepatocellular carcinoma. Our results indicate that 100% of viral and non-viral related HCC samples have positive expression of p62, and the adjacent non-tumor areas are negative for p62. We also found that an impaired expression of glypican-3 in HCC cells is related to autophagy defect, since activation of cellular autophagy by mTOR inhibitor clears the expression of p62 and glypican-3.
Materials and Methods
Tissue Specimens
Paraffin blocks of 20 HCCs from patients with and without hepatitis C virus infection were obtained from the Department of Pathology, The Mount Sinai Medical Center, New York. Hematoxylin and eosin (H & E)-stained sections of all specimens including cancer and non-cancer areas of the liver tissue were examined by three Pathologists (SNT, TW and KM)
Antigen retrieval and immunohistological staining
Five-micron tissue sections were prepared and the slides were deparaffinized for 15 minutes at 50–60°C followed by treatment with xylene twice for 5 minutes. The tissue sections were rehydrated by sequential treatment with 100%, 95% and 80% alcohol. Peroxidase quenching was carried out by incubation with 3% hydrogen peroxide and 100% methanol for 5 minutes. The slides were placed in a plastic Coplin jar with Reveal Deckloaker RTU (Biocare Medical) for 25 minutes at 95°C in a steamer for heated antigen retrieval. Following this step, the slides were allowed to cool down at room temperature for 20 minutes. The tissue sections were rinsed in deionized distilled water and marked using a PAP pen. The slides were incubated with a blocking sniper (Biocare Medical) for 10 minutes and incubated with a primary antibody for 1 hour at room temperature. The primary antibodies we used were p62 mouse monoclonal antibody (Cell signaling) (1:200 dilution) and pre-diluted antibody to glypican-3 (Biocare Medical). After the primary antibody incubation, slides were washed 3 times in Tris Buffer Saline (TBS) (pH 8.0), and incubated with a MACH 4 mouse probe (Biocare Medical, UP534) for 20 minutes and MACH 4 HRP Polymer (Biocare Medical, MRH534) for 30 minutes each, then washed 3 times using TBS. Finally, tissue sections were treated with diaminobenzidine (DAB) chromogen (Dako Cytomation, Carpinteria, CA) for 1–5 minutes. The slides were then counterstained with hematoxylin for 30 seconds and Tacha’s bluing Solution (Biocare Medical, HTBLU) for 30 seconds, dehydrated with 95% and 100% alcohol, mounted and observed by light microscopy.
Evaluation of immunohistochemical Staining
Immunohistochemical staining of HCC tissue sections were examined by two pathologists (TW and KM). Scores were assigned to the intensity and percentage of positive staining of the all the slides used in this study. Score 0 means negative staining, score (+) when 1–10% of cells were positive, score (++) when 10–50% of cells were positive and score (+++) when 50–100% cells were positive. Discrepancies were resolved by a consensus between the two pathologists using a multiheaded microscope in the Pathology Department, Tulane University Health Sciences Center. H&E-stained sections of all specimens including cancer and non-cancer cases were examined by the same two pathologists following the immunohistichemical evaluation.
Immunostainings of Cultured Hepatoma Cells and Primary Human Hepatocytes
Cultured Huh-7 cells and primary human hepatocytes (Xenotech) were mounted onto glass slides via cytospin. The cells were washed twice with 10mM PBS pH 7.4 (Sigma-Aldrich, St Louis, MO) for 5 minutes, fixed in chilled acetone for 15 minutes and then permeabilized by treatment with Reveal Decloaker RTU (Biocare Medical, RV 100) for 25 minutes at boiling point. Slides were then cooled down to room temperature for 20 minutes. Blocking was performed utilizing Background Sniper (Biocare Medical, BS966) for 10 minute at room temperature. The cells were incubated with monoclonal anti-p62 antibody (Cell Signaling) at 1:200 diluted with Da Vinci Green Diluent (Biocare Medical, PD900) for 1 hour at room temperature. Following the primary antibody incubation, the cells were washed 3 times in Tris Buffered Saline (pH 8.0), and incubated with MACH 4 mouse probe (Biocare Medical, UP534) for 20 minutes. After the mouse probe treatment, the cells were incubated with MACH 4 HRP Polymer (Biocare Medical, MRH534) for 30 minutes, and cells were washed with TBS 3 times. Next, the cells were treated with diaminobenzidine (DAB) chromogen (Dako Cytomation, Carpinteria, CA) for 5 minutes. The slides were counterstained with hematoxylin for 30 seconds and Tacha’s bluing Solution (Biocare Medical, HTBLU) for 30 seconds, dehydrated with 95% and 100% alcohol, mounted and observed by light microscopy.
Results
Expression of p62 in human hepatocellular carcinomas
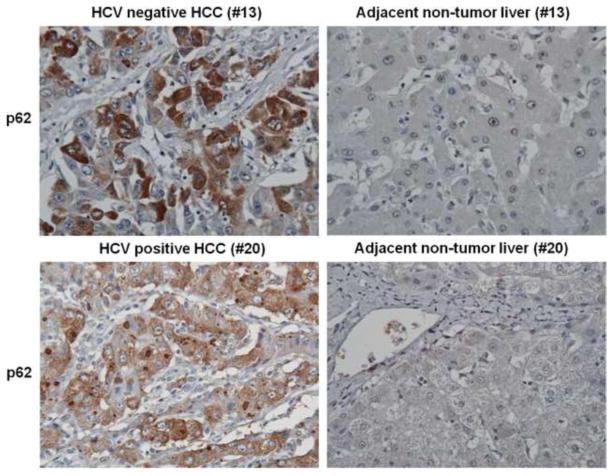
The expression of p62 was examined following immunocytochemical staining of 20 archival formalin-fixed, paraffin-embedded HCC samples and their surrounding non-tumorous liver tissue. Most of the specimens used in our study have liver cirrhosis and all have HCC with or without HCV infection (Table 1). The expression of p62 was absent in the control livers and cirrhotic nodules. Most of the HCCs showed positive expression of p62 with a variation in the staining intensity. The expression of p62 in HCC was localized mostly in the cytoplasmic vacuoles and also in perinuclear in some samples (Figure 1). The specimens with high expression of p62 and low expression of p62 are shown in Table 1. P62 expression was not detectable in non-tumor areas and cirrhotic nodules. The expression of p62 was detected in 20 out of 20 tumor specimens. There was no correlation between the amount of p62 expression and the degree of differentiation, i.e. well differentiated versus poorly differentiated HCC. The expression of p62 between HCV positive and HCV negative HCC was not different. The expression of p62 was weak or negative in six-cholangiocarcinoma tissues tested.
Table 1.
Summary of glypican-3 and p62 expression in HCC
| SAMPLES | HCV | Diagnosis | P62 Staining | Glypican 3 |
|---|---|---|---|---|
| 1. | (+) | Cirrhosis/HCC | + | 0 |
| 2. | (−) | Cirrhosis/HCC | + | 0 |
| 3. | (−) | Cirrhosis/HCC | ++ | 0 |
| 4. | (−) | Cirrhosis/HCC | ++ | +++ |
| 5. | (+) | Cirrhosis/HCC | ++ | 0 |
| 6. | (+) | Cirrhosis/HCC | ++ | +++ |
| 7. | (−) | Cirrhosis/HCC | + | + |
| 8. | (−) | Cirrhosis/HCC | + | 0 |
| 9. | (+) | Cirrhosis/HCC | +++ | ++ |
| 10. | (+) | Cirrhosis/HCC | ++ | ++ |
| 11. | (−) | Cirrhosis/HCC | + | +++ |
| 12. | (−) | Cirrhosis/HCC | + | 0 |
| 13. | (−) | Cirrhosis/HCC | ++ | +++ |
| 14. | (+) | Cirrhosis/HCC | +++ | +++ |
| 15. | (+) | Cirrhosis/HCC | + | +++ |
| 16. | (−) | Cirrhosis/HCC | +++ | +++ |
| 17. | (−) | Cirrhosis/HCC | + | ++ |
| 18. | (+) | Cirrhosis/HCC | + | ++ |
| 19. | (+) | Cirrhosis/HCC | ++ | +++ |
| 20. | (+) | Cirrhosis/HCC | ++ | +++ |
0 = No Staining
+ = 1–10% cells show positive staining
++= 10–50% cells show positive staining
+++ =50–100% cells show positive staining
Figure 1.
Immunohistochemical staining of p62 in hepatocellular carcinoma and adjacent non-tumorous tissues samples. Representative staining of tissue section of case 13 and 20 are shown for comparison.
Glypican-3 expression in HCC
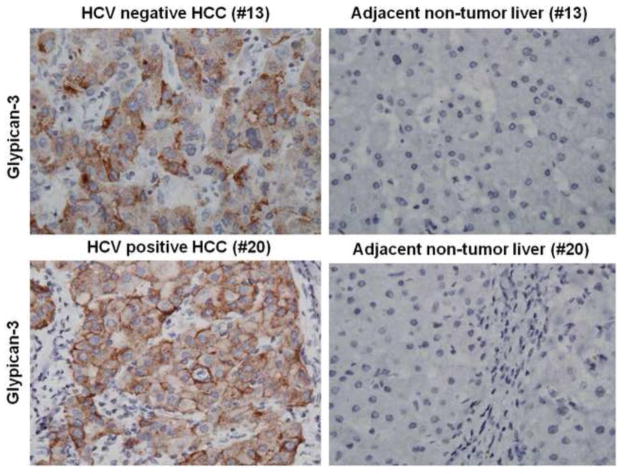
A number of reports have shown that glypican-3 is a specific and sensitive biomarker for the diagnosis of HCC by immunohistochemistry. Abnormal expression of glypican-3 has been seen in the majority of HCCs, while normal livers and benign liver lesions show no expression. Immunohistochemical analysis using monoclonal antibodies specific to glypcan-3 demonstrated strong membranous and cytoplasmic staining, whereas the non-tumorous liver was negative. To compare the sensitivity and specificity of p62 expression with glypican-3, these two markers were examined in our 20-HCC specimens. Among the 20 HCC specimen used in our study, 14 of them showed cytoplasmic and membranous expression of glypican-3 and in the remaining six HCC specimens, the glypican-3 expression was negative. The expression of p62 expression showed 100% correlation in 20 HCC samples as compared to only 70% showing the glypican-3 positive staining (Table 1). The expression of p62 was mostly cytoplasmic; where as the expression of glypican-3 was both cytoplasmic and membranous. The expression of p62 and glypican-3 was seen only in HCC cell line (Huh-7 cells) but not in the primary human hepatocytes. Representative pictures showing cytoplasmic staining of two HCCs with glypican-3 antibody are illustrated in Figure 2. The normal liver and cirrhotic liver were negative for glypican-3. There were no differences in the expression of p62 and glypican-3 in the HCC samples with or without hepatitis C virus infection, which indicate that HCC development due to autophagy defect is not only specific to viral liver diseases.
Figure 2.
Glypican-3 expression and localization in human hepatocellular carcinoma and adjacent non-tumorous tissue samples. Representative staining of case 13 and 20 shows membranous and cytoplasmic staining of tumor cells.
Cell culture study shows that mTOR inhibitor abolished the expression of glypican-3 and p62 in human hepatocellular carcinoma cells in vitro
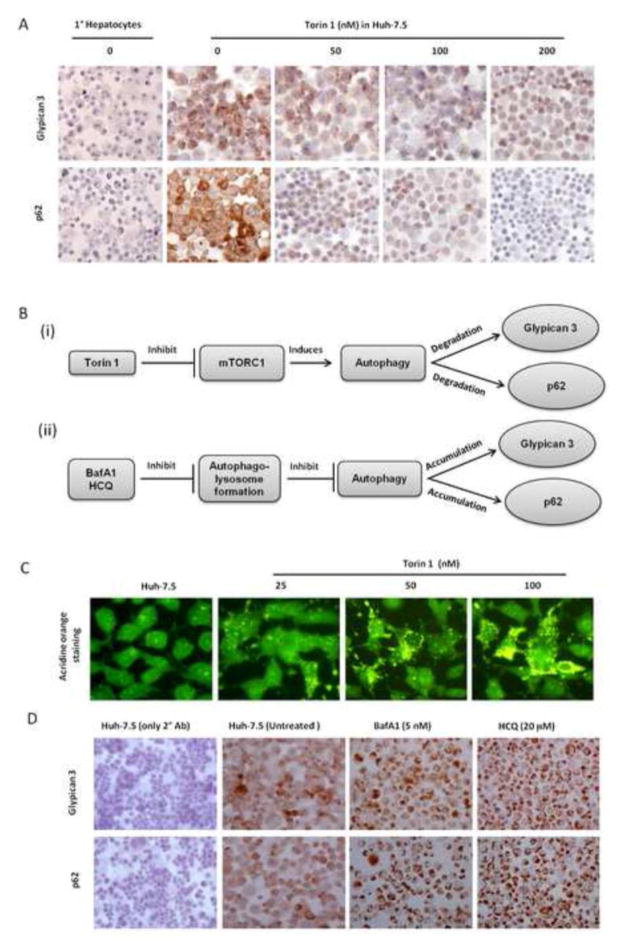
Several studies including our own have shown that glypican-3 is overexpressed in HCC but not in the surrounding non-tumorous livers. The mechanisms as to why the cell membrane of HCC over-expresses heparin sulfate proteoglycan molecule is not known. First, we determined whether the expression of p62 and glypican-3 could also show a similar pattern between non-tumorous primary human hepatocytes and human hepatocellular carcinoma cell lines. The expression of p62 and glypican-3 were examined using immunocytochemical staining. Our results show that the expression of p62 and glypican-3 was specifically upregulated in the human hepatocellular carcinoma cell line (Huh-7.5) but not in primary human hepatocytes (Figure 3A). Since the expression of p62 in HCC cells is due to a defect in autophagy, we further investigated whether autophagy defect mechanisms could also explain the expression of glypican-3 in HCC. For this purpose, the expression of p62 and glypican-3 in Huh-7.5 cells were examined after treatment with a known autophagy inducer, Torin. Recent studies show that Torin1 is a better substrate for mTOR than rapamycin (Settembre et al., 2012; Thoreen et al., 2009; Feldman et al., 2009). Torin 1 efficiently inhibits mTOR activity and induces cellular autophagy (Figure 3B). The process of autophagy starts with autophagosome formation, which then progresses to autophagolysosomes through the fusion of acidic lysosomes. Huh-7.5 cells were treated with different concentrations of Torin1 and activation of autophagy was monitored after 24 hours by acridine orange staining. The green dye changes to an orange color when autophagysome is fused with acidic lysosome (Wang et al., 2008). Results of this experiment indicate there is a dose dependent increase in the number of orange-colored autophagolysomes in the cytoplasm in Torin 1 treated cells (Figure 3C). The untreated Huh-7.5 cells show cytoplasmic and nuclear green fluorescence without autophagy induction. Immunoperoxidase staining results shows that p62 and glypican-3 were expressed only in Huh-7.5 cells but not in the non-cancerous primary human hepatocytes. Immunostaining results show Torin 1 treatment induced clearance of p62 and glypican-3 expression in the hepatocellular carcinoma cell line in a dose dependent manner (Figure 3A). To verify that autophagy defective mechanism prevented degradation of p62 and glypican-3 expression, Huh-7.5 cells were treated with autophagy inhibitors (Bafilomycin and hydroxychloroquine) and then the levels of p62 and glypican-3 expression were examined. Autophagy inhibition indeed induced the expression of glypican-3 and p62 (Figure 3D). These results suggest that impaired autophagy response in HCC cells may be responsible for glypican-3 expression.
Figure 3.
Mechanistic studies to show that impaired autophagy leads to accumulation of p62 and glypican-3. Panel A: Shows negative expression of glypican-3 and p62 in primary human hepatocytes and positive expression in HCC. Cell culture studies show that expression of glupican-3 and p62 was reduced in a concentration dependent manner by inducing autophagy response in Huh-7.5 cell line by Torin-1 (mTOR inhibitor). Panel B: (i) Show the molecular target how autophagy response in Huh-7.5 cells was induced by treatment with mTOR inhibitor (Torin1). (ii) Show molecular target how inhibiting autophagy response by Bafilomycin A1 (BafA1) and Hydroxychloroquine (HCQ) induced expression of glypican-3 and p62. Panel C: Huh-7.5 cells were treated with different concentrations of Torin1 for 24 hours stained with acridine orange dye and then examined under a fluorescence microscope. Acridine orange is a lysotropic dye that accumulates in acidic organelles in a pH-dependent manner. At neutral pH, acridine orange show green fluorescence. Within acidic environment of autophagolysosome vesicles, acridine orange becomes protonated and trapped within the organelle. Protonated acridine orange forms aggregates that emit bright red fluorescence due to autophagosome/lysosome fusion. This shows dose dependent increase in orange fluorescence in the acidic vacuoles due to Torin1 treatment. Panel D: Induced expression of glypican-3 and p62 seen in Huh-7.5 cells after treatment with either Bafilomycin A1 (BafA1) or Hydroxychloroquine (HCQ) (authophagy blocker).
Discussion
The results obtained from twenty paraffin embedded HCC samples confirm the defective autophagy response in human hepatocellular carcinoma tissue samples by immunohistochemical detection of p62 expression. All HCC samples used in our study showed positive expression of human hepatocellular carcinoma as compared to the surrounding non-tumorous liver as well as cirrhotic nodules. Our study shows that both of the markers are expressed in HCC probably because of an autophagy defect. It is well known that HCC is usually associated secondary to chronic liver diseases of viral and non-viral etiology. The majority of HCC develop in the background of long-standing chronic liver diseases and cirrhosis. Liver cirrhosis of any etiology has been found to be strongest risk factor for the development of HCC (Et-Serag et al., 2007). The molecular mechanisms of how the microenvironment of a cirrhotic liver drives hepatocytes to develop into a malignant tumor has been widely studied for a number of years and appears to be complex (Thorgeirsson et al., 2002). There is a demand to develop specific molecular markers that could facilitate early detection and monitoring treatment response for HCC. Glypican-3 is widely used as marker for HCC diagnosis and is expressed in 72% of HCC and undetectable in hepatocytes of benign liver diseases, cirrhotic livers and normal liver. There is a need to develop an improved molecular marker which can detect 100% HCC with high specificity.
Autophagy is a cellular lysosomal degradation mechanism that plays an important role in cellular homeostasis and cell survival mechanisms. Evidence has been presented in a number of studies that autophagy in the liver is induced during chronic viral and non-viral liver diseases indicating that cellular autophagy may be important in the pathogenesis of liver diseases (Sasaki et al. 2012, Rautou et al., 2011; Ni et al., 2012). Some recent studies provide evidence indicating that a defect in cellular autophagy may lead to a number of diseases, such as Crohn’s diseases, stroke, neurodegenerative disorders, pancreatitis and cancer (Ni et al., 2012; Ding et al., 2008, Nagalska et al., 2010). Recent studies have shown that a transgenic mouse with an autophagy defect develops multiple liver tumors (Takamura et al. 2011). Also, activation of mTOR pathways lead to HCC development in a mouse model has been documented (Menon et al., 2012). The accumulation of p62 in hepatocytes is also seen in the animal model of autophagy deficiency transgenic mice. The p62 protein is an adaptor protein involved in the delivery of ubiquitin bound cargo to the autophagysome and lysosomal degradation. When cellular autophagy is disrupted, cytoplasmic protein aggregates, including ubiquitin and p62, can be detected in a number of disease models. The impaired expression of p62 in liver cancer and liver cirrhosis has been initially observed by Eng M Tan laboratory as a fetal RNA binding protein. These authors have detected autoantibodies against p62 proteins among HCC patients (Zhang et al., 1999; Lu et al., 2001). The accumulation of p62 due to impaired autophagy response reported here is novel. These studies including ours, have now provided strong evidence that p62 can be used as a liable marker for HCC detection in human samples. We confirmed that an accumulation of p62 protein expression in the hepatocellular carcinoma, but not in the surrounding non-tumor cells, is an indication of an autophagy defect in HCC. Our results show p62 staining was more sensitive than glypican-3 in our tissue samples, suggesting that this can be a better and more reliable marker for diagnosis of HCC of viral and non-viral etiology. The expression of p62 was weak in the six cholangiocarcinomas tested. The significance of strong staining in HCC and weak staining in cholangiocarcinoma needs further investigation. In summary, our results confirm that human HCC shows high-level expression of p62 indicating that HCC is derived from an autophagy defect.
Acknowledgments
We thank Daniel Hoskins for critically reading this manuscript. This work was supported from NIH grants CA127481, CA089121, AI103106 (SD).
Footnotes
Conflict of interest disclosures
The authors made no disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609–1619. doi: 10.1053/gast.2002.33411. [DOI] [PubMed] [Google Scholar]
- Collier J, Sherman M. Elevated alphafetoprotein in benign liver disease. Viral Hepatitis review. 1998;4:31–41. [Google Scholar]
- Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: A novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterol. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim Biophys Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: Molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836:15–26. doi: 10.1016/j.bbcan.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Ding ZB, Shi YH, Zhou J, Qiu SJ, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, Fan J. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Research. 2008;68:9167–9175. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Saftig P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Bioch Biophy Acta. 2009;1793:664–474. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, Tokita S, Iwanari H, Ito Y, Nakano K, Nezu J, Tsunoda H, Yoshino T, Ohizumi I, Tsuchiya M, Ohnishi S, Makuuchi M, Hamakubo T, Kodama T, Aburatani H. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Research. 2004;64:2418–2423. doi: 10.1158/0008-5472.can-03-2191. [DOI] [PubMed] [Google Scholar]
- Jakubovic BD, Jothy S. Glypican-3: From the mutations of Simpson-Golabi Genetic Syndrome to a tumor marker for human hepatocellular carcinoma. Exp Mol Path. 2007;82:184–189. doi: 10.1016/j.yexmp.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Disease. 2001;5:145–159. doi: 10.1016/s1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- Kandil D, Leiman G, Allegretta M, Trotman W, Pantanowitz L, Goulart R, Evans M. Glypican-3 immunohistochemistry in Liver fine needle aspirates. Cancer (Cancer cytopathology) 2007;111:316–322. doi: 10.1002/cncr.22954. [DOI] [PubMed] [Google Scholar]
- Kotsafti A, Farinati F, Cardin R, Cillo U, Nitti D, Bortolami M. Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterol. 2012;12:118. doi: 10.1186/1471-230X-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Li L, Jin R, Zhang X, Lv F, Liu L, Liu D, Liu K, Li N, Chen D. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology. 2012;56:1380–1390. doi: 10.1002/hep.25891. [DOI] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Yu C, Han T, Moser SD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR. Sulfatase 2 up-regulates glypican-3 promotes fibroblast growth factor signaling and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Nakamura RM, Dent ED, Zhang JY, Nielson FC, Christiansen J, Chan EKL, Tan EM. Aberrant expression of fetal RNA-binding protein p62 in liver cancer and liver cirrhosis. Am J Pathology. 2001;159:945–953. doi: 10.1016/S0002-9440(10)61770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Yecies J, Zhang HH, Howell JJ, Nicholatos J, Harputiugil E, Bronson RT, Kwiatkowski DJ, Manning BD. Chronic activation of mTOR complex 1 is sufficient to cause hepatocellular carcinoma in Mice. Science Signaling. 2012;5:ra24. doi: 10.1126/scisignal.2002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M, Jalvy S, Ladeiro Y, Combe C, Vachet L, Sagliocco F, Bioulac-Sage P, Pitard V, Jacuemin-Sablon H, Zucman-Rossi J, Laloo B, Grosset CF. A functional screening identified five microRNAs controlling glypican-3: role of miR-1271 down-regulation in hepatocellular carcinoma. Hepatology. 2013;57:195–204. doi: 10.1002/hep.25994. [DOI] [PubMed] [Google Scholar]
- Mounajjed T, Zhang L, Wu TT. Glypican-3 expression in gastrointestinal and pancreatic epithelial neoplasms. Human Pathol. 2013;44:542–550. doi: 10.1016/j.humpath.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Man XB, Tang L, Zhang BH, Li SJ, Qiu XH, Wu MC, Wang HY. Upregulation of glypican-3 expression in hepatocellular carcinoma but down regulation in cholangiocarcinoma indicates its differential diagnosis value in primary liver cancers. Liver International. 2005;25:962–966. doi: 10.1111/j.1478-3231.2005.01100.x. [DOI] [PubMed] [Google Scholar]
- Ni HM, Williams JA, Yang H, Shi YH, Fan J, Ding WX. Targeting autophagy for the treatment of liver diseases. Pharmaceutical Research. 2012;66:463–474. doi: 10.1016/j.phrs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalska A, D’Agostino C, Terracciano C, Engel WK, Askanas V. Impaired autophagy in sporadic inclusion body myositis and in endoplasmic reticulum stress provoked cultured human muscle fibers. Am J Pathology. 2010;177:1377–1387. doi: 10.2353/ajpath.2010.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Fenouille N, Auberger P. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2:397–413. [PMC free article] [PubMed] [Google Scholar]
- Rustgi VK. Epidemiology of hepatocellular carcinoma. Gastroenterol Clin North America. 1987;16:545–551. [PubMed] [Google Scholar]
- Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in Liver Diseases. J Hepatology. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Rautou PE, Cazals-Hatem D, Feldmann G, Mansouri A, Grodet A, Barge S, Martinot-Peignoux M, Duces A, Bièche I, Lebrec D, Bedossa P, Paradis V, Marcellin P, Valla D, Asselah T, Moreau R. Changes in autophagic response in patients with chronic HCV infection. Am J Path. 2011;178:2708–2715. doi: 10.1016/j.ajpath.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signaling mechanism senses and regulates the lysosomes via mTOR and TFEB. The EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Miyakoshi M, Sato Y, Nakanuma Y. A possible involvement of p62/sequestosome-1 in the process of biliary epithelial autophagy and senescence in primary biliary cirrhosis. Liver International. 2012;32:660–666. doi: 10.1111/j.1478-3231.2011.02656.x. [DOI] [PubMed] [Google Scholar]
- Sherman M. Alpha-fetoprotein: an abituary. J Hepatology. 2001;34:603–605. doi: 10.1016/s0168-8278(01)00025-3. [DOI] [PubMed] [Google Scholar]
- Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, Nakagohri T, Konishi M, Kobayashi N, Kinoshita T, Nakatsura T. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100:1403–1407. doi: 10.1111/j.1349-7006.2009.01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri A, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Development. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nature Genetics. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Cgang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant function of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Degos F, Dubois S, Tessiore S, Allegretta M, Guttmann RD, Jothy S, Belghiti J, Bedossa P, Paradis V. Glypican-3 expression in hepatocellular tumors: diagnostic value for preneoplastic lesions and hepatocellular carcinomas. Hum Pathology. 2006;37:1435–1441. doi: 10.1016/j.humpath.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, Casey PJ. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC2 prostate cancer cells. J Biol Chem. 2008;283:18678–18684. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- Yamauchi N, Watanabe A, Hishinuma M, Ohashi K, Midorikawa Y, Morishita Y, Niki T, Shibahara J, Mori M, Makuuchi M, Hippo Y, Kodama T, Iwanari H, Aburatani H, Fukayama M. The glypican-3 oncofetal protein is a promising diagnostic marker for hepatocellular carcinoma. Mod Pathology. 2005;18:1591–1598. doi: 10.1038/modpathol.3800436. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Chan EKL, Peng XX, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Experimental Medicine. 1999;189:1101–1110. doi: 10.1084/jem.189.7.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]