Abstract
Memory T cells are primed for rapid responses to antigen; however, the molecular mechanisms responsible for priming remain incompletely defined. CpG methylation in promoters is an epigenetic modification, which regulates gene transcription. Using targeted bisulfite sequencing, we examined methylation of 2100 genes (56,000 CpG) mapped by deep sequencing of T cell activation in human naïve and memory CD4 T cells. 466 CpGs (132 genes) displayed differential methylation between naïve and memory cells. 21 genes exhibited both differential methylation and gene expression before activation, linking promoter DNA methylation states to gene regulation; 6 of 21 genes encode proteins closely studied in T cells, while 15 genes represent novel targets for further study. 84 genes demonstrated differential methylation between memory and naïve cells that correlated to differential gene expression following activation, of which 39 exhibited reduced methylation in memory cells coupled with increased gene expression upon activation compared to naïve cells. These reveal a class of primed genes more rapidly expressed in memory compared to naïve cells and putatively regulated by DNA methylation. These findings define a DNA methylation signature unique to memory CD4 T cells that correlates with activation-induced gene expression.
Keywords: DNA Methylation, Epigenetics, T Cell
Introduction
Differentiation into fast acting memory cells following antigen recognition is a central tenet of T cell immunology. Memory T cells are often described as being “primed” for rapid responses to antigen; however, the molecular mechanisms responsible for this “priming” remain incompletely defined. Methylation of CpG dinucleotides in promoter regions is one mechanism of epigenetic regulation of gene transcription (1-3). CpG methylation is maintained during cell division, but can be altered by aging or environmental stimuli such as disease (1, 4) and T cell activation (5-7). Many genes corresponding to immune function have been identified as being regulated by CpG methylation, implicating a role for CpG methylation in T cell function and differentiation. Clear examples of this phenomenon include the patterns of DNA methylation and gene expression for IFNγ and IL-4 in the development of Th1 and Th2 lineages. Differentiated Th1 cells express IFNγ and exhibit a demethylated IFNγ promoter (8, 9). These same cells do not express IL-4 and have a methylated IL-4 promoter. Conversely, Th2 cells express IL-4, have a demethylated IL-4 gene promoter (10), do not express IFNγ and have a methylated IFNγ promoter (11, 12). Similarly, FoxP3 (13-15), IL-2 (6, 16-18), IL-17A (19) and other immune genes have been shown to be regulated by DNA methylation.
While these studies have been performed to look at the impact of DNA methylation on the expression of single genes, few have employed a more global examination of DNA methylation in CD4 T cells (20). Indeed, many candidate genes have been screened for promoter CpG methylation in CD4 T cells profiled at rest and following activation (9, 17, 18, 21), during development (3, 22), or comparing conventional T cells to regulatory T cells (23). CD4 T cells have also been studied in disease contexts such as latent autoimmune diabetes in adults (24), bronchial asthma (8), or systemic lupus erythematosus (25). Recently, Hashimoto et. al. conducted a global DNA methylation analysis in murine naïve, effector and memory CD4 T cells (7). The authors found that the majority of differential methylation between naïve and memory cells occurred in introns and intergenic regions. Interestingly, the methylation changes occurring following activation of memory CD4 T cells localized to enhancer regions.
To fully appreciate the impact of epigenetic changes in disease states it is important to understand how CpG methylation regulates function and activation-dependent lineage commitments of human naïve and memory CD4 T cells in healthy individuals. We used deep RNA sequencing of naïve and memory cells activated by CD3/CD28 crosslinking to identify a set of high value candidate genes for epigenetic regulation. Then, using high-throughput targeted microdroplet PCR (26), we successfully mapped 57,706 CpGs across 1,946 selected genes.
This study shows an inverse association between promoter CpG methylation and RNA expression, particularly in genes without a promoter CpG island (CGI). We identified 132 genes that were differentially methylated between CD4 naïve and memory subsets. In contrast, 48 hours following activation, there was surprisingly little variation in CpG methylation from resting to activated cells. These 132 genes mapped to pathways involved in cellular migration, hematological system development and function, and inflammatory responses, consistent with the conclusion that DNA methylation is important for regulation of CD4 T cell function. Moreover, 21 genes exhibited differential methylation between naïve and memory CD4 T cells that correlated with differential gene expression at rest, and 84 genes demonstrated differential methylation between memory and naïve cells that correlated to differential gene expression following activation. Ultimately, we mapped the majority of these genes to three pathways involved in delivering a number of critical inflammatory cytokine signals, many with established biological significance in CD4 T cell activation and innate immunity and many as novel candidates for additional research.
Materials and Methods
Ethics Statement
All the studies in this manuscript were covered by Human Subjects Research Protocols approved by the Institutional Review Board of The Scripps Research Institute. Informed written consent was obtained from all study subjects in the study.
Isolation and activation of human lymphocytes
Peripheral blood was collected from healthy donors, and peripheral blood mononuclear cells (PBMC) were collected by centrifugation through a histopaque (Sigma) gradient. CD4 T cells were negatively selected from PBMC using the Naïve CD4 T cell isolation kit II (Miltenyi Biotec) or the Memory CD4 T cell isolation kit (Miltenyi Biotec) from 6 donors. CD8 T cells and B cells were positively selected from PBMC using CD8 or CD19 microbeads (Miltenyi Biotec), respectively. CD8 T cells and CD19 B cells were isolated at a later time from 2 donors included in the CD4 T cell sampling. Cell purity was assessed by flow cytometry staining with antibodies specific for CD4 (SK3, eBioscience), CD45RA (HI100, eBioscience), CD45RO (UCHL1, eBioscience), CD8 (BW135/80, Miltenyi Biotec) and CD19 (LT19, Miltenyi Biotec). Live cells were gated based on forward by side scatter area. Doublets were excluded based on forward scatter height by forward scatter width and side scatter height by side scatter width. Live cells were then gated on CD4 staining and cell purity following isolation was determined by CD45RA vs. CD45RO staining of the CD4+ population. Cell purity for all donors was >95%. T and B cells were cultured in RPMI 1640 (Mediatech) supplemented with 100 U/ml Penicillin, 100 μg/ml Streptomycin and 10% FBS at 37 °C and 5% CO2. T cells were activated with DynaBead Human T-Activator CD3/CD28 (Invitrogen) for 48 h. DNA and RNA for methylSeq and microarrays were isolated from purified cells using an All Prep kit (QIAGEN) following the manufacturer's instructions. RNA for RNAseq was purified with Trizol (Invitrogen) following the manufacturer's instructions.
Preparation of sequencing libraries and deep RNA sequencing
Purified total RNA was converted to cDNA using the Ovation RNA-seq system (NuGEN) followed by S1 endonuclease digestion (Promega) as previously described (27). Digested cDNA libraries were then end-repaired and A-tailed. Indexed adapters were ligated, and ligation product was purified on Agencourt AMPure XP beads (Beckman Coulter Genomics) followed by size selection from 2% agarose. Purified product was amplified with 15 cycles of PCR followed by size selection from 2% agarose. Libraries were assessed on an Agilent Bioanalyzer using a DNA chip and quantitated using the Quant-iT ds DNA BR Assay kit (Invitrogen) and a Qubit Fluorimeter (Invitrogen). Cluster generation and sequencing on an Illumina GAIIx system was conducted directly with purified libraries following manufacturer's instructions. 100 bp single-end reads were generated for naïve and memory CD4 T cells from 3 donors with 2 samples per lane.
RNA sequencing and analysis
For RNA-seq, reads were mapped to hg18 using Tophat (28) with default parameters and without specifying splice junctions or transcript definitions. Transcript quantitation was performed using Cufflinks (29) in quantitation-only mode (-G) with RefSeq gene models downloaded from the Human Genome Browser in GTF format. Genes with expression levels of log2(FPKM) < −2 were considered non-expressed. Differential expression between sample classes was measured by DESeq (30).
Preparation of bisulfite converted DNA
Bisulfite converted DNA was prepared as previously described (26). Briefly, DNA was bisulfite treated using the Epitect Bisulfite kit (QIAGEN) following the manufacturer's protocol. Converted DNA was concentrated on Agencourt AMPure XP beads and quantitated using the Quant-iT ssDNA Assay Kit (Invitrogen) and a Qubit fluorimeter (Invitrogen).
RainDance microdroplet PCR
Microdroplet PCR was conducted as previously described (26). Briefly, 2 μg bisulfite converted DNA was merged with the CD4 specific 2100 gene droplet library and amplified with 55 cycles of PCR. Following amplification, the droplet emulsion was broken and the amplified DNA was purified using a MinElute PCR purification kit (QIAGEN) following standard protocols.
Preparation of sequencing libraries and deep bisulfite sequencing
RainDance PCR products were concatenated as previously described (26). Briefly, 400 ng microdroplet PCR product was end repaired and concatenated. Concatenated products were then fragmented to 200 bp using a Covaris S2. Fragmentation was confirmed on an Agilent Bioanalyzer using a HS DNA chip. 10-100 ng of fragmented PCR product was end-repaired and A-tailed. Indexed adapters were ligated, and ligation product was purified on Agencourt AMPure XP beads followed by size selection from 2% agarose. Purified product was amplified with 18 cycles of PCR followed by size selection from 2% agarose. Libraries were assessed on an Agilent Bioanalyzer using a DNA chip and quantitated using the Quant-iT ds DNA BR Assay kit and a Qubit Fluorimeter. Cluster generation and sequencing on an Illumina HiSeq 2000 system was conducted directly with purified libraries following manufacturer's instructions. 100 bp single-end reads were generated for 5 naïve and 6 memory CD4 T cell, 4 CD8 T cell, and 4 B cell with 10 samples per lane. The CD4 T cells used for targeted bisulfite sequencing and RNAseq were from different donors, except for one that was used for both RNAseq and targeted bisulfite sequencing. The samples used from a single donor for both methods were collected at different times but even so there was ~70% concurrence with differential gene expression using the two different technologies (data not shown).
Bisulfite sequencing alignment and analysis
Reads were mapped to hg18, the reference genome from which the amplicons were designed, using Novoalign version 2.7 (primer coordinates for hg18 or hg19 available upon request). In bisulfite mode, Novoalign assembles a four-strand index and allows mapping to ambiguous bases in the reference genome without penalty; e.g. both CpG and TpG reads will map equally to a CpG locus. Reads that overlap an amplicon concatenation junction are not expected to map properly; we used the -s parameter to truncate unmapped sequence tags to rescue a portion of these reads. Other runtime parameters were set in consultation with the vendor; the novoalign command line was:
novoalign -d hg18.bis.nbx -f [fastq-file] -F ILMFQ -b 4 -c 8 -a -h 120 -t 240 -s 50 -o SAM > [output-file.sam]
The samtools suite and custom perl scripts were used to calculate read depth and percent methylation. Percent methylation is defined as # C reads / (#C + #T reads) at each CpG locus, determined at the C locus for forward-strand amplicons and the G locus for reverse-strand amplicons. Where both forward- and reverse-strand amplicons overlapped the same CpG locus, excluding primer sequences, both sets of reads were included in the calculation. As there was no significant change in the promoter methylation (± 0.1, FDR ≤ 0.1) upon activation of CD4 T cells with anti-CD3/CD28 stimulation for 48 h, the activated methylation profiles were combined with the resting profiles to take advantage of higher sample numbers to determine average CpG methylation across the promoter regions. Mapping to functional pathways was conducted using Ingenuity Pathways Analysis (Ingenuity Systems).
Microarray sample preparation
Total RNA was amplified and labeled using the Applause WT Amp ST kit (NuGen). 2.5μg labeled cDNA was hybridized to Human Gene 1.1 ST arrays (Affymetrix). Raw data was Robust Multi-array Average (RMA) normalized and analyzed with Partek Genomics Suite (Partek Incorporated).
Cytokine ELISAs
Naïve and memory CD4 T cells were activated as described above for 48 h. Supernatants were collected and stored at −80 °C. Cytokine expression was determined using the QPlex Human Cytokine Screen IR (16-plex, Quansys) following manufacturers instructions. Plates were read on a LI-COR Odyssey imaging system at multiple scanning intensities. Data was analyzed using Q-View Software (Quansys). Cytokine expression was background corrected by subtracting any signal from media only samples. ELISAs were performed for six donors.
Luciferase reporter assay
Differentially methylated promoter regions for CCL3, IL17A, TOX, AIM2, CD4, and a differentially methylated fragment of the CD4 intron 1 were amplified by PCR from genomic DNA and the primers listed in Supplemental Table I. The ~1 kb fragments were purified using a QIAquick Gel Extraction kit (QIAGEN) and cloned into the pCR 2.1-TOPO vector (Life Technologies) following manufacturers instructions. The promoter fragments were digested from pCR2.1-TOPO and inserted into the CpG free vector pCpGfree-Lucia (Invivogen), replacing the EF1 promoter with the cloned fragments. The CD4 intron fragment was inserted into pCpGfree-Lucia, replacing the CMV enhancer. Purified vectors were methylated in vitro using the methylase SssI (New England Biolabs) for 2 hours at 37 °C followed by purification on a DNA Clean & Concentrate Column (Zymo Research). Methylation was assessed by digestion with the methyl-CpG sensitive enzyme HpaII (New England Biolabs) and the methyl-CpG insensitive enzyme MspI (New England Biolabs).
Jurkats were transfected with either 0.4 μg methylated or unmethylated vector in triplicate. The unmodified pCpGfree-Lucia vector containing the EF1 promoter and CMV enhancer was used as a control. Cells were co-transfected with 0.4 μg of the pGL4.13[lucZ/SV40] vector (Promega), which contains a firefly luciferase. Cells were allowed to rest overnight following transfection followed by stimulation with and without 0.1 μg/ml PMA (Sigma) and 0.1 μg/ml ionomycin (Sigma) for 24 h. Supernatant was collected and secreted synthetic Renilla luciferase was detected using QuantiLuc (Invivogen). Intracellular firefly luciferase was measured with the Bright-Glo Luciferase Assay System (Promega) following manufacturer's instructions. Renilla luciferase signals were normalized to the internal firely luciferase signal, and this signal was further normalized to the unmethylated vector signal. These experiments were performed at least 3 times for each differentially methylated region. Significance was determined using a paired 2-tailed Student's t-Test.
Results
Selection of the candidate genes for CpG methylation profiling
To fully understand the role of CpG methylation in differentiation of CD4 T cells, it would be optimal to assess the methylation status of all CpGs using whole genome bisulfite sequencing. However, that approach is cost prohibitive and bioinformatically challenging. To reduce both cost and complexity, we interrogated the promoter CpG methylation status of ~2,100 genes in a targeted fashion using microdroplet PCR coupled with bisulfite sequencing (methylSeq) (26, 31). The microdroplet PCR system allows for 1.5 × 106 separate amplifications in less than an hour in a single reaction (32). Moreover, microdroplet PCR significantly reduces amplification bias (32, 33) creating an ideal platform for designing a primer library for targeted CpG studies. At the time these studies were designed, we could optimally target ~3,500 amplicons (~2,000 genes) in one library based on the primer selection guidelines we previously developed for bisulfite converted DNA (26).
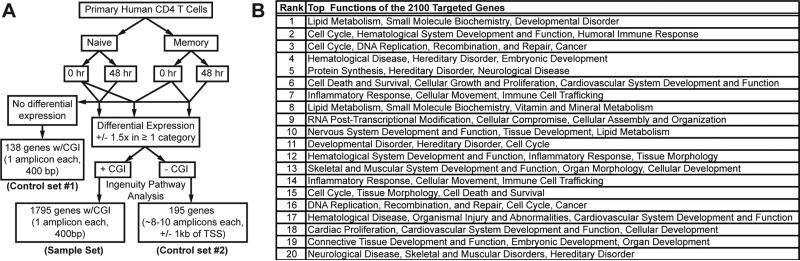
As we could only target approximately 2,000 genes, it was critical that the selection process was informed by function and differential expression in naïve and memory CD4 T cells at rest and following 48 h of activation as outlined in Figure 1a. To select genes for promoter methylation study, RNAseq expression data from memory and naïve CD4 T-cells at rest (T0) and 48 h following activation (T48) were filtered and sorted according to the normalized log fold-change, false discovery rate (FDR, (34)), and promoter CGI status. All genes were filtered to those with a FDR ≤ 0.01 for consideration. For each subset, genes with a minimum ± 1.5-fold change in expression were considered to be up- or down-regulated. Taking three contrasts (naïve vs. memory at T0, naïve at T0 vs. naïve at T48, and memory at T0 vs. memory at T48) into consideration, 7,987 genes were found to be differentially expressed in one or more categories. These genes were mapped to literature-based functional networks. To enrich our analysis for functionally important molecular networks during T cell activation, all genes corresponding to pathways with 10 or more molecules per network were chosen. While many networks identified were linked directly to immune function and inflammation, others were centered upon cell cycle, proliferation and cell signaling (data not shown). Networks were not selected solely based upon documented involvement in T cell regulation or function, but instead were selected based on the differential expression patterns revealed in our RNAseq experiments in order to avoid bias.
Figure 1.
Selection and curating of 2100 genes for targeted microdroplet PCR. (A) Criteria for selection of the 2100 genes for targeted microdroplet PCR. (B) Top functions of the selected 2100 genes. RNAseq was performed on samples from 6 donors.
At the time we designed these studies, the general principle in the field was that CGI were the dominant target for CpG methylation-induced regulation of gene transcription (20, 23, 35). Indeed, ~70% of gene promoters contain CGI (36) and 82% of the 7,987 genes contained promoter CGI. Considering this result and the fact that our first studies demonstrated the expected strong correlation between methylation status and gene expression of CGI-containing promoters (26), the primer library developed for this study was purposely skewed towards CGI-containing genes (90% CGI and 10% no CGI). Ultimately, 1,795 differentially expressed genes with promoter CGI and 195 differentially expressed genes without a promoter CGI were selected for targeted bisulfite sequencing to determine if their differential expression in naïve and memory cells was regulated by promoter methylation. Additionally, 138 genes with a promoter CGI but not differentially expressed between any of the contrasts were selected for inclusion in the targeted primer library as a negative control to assess the impact of promoter methylation. In the case of the 138 genes without differential expression, any differential methylation would have no impact on gene expression. As our initial studies showed that CGI-containing promoters were either 0-20% or 80-100% methylated (26) we chose to target only up to 200 bp of the promoters containing CGI even if the mapped CGI was larger. For genes without a promoter CGI, regions 1 kb up- and downstream of the transcriptional start site (TSS) were targeted for amplification. The final primer library consisted of 3,519 primer pairs targeting 2,128 genes and 77,674 individual CpGs. Network analysis mapped the 2,128 targeted genes to over 50 networks, the top 20 of which are shown in Figure 1B.
Analysis as a function of mean promoter CpG methylation
Following bisulfite conversion, targeted amplification, and sequencing, we successfully mapped 76 ± 21% of the 8.3 ± 2.9 million raw reads to the genome. 64 ± 10% of these mapped to the specific regions targeted by our microdroplet library. 55,707 of 77,674 (72%) of the targeted CpGs had a minimum read depth of 25 reads across all samples, which was the minimum number of reads chosen to allow for both confident calling of the methylation status and broad coverage across the chosen amplicons. As the targeted genes were selected based solely upon expression in CD4 T cells, it was of interest to determine if these genes are specific to CD4 T cells or if they were more broadly expressed in other immune subsets. CpG methylation status was averaged across the targeted regions and compared between resting and activated CD4 naïve and memory T cells as well as resting B cells and resting CD8 T cells.
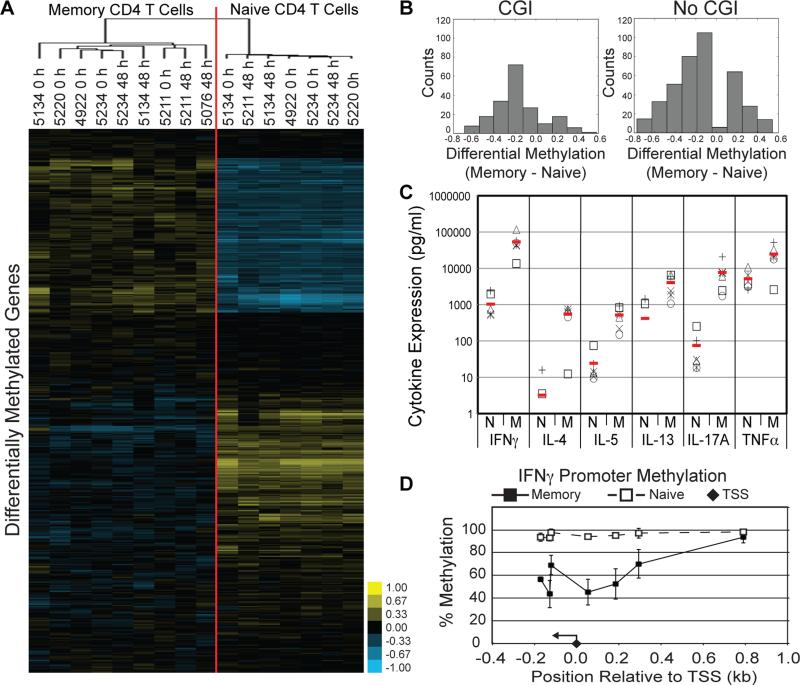
By pooling the data for samples, the majority of genes demonstrated either 0-20% or 80-100% methylation consistent with our original data using this approach (26) or they demonstrated no significant differences as a function of cell type (change in methylation < ± 0.1; FDR > 0.1; Fig. 2A). Both of these classes of genes were excluded from further analysis, as we were only interested in identifying genes with CD4 memory-specific CpG methylation. The remaining 188 genes allowed us to discriminate all four cell types including naïve vs. memory CD4 (Fig. 2B). Even though there was clearly an epigenetic signature for naïve vs. memory CD4 T cells, the next important observation was that within each CD4 cell type methylation did not change within 48 hours of activation. Mapping the function of these genes revealed that the differential methylation of memory CD4 T cells is linked to genes with immune function rather than genes with general functions such as cell cycle (Fig. 2C). In particular, these genes populated networks involved in cell activation, chemokines and leukocyte trafficking.
Figure 2.
Average CpG methylation across multiple immune cell subsets. (A) Variation in average methylation across naïve and memory CD4 T cells, CD8 T cells and B cells. Red line indicates a standard deviation of 0.05 across all samples. (B) Hierarchical clustering of methylation status. Samples are labeled with cell subset, donor identifier (numerical value), and time point following activation (0 or 48 h). (C) Network analysis of the genes with differential methylation between naïve and memory CD4 T cells. (D) Distribution of average promoter methylation in memory CD4 T cells with a CGI. (E) Distribution of average promoter methylation in memory CD4 T cells without a CGI. Data are representative of ≥ 3 donors.
A closer look at the methylation patterns specifically in the CD4 T cell populations demonstrated that 40 genes (Fig. S1A) are differentially methylated (change in methylation ≥ ± 0.1, FDR ≤ 0.1) and define the epigenetic differences between naïve and memory CD4 T cells in the context of the genes we profiled. These genes mapped to only two networks involved in inflammatory responses, cell-to-cell signaling, cellular movement, and development (Fig. S1B).
In our initial study based on a 50 gene library, genes containing a promoter CGI displayed a bimodal methylation profile of being either 80-100% or 0-20% methylated, with the majority of CGI-containing genes being 0-20% methylated (26). While the majority of CGI-containing genes interrogated with the new 2100 gene library were unmethylated in memory CD4 T cells, the distribution was not bimodal, as there were many genes displaying intermediate levels of methylation (Fig. 2D). In the case of the genes without promoter CGI, where a more inclusive region surrounding the TSS was targeted, the distribution of average methylation is even more diffuse with many genes displaying an intermediate level of methylation (Fig. 2E). Therefore, the key point is that methylation of these regulatory regions is not a dichotomous state. Thus, we next determined how different degrees of promoter methylation mapped to the individual CpGs and how that impacted gene expression.
Individual CpG Methylation Analysis
While assays such as DNA methylation arrays and methylation-sensitive restriction enzyme digests have provided invaluable insight into the role of DNA methylation in controlling gene expression, they are limited in that they lack single base resolution. The sequencing based method employed in our study can examine the roles of individual CpGs in each of the targeted regions. When methylation at individual CpGs is considered, 464 CpGs representing 132 genes are differentially methylated between memory and naïve CD4 T cells. Again, samples clustered by cell type (naïve vs. memory, Fig. 3A). There is also no significant change in methylation after 48 h of activation as demonstrated by the failure to cluster on this basis. The majority of these genes mapped to 6 networks (Table I) linked to cell-to-cell signaling, cellular movement and immune/inflammatory responses. These are basically the same functions mapped above to the 40 differentially methylated genes defined by average CpG methylation, though we tripled the number of genes to study by analyzing individual CpG status. Of the 132 differentially methylated genes, 33 had increased methylation in memory cells, 87 had decreased methylation, and 12 had promoters with CpGs demonstrating both increased and decreased methylation. As shown in Fig. 3B, the majority of these CpG are relatively less methylated in memory cells.
Figure 3.
Analysis of methylation at individual CpGs in CD4 T cells. (A) Hierachical clustering of individual CpG methylation in CD4 T cells. Samples are labeled with cell subset, donor identifier (numerical value), and time point following activation (0 or 48 h). (B) Differential methylation between memory and naïve CD4 T cells. (C) Cytokine expression by naïve and memory cells activated for 48 h. Data are representative of 6 donors (symbols represent different donors, red lines represent average signal). (D) Promoter methylation of IFNγ for naïve (open circles, dashed line) and memory (filled squares, solid line) CD4 T cells. The filled diamond marks the TSS and the arrow indicates directionality of transcription. Data are representative of 2 donors.
Table I.
Top networks (> 10 focus molecules) identified by IPA for 132 genes with differential individual CpG methylation between naïve and memory CD4 T cells. Bold genes are differentially methylated. Data is representative of ≥ 3 donors.
| Molecules in Network | Focus Molecules | Top Functions |
|---|---|---|
| CAMP, CCL2, CCL3, CCL5, CCL20, CCL22, Fcer1, FCER1A, FCER1G, Fcgr3, G0S2, ICOS, IL1, IL13, IL22, IL23,IL12 (complex), IL12 (family), IL17A, LSP1, MAP2K6, NFkB (complex), NOD2, P38 MAPK, PRF1, RORC, SELE, STAT4, TLR6,T NFRSF25, TNFSF4, TNFSF15, TNIP3, TRIM69, TRIP6 | 27 | Cell-To-Cell Signaling and Interaction, Cellular Movement, Hematological System Development and Function |
| A2M, AIM2, CD3, CD4, CTLA4, ERK1/2, Fibrinogen, FLT1, Focal adhesion kinase, GBP4, GZMB, HAVCR1, IFIT1, IFN Beta, IKBKE, Interferon alpha, ISG20, ITGAX, ITGB2, ITGB7, LCK, LGALS1, Mapk, MUC4, OASL, PI3K (complex), Ras, RSAD2, SEMA4A, SH2D2A, TCR, TLR3, TNC, TRAF3, Vegf | 24 | Dermatological Diseases and Conditions, Cellular Development, Cellular Growth and Proliferation |
| ABCA1, ACPP, Akt, Ap1, BCR (complex), CD7, CD97, Cg, CIITA, DIO1, EHMT1, EMP1, FLNB, Histone h3, IFITM3, IgG, IL16, IL24, IL1R2, Immunoglobulin, Jnk, LDL, Nr1h, PALLD, PLAT, RAC2, Ras homolog, RUNX3, S100A4, S100A8, TNFRSF1A, TNFSF14, TNS1, TOB1, TP63 | 24 | Inflammatory Response, Cellular Movement, Cellular Function and Maintenance |
| ANK3, CDH1, CFB, EFNA1, EHD1, ELF3, EZH2, GRIP1, GSTM2, HOXA11, ICOSLG, IFNE, IL32, ITGB6, KRT34, LAMP3, LYZ, MAPK1, MAPK13, MAST3, MEOX1, MPZL2, NCOA2, OAS2, PPBP, PSMB8, PSME2, PTPN7, RAP1GAP, RARRES3, RNASE2, TDRD7, TNF, UBE2I, ZNF318 | 13 | Cellular Development, Hematopoiesis, Infectious Disease |
| ASPM, ATAD2, BRWD1, CALML3, CCND1, CD226, CDCA7L, E2F1, EME1, FOXO1, GLIPR1, GLUL, IKZF1, IL15, IL18R1, ITGB1, KCNA3, MYO10, NEFH, NR3C1, PARVG, PDCD1LG2, PIWIL2, PRIM2, PXN, RASGRP2, RBMS3, SATB1, SCG5, SPC25, STAT3, TCF/LEF, TFDP1, TGFB1, TGFBI | 13 | Cell-To-Cell Signaling and Interaction, Tissue Development, Cell-mediated Immune Response |
| ABLIM, ADCY7, ARL4C, ARPC1A, ATP9A, CASP4, DCHS1, DUSP3, DYRK3, ERK,estrogen receptor, FSH, GNLY, GNRH, HNF1A, IL13, ITGAM, ITIH4, LEP, Lh, LOC81691, MPO, MT3, MT1X, NFkB (complex), P4HA2, PDXK, PLCL1, POP5, PPFIA4, RGS5, SLC43A3, UCP3, USPL1, ZNF331 | 12 | Hematological Disease, Immunological Disease, Infectious Disease |
Seventy-four of the 132 differentially methylated genes (56%) do not contain promoter CGIs and 58 (44%) have promoter CGIs (Table II). Of 1,954 genes profiled with CGI-containing promoters, only 156 (8%) demonstrated any significant CpG methylation. However, of the CGIs differentially methylated in memory cells, we observed increased methylation in 23 of the 58 genes (40%) and decreased methylation in 32 (55%). In contrast, of the 206 genes with promoters without a CGI, nearly all are relatively less methylated in memory cells. These differences between CGI and non-CGI methylation reveals the importance of examining these two classes of CpG-containing promoters separately in the context of mapping epigenetic regulation of memory T cell agendas.
Table II.
132 genes have differential methylation at individual CpGs between naïve and memory cells. 21 of these have differential gene expression between resting naïve and memory CD4 cells and 84 have differential gene expression following activation with anti-CD3/CD28 beads for 48 hours. Genes without significant differential expression (± 1.5× fold-change, P < 0.005) indicated as n.s. Data is representative of ≥ 3 donors.
| Methylation | Gene Expression (Log2 Fold Change) | |||||
|---|---|---|---|---|---|---|
| Gene | CGI | M v N | M0 v N0 | N48 v N0 | M48 v M0 | M48 v N48 |
| ARL4C | CGI | ↓ | n.s. | −1.855 | −1.414 | n.s. |
| DNAJA4 | CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| EME1 | CGI | ↓ | n.s. | 3.570 | 2.320 | n.s. |
| IL18R1 | CGI | ↓ | n.s. | n.s. | n.s. | 1.398 |
| LSP1 | CGI | ↓ | n.s. | n.s. | n.s. | 0.506 |
| MAP2K6 | CGI | ↓ | n.s. | −2.280 | −2.592 | n.s. |
| MEOX1 | CGI | ↓ | n.s. | n.s. | −0.545 | 0.439 |
| MPEG1 | CGI | ↓ | 3.605 | n.s. | −4.740 | n.s. |
| MYO10 | CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| NOD2 | CGI | ↓ | 1.899 | n.s. | 1.579 | 3.091 |
| OASL | CGI | ↓ | n.s. | n.s. | 1.746 | 2.515 |
| PALLD | CGI | ↓ | n.s. | n.s. | 2.149 | n.s. |
| PITPNM2 | CGI | ↓ | n.s. | n.s. | −0.652 | n.s. |
| PLCL1 | CGI | ↓ | n.s. | −2.429 | −2.548 | n.s. |
| PPBP | CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| PRF1 | CGI | ↓ | 1.512 | n.s. | n.s. | 2.220 |
| PRIM2 | CGI | ↓ | n.s. | 2.138 | 1.465 | n.s. |
| PSTPIP1 | CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| PTPN7 | CGI | ↓ | n.s. | 1.083 | 0.882 | n.s. |
| RUNX3 | CGI | ↓ | n.s. | n.s. | n.s. | 0.541 |
| SLC43A3 | CGI | ↓ | n.s. | 3.198 | 2.391 | n.s. |
| SMCR7L | CGI | ↓ | n.s. | 0.638 | n.s. | n.s. |
| SPC25 | CGI | ↓ | n.s. | 5.399 | 3.815 | −1.012 |
| TFDP1 | CGI | ↓ | n.s. | 1.359 | n.s. | n.s. |
| TIMD4 | CGI | ↓ | n.s. | −2.100 | n.s. | n.s. |
| TNFRSF1A | CGI | ↓ | n.s. | n.s. | n.s. | 0.403 |
| TNFRSF25 | CGI | ↓ | n.s. | n.s. | −0.783 | n.s. |
| TNS1 | CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| TOB1 | CGI | ↓ | n.s. | −1.954 | n.s. | n.s. |
| VDAC1 | CGI | ↓ | n.s. | 2.054 | 1.552 | n.s. |
| MPO | CGI | ↑ ↓ | n.s. | n.s. | −2.717 | n.s. |
| PLAT | CGI | ↑ ↓ | n.s. | n.s. | 1.411 | 1.125 |
| PRKCH | CGI | ↑ ↓ | n.s. | −0.906 | −0.470 | n.s. |
| TP63 | CGI | ↑ ↓ | 0.651 | n.s. | n.s. | n.s. |
| ZNF331 | CGI | ↑ ↓ | n.s. | n.s. | n.s. | n.s. |
| ABCA1 | CGI | ↑ | n.s. | n.s. | 1.885 | n.s. |
| CTSA | CGI | ↑ | 0.899 | n.s. | n.s. | 0.668 |
| DCHS1 | CGI | ↑ | n.s. | −1.707 | −1.007 | n.s. |
| FLNB | CGI | ↑ | −0.931 | n.s. | n.s. | −0.469 |
| FLT1 | CGI | ↑ | −1.115 | 2.285 | 4.359 | n.s. |
| G0S2 | CGI | ↑ | n.s. | 1.680 | 1.926 | n.s. |
| GLUL | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| GRIK1 | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| GSTM2 | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| KCNJ10 | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| LCK | CGI | ↑ | n.s. | −0.697 | −0.339 | n.s. |
| MAST3 | CGI | ↑ | n.s. | −0.790 | n.s. | 0.611 |
| MPZL2 | CGI | ↑ | n.s. | 4.907 | 3.852 | −0.727 |
| MT1X | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| NCOA2 | CGI | ↑ | n.s. | n.s. | −0.554 | n.s. |
| PARVG | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| RASGRP2 | CGI | ↑ | n.s. | −4.143 | −3.523 | n.s. |
| SATB1 | CGI | ↑ | −0.721 | −1.133 | 0.752 | 1.165 |
| SLFN13 | CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| TGFBI | CGI | ↑ | 2.302 | n.s. | −2.735 | n.s. |
| TNIP3 | CGI | ↑ | n.s. | n.s. | 2.910 | 1.910 |
| TOX | CGI | ↑ | n.s. | −1.667 | −1.455 | 1.279 |
| TRIP6 | CGI | ↑ | n.s. | 1.258 | 0.909 | n.s. |
| Methylation | Gene Expression (Log2 Fold Change) | |||||
|---|---|---|---|---|---|---|
| Gene | CGI | M v N | M0 v N0 | N48 v N0 | M48 v M0 | M48 v N48 |
| A2M | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| ACPP | No CGI | ↓ | n.s. | n.s. | −2.972 | n.s. |
| AIM2 | No CGI | ↓ | 2.628 | n.s. | 4.406 | n.s. |
| ANK2 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| CAMP | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| CASP4 | No CGI | ↓ | n.s. | −0.606 | −0.639 | n.s. |
| CCL2 | No CGI | ↓ | n.s. | n.s. | 3.104 | 2.830 |
| CCL20 | No CGI | ↓ | n.s. | n.s. | 5.159 | n.s. |
| CCL22 | No CGI | ↓ | n.s. | n.s. | 1.718 | n.s. |
| CCL3 | No CGI | ↓ | n.s. | n.s. | 5.916 | 4.030 |
| CCL5 | No CGI | ↓ | 2.945 | n.s. | n.s. | 3.619 |
| CD4 | No CGI | ↓ | n.s. | n.s. | 0.353 | 0.721 |
| CD97 | No CGI | ↓ | n.s. | n.s. | −0.274 | n.s. |
| CIITA | No CGI | ↓ | 1.144 | n.s. | −0.977 | 0.507 |
| CTLA4 | No CGI | ↓ | n.s. | n.s. | 1.479 | 1.664 |
| DIO1 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| EHMT1 | No CGI | ↓ | n.s. | −0.521 | −0.444 | n.s. |
| EMP1 | No CGI | ↓ | n.s. | n.s. | 2.995 | 3.529 |
| FCER1A | No CGI | ↓ | 2.310 | n.s. | −2.785 | n.s. |
| GRIP1 | No CGI | ↓ | n.s. | n.s. | −0.842 | n.s. |
| GZMB | No CGI | ↓ | n.s. | n.s. | 5.473 | n.s. |
| HAVCR1 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| HSD17B11 | No CGI | ↓ | n.s. | −1.171 | −1.145 | n.s. |
| ICOS | No CGI | ↓ | n.s. | n.s. | n.s. | 0.604 |
| IKBKE | No CGI | ↓ | n.s. | −1.481 | −1.404 | n.s. |
| IL13 | No CGI | ↓ | n.s. | 1.564 | 5.898 | 4.302 |
| IL16 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| IL17A | No CGI | ↓ | n.s. | n.s. | 5.949 | 5.897 |
| IL22 | No CGI | ↓ | n.s. | n.s. | 5.666 | 5.408 |
| IL24 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| IL32 | No CGI | ↓ | n.s. | n.s. | 0.801 | 0.863 |
| ITGAM | No CGI | ↓ | n.s. | n.s. | −1.558 | 1.487 |
| ITGAX | No CGI | ↓ | n.s. | n.s. | n.s. | 3.014 |
| ITGB2 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| ITGB7 | No CGI | ↓ | n.s. | −1.581 | n.s. | 1.399 |
| ITIH4 | No CGI | ↓ | n.s. | −1.071 | −1.117 | n.s. |
| LGALS1 | No CGI | ↓ | 1.831 | n.s. | 2.080 | 2.429 |
| LYZ | No CGI | ↓ | n.s. | n.s. | −6.131 | n.s. |
| MUC4 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| OAS2 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| RARRES3 | No CGI | ↓ | n.s. | −1.775 | −2.095 | n.s. |
| RORC | No CGI | ↓ | 2.572 | n.s. | n.s. | 2.578 |
| S100A4 | No CGI | ↓ | n.s. | n.s. | n.s. | 2.031 |
| S100A8 | No CGI | ↓ | n.s. | n.s. | −6.223 | n.s. |
| SELE | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| SEMA4A | No CGI | ↓ | 2.027 | n.s. | 2.101 | 2.672 |
| SH2D2A | No CGI | ↓ | n.s. | 2.360 | 2.417 | 0.961 |
| SLC3A1 | No CGI | ↓ | n.s. | −0.579 | n.s. | n.s. |
| SRGN | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| STAT4 | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| TLR6 | No CGI | ↓ | 1.445 | n.s. | −0.738 | n.s. |
| TNC | No CGI | ↓ | n.s. | n.s. | n.s. | n.s. |
| TNFSF14 | No CGI | ↓ | 0.747 | 1.880 | 2.027 | 0.893 |
| TPH1 | No CGI | ↓ | n.s. | n.s. | −0.638 | n.s. |
| TRAF3 | No CGI | ↓ | n.s. | 0.688 | 1.120 | 0.537 |
| TRIM69 | No CGI | ↓ | n.s. | 1.857 | 1.861 | n.s. |
| UCP3 | No CGI | ↓ | n.s. | −1.403 | n.s. | n.s. |
| ANK3 | No CGI | ↑ ↓ | n.s. | −1.446 | −1.091 | 0.445 |
| CD226 | No CGI | ↑ ↓ | 0.838 | −1.804 | n.s. | 2.135 |
| FCER1G | No CGI | ↑ ↓ | n.s. | n.s. | n.s. | n.s. |
| IL1R2 | No CGI | ↑ ↓ | n.s. | n.s. | 4.422 | 3.276 |
| ISG20 | No CGI | ↑ ↓ | n.s. | n.s. | n.s. | n.s. |
| TLR3 | No CGI | ↑ ↓ | 2.044 | n.s. | −3.045 | n.s. |
| TXN2 | No CGI | ↑ ↓ | n.s. | 1.032 | 0.859 | n.s. |
| AEBP1 | No CGI | ↑ | n.s. | n.s. | n.s. | −0.974 |
| CD7 | No CGI | ↑ | −0.675 | n.s. | 1.852 | n.s. |
| GBP4 | No CGI | ↑ | n.s. | n.s. | −1.695 | n.s. |
| IFIT1 | No CGI | ↑ | n.s. | −1.213 | n.s. | n.s. |
| IFITM3 | No CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| NEFH | No CGI | ↑ | n.s. | 0.875 | n.s. | n.s. |
| RAC2 | No CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| RSAD2 | No CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| TNFSF15 | No CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
| TNFSF4 | No CGI | ↑ | n.s. | n.s. | n.s. | n.s. |
Memory cells demonstrate more variable methylation and functional heterogeneity
Regardless of CGI content, the changes in methylation between memory and naïve CD4 T cells are not dichotomous, i.e. either 0% methylated in one subset and 100% methylated in the other subset (Fig. 3B). Such intermediate levels of methylation could be explained by the fact that the memory pool is composed of multiple CD4 T cell subsets (Th1, Th2, etc). Thus, multi-analyte cytokine ELISAs were performed to determine the cytokine milieu of the activated naïve and memory CD4 T cells that we profiled. Activation of these cells with anti-CD3/CD28 beads for 48 h resulted in the naïve and memory cells expressing IFNγ, IL-4, IL-5, IL-13, IL-17A and TNFα (Fig. 3C). Both subsets of cells also produced levels of IL-2 that saturated the assay (data not shown). These findings demonstrate that by 48 h post-activation the primary naïve and memory cells are both making a diverse set of cytokines and interestingly, the same cytokines. Thus, naïve cells when robustly activated can make many different cytokines but the maturation to memory phenotypes subsequently creates the currently known CD4 T cell subsets.
IL-13 and IL-17A were found to have decreased methylation in at least one CpG (4/18, and 3/3 respectively) in memory cells compared to naïve, suggesting that promoter hypomethylation may be linked to a faster induction of expression of IL-13 and IL-17A in memory CD4 T cells. The minimum threshold for read depth for the promoter regions of IL-5, TNFα and IFNγ was reached for 2 donors sequenced at higher depth. These donors demonstrated that IL-5 is differentially methylated between naïve and memory cells at 4/7 CpGs and that the single covered CpG for TNFα was 40% methylated in resting naïve cells 13% methylated in resting memory cells (data not shown). For IFNγ, resting memory cells were 50% methylated at 6/7 total CpGs compared to nearly 100% methylated in naïve cells (Fig. 3D). Consistent with previous studies demonstrating a strong correlation between hypomethylation of the IFNγ promoter and expression of IFNγ (8, 9, 21), our results support the conclusion that CpG methylation accounts for the ability of memory CD4 T cells to induce IFNγ, the key Th1 cytokine, faster than naïve cells.
Previous studies have shown rapid demethylation (within 6 hours of stimulation) of cytokine promoters following T cell activation (5, 6, 17). We examined 17-targeted cytokine and chemokine promoters comprised of 165 individual CpGs that were covered with sufficient read depth in two donors for naïve CD4 T cells. This analysis revealed that only 21 of 165 (13%) individual CpGs demonstrated a change in methylation >10% following activation for 48 h, with the largest change being ~20%. These individual CpGs mapped to 7 genes (CCL4, CCL22, CCL3, IL16, IL17F, IL32 and IL8) with the majority of differential methylation being decreased at 48 h post-activation (Supplemental Table II). In some cases, such as IL16 and IL17F, a change in methylation is only seen for a single CpG while there were many CpGs that did not change in the promoter region. The impact of a change in methylation of a single CpG is unclear but there are examples where such a change is biologically significant (18). Moreover, mRNA levels for 6 of 7 were significantly upregulated with activation (exception IL16, Table II and data not shown). Thus, the impact likely depends on the precise location of the CpG in relation to transcription factor/repressor binding sites and also on the context of histone modifications. These findings suggest that while some cytokines may rely on rapid demethylation of promoter CpGs, others do not. A more thorough study of cytokine promoters at various times following T cell activation is required to determine which cytokines rely on CpG demethylation for rapid activation and gene transcription. Indeed this is exactly the kind of study that our targeted CpG methylation sequencing is ideally suited to do.
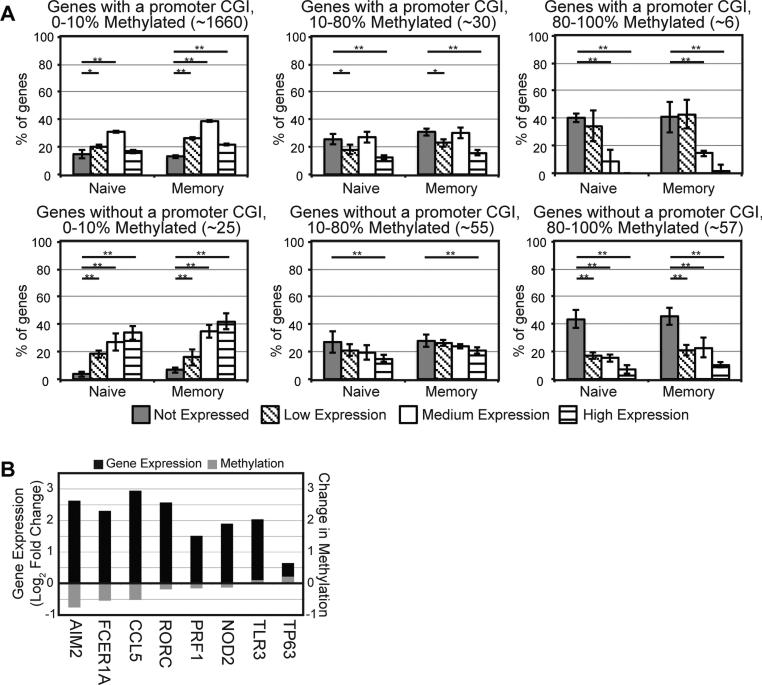
CpG methylation correlates with differential gene expression between naïve and memory CD4 T cells
It cannot be assumed that CpG methylation explains all the differences in gene expression defining naïve vs. memory CD4 T cells. Thus, it is important to identify which genes are regulated by DNA methylation. To this end, global gene expression profiling was conducted by microarray for all samples. In agreement with previous studies (3), an inverse association was found between average promoter methylation and gene expression (Fig. 4A). This relationship was true for both classes of genes profiled (e.g. CGI-containing and no CGI). The negative association seen between methylation of CGI-containing genes and low expression also suggests that we are accurately profiling the impact of CGI methylation. However, even with our selection for differentially expressed genes, we discovered that the vast majority of the CGI-containing promoters are unmethylated in CD4 T cells. Therefore, the expression of all these genes cannot be explained by CpG methylation in the regions that we studied. With respect to non-CGI regulation, there are 10 genes that demonstrate both high methylation and high gene expression in at least one condition (naïve resting, naïve activated, memory resting, memory activated), suggesting that promoter methylation does not regulate expression for these genes. In particular, IL1A, IL-16, IL-17F, IL-23A, IL1R2, and IL1RN (all non-CGI genes) were found to be highly methylated but highly expressed in at least one condition (naïve resting, naïve activated, memory resting, or memory activated). These genes represent >25% of the targeted cytokine related genes.
Figure 4.
Promoter CpG methylation is inversely associated to gene expression. (A) Relationship between average promoter methylation and gene expression. Genes were sorted by log2 normalized expression data (RMA) values into 4 bins: not expressed (RMA 0-6, grey), low expression (RMA 6-8, diagonal stripes), medium expression (RMA 8-10, white) and high expression (RMA 10-14, horizontal stripes). Samples were then separated into three different levels of CpG methylation: 0-10%, 10-80% and 80-100% and the results for the CGI-contained and non-CGI genes are shown separately. Student's t-test p-values: **, P < 0.01; *, P < 0.05. (B) Genes with both differential average methylation (grey) and RNA expression (black) between naïve and memory cells at rest. Data are representative of ≥ 3 donors.
Next, we evaluated how many genes that differ between naïve and memory CD4 T cells are regulated by CpG methylation. By average methylation, 8 genes (3 CGI and 5 no CGI) showed differential methylation (± 0.1, FDR < 0.1) and differential gene expression (± 1.5x fold change, p < 0.005) between memory and naïve CD4 T cells at rest (Fig. 4B). The majority of these genes (6/8, 75%) demonstrated decreased methylation and increased gene expression in memory cells.
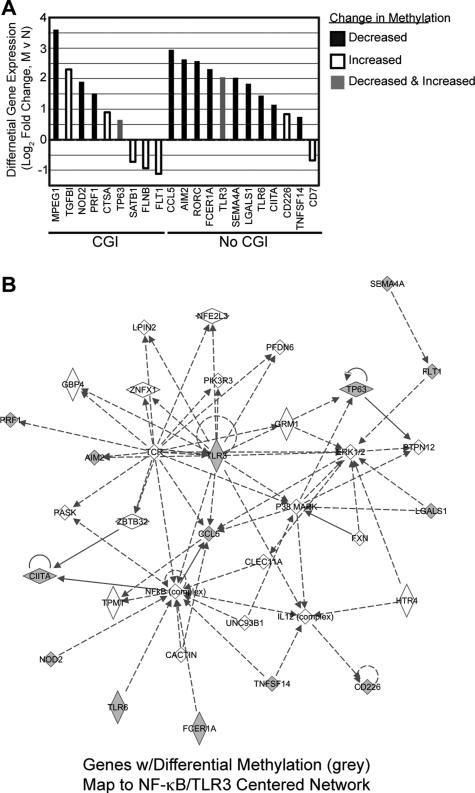
Taking methylation of individual CpGs into account, 21 genes were identified to have both differential methylation and gene expression between memory and naïve CD4 T cells at rest (Fig. 5A and Table II). This is over twice the number of genes identified by average methylation across the promoter, demonstrating many genes that are potentially regulated by DNA methylation are missed by technologies that measure average methylation. An average of 20 CpG (range 2-59) were assayed for each of these 21 genes. The majority (18/21) conformed to the negative relationship predicted between methylation and mRNA expression. Note that 2 genes demonstrated a mix of CpG methylation changes (up and down) in their promoters and in both cases demonstrated increased gene expression in memory cells. The top biological functions associated with these genes are all linked to inflammatory signaling. In particular, 14 of these genes mapped to a single network centered on TCR, TLR3 and NFκB (Fig. 5B).
Figure 5.
21 genes have differential methylation at individual CpGs and differential gene expression. (A) Promoter methylation is grouped into three categories, decreased methylation (black), increased methylation (white) or promoters containing at least one CpG with decreased methylation and one CpG with increased methylation (grey). (B) 14/21 genes populate a single network identified by IPA. Grey molecules are those identified to have differential methylation. Data is representative of ≥ 3 donors.
Of the 132 genes with differential methylation between naïve and memory cells for at least one CpG, 84 genes demonstrated differential gene expression (p < 0.005, FC ± 1.5X) between resting and activated states or between activated memory and activated naïve cells (Table II). For example, a number of chemokines (CCL5, CCL2, CCL3, CCL20, and CCL22) and cytokines (IL-13, IL-17A, IL-22) have decreased methylation in memory CD4 T cells and increased gene expression upon activation (Fig. S2). None of these genes contain a promoter CGI. While expression of IL-13 (37, 38), IL-17A (19), CCL2 (39) and CCL20 (40) have been previously shown to be negatively impacted by promoter CpG methylation, to our knowledge, this is the first data linking DNA methylation to the regulation of expression of IL-22, CCL3, CCL22, TLR3 and TLR6. TNFSF14 (LIGHT) is another interesting molecule revealed in this analysis to be less methylated in memory and correlated with significantly increased gene expression. These results are consistent with the pivotal role proposed recently for TNFSF14 in maintaining antigen-specific memory T cells after re-exposure to antigen in a mouse model (41).
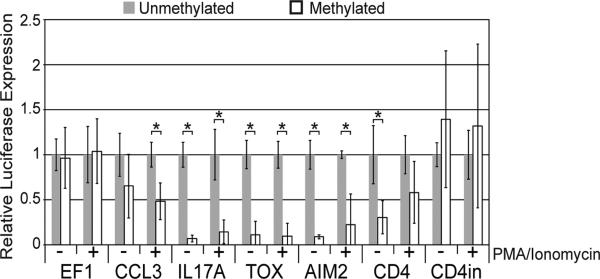
To validate the effect of promoter methylation on gene expression, luciferase reporter constructs were developed for 5 genes (AIM2, CCL3, CD4, IL17A and TOX) that demonstrated clear correlations between promoter methylation and gene expression. We also chose to look at the effect of methylation in the first intron of CD4 as we saw differences in methylation in this region between naïve and memory CD4 T cells. Unmethylated or in vitro SssI-methylated reporter vectors were transiently transfected into Jurkat cells, and luciferase expression was measured with and without activation by CD3/CD28 crosslinking. Methylation of the CD4 intron had no impact on luciferase expression, suggesting that methylation in this region does not affect gene expression. Luciferase expression driven by methylated IL-17A, TOX, and AIM2 promoters was reduced by at least 80% at rest and following activation compared to the unmethylated promoters (Fig. 6). Similarly, CCL3 promoter-driven luciferase expression was significantly reduced upon methylation following activation, and the methylated CD4 promoter driven expression was reduced at rest (Fig. 6). These findings further validate the impact of promoter methylation on gene expression.
Figure 6.
Effect of methylation on luciferase expression for 5 genes with differential methylation. Jurkats were transiently transfected with methylated and unmethylated reporter vectors with and without activation with anti-CD3/CD28 beads. EF1 represents a control promoter that contains no CpG sites and therefore cannot be methylated. * indicates a Student's t-test p-value < 0.01. Data is representative of ≥ 3 experiments.
Revealing the landscape of individual CpG methylation
The single base resolution achieved by high throughput bisulfite sequencing allowed us to assess the distribution of CpG methylation in relationship to the TSS. To further address the question of an epigenetic methylation landscape, we focused on genes without a promoter CGI for which we interrogated methylation status of CpGs within 1 kb up- and downstream of the TSS. Methylation status was measured at 1,421 CpG sites for the 74 non-CGI genes with differential methylation between memory and naïve CD4 T cells (Fig 7A). As shown in Table II, 53 (72%) of these genes also show differential gene expression either in the resting or activated states. In landscape terms, the CpGs with decreased methylation were fairly evenly distributed along the entire 2 kb region (Fig. 7B), while CpGs with increased methylation were predominantly distributed upstream of the TSS (Fig 7C).
Figure 7.
Differential methylation is evenly distributed across the 2 kb of sequence targeted for no CGI genes. (A) Distribution of CpG and methylation states (memory vs naïve) in relation to the TSS. Red indicates CpGs with differential expression (± 0.1 or greater). (B) Histogram of the distribution of CpG with significantly decreased methylation in memory cells compared to naïve. (C) Histogram of the distribution of CpG with significantly increased in memory cells compared to naïve. Data is representative of ≥ 3 donors.
Discussion
Here, we report targeted CpG DNA methylation profiling in human naïve and memory CD4 T cells from healthy, normal donors. This study focused on promoter regions from 2100 genes with established biological functions and for which we had demonstrated differential, activation-induced expression by RNAseq comparing naïve and memory cells. Using targeted microdroplet PCR coupled with methylSeq, we identified over 400 unique DNA methylation differences defining naïve and memory CD4 T cells. We have successfully validated our first report of this method (26) and expanded the targeted CpG region sequencing from 50 to 2100 genes. Thus, any investigator can take advantage of this protocol to interrogate a model system and/or selection of genes mapping to biologically relevant pathways.
In agreement with previous studies, a negative relationship between average methylation and gene expression was observed. Moreover, concurring with a recent report in neonatal CD4 T cells (22), immune activation with anti-CD3/CD28 did not reveal any immediate changes in DNA methylation within 48 h of activation despite literature suggesting such changes occur in several genes including IL-2 (6, 17) and IFNγ (21). In the case of human IFNγ, modest demethylation of a few CpG in the CNS-1 and promoter regions was observed within 16 h of stimulation of naïve CD4 cells, with more robust demethylation apparent by 48-70 h concurrent with cell division (21). While we are unable to address the question of demethylation of the IFNG promoter as the targeted regions did not reach sufficient read depth in our analysis, high depth analysis of unstimulated naïve and memory cells from two donors showed that CD4 memory T cells are hypomethylated compared to naïve, suggesting that the demethylation of the IFNG promoter upon differentiation into effector cells is maintained following development of a CD4 memory phenotype. Interestingly, studies in murine CD8 T cells have demonstrated that the IFNG promoter and enhancer is hypermethylated in memory cells compared to effector cells, but is rapidly demethylated in response to antigenic stimulation (42, 43). Such differences between epigenetic regulation of single genes in CD4 and CD8 T cells highlights the importance of cell specific transcriptional regulation. Moreover, many studies have identified dynamic methylation changes which occur during T cell differentiation into effector and memory populations in both CD4 and CD8 T cells, including changes in regulatory regions of PD-1, CCR6, and RORC, GZMB, perforin and many cytokines (44-47). Our study confirms many of these observations including the hypomethylation of RORC (48) in memory CD4 T cells and GZMB (49) in effector CD8 T cells.
Other studies demonstrated that antigen specific stimulation of murine CD8 T cells (50) and memory CD4 T cells had little effect on promoter methylation (7), consistent with our results. In the CD8 T cell study, it was shown that IL-7Rα expression was linked to promoter hypomethylation in naïve cells, while IL-7Rα negative memory cells have promoter hypermethylation. However, downregulation of IL-7R transcription occurring rapidly after TCR signaling did not correspond to immediate changes in promoter methylation. In the memory CD4 T cell study, Hashimoto et al. observe significant activation-induced differential methylation in enhancer regions, areas outside the design of our targeted sequencing. Thus, remodeling of the methylation landscape appears to occur following TCR ligation in this mouse model. Similarly, comparing conventional CD4 T cells to regulatory T cells also demonstrates differentially methylated regions localize to promoter distal sites such as enhancers (23) and a genome-wide methylome study in CD8 T cells found enrichment of differentially methylated regions in cis-elements such as active enhancers (49). One advantage of our targeted sequencing method is that we can easily redesign our primer library to include enhancer regions and test this hypothesis in human CD4 T cells. Despite our original assumption that profiling of CGI would be most revealing, we discovered that the majority of CGI were not methylated in CD4 T cells and many of the significant methylation differences appear to be in the 2 kb regions up and downstream of the TSS of non-CGI-containing genes. Moreover, differential methylation associated with gene expression during lymphoid differentiation is more strongly correlated with CGI shores (+/− 2 kb of an island) than with CGI themselves (51). As already noted, extending our design to include additional regions such as enhancers and CGI shores further upstream of the TSS might significantly expand our findings.
Bisulfite sequencing has the major advantage of providing single base resolution, and as such, is the gold standard for methylation studies. When average methylation across the entire targeted region was considered, only 40 genes were identified as being differentially methylated between naïve and memory CD4 T cells. However, 132 genes had differential methylation of at least one targeted CpG between naïve and memory CD4 T cells. These results suggest that methods that account for only average methylation (MeDIP, methylArrays, etc) are potentially underreporting the number of genes with changes in methylation. It also underlines the value of thinking in terms of epigenetic landscapes rather than simply as a dichotomous view of a whole region being in one methylation state. However, the impact of differential methylation from a few single CpGs upon gene expression is still unclear, and we believe it is likely to be highly dependent on the location of the CpG in relation to the TSS, as well as transcription factor and enhancer binding sites. In this context, our analysis in Figure 6 shows that the majority of increased methylation changes are upstream of the TSS.
While several of the genes demonstrating both differential methylation and differential gene expression between naïve and memory CD4 T cells have been ascribed to having higher expression or function in memory or activated CD4 T cells (i.e. IL18R1 (52), NOD2 (53), FLT1 (54), CCL3, CCL5 (55), ITGB7 (56)), many have not been linked specifically to either subset in the literature (AIM2, EMP1, ANK3, MPEG1; Table II). To obtain a more complete understanding of which functions might be regulated by CpG methylation as revealed in our data, functional pathway mapping was conducted. Most of the 132 genes with differential methylation by CD4 subset mapped to three significant literature and function-based networks centered on: 1) TCR and TLR3 signaling (Fig. 5), 2) NFκB and p38 MAPK (Fig. S2) and 3) JNK and Akt (Table I).
TLR3 recognizes dsRNA and is located on endosomal membranes (57). Previous studies have shown that TLR3 is costimulatory for CD4 T cells (58, 59) and activation through both CD3 and TLR3 leads to expression of IL-17A, IL-21 (59), CCL3, CCL4, CCL5 and granzyme B (58). While TLR3 signaling is not sufficient to induce proliferation of T cells with anti-CD3, the addition of a TLR3 stimulus enhances CD3/CD28 costimulation-induced proliferation (58). While no exogenous TLR3 stimulus was added in these studies, we observed induction of IL-17A, CCL3, CCL5 and granzyme B upon activation with anti-CD3/CD28, similar to what has been previously reported following costimulation with TLR3 ligands. These findings suggest that the downstream TLR3 signaling pathway may be activated, possibly by recognition of an unidentified endogenous ligand that is induced upon activation such as duplexed RNA.
One of the genes with the greatest differential methylation and the greatest differential gene expression between naïve and memory T cells at rest is AIM2 (absent in melanoma 2). AIM2 is found in the cytoplasm and forms a caspase 1-activating inflammasome upon recognition of dsDNA in macrophages (60, 61). While AIM2 has been characterized in macrophages (61, 62), epidermal cells (63), and numerous cancers (64-66), its function in T cells has not been recognized. IFNγ is known to induce AIM2 expression (67) and could be responsible for the activation of AIM2 in CD4 T cells. As both TLR3 and AIM2 are nucleic acid sensing receptors, it is intriguing to consider the impact that fragments of DNA and RNA duplexes may be having on T cell activation. Recent studies showed that human CD4 T cells undergo inflammasome driven pyroptosis during non-productive HIV infection (68). While AIM2 is not responsible for inflammasome formation with HIV, these studies demonstrate that inflammasomes, typically associated with innate immunity, are intact and functional in CD4 T cells (69). During activation of T cells in vitro there is a large amount of cell death. This cell death would result in release of endogenous dsRNA in the form of stem loop duplexes and DNA being accessible to contribute to T cell activation through TLR3 and AIM2. In vivo, there is also abundant cell death at sites of inflammation and cell injury as well as during the contraction phase following activation of T cells. Thus, further study of the role of TLR3 and AIM2 in controlling T cell proliferation and apoptosis under these conditions is warranted and underway.
While the majority of genes with differential methylation between memory and naïve cells did not contain a CGI, we identified 46 genes that contained a CGI and demonstrated differential methylation. The definition of a CGI is a region dense in CpG and these are largely unmethylated (70). Other recent studies have demonstrated CGIs that are constitutively or differentially methylated (71-73). As more work analyzing CpG methylation at genome scale has been conducted, the importance of CpG outside of classically defined CGI in regulating gene expression has been recognized (7, 23, 51, 73). Moreover, our analysis of the CpG methylation landscape around the TSS for non-CGI genes demonstrated that there is a complex landscape of differentially methylated CpGs. This raises the question of the impact of both non-CGI and intragenic CpG methylation on gene expression. Several studies have been conducted that examine CpGs across the entire genome (73-75) or that classify DNA by CpG content (76). Additionally, many studies have demonstrated CpG methylation driven regulation of many immune genes that do not contain CGI, including IL-2 (6, 18), IL-4 (9, 10), IL-7R (50), IL-17A (1, 19) and IFNγ (8, 9). Coupled with our findings, such studies emphasize the important role of DNA methylation in non-CGI containing regions in regulating gene expression, suggesting that one must look beyond CGI to fully understand the impact of CpG methylation on transcriptional regulation in any cell.
While the precise mechanism of methylation induced transcriptional regulation is unclear, one hypothesis is that DNA methylation in promoter regions may inhibit initiation of transcription through various mechanisms (3). While small-scale gene studies provide important insights into transcriptional regulation, they cannot address how transcriptional networks act in concert with epigenetic regulation to control complex processes such as differentiation, activation and disease progression. The advent of complementary high throughput technologies (ChIPseq, MethylSeq, etc (77)) has allowed investigators to begin to address such sophisticated biological questions. Genome wide studies of histone modifications have led to the hypothesis that histone modifications play a larger role in repression of CGI containing genes (78), suggesting that CGI and non-CGI containing genes are regulated by different epigenetic mechanisms. The strong negative correlation we observed between DNA methylation and gene expression in non-CGI containing promoters suggests that a high density of CpG methylation is not required to accomplish such regulatory function. For example, a methylated CpG in a transcription factor binding motif may be sufficient to inhibit transcription factor binding thereby inhibiting mRNA synthesis. Many studies, including the ENCODE project (79, 80), have mapped transcription factor binding sites and histone modifications for many cell types (47, 81-85) and this data can now be used to align dynamic DNA methylation changes with other regulatory mechanisms to better understand the complex landscape of transcriptional control.
While our current primer library was focused primarily on CGI-containing genes, a new study with a primer library spanning a greater breadth of non-CGI genes and enhancer regions will further refine the roles of CpG methylation landscapes in memory CD4 T cells. We are paralleling this targeted profiling in the next work by also expanding to global genome-wide CpG methylation of CGI and their shores and shelves using a new generation of methyl capture protocols.
The objective of this study in targeted CpG methylation profiling of naïve and memory CD4 T cells was to understand the role of CpG methylation in determining the activation-induced gene expression repertoire of these two primary subsets of human CD4 T cells. Together with previous single gene and high throughput studies in mouse and human CD4 and CD8 T cell differentiation, this work confirms the importance of DNA methylation in regulating gene transcription and reveals a set of unique epigenetic signatures that map to a relatively small number of defined networks that define naïve vs. memory CD4 T cells. Differential gene expression of the majority of CGI-containing genes is not directly determined by the methylation status of the CGI, the majority of which are unmethylated. In contrast, the methylation status for 28% of the non-CGI genes we selected did correlate. These results suggest that there is an evolutionary bias to regulating immune pathway genes without CGI in CD4 T cells by CpG methylation. Interestingly, profiling these same genes in CD8 T cells and B cells reveals unique differences for each cell type, demonstrating that differentiation of lymphoid progenitors to mature subsets involves epigenetic choices that can be studied by targeted CpG profiling. Finally, our targeted, function-based approach clearly links differential CpG methylation between naïve and memory CD4 T cells to a large number of critically important signaling networks underlining the immunological significance of epigenetics.
Supplementary Material
Acknowledgements
We thank A. Feeny, D. Kono and K. Podshivalova for critical reading of the manuscript and E. Wang for technical support. This is manuscript #23060 from The Scripps Research Institute.
This research was supported by funds from the National Institutes of Health: U19 A1063603 (DRS, GTH), UL1 RR025774 (SAL), T32DK007022-30 and a Postdoctoral Juvenile Diabetes Research Foundation fellowship (HKK), and the Molly Baber and Vernah Harrah Research Funds supporting the Salomon laboratory.
Footnotes
The sequences presented in this article have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE59860
References
- 1.Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. J Autoimmun. 2010;34:J207–219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Olkhov-Mitsel E, Bapat B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 2012;1:237–260. doi: 10.1002/cam4.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Chen G, Ma L, Ohms SJ, Sun C, Shannon MF, Fan JY. Plasticity of DNA methylation in mouse T cell activation and differentiation. BMC Mol Biol. 2012;13:16. doi: 10.1186/1471-2199-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Ohms SJ, Shannon FM, Sun C, Fan JY. IL-2 and GMCSF are regulated by DNA demethylation during activation of T cells, B cells and macrophages. Biochem Biophys Res Commun. 2012;419:748–753. doi: 10.1016/j.bbrc.2012.02.094. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto S, Ogoshi K, Sasaki A, Abe J, Qu W, Nakatani Y, Ahsan B, Oshima K, Shand FH, Ametani A, Suzuki Y, Kaneko S, Wada T, Hattori M, Sugano S, Morishita S, Matsushima K. Coordinated changes in DNA methylation in antigen-specific memory CD4 T cells. J Immunol. 2013;190:4076–4091. doi: 10.4049/jimmunol.1202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol. 2008;28:139–146. doi: 10.1007/s10875-007-9148-1. [DOI] [PubMed] [Google Scholar]
- 9.Melvin AJ, McGurn ME, Bort SJ, Gibson C, Lewis DB. Hypomethylation of the interferon-gamma gene correlates with its expression by primary T-lineage cells. Eur J Immunol. 1995;25:426–430. doi: 10.1002/eji.1830250218. [DOI] [PubMed] [Google Scholar]
- 10.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–1903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- 11.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janson PC, Winerdal ME, Marits P, Thorn M, Ohlsson R, Winqvist O. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3:e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Ballas ZK. The use of 5-azacytidine to establish constitutive interleukin 2-producing clones of the EL4 thymoma. J Immunol. 1984;133:7–9. [PubMed] [Google Scholar]
- 17.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 18.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, Shibuya A, Kawabe Y, Yanagisawa J. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas RM, Sai H, Wells AD. Conserved intergenic elements and DNA methylation cooperate to regulate transcription at the il17 locus. J Biol Chem. 2012;287:25049–25059. doi: 10.1074/jbc.M112.351916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes T, Webb R, Fei Y, Wren JD, Sawalha AH. DNA methylome in human CD4+ T cells identifies transcriptionally repressive and non-repressive methylation peaks. Genes Immun. 2010;11:554–560. doi: 10.1038/gene.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong J, Chang HD, Ivascu C, Qian Y, Rezai S, Okhrimenko A, Cosmi L, Maggi L, Eckhardt F, Wu P, Sieper J, Alexander T, Annunziato F, Gossen M, Li J, Radbruch A, Thiel A. Loss of methylation at the IFNG promoter and CNS-1 associates with the development of functional IFN-gamma memory in human CD4(+) T lymphocytes. Eur J Immunol. 2013;43:793–804. doi: 10.1002/eji.201242858. [DOI] [PubMed] [Google Scholar]
- 22.Martino D, Maksimovic J, Joo JH, Prescott SL, Saffery R. Genome-scale profiling reveals a subset of genes regulated by DNA methylation that program somatic T-cell phenotypes in humans. Genes Immun. 2012;13:388–398. doi: 10.1038/gene.2012.7. [DOI] [PubMed] [Google Scholar]
- 23.Schmidl C, Klug M, Boeld TJ, Andreesen R, Hoffmann P, Edinger M, Rehli M. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zhao M, Hou C, Liang G, Yang L, Tan Y, Wang Z, Yin H, Zhou Z, Lu Q. Abnormal DNA methylation in CD4+ T cells from people with latent autoimmune diabetes in adults. Diabetes Res Clin Pract. 2011;94:242–248. doi: 10.1016/j.diabres.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komori HK, Lamere SA, Torkamani A, Hart GT, Kotsopoulos S, Warner J, Samuels ML, Olson J, Head SR, Ordoukhanian P, Lee PL, Link DR, Salomon DR. Application of microdroplet PCR for large-scale targeted bisulfite sequencing. Genome Res. 2011 doi: 10.1101/gr.116863.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, Ordoukhanian PT. Method for improved Illumina sequencing library preparation using NuGEN Ovation RNA-Seq System. Biotechniques. 2011;50:177–180. doi: 10.2144/000113613. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrmann A, Haake A, Ammerpohl O, Martin-Guerrero I, Szafranski K, Stemshorn K, Nothnagel M, Kotsopoulos SK, Richter J, Warner J, Olson J, Link DR, Schreiber S, Krawczak M, Platzer M, Nurnberg P, Siebert R, Hampe J. Pipeline for large-scale microdroplet bisulfite PCR-based sequencing allows the tracking of hepitype evolution in tumors. PLoS One. 2011;6:e21332. doi: 10.1371/journal.pone.0021332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB, Larson JW, Topol EJ, Weiner MP, Harismendy O, Olson J, Link DR, Frazer KA. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nature biotechnology. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hori M, Fukano H, Suzuki Y. Uniform amplification of multiple DNAs by emulsion PCR. Biochem Biophys Res Commun. 2007;352:323–328. doi: 10.1016/j.bbrc.2006.11.037. [DOI] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 35.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, McCombie WR, Wigler M, Hannon GJ, Hicks JB. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Q, Zhou B, Zhang Y, Nguyen ET, Du J, Glosson NL, Kaplan MH. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:541–546. doi: 10.1073/pnas.1103803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao M, Tang J, Gao F, Wu X, Liang Y, Yin H, Lu Q. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. Journal of biomedicine & biotechnology. 2010;2010:931018. doi: 10.1155/2010/931018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aoi Y, Nakahama K, Morita I, Safronova O. The involvement of DNA and histone methylation in the repression of IL-1beta-induced MCP-1 production by hypoxia. Biochem Biophys Res Commun. 2011;414:252–258. doi: 10.1016/j.bbrc.2011.09.066. [DOI] [PubMed] [Google Scholar]
- 40.Yin L, Chung WO. Epigenetic regulation of human beta-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal immunology. 2011;4:409–419. doi: 10.1038/mi.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soroosh P, Doherty TA, So T, Mehta AK, Khorram N, Norris PS, Scheu S, Pfeffer K, Ware C, Croft M. Herpesvirus entry mediator (TNFRSF14) regulates the persistence of T helper memory cell populations. The Journal of experimental medicine. 2011;208:797–809. doi: 10.1084/jem.20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kersh EN. Impaired memory CD8 T cell development in the absence of methyl-CpG-binding domain protein 2. J Immunol. 2006;177:3821–3826. doi: 10.4049/jimmunol.177.6.3821. [DOI] [PubMed] [Google Scholar]
- 43.Northrop JK, Thomas RM, Wells AD, Shen H. Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol. 2006;177:1062–1069. doi: 10.4049/jimmunol.177.2.1062. [DOI] [PubMed] [Google Scholar]
- 44.Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139:277–284. doi: 10.1111/imm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nature reviews. Immunology. 2012;12:306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zediak VP, Wherry EJ, Berger SL. The contribution of epigenetic memory to immunologic memory. Current opinion in genetics & development. 2011;21:154–159. doi: 10.1016/j.gde.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 47.Gray SM, Kaech SM, Staron MM. The interface between transcriptional and epigenetic control of effector and memory CD8(+) T-cell differentiation. Immunological reviews. 2014;261:157–168. doi: 10.1111/imr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidl C, Hansmann L, Andreesen R, Edinger M, Hoffmann P, Rehli M. Epigenetic reprogramming of the RORC locus during in vitro expansion is a distinctive feature of human memory but not naive Treg. Eur J Immunol. 2011;41:1491–1498. doi: 10.1002/eji.201041067. [DOI] [PubMed] [Google Scholar]
- 49.Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. J Immunol. 2013;191:3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HR, Hwang KA, Kim KC, Kang I. Down-regulation of IL-7Ralpha expression in human T cells via DNA methylation. J Immunol. 2007;178:5473–5479. doi: 10.4049/jimmunol.178.9.5473. [DOI] [PubMed] [Google Scholar]
- 51.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bofill M, Almirall E, McQuaid A, Pena R, Ruiz-Hernandez R, Naranjo M, Ruiz L, Clotet B, Borras FE. Differential expression of the cytokine receptors for human interleukin (IL)-12 and IL-18 on lymphocytes of both CD45RA and CD45RO phenotype from tonsils, cord and adult peripheral blood. Clinical and experimental immunology. 2004;138:460–465. doi: 10.1111/j.1365-2249.2004.02651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanello G, Goethel A, Forster K, Geddes K, Philpott DJ, Croitoru K. Nod2 activates NF-kB in CD4+ T cells but its expression is dispensable for T cell-induced colitis. PLoS One. 2013;8:e82623. doi: 10.1371/journal.pone.0082623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu A, Hoerning A, Datta D, Edelbauer M, Stack MP, Calzadilla K, Pal S, Briscoe DM. Cutting edge: Vascular endothelial growth factor-mediated signaling in human CD45RO+ CD4+ T cells promotes Akt and ERK activation and costimulates IFN-gamma production. J Immunol. 2010;184:545–549. doi: 10.4049/jimmunol.0900397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorner BG, Steinbach S, Huser MB, Kroczek RA, Scheffold A. Single-cell analysis of the murine chemokines MIP-1alpha, MIP-1beta, RANTES and ATAC/lymphotactin by flow cytometry. Journal of immunological methods. 2003;274:83–91. doi: 10.1016/s0022-1759(02)00498-2. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez MW, Paquet AC, Yang YH, Erle DJ. Differential gene expression by integrin beta 7+ and beta 7- memory T helper cells. BMC immunology. 2004;5:13. doi: 10.1186/1471-2172-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 58.Meyer T, Oberg HH, Peters C, Martens I, Adam-Klages S, Kabelitz D, Wesch D. poly(I:C) costimulation induces a stronger antiviral chemokine and granzyme B release in human CD4 T cells than CD28 costimulation. Journal of leukocyte biology. 2012;92:765–774. doi: 10.1189/jlb.0811407. [DOI] [PubMed] [Google Scholar]
- 59.Holm CK, Petersen CC, Hvid M, Petersen L, Paludan SR, Deleuran B, Hokland M. TLR3 ligand polyinosinic:polycytidylic acid induces IL-17A and IL-21 synthesis in human Th cells. J Immunol. 2009;183:4422–4431. doi: 10.4049/jimmunol.0804318. [DOI] [PubMed] [Google Scholar]
- 60.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 63.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, Simanski M, Glaser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Science translational medicine. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 65.Liu G, Yu JS, Zeng G, Yin D, Xie D, Black KL, Ying H. AIM-2: a novel tumor antigen is expressed and presented by human glioma cells. Journal of immunotherapy. 2004;27:220–226. doi: 10.1097/00002371-200405000-00006. [DOI] [PubMed] [Google Scholar]
- 66.Woerner SM, Kloor M, Schwitalle Y, Youmans H, Doeberitz M, Gebert J, Dihlmann S. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Genes, chromosomes & cancer. 2007;46:1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- 67.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cross SH, Bird AP. CpG islands and genes. Current opinion in genetics & development. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 71.Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–1383. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 75.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, Gnirke A, Lander ES, Bernstein BE, Meissner A. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annual review of immunology. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 79.Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O'Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nature reviews. Immunology. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nature reviews. Immunology. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 84.Vahedi G, Hand CPA,TW, Laurence A, Kanno Y, O'Shea JJ, Hirahara K. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunological reviews. 2013;252:24–40. doi: 10.1111/imr.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim PS, Shannon MF, Hardy K. Epigenetic control of inducible gene expression in the immune system. Epigenomics. 2010;2:775–795. doi: 10.2217/epi.10.55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.