Abstract
Chemotaxis, the directed migration of cells in chemical gradients, is a vital process in normal physiology and in the pathogenesis of many diseases. Chemotactic cells display motility, directional sensing, and polarity. Motility refers to the random extension of pseudopodia, which may be driven by spontaneous actin waves that propagate through the cytoskeleton. Directional sensing is mediated by a system that detects temporal and spatial stimuli and biases motility toward the gradient. Polarity gives cells morphologically and functionally distinct leading and lagging edges by relocating proteins or their activities selectively to the poles. By exploiting the genetic advantages of Dictyostelium, investigators are working out the complex network of interactions between the proteins that have been implicated in the chemotactic processes of motility, directional sensing, and polarity.
Keywords: Dictyostelium, adaptation, Local Excitation Global Inhibition (LEGI) model, protein localization
INTRODUCTION: CHEMOTAXIS OCCURS IN MANY CELL TYPES
Many cells have an internal compass that allows them to detect extracellular chemical gradients and move toward or away from higher concentrations. This process is referred to as chemotaxis or directed cell migration. During embryogenesis, chemotaxis is important for individual and group cell migration events, organ formation, and wiring of the nervous system. In the adult, chemotaxis is critical for the trafficking of immune cells and in inflammation, regenerative processes such as wound healing, and maintenance of tissue architecture. Evidence suggests that chemotaxis allows stem cells to target to and persist in their niches. See Supplemental Sidebar 1 for examples of the importance of chemotaxis in disease (follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
Although an increasing number of cell types that carry out chemotaxis are being discovered, the signal transduction events mediating directed migration have been most thoroughly studied in Dictyostelium, neutrophils, and a variety of transformed mammalian cells (100, 105, 119; see also Related Resources). The amoeboid movements of Dictyostelium toward 3′,5′-cyclic adenosine monophosphate (cAMP) and of neutrophils toward a variety of chemokines are based on the extension of pseudopodia. In both cell types, chemoattractants activate G-protein coupled receptors (GPCRs), resulting in the localized accumulation of signaling molecules, such as phosphatidylinositol 3,4,5-trisphosphate (PIP3), toward the high side of the gradient. PIP3 accumulation leads to pseudopodia extension at the leading edge of cells, which is thought to be driven by localized Rac-mediated actin polymerization. Primordial germ cells (PGCs) also use GPCRs to sense chemoattractants, but these cells maintain uniform PIP3 levels throughout the membrane and migrate by extending actin-free blebs (3, 30). These blebs may be generated by myosin-based contraction, which is also important at the lagging edge for migration in Dictyostelium and neutrophils. These contractions are generally directed by Rho, and in Dictyostelium, myosin regulation also involves 3′,5′-cyclic guanosine monophosphate (cGMP). The combination of forces at the leading and lagging edges generates rapid movement. Adaptation to persistent stimulation in Dictyostelium and neutrophils allows for enhanced sensitivity to differences in chemoattractant concentrations across the cell.
Stimulation of fibroblasts or breast carcinoma cells with growth factors that bind to Receptor Tyrosine Kinases (RTKs) also results in PIP3 accumulation and Rac-mediated actin polymerization at the leading edge (reviewed in References 64, 100, and 119). In carcinoma cells, the activation of Cofilin through its release from the membrane, which is mediated by chemoattractant-induced reductions in phosphatidylinositol 4,5-bisphosphate (PIP2) levels, generates actin barbed ends and contributes to actin polymerization (114). As with amoeboid cells, the actin-mediated events of fibroblasts and carcinoma cells are coordinated with myosin-based contraction at the lagging edge, which is regulated by Rho and calcium signaling. Together, these result in migration that occurs much more slowly than that of amoeboid cells. Because there is no adaptation in fibroblasts, these cells respond only to absolute concentrations of chemoattractant.
Despite slight differences in the migratory behaviors and specific signaling components of Dictyostelium, neutrophils, and the other cell types, the overall pathways that regulate chemotaxis are similar. Furthermore, many of the guidance mechanisms in these cell types are evolutionarily conserved among most migratory eukaryotic cells, including neurons (reviewed in References 100 and 119). For the remainder of this review, we focus primarily on chemoattractant-mediated signaling events in Dictyostelium, in which genetic analysis has facilitated a thorough assessment of chemotactic behavior (Supplemental Sidebar 2).
MOTILITY, DIRECTIONAL SENSING, AND POLARITY
Chemotaxis can be conceptually divided into the processes of motility, directional sensing, and polarity (Figure 1). In the absence of a stimulus, cells provided with a suitable surface will crawl about, a process referred to as motility. Amoeboid cells, such as Dictyostelium and neutrophils, extend pseudopodia rhythmically, propelling the cell in random directions. When the cells are exposed to a gradient of chemoattractant or chemorepellent, their motility is biased toward or away from, respectively, higher concentrations. The molecular mechanisms that read the gradient and provide this chemotactic bias are referred to as directional sensing and correspond to the cells’ internal compass described above. However, motility and directional sensing are separable, since molecules within immobilized cells can move toward external stimuli and can dynamically track changes in gradient direction. Finally, chemotactic cells often display a relatively stable axis of polarity, which restricts pseudopodia extension to the cell anterior. Polarity is also separable from directional sensing, as cells in uniform chemoattractant can be polarized. Although polarized cells move with more persistence than unpolarized cells, they do not move in a specific direction. Chemotaxis typically incorporates motility, directional sensing, and polarity and should not be confused with any one of these processes alone.
Figure 1.
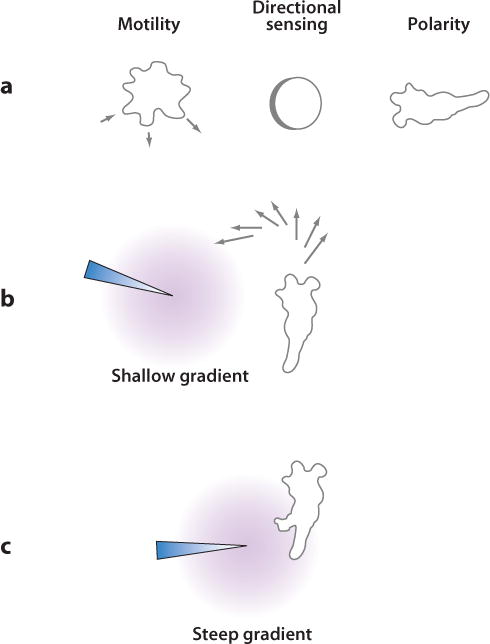
Chemotaxis is composed of motility, polarity, and directional sensing. In the presence of a chemoattractant (or chemorepellent) gradient, cells move toward (or away from) higher concentrations. (a) Left: Free amoeboid cells rhythmically extend pseudopodia and move in random directions. Middle: Spatial sensing, a means of directional sensing, can be demonstrated by the gradient-mediated relocalization of proteins in cells immobilized by actin inhibitors. Right: Chemotactic cells are often polarized, with a stable leading edge from which pseudopodia are extended. (b) In a shallow gradient, polarized cells display biased patterns of pseudopodia extension at the leading edge that cause cells to turn gradually toward higher concentrations of chemoattractant. (c) Sufficiently steep gradients can trigger new projections anywhere along the cell periphery.
Motility: Pseudopod Extension in Migrating Cells
Several recent reports analyze the motile behavior of Dictyostelium cells through observations of pseudopod extension in the absence or presence of shallow cAMP gradients (1, 8). These studies conclude that gradients modify the basal behavior that unstimulated cells already display. In the absence of chemoattractant, polarized cells extend pseudopodia of uniform size and duration alternately from either side of the axis of motion in a behavior reminiscent of “ice skating.” Occasionally, the alternation is skipped and several subsequent pseudopodia are extended from the same side. Chemotactic gradients cause more pseudopodia to be extended toward the “correct” direction. In one model, this is achieved by more often choosing to retract pseudopodia extended in the “wrong” direction (1). In another model, the probability of extending pseudopodia toward the gradient is higher; in addition, the angle at which pseudopodia are extended is altered to favor movement in the correct direction (8). In both models, the bias causes cells to turn toward and remain facing the source of chemoattractant (Figure 1). The ice skating behavior is less obvious in neutrophils, in which individual pseudopodia are not as readily separable, and an alternative mechanism may exist for lamellipod extension in fibroblasts.
Analyses of cells in shallow gradients suggest that generation of pseudopodia is autonomous and that the gradient can only bias this behavior; however, a strong chemotactic stimulus can also directly elicit de novo production of a pseu-dopod (106). Whether a chemotactic stimulus causes turning or triggers a new projection depends on the relative polarity of the cell versus the strength of the stimulus. In a weakly polarized cell, a chemotactic stimulus applied anywhere around the perimeter often triggers the formation of a new front. In a highly polarized cell, the side and rear are less sensitive than the front, resulting in the turning behavior described above. However, even in a highly polarized cell, a sufficiently steep gradient applied to the side or back can break the polarity and create a new front (Figure 1).
Recent observations of fluorescent cytoskeletal proteins on the basal surfaces of migrating neutrophils or Dictyostelium cells show wave-like propagation through the cytoskeleton (10, 118, 123; see also Supplemental Movie 1 and references therein). Actin-binding proteins or Scar/WAVE complex components are recruited sequentially from the cytosol to adjacent points on the basal surface, giving rise to a propagated wave. Basic wave propagation has been modeled and requires the mechanisms of signal relay, positive feedback, and reversible inhibition. Waves can originate at random on the basal surface, although in polarized cells they arise more often toward the anterior and move outward toward the edge. The arrival of waves at the perimeter coincides with the onset of a protrusion and may underlie the spontaneous generation of pseudopodia. The cytoskeletal events that mediate actin polymerization for the formation of cell protrusions have been studied extensively and are beyond the scope of this review; however, little is known about how these events link to receptor signaling (27, 50). Recent observations suggest that chemoattractants influence wave propagation, which may provide insight into the mechanisms by which signaling pathways regulate the cytoskeleton and motility.
Directional Sensing: Temporal and Spatial Sensing of Chemoattractants
Physiological responses triggered by chemoattractants
When differentiated Dictyostelium cells are exposed to the chemoattractant cAMP, a series of morphological changes and physiological responses is triggered (Figure 2a,b; see also Related Resources). The network of signaling pathways mediating these responses is discussed in detail below. In Supplemental Table 1, we list the genes implicated in chemotaxis or in the transduction of chemotactic signals. Within seconds of cAMP stimulation, the heterotrimeric G-protein G2 linked to the cAMP receptor cAR1 is activated, as evidenced by a rapid decrease in the fluorescence resonance energy transfer (FRET) signal between the Gα2- and Gβ-subunits (54, 129). In addition, cAR1 becomes phosphorylated, the mitogen-activated protein kinase (MAPK) Erk2 is activated, and a calcium influx is triggered (11, 86). These responses persist during continuous stimulation. However, many other responses, listed below, are transiently activated upon chemoattractant stimulation. These transient responses typically display an initial rapid activation and a secondary, delayed peak. For example, multiple Ras proteins with similar kinetics are activated (57, 61, 99). The adenylyl cyclase ACA and both soluble and membrane-bound guanylyl cyclases (sGC and GCA, respectively) are activated, resulting in cAMP and cGMP production (93, 98). Phosphoinositide 3-kinases (PI3Ks) are activated, resulting in PIP3 production (34, 42). Many proteins are phosphorylated, including the Gα2-subunit, a series of protein kinase B (PKB) substrates, and Myosin II heavy and light chains (36, 62, 104, 131). Actin is polymerized and actin-binding proteins are recruited to the cortex (25, 59, 101). The phosphoinositide 3-phosphatase PTEN (phosphatase and Tensin homolog on chromosome ten) dissociates from the membrane, and Myosin II is lost from the cortex (45; C. Janetopoulos & P. Devreotes, unpublished observations). In growing undifferentiated cells, folic acid and pterines trigger many of these same responses (37).
Figure 2.
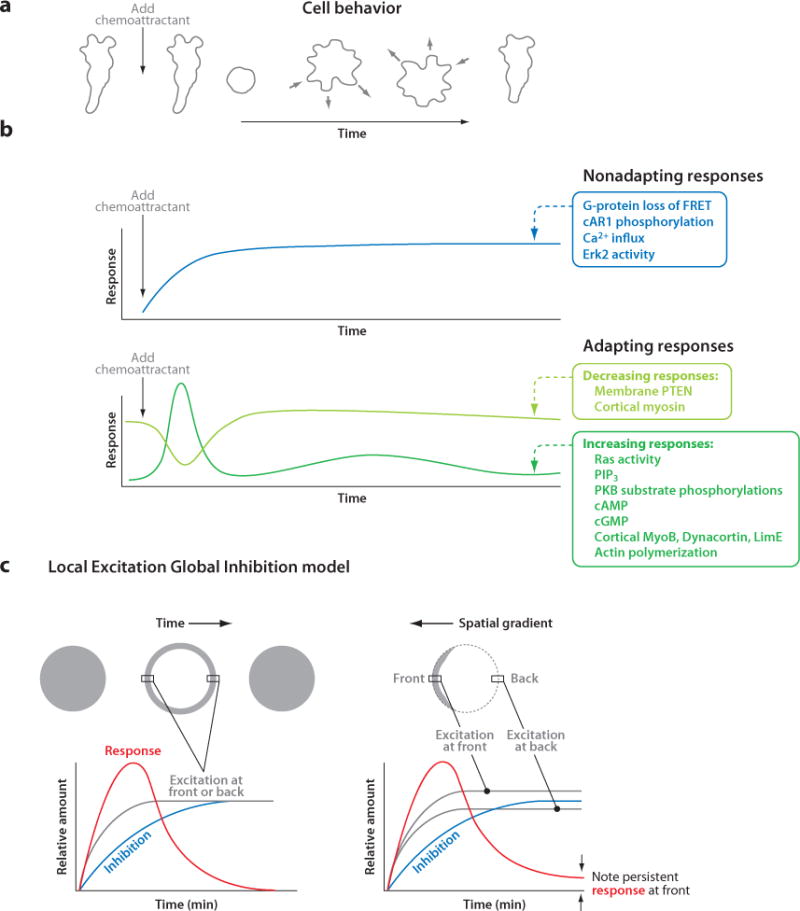
Temporal and spatial responses triggered by chemoattractants and the Local Excitation Global Inhibition (LEGI) model. (a) When exposed to a sudden increase in cAMP, cells retract projections or cringe; then, they periodically extend and retract projections at random sites on the periphery until, after several minutes, they regain their polarized morphology. (b) The biochemical responses triggered by cAMP can be divided into two groups on the basis of whether or not they adapt to constant stimuli. Some responses, such as G-protein activation, are nonadapting and persist as long as the stimuli are maintained. Of the adapting responses, most, such as PIP3 production, transiently increase, whereas others, such as membrane-localized PTEN, transiently decrease. The timescales shown in panels a and b are the same so that the cell behavior in panel a can be directly compared to the response curves in panel b. (c) To explain the temporal and spatial adapting responses of immobilized cells, the LEGI model proposes that chemotactic stimuli elicit an excitor that reflects local receptor occupancy, as well as an inhibitor that is broader and more closely reflects the mean receptor occupancy. Excitation rises faster than inhibition, resulting in an initial response. Left: In a uniform stimulus at steady state, excitation equals inhibition throughout the cell, which explains the experimentally observed disappearance of the initial response. Right: In a gradient at steady state, excitation exceeds inhibition at the high side and vice versa at the low side; therefore, the response persists only at the high side of the gradient, as seen experimentally and indicated by arrows. Abbreviations: cAMP, 3′,5′-cyclic adenosine monophosphate; cGMP, 3′5′-cyclic guanosine monophosphate; FRET, fluorescence resonance energy transfer; PIP3, phosphatidylinositol 3,4,5-trisphosphate; PKB, protein kinase B; PTEN, phosphatase and Tensin homolog on chromosome ten.
The tendency of cellular responses to subside during constant receptor occupancy is known as adaptation. The responses triggered by cAMP can be broadly divided into two groups on the basis of whether or not they adapt to continuous stimulation (Figure 2b). The persistent responses, including receptor phosphorylation, G-protein and Erk2 activation, and the calcium influx, are nonadapting, whereas most of the other responses, which are transient, are adapting (12, 54, 61, 86). The molecular level at which adaptation occurs is an important open question that is addressed below.
Temporal versus spatial sensing
For directional sensing, cells need to detect nonuniform distributions of chemoattractants. To this end, cells must be able to sense changes in receptor occupancy over time, referred to as temporal sensing, and/or space, referred to as spatial sensing. Moving cells can detect spatial gradients using a temporal sensing mechanism because their receptor occupancy changes as a function of time. In fact, chemotactic bacteria rely on temporal sensing to bias their random walks and to detect spatial gradients (120). However, here, the term spatial sensing is reserved for the detection of differences in receptor occupancy across the cell length.
Observations of immobilized cells show that eukaryotic cells are capable of both temporal and spatial sensing (Supplemental Movies 2 and 3 and references therein). In cells immobilized with Latrunculin, an inhibitor of actin polymerization, responses that have been examined, such as Ras activation or PIP3 production, adapt normally to constant uniform stimuli (Supplemental Movie 2). If the stimuli are further increased, these responses can be reactivated but again adapt over time. Therefore, cells respond to temporal changes in receptor occupancy rather than the absolute level. In contrast, immobilized cells placed in stable gradients show persistent Ras and PIP3 responses toward the high side of the gradient, as detected by recruitment of Ras-binding domains (RBDs) and specific Pleckstrin homology (PH) domains, respectively (Supplemental Movie 3). Because these cells are not moving, the steady-state level of receptor occupancy at each position on the membrane is constant over time. Therefore, cells are able to compare receptoroccupancy across their lengths and selectively maintain responses only at the high side.
The temporal phenomenon of adaptation leads to spatial sensing, as illustrated by considering how the steady-state response evolves. When cells are first exposed to a gradient, receptor occupancy increases everywhere and cells show initial global responses similar to those induced by uniform stimuli. Over time, responses at the high side of the gradient arrive at a steady, nonzero level, whereas those at the low side eventually vanish. Thus, adaptation still occurs, but instead of eliminating the signal, it allows cells to respond to the difference in receptor occupancy rather than the absolute level, resulting in an internal amplification of the gradient. So far, mutants that fail to adapt have not been identified. However, defects in adaptation can be mimicked by removing negative regulators such as PTEN or the Ras GTPase activating protein (GAP) NF1 (45, 134). These cells have prolonged and poorly localized PIP3 responses, resulting in impaired chemotaxis.
Adaptation and the Local Excitation Global Inhibition model
If responses such as Ras activation and PIP3 accumulation adapt to constant receptor occupancy, how can they persist locally in an immobilized cell in a stable gradient? The apparent paradox posed by this question can be explained by the Local Excitation Global Inhibition (LEGI) model (Figure 2c) (78, 91; reviewed in Reference 44). According to the LEGI model, two processes are elicited by chemotactic stimuli: a fast excitation that reflects local receptor occupancy and a delayed inhibition that is broader and more closely reflects the mean receptor occupancy. Neither process adapts; both persist as long as stimuli are maintained. The balance between excitation and inhibition determines the magnitude of the observed responses, such as Ras activation and PIP3 production. Because excitation is faster than inhibition, there is a positive response immediately after stimuli are applied. However, with a uniform stimulus at steady state, inhibition eventually equals excitation all over the cell, and there is no response. That is, the cells adapt. In a chemoattractant gradient at steady state, local excitation is higher toward the gradient, whereas inhibition is nearly uniform and intermediate in strength. Therefore, excitation exceeds inhibition at the high side, whereas the opposite occurs at the low side. In this case, adaptation results in a persistent response toward the gradient. The model therefore recapitulates the observed temporal and spatial behaviors of immobilized cells as outlined above. Two predictions of the LEGI model have been verified experimentally (55). For example, when gradients change direction, the responses reorient to the new high side. In addition, localized responses can be induced simultaneously at two points on the membrane by applying two steep gradients.
Four points about the LEGI model need to be clarified. First, the nonadapting property of excitation and inhibition does not imply that the molecules involved are stable. Because the excitatory molecules are constantly diffusing, they must be continually inactivated for excitation to remain localized and adjust to directional changes in the gradient (94). Second, local and global are relative terms. The LEGI model predictions hold true as long as the inhibitor has a longer range of action, or dispersion length, than the excitor. The dispersion length of a molecule is defined as , where D is its diffusion coefficient and k is its rate constant for inactivation. Thus, the inhibitor should have either a faster diffusion rate or a longer half-life (or both) than the excitor. Third, the LEGI model can only fully explain responses in unpolarized cells such as those immobilized by Latrunculin treatment. In polarized cells, the differential sensitivities at the opposing ends of the cell must be taken into account (see below). Fourth, although the inhibitor is often assumed to be a different molecule than the excitor, this does not have to be the case. Hypothetically, if cAMP stimulation resulted in rapid activation and then slow inactivation of the receptor, the receptor itself could act as the excitor when first stimulated and become the inhibitor as adaptation ensued. In this case, the inhibitor would have a longer half-life and therefore dispersion length than the excitor, thus satisfying the requirements of the LEGI model.
While the LEGI model successfully explains the responses of cells to uniform stimuli and gradients, it is not known where adaptation, the balance between excitation and inhibition, occurs along the signaling pathway. FRET experiments of the heterotrimeric G-protein indicate that dissociation of the α- and β-subunits, a readout for G-protein activation, does not adapt to persistent stimulation, whereas the activation of the Ras proteins does adapt (Figure 2b; Supplemental Movie 4 and references therein). Furthermore, prolonged Ras activation leads to prolonged downstream responses (134). Together, these results imply that the major site of adaptation occurs after the G-protein dissociates and before the signal reaches the Ras proteins. The molecular links from the heterotrimeric G-protein to the Ras guanine nucleotide exchange factors (GEFs) are unknown, and exactly where and how adaptation occurs are still open questions. One possible link is an extracellular superoxide dismutase (SodC) that was identified in a restriction enzyme-mediated insertional (REMI) mutagenesis screen (Supplemental Sidebar 2) as having elevated PIP3 levels on the membrane (117). Disruption of SodC leads to continuous recruitment of RBD to the membrane, indicating persistent activation of Ras proteins. The G-protein α9-subunit has also been implicated as a potential negative regulator of the pathway, as disruption of the α9-subunit causes prolonged activation of ACA and increases the size of multicellular aggregates (13). However, there is still no direct evidence that Gα9 regulates Ras activity.
Models based on positive feedback mechanisms have also been proposed to explain the asymmetric responses of cells (48, 82, 94, 124). These models describe the observed responses of polarized cells, which cannot be readily explained by the LEGI model. However, the positive feedback models cannot easily account for the rapid reorientation of responses to changes in the direction of gradients or the dual responses induced by two sharply localized stimuli observed in Latrunculin-treated cells. It is possible that some combination of the LEGI and the positive feedback models is necessary to account for the entire set of physiological responses of chemotactic cells.
Polarity: Selective Localization of Molecules and Reactions to the Front or Back of Cells
The polarization component of chemotaxis manifests itself in two ways. First, cells take on an elongated morphology that becomes increasingly pronounced as they differentiate. Mutants that cannot maintain this morphology have reduced directional persistence, resulting in poor chemotaxis (see below). Second, in polarized cells, certain molecules are spatially restricted to the leading or lagging edge. These localizations are maintained even in the absence of a gradient. The clustering of signaling molecules generates regions at opposing ends of the cell that have distinct functions and different sensitivities to chemoattractant, which alters the way the cell responds to a gradient (Figure 1). Less differentiated cells, which have low or moderate polarity, can easily form new fronts, whereas fully differentiated cells, which are extremely polarized, tend to persist along a constant path with increased velocity.
Although it is thought that positive feedback must be involved, the molecules that are responsible for initiating polarity are still unknown. In neutrophils, a positive feedback loop involving PIP3 and actin has been described (48, 124). However, the factors mediating polarization must be redundant with the PIP3 pathway, because neutrophils or Dictyostelium cells lacking PI3K activity can initiate and maintain relatively normal polarity (33, 40). cGMP signaling may contribute to polarity as discussed below (9, 88). It is also apparent from studies of mutants and chemical inhibitors that the cytoskeleton is required for polarity. Evidence for the requirement of microtubules comes from overexpression of Lissencephaly protein 1 (Lis1) or fragments of Dynein, as well as Lis1 hypomorphic mutants (97). In all three cases, microtubules are unable to attach to the cortical shell, resulting in a flattened morphology with increased lateral protrusions and reduced polarity. Furthermore, cells lacking Tsunami (TsuA) have normal motility and directional sensing capabilities but cannot properly orient their microtubules, which causes defects in polarity and chemotaxis (107). In addition, drugs, such as Benomyl, that depolymerize microtubules eliminate polarity (107). The actin cytoskeleton is also required for polarity. Treatment with Latrunculin not only immobilizes cells, but also disrupts their stable axis of polarity and abolishes spatially restricted protein localizations (55, 73). Furthermore, pten-and tsuA- cells have high levels of F-actin, resulting in the formation of lateral pseudopodia, reduced polarity, and impaired chemotaxis (45, 107). From these and other mutants, including disruptions in the MAPKK MEK1, Tortoise (TorA), and the Na-H exchanger Nhe1, which also cause rather specific defects in polarity, it is apparent that an increase in the number of lateral pseudopodia is correlated with impaired chemotaxis (Supplemental Table 1 and references therein).
Many molecules involved in chemotaxis, including both lipids and proteins, are localized on the membrane or in the cortex specifically at either the leading or lagging edge of polarized cells, and examples of spatially restricted proteins are continuously being identified (Supplemental Table 2 and references therein). In polarized cells, PI3Ks are found at the leading edge, whereas PTEN is found at the lagging edge, creating a localized accumulation of PIP3 and PH domain-containing effectors at the front (Figure 3). In addition, actin and many actin-binding proteins have been identified in the cortex at the front of the cell. Interestingly, S-adenosylhomocysteine hydrolase (SAHH) and the Shwachman-Bodian-Diamond syndrome protein (SBDS) localize to the entire pseudopod at the anterior of the cell. Several other proteins are localized uniformly on the membrane or in the cytosol but are enzymatically active specifically at the leading edge, including Ras, Target of Rapamycin Complex 2 (TorC2) subunits, and the PKB-related protein PKBR1. In contrast, both cAR1 and the heterotrimeric G-proteins, as well as their activities, are uniformly localized throughout the membrane in polarized cells (58, 128).
Figure 3.
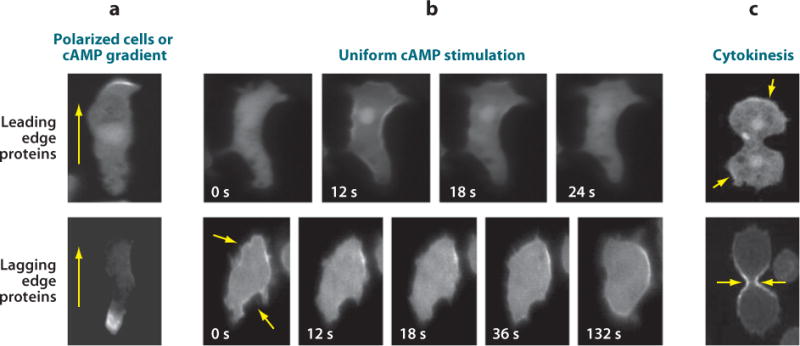
Localization of signaling components to the leading or lagging edge. The distributions of leading edge proteins are represented by a PIP3-specific PH domain tagged with GFP, and those of lagging edge proteins are represented by PTEN-GFP (a) In polarized or chemotaxing cells, many proteins are recruited to the leading or lagging edge. Arrows reflect the direction of migration. (b) When unpolarized cells are stimulated globally with cAMP, “leading edge” proteins, such as PI3Ks and several actin-associated proteins, translocate uniformly to the plasma membrane or cortex and then return to the cytosol. Conversely, “lagging edge” proteins, such as PTEN or Myosin II, transiently fall off the membrane or cortex (arrows) and then return to the periphery. Time in seconds after the addition of chemoattractant is indicated for each frame. (c) During cytokinesis, “leading edge” proteins localize to the poles, whereas “lagging edge” proteins are targeted to the cleavage furrow (arrows). Images in panel c are reproduced from Reference 53. Abbreviations: cAMP, 3′,5′-cyclic adenosine monophosphate; GFP, green fluorescent protein; PIP3, phosphatidylinositol 3,4,5-trisphosphate; PTEN, phosphatase and Tensin homolog on chromosome ten.
Although most asymmetrically localized proteins are found at the leading edge, several have also been identified at the lagging edge (Figure 3; Supplemental Table 2). These proteins are found at the lateral and rear edges of the cell at the outset of differentiation but become more tightly localized specifically to the back as differentiation progresses (K. Swaney & P. Devreotes, unpublished observations). These proteins include PTEN, as described above, as well as Myosin II and the actin-binding protein Cortexillin I, which localize in the cortex and mediate rear contractility. Furthermore, p21-activated protein kinase A (PakA) is at the lagging edge and is proposed to prevent Myosin II dissociation by inhibiting Myosin II Heavy Chain (MHC) kinases (MHCKs). ACA is the only known integral membrane protein in the group of lagging edge proteins. The distribution of ACA has been proposed to create a localized synthesis and release of cAMP at the lagging edge, which attracts other nearby cells and is important for generating the head-to-tail migration, or streaming, characteristic of polarized cells.
Proteins that localize asymmetrically in polarized cells display characteristic behaviors in cells stimulated by chemoattractants (Figure 3; Supplemental Table 2 and Supplemental Movies 5 and 6). In less polarized cells, “leading edge” proteins such as PI3Ks accumulate in the cytosol and on membrane protrusions, whereas “lagging edge” proteins such as PTEN are localized uniformly on the plasma membrane with the exception of membrane protrusions. Upon the addition of cAMP, both sets of proteins respond by transiently relocalizing with respect to the plasma membrane or cytoskeletal cortex. The leading edge proteins translocate uniformly to the periphery within 10 s and then return to the cytosol roughly 30 s after stimulation (Supplemental Movie 5 and references therein). With the same kinetics, most of the lagging edge proteins transiently fall off the cell periphery and into the cytosol before returning to the membrane or cortex (Supplemental Movie 6 and references therein). These redistribution patterns change as the cells become more polarized. Leading edge proteins are found at the front of polarized cells but are additionally and transiently recruited to the membrane globally with uniform stimuli. Lagging edge proteins, already sharply localized to the back of cells, display limited redistribution in response to uniform stimuli in polarized cells. Protein localizations at the leading or lagging edge are maintained in polarized cells even in the absence of a gradient, but, as noted above, treatment with Latrunculin eliminates these spatially restricted distributions (55, 73). In Latrunculin-treated cells, many of these proteins translocate in response to uniform stimuli as they do during early differentiation and localize toward or away from the source of chemoattractant gradients (47, 55, 90, 99).
Current evidence, although limited, implies that the preferential targeting of proteins to the leading or lagging edge of migrating cells is important for polarity and chemotaxis. For example, removing the PIP2-binding domain of PTEN causes mislocalization of the protein to the cytosol, resulting in a broader distribution of PIP3, loss of polarity, and chemotactic impairment that resemble the phenotype of pten- cells (see below) (47). Similarly, expression of PI3Ks tagged with CAAX or myristoylation motifs, which target the proteins uniformly to the plasma membrane, in wild-type cells mimics the pten- cell phenotype (34, 41). Furthermore, the mislocalization of Myosin II and its regulatory proteins causes cells to lose polarity and impairs chemotaxis. For example, excessive recruitment of MHCKA to the membrane results in the overall cortical loss of Myosin II and the overproduction of pseudopodia from the lateral edges of the cell (9, 87). In the future, it will be important to disrupt the localizations of other spatially restricted proteins and to characterize the effects of these changes on polarity and chemotaxis. However, the mechanisms that target many of these asymmetrically localized proteins, including PI3Ks and PTEN, to the leading or lagging edge are still unclear, which currently hinders the investigation of protein mislocalizations.
Several intriguing observations suggest that different components of the same signaling pathway sometimes have different localizations. For example, consider the Ras-TorC2-PKB pathway. The RasGEF Aimless (AleA) activates RasC, which activates TorC2, which in turn mediates the phosphorylation and activation of PKBs, which then phosphorylate a number of substrates (49, 60, 62, 75). The AleA, Ras, TorC2, and PKB proteins are localized globally on the membrane or in the cytosol, but their activities appear to be localized specifically at the leading edge (62, 99). The dispersion lengths of the active forms of these molecules must be sufficiently short to maintain localized downstream responses, implying that these proteins are deactivated before diffusing away from the cell anterior. More puzzling, there are cases in which an upstream activator and its downstream effector are localized at opposite ends of the cell. For example, the PKB substrate PakA resides at the lagging edge despite the restriction of PKB kinase activity to the leading edge (21). It is possible that phosphorylation by PKBs is not required for or related to PakA function at the lagging edge, but this apparent paradox requires further study. Similarly, the activation of the lagging edge integral membrane protein ACA requires both Cytosolic Regulator of Adenylyl Cyclase (Crac), which is recruited by its PH domain to PIP3 at the leading edge, and Pianissimo (PiaA), a TorC2 component (18, 90). The mechanism of ACA activation by two proteins that are active at the leading edge is still unclear.
The asymmetric distribution of proteins has implications beyond polarity and chemotaxis, because the same proteins display characteristic localization patterns when cells undergo morphological changes in general (Figure 3; Supplemental Table 2). For example, in growing cells, both “leading edge” and “lagging edge” proteins have localizations that resemble those of early differentiation discussed above. During phagocytosis, many of the leading edge proteins, such as Crac and the LIM domain-containing protein, LimE, localize to the phagocytic cup and phagosome, whereas lagging edge proteins, such as PTEN, are excluded from these structures (22, 29). During cytokinesis, PI3Ks and other leading edge proteins are localized to the poles, whereas lagging edge proteins, such as PTEN and Myosin II, are restricted to the cleavage furrow (53; see also Supplemental Table 2). It will be interesting to determine how the signals that target these proteins change during these different morphological states.
A NETWORK OF SIGNALING PATHWAYS CONTROLS CHEMOTAXIS
Studies of numerous Dictyostelium mutants with single or multiple gene deletions, or expressing various mutant proteins, have facilitated a molecular analysis of chemotaxis (Supplemental Sidebar 2). With current techniques, it appears that most of the isolated mutants display general chemotactic impairments rather than defects specific to motility, directional sensing, or polarity. In Figure 4, a subset of the genes listed in Supplemental Table 1 is organized into an internally consistent network of pathways that can account for many experimental observations. This network is characterized by redundancy and cross-talk among the pathways. For ease of discussion, the network is divided into a series of overlapping modules where, to a rough approximation, the output of one module serves as an input for the next.
Figure 4.
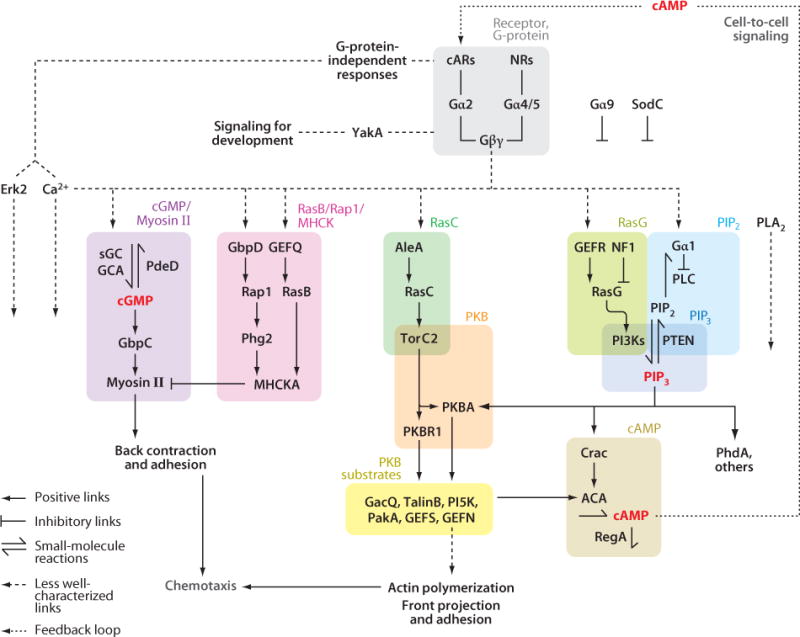
A network of signal responses controls chemotaxis. The interactions among the signaling components that generate chemotactic responses in Dicytostelium cells are shown. Symbols used to indicate positive or inhibitory links, small molecule reactions, and less-well characterized connections are shown in the key located in the lower left corner of the figure. The network is divided into several modules, which are contained in shaded boxes of different colors. Experimental data supporting the links between different components are discussed in detail in the main text. Abbreviations: AleA, RasGEF Aimless; cAMP, 3′,5′-cyclic adenosine monophosphate; cARs, cAMP receptors; cGMP, 3′5′-cyclic guanosine monophosphate; FRET, fluorescence resonance energy transfer; GEF, guanonucleotide exchange factors; PIP3, phosphatidylinositol 3,4,5-trisphosphate; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; PLA2, phospholipase A2; PTEN, phosphatase and Tensin homolog on chromosome ten; TorC2, Target of Rapamycin Complex 2.
Receptor and G-Protein Module
All responses to cAMP are initiated by a set of similar cAMP receptors, cAR1–4, that are expressed at different stages of differentiation (reviewed in Reference 85; see also Supplemental Table 1). When the cAMP receptors are deleted and cells are assessed in early development, car1- cells respond weakly to high doses of cAMP, whereas car1-/car3- cells do not respond at all (51). When cAR1, cAR3, or cAR2 is expressed ectopically in car1-/car3- cells, each receptor can mediate the same set of physiological responses but requires a proportionally higher stimulus concentration corresponding to its respective affinity for cAMP (67). Differences in affinity have been traced to sequence differences in the extracellular loop linking transmembrane domains four and five (69).
The properties of cAR1 have been extensively characterized. As with most GPCRs, there are low- and high-affinity cAMP binding sites, the latter of which are sensitive to GTP in isolated membranes (111). The dissociation rate of cAMP from the receptor is on the order of a few seconds (108, 111). cAR1 undergoes robust agonist-induced phosphorylation, which rises to a steady-state level proportional to the level of receptor occupancy, with a half-time of about 45 s (39). Upon removal of the stimulus, dephosphorylation ensues with a half-time of about 2 min (115). A phosphorylated receptor displays an average fivefold-lowered affinity for cAMP compared with its native counterpart (14). The shift in affinity does not occur if specific phosphorylated serines are removed or replaced by alanines. However, the remainder of downstream responses still adapt to constant stimuli, suggesting that receptor phosphorylation is not the mechanism controlling adaptation (70). A series of point mutations in cAR1 have been described that alter its affinity or lock it in constitutively active or various intermediate functional states. All of these point mutations result in corresponding chemotactic defects that are consistent with the biochemical properties of the receptors (68, 132).
Evidence suggests that the cARs are directly linked to the heterotrimeric G-protein, G2, consisting of α2-, β-, and γ-subunits (reviewed in Reference 17). The β- and γ-subunits have been shown experimentally to directly interact, and cAMP triggers a rapid loss of FRET between the α2-subunit and the βγ-complex (43, 54, 129). In gα2- or gβ- cells, cAMP does not activate actin polymerization, ACA, sGC or GCA, or PI3Ks, nor does it mediate chemotaxis (42, 74, 127, 135). Cells expressing a dominant negative γ-subunit or gγ- cells display a similar phenotype (133; M. Ueda, personal communication). Observations suggest that the βγ-complex mediates downstream signaling, whereas the α2-subunit is required to link the heterotrimeric G-protein to the cARs (127). The Dictyostelium genome contains 13 additional Gα-subunits, and evidence suggests that heterotrimeric G-proteins, consisting of either α4- or α5-subunits and sharing the βγ-complex, link to a set of undefined nutrient receptors (32, 37). Consistently, in growth-stage gα2- cells, but not gβ- cells, folic acid activates cGMP production and mediates chemotaxis (74, 127). The Dictyostelium genome has at least seven Regulator of G-protein Signaling (RGS) domain-containing proteins, which are typically negative regulators of G-protein signaling, although none has been connected to specific Gα-subunits so far (32). Disruption or overexpression of one of these, RGS domain-containing protein kinase 1 (RCK1), results in a corresponding enhancement or reduction of chemotactic speed (Supplemental Table 1).
Expression of genes in early, but not late, development and progression through the entire developmental program require oscillatory cAMP signaling through the receptor (Supplemental Sidebar 2 and references therein). Cells that do not oscillate, such as those with disruptions in ACA, PiaA, or Crac, can still differentiate when pulsed with exogenous cAMP However, cells lacking cARs or G2 fail to enter the developmental program even when supplemented with exogenous cAMP (102, 127). The initiation and progression of the developmental program also require the protein kinase YakA, which appears to act immediately downstream of the heterotrimeric G-protein in the mediation of signaling events (110; see also Supplemental Table 1). Disruption of YakA results in cells that cannot transduce signals in response to cAMP or folic acid stimulation, the latter of which does not require differentiation. Constitutive activation of cAMP-dependent protein kinase A (PKA) can partially bypass the requirement for cAMP oscillations in the developmental program (121).
The cAMP receptors are capable of triggering certain physiological responses in the absence of functional G-proteins (reviewed in Reference 11). In gα2- or gβ- cells, chemoattractant-induced phosphorylation of cAR1 occurs normally and calcium influx and Erk2 activation are reduced by only 50% (84, 86). Abrogation of calcium influx by disrupting the putative inositol 1,4,5-trisphosphate (IP3) receptor (IplA) has little effect on chemotaxis under normal conditions, possibly owing to redundancy (Supplemental Table 1). Further study is necessary to determine the physiological importance of this response.
RasC and RasG Modules
The receptor- and G-protein-mediated transient activations of Ras proteins appear to be significant early steps in the network (reviewed in Reference 122). Although genetic analysis of these proteins is complicated by apparent redundancy and/or compensation among the pathways they trigger, some of the unique roles have been established. For example, AleA and GEFR appear to be the predominant exchange factors for RasC and RasG, respectively (60). In addition, studies of rasC-, rasG-, and rasC-/G- cells suggest that RasC and RasG activate TorC2 and PI3Ks, respectively, with possible overlaps in specificity (62, 99). In rasC- or rasC-/G- cells, phosphorylation of PKBR1, a readout of TorC2 activation, is reduced by 70% (62). The residual activation may be attributable to the upregulation of RasD in rasG- and rasC-/G- cells (66). Ithas been reported that PKBA is not activated by chemoattractant in rasG- cells, suggesting that PIP3 levels are not elevated (4). However, PIP3-binding PH domains are still recruited to the membrane (C. Janetopoulos, personal communication). These apparent discrepancies could be explained by variations in experimental conditions or compensation by RasD. A link between RasG and PI3Ks is also supported by the disruption of the RasGAP NF1, which coincidently prolongs the activation of both proteins (134). The resulting excessive PIP3 levels cause a chemotactic phenotype resembling that of pten- cells (see below). Disruption of RasG or inhibition of PI3K activity partially rescues the nf1- cell phenotype. The prolonged PIP3 production in nf1- cells is consistent with adaptation occurring upstream of the Ras proteins. Both TorC2 and PI3Ks lead to multiple downstream responses, including chemotaxis and the activation of ACA. This can be partially reconciled with early studies, which suggested that RasC regulates ACA activation, whereas RasG regulates chemotaxis, although the actual roles of the Ras proteins are probably more complex than originally thought (4).
PIP3 and PIP2 Modules
The activation of Ras proteins leads to increases in PIP3, mediated by a burst in PI3K activity and a kinetically similar loss of PTEN from the membrane (28, 42, 45, 46, 83). These regulations are independent of PIP3 levels (47). Evidence suggests that PI3K activation requires both recruitment to the membrane and interaction with Ras (34, 42). N-terminal regions of PI3Ks, lacking the RBD and kinase domain, are sufficient for recruitment of GFP to the membrane in a chemoattractant-dependent manner. PI3Ks lacking the N-terminal regions cannot rescue the pi3k1-/pi3k2- cell phenotype, but addition of a CAAX motif to a truncated protein can produce a membrane-associated enzyme that is active in the absence of chemoattractant (34, 41, 42). For PTEN, an N-terminal, 15-residue PIP2-binding motif is required for membrane association, activity in cells, and reversal of the pten- phenotype (47). The essential function of this motif is conserved in human PTEN (hPTEN), which incidentally can also rescue Dictyostelium pten- cells (77, 116). In HeLa cells, rapid depletion of PIP2 causes hPTEN to immediately dissociate from the membrane, which further suggests that PIP2 anchors PTEN to the membrane (96). It is speculated that temporal and spatial regulation of PIP2 levels by chemoattractant occur primarily through Phospholipase C (PLC) in Dictyostelium (71). In theory, activation of PLC leads to a decrease in PIP2 levels, dissociation of PTEN from the membrane, and an enhanced increase in PIP3. In chemotaxing plc- cells, PTEN is not dissociated from the membrane at the front and PIP3 increases at the leading edge are reduced. The chemorepellent 8-CPT-cAMP, a cAMP analog, acts through the G-protein α1-subunit to inhibit PLC activity, which is presumed to increase PIP2 and recruit PTEN toward the high side of the gradient, flipping the axis of polarization (65).
Local accumulations of PIP3 lead to the recruitment of multiple PH domain–containing proteins to the membrane (Supplemental Table 2). The PH domain–containing proteins that translocate to the membrane in response to chemoattractant stimulation in Dictyostelium include Crac, PKBA, and PH domain protein A (PhdA), and cells lacking these proteins have been reported to display weak defects in chemotaxis (23, 90; see also Supplemental Table 2). Characterization of the other translocating PH domain-containing proteins is in progress, and many more that have yet to be studied are currently being evaluated for responsiveness to PIP3 signaling.
Chemoattractant-induced PIP3 production is a highly conserved signature of chemotactic signaling in many cell types, yet inhibiting this response alone does not always block chemotaxis (16, 33, 40, 63, 105). Dictyostelium cells or neutrophils lacking PI3Ks still carry out essentially normal chemotaxis in steep gradients or under specific adhesive conditions. In contrast, pten- cells have high basal PIP3 levels and exaggerated PIP3 increases with chemoattractant stimulation, causing defects in cell morphology, directed migration, and cell-cell signaling (45). Taken together, these results suggest that partially redundant pathways act in parallel with PIP3 signaling to mediate chemotaxis. One such pathway appears to involve Phospholipase A2 (PLA2), because the simultaneous loss or inhibition of PI3K and PLA2 activities causes a stronger chemotactic defect than does the loss of either activity alone (15, 112). Another parallel pathway described in detail below involves TorC2- and PKB-mediated phosphorylation events.
PKB and PKB Substrate Modules
Activated Ras proteins and PIP3 accumulations are involved in the regulation of the chemoattractant-induced activities of two major kinases, PKBA and PKBR1, and the phosphorylation of PKB substrates (Supplemental Table 1). Like mammalian PKBs, each kinase is phosphorylated within a hydrophobic motif (HM) by TorC2 and within an activation loop (AL), presumably by phosphoinositide-dependent protein kinases (PDKs). In cells lacking putative TorC2 subunits PiaA or Ras-interacting protein 3 (Rip3), chemoattractants fail to elicit phosphorylation of the HM of either PKB (62). The absence of HM phosphorylation prevents phosphorylation of the AL of PKBR1 and substantially decreases phosphorylation of the AL of PKBA. Unlike typical PKBs, including PKBA, PKBR1 lacks a PIP3-sensitive PH domain and is tethered constitutively to the membrane by myristoylation, and therefore its activation is independent of PIP3.
The two PKBs act somewhat redundantly to phosphorylate a series of substrates, including TalinB, the RacGAP GacQ, the RasGEFs GEFS and GEFN, PakA, and a putative phosphoinositide 5-kinase (PI5K) (21, 62). The bulk of chemoattractant-mediated PKB-specific phosphorylation events are insensitive to inhibition or disruption of PI3Ks, indicating that PKBR1 is the predominate kinase. The phosphorylation of substrates is substantially reduced in pkbR1- cells and nearly abolished in piaA- cells. Consistently, pkbR1- and piaA- cells have impaired chemotaxis (18, 62). Furthermore, phosphorylation of specific PKB substrates is exaggerated and prolonged in pten- cells, which have extraneous projections that interfere with chemotaxis. These defects are suppressed by simultaneous disruption of PKBA, indicating that the effects of PIP3 are mediated by this protein (M. Tang, M. Iijima, Y. Kamimura & P. Devreotes, manuscript in preparation). Thus, in this series of mutants, the level of PKB substrate phosphorylation correlates well with the observed behavior of the cells. Consistently, inhibition of PKBs in mammalian cells has been reported to interfere with chemotaxis (35, 130). The functions of specific PKB substrates in Dictyostelium are currently being investigated; it is expected that each has a distinct role in mediating chemotaxis.
cAMP Module
The oscillatory production and secretion of cAMP by ACA mediate cell-cell signal relay, and, although not necessary for chemotaxis in differentiated cells, the regulation of this response provides important insights into the chemotactic signaling networks (93; see also Supplemental Sidebar 2). Activation of ACA requires PIP3 accumulation and PKB activation. An early study showed that, in vitro, addition of supernatants from piaA- or crac- cells restored nonhydrolyzable GTP (GTPγS) stimulation of ACA to extracts from crac- or piaA- cells, respectively, but only wild-type supernatants restored activation to extracts of crac-/piaA- cells (18). For ACA activation, Crac requires an intact PH domain, which mediates the recruitment of Crac to PIP3 at the leading edge of the cell (90). Consistently, inhibitors of PI3Ks block activation of ACA in vitro, and pten- cells display excessive ACA activation (24, 45). The involvement of TorC2 strongly suggests that activation of ACA also requires PKB activity and probably phosphorylation of one or more PKB substrates. Preliminary evidence shows that ACA is not activated in pkbR1-/pkba- cells (H. Cai, Y. Kamimura, C. Parent, F. Comer, S. Das & P. Devreotes, manuscript in preparation).
cAMP is synthesized by ACA and degraded both intracellularly by the phosphodiesterase RegA and extracellularly by membrane-bound and secreted phosphodiesterases (2). A circuit involving Erk2, RegA, ACA, and PKA has been proposed to contribute to the spontaneous oscillations of cAMP levels that occur during development (80; see also Supplemental Sidebar 2). In addition to activating ACA, a receptor also activates Erk2, which inhibits RegA, thus preventing the degradation of cAMP. The resulting elevated levels of cAMP activate PKA, which in turn deactivates ACA. In this model, the oscillations arise from a combination of this inhibition and a positive feedback loop in which cAMP stimulates the receptor.
cGMP/Myosin II Module
Activation of the receptor and heterotrimeric G-protein triggers a transient burst of cGMP that is closely associated with the chemotactic response in Dictyostelium (reviewed in Reference 113). The cGMP increase is mediated by membrane-bound and soluble guanylyl cyclases (GCA and sGC, respectively) (98). Curiously, GCA has 12 transmembrane domains, as do adenylyl cyclases; both GCs have catalytic domains that are similar to those of the ACs. Unlike mammalian GCs, neither Dictyostelium GC contains a heme group. Simultaneous disruption of GCA and sGC prevents cGMP accumulation and leads to a defect in chemotaxis. Mutations in the complementation group referred to as streamer F, which maps to the gene for a cGMP-specific phosphodiesterase, PdeD, were some of the earliest described chemotactic defects (26). Streamer F mutants have excessive and prolonged stimulus-mediated cGMP accumulation, have excessive Myosin II association with the cortex, and are hyperpolarized (reviewed in Reference 88). These findings led to the concept that cGMP positively regulates cell polarity.
The Roco protein kinase family member cGMP-binding protein C (GbpC) appears to bind to and mediate the effects of cGMP (9). GbpC contains Leucine-rich repeat (LRR), Ras, and MAPKKK domains, like other Roco family members, and also a unique C-terminal extension with cyclic nucleotide binding and GEF domains (reviewed in Reference 81). In vitro, cGMP activates the GbpC GEF domain, which in turn activates the Ras domain and leads to the subsequent activation of the MAPKKK domain (109). GbpC activity mediates the recruitment of Myosin II to the cortex at the rear of the cell, which is important for generating the tension and contraction that facilitate chemotaxis (6, 9). Disruption of the gbpC gene causes defects in polarity and chemotaxis that resemble the loss of GCs or Myosin II (6, 9, 98, 126). Recent evidence suggests that PIP3 may also be involved in restricting Myosin II to the lagging edge, because Myosin II localization is abnormal in pten- cells during cytokinesis and other growth stages (53, 95, 125).
RasB/Rap1/MHCK Module
The activation of receptor and G-protein triggers the activation of RasB and Rap1, which regulate Myosin Heavy Chain Kinases (MHCKs) and Myosin II. The phosphorylation of Myosin II Heavy Chain (MHCA) by MHCKs promotes the disassembly and release of Myosin II from the cortex and opposes cGMP-mediated Myosin IIassembly (reviewed in Reference 7). RasB and Rap1 are thought to mediate the chemoattractant-induced recruitment and activation of MHCKA at the front of cells, resulting in the restriction of Myosin II to the rear (31, 79). Furthermore, it has been proposed that PakA, which is localized at the lagging edge, inhibits MHCK activity at the rear (19).
Several studies support the involvement of RasB or Rap1 in the activation of MHCKs (56, 57, 72, 87). Excessive activation of RasB, caused by the overexpression of the GEF domain from GEFQ, recruits MHCKA to the membrane and phenocopies mhcA- cells (87, 126). Conversely, gefQ- cells underphosphorylate and overassemble Myosin II, which also impairs chemotaxis (87). Rap1 and its putative effector, tyrosine kinase-like protein Phg2, are also speculated to activate MHCK at the leading edge of the cell (56, 57, 72). Rap1 is activated by the GEF domain of cGMP-binding protein D (GbpD), and although GbpD contains cyclic nucleotide-binding domains, it is not stimulated by cGMP or cAMP (6, 72). Disruption of GbpD or expression of dominant-negative Rap1 leads to excessive polarization and reduced lateral pseudopodia extension, whereas overexpression of GbpD, expression of constitutive-active Rap1, or disruption of RapGAP1 leads to enhanced adhesion, many protrusions, and inhibition of chemotaxis (56, 57, 72). This phenotype is partially suppressed when GbpD is expressed in phg2- cells, suggesting that Phg2 mediates some of the effects of GbpD and Rap1 (72). As expected from the restriction of MHCK to the leading edge, Phg2, RapGAP1, and markers that should be specific for Rap1-GTP are all localized to the front of cells (56, 57). Recent evidence suggests that Rap1 activity is regulated by RasG, which is consistent with the slower chemoattractant-mediated activation of Rap1 compared with the other Ras proteins (5, 57). However, other reports state that prolonged activation of RasG does not appear to increase Rap1 activation (134).
Linking the Signaling Network to the Cytoskeleton
Chemotaxis depends on both myosin-mediated contraction at the rear of the cell and actin polymerization at the front. It is thought that signaling through the Rac proteins mediates actin polymerization in Dictyostelium as in neutrophils and other cell types. Disruption of some of the individual Dictyostelium Rac proteins or putative Rac exchange factors and effectors results in impaired chemotaxis (20, 38, 52, 76, 89, 92, 103). However, a definitive role for the Racs has been elusive owing to redundancy between the many Rac proteins. Indeed, although there has been intense investigation of the cytoskeletal events involved in actin polymerization, little is known about the mechanisms by which signaling events regulate the actin cytoskeleton, but there are some clues. First, excessive or prolonged levels of PIP3, as found in pten- and nf1- cells, elevate the actin polymerization response and interfere with chemotaxis (16, 45, 134). The effects of elevated PIP3 in these mutants may be a result of excessive PKB substrate phosphorylation, because disruption of PKBA in pten- cells restores polarity, suppresses extraneous pseudopodia, and enhances chemotaxis (M. Tang, M. Iijima, Y. Kamimura & P. Devreotes, manuscript in preparation). Therefore, phosphorylation of certain PKB substrates may be an important link between receptor signaling and actin polymerization. Second, signals mediated by contractive forces at the rear may regulate actin polymerization as part of a negative feedback loop. For example, mhcA- cells have an increased number of lateral pseudopodia and chemotactic defects, similar to pten- cells (45, 126). By reading extracellular gradients and integrating the regulation of actin polymerization and myosin-based contraction, the signaling network can interpret the intracellular compass and translate inputs from chemical stimuli into directed migration.
SUMMARY POINTS.
A conserved process referred to as chemotaxis guides the migration of cells, such as neurons, leukocytes, stem cells, and simple amoeba, along chemical gradients.
Chemotaxis can be conceptually divided into the processes of motility, directional sensing, and polarity.
Eukaryotic cells can compare and react to the small concentration differences across their dimensions. In some cells, adaptation to constant chemotactic stimulation allows subtraction of ambient chemoattractant concentrations and greatly increases the accuracy of gradient sensing.
Genetic analysis of Dictyostelium is revealing that chemotaxis depends on a network of overlapping interactions among receptors, G-proteins, Ras proteins, PI3Ks, protein kinases, and phosphatases.
FUTURE ISSUES.
What are the components involved in the generation and propagation of waves of cytoskeletal proteins that are seen on the basal cortex of migrating cells? What is the causal relationship between these waves and cell motility, and how are they affected by chemotactic stimuli and gradients?
What is the molecular mechanism of adaptation? Evidence suggests that it occurs between G-proteins and Ras proteins. How are the Ras proteins activated and inactivated by G-protein signaling?
What is the nature of the positive feedback mechanisms that drive polarity, and how are proteins restricted to the front or back of polarized cells? How is it that different components of the same signaling pathway sometimes show widely different localization patterns?
What is the relative importance of the parallel pathways in the chemotactic signaling network, and what is the purpose of the apparent redundancy?
How is actin polymerization activated and regulated by the chemotactic signaling network?
To what extent is the chemotactic signaling network described in Dictyostelium applicable to other systems, such as leukocytes, fibroblasts, and cancer cells?
Acknowledgments
Kristen F. Swaney and Chuan-Hsiang Huang contributed equally to this review. The authors wish to thank Pablo Iglesias and Yulia Artemenko for critical reading of the manuscript and Peter van Haastert for scientific suggestions. KFS is supported by the American Heart Association. CHH is a Harold L. Plotnick Fellow of the Damon Runyon Cancer Research Foundation. This work was supported by NIH grants GM28007 and GM34933 to PND.
Glossary
- Chemotaxis
the directed migration of cells toward higher or lower concentrations of chemical stimuli
- cAMP
3′,5′-cyclic adenosine monophosphate
- GPCR
G-protein coupled receptor
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- cGMP
3′5′-cyclic guanosine monophosphate
- Adaptation
the tendency of responses to subside when receptor occupancy is held constant
- Directional sensing
the molecular mechanisms that read the direction of chemoattractant gradients and provide a bias to guide the motility of chemotactic cells
- Polarity
a morphological state with stable, functionally distinct leading and lagging edges that are characterized by the preferential localization of specific molecules
- cAR
cAMP receptor
- FRET
fluorescence resonance energy transfer
- PTEN
phosphatase and Tensin homolog on chromosome ten
- Spatial versus temporal sensing
the ability of cells to detect differences in receptor occupancy across the cell length versus over time
- Local Excitation Global Inhibition (LEGI) model
a model, involving a balance between local excitatory and global inhibitory processes, that can explain the temporal and spatial responses of immobilized cells to chemoattractant stimulation
- Dispersion length
the effective range of a signaling molecule, determined by the diffusion coefficient and half-life of the molecule
- TorC2
Tor (Target of Rapamycin) Complex 2
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 2.Bader S, Kortholt A, Van Haastert PJ. Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP. Biochem J. 2007;402:153–61. doi: 10.1042/BJ20061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, et al. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell. 2006;11:613–27. doi: 10.1016/j.devcel.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol Biol Cell. 2006;17:4543–50. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolourani P, Spiegelman GB, Weeks G. Rap1 activation in response to cAMP occurs downstream of Ras activation during Dictyostelium aggregation. J Biol Chem. 2008;283:10232–40. doi: 10.1074/jbc.M707459200. [DOI] [PubMed] [Google Scholar]
- 6.Bosgraaf L, Russcher H, Smith JL, Wessels D, Soll DR, Van Haastert PJ. A novel cGMP signaling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. EMBO J. 2002;21:4560–70. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosgraaf L, van Haastert PJ. The regulation of myosin II in Dictyostelium. Eur J Cell Biol. 2006;85:969–79. doi: 10.1016/j.ejcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Bosgraaf L, van Haastert PJ. The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS One. 2009;4:e5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosgraaf L, Waijer A, Engel R, Visser AJ, Wessels D, et al. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J Cell Sci. 2005;118:1899–910. doi: 10.1242/jcs.02317. [DOI] [PubMed] [Google Scholar]
- 10.Bretschneider T, Jonkman J, Kohler J, Medalia O, Barisic K, et al. Dynamic organization of the actin system in the motile cells of Dictyostelium. J Muscle Res Cell Motil. 2002;23:639–49. doi: 10.1023/a:1024455023518. [DOI] [PubMed] [Google Scholar]
- 11.Brzostowski JA, Kimmel AR. Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem Sci. 2001;26:291–97. doi: 10.1016/s0968-0004(01)01804-7. [DOI] [PubMed] [Google Scholar]
- 12.Brzostowski JA, Kimmel AR. Nonadaptive regulation of ERK2 in Dictyostelium: implications for mechanisms of cAMP relay. Mol Biol Cell. 2006;17:4220–27. doi: 10.1091/mbc.E06-05-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brzostowski JA, Parent CA, Kimmel AR. A G alpha-dependent pathway that antagonizes multiple chemoattractant responses that regulate directional cell movement. Genes Dev. 2004;18:805–15. doi: 10.1101/gad.1173404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caterina MJ, Devreotes PN, Borleis J, Hereld D. Agonist-induced loss of ligand binding is correlated with phosphorylation of cAR1, a G protein-coupled chemoattractant receptor from Dictyostelium. J Biol Chem. 1995;270:8667–72. doi: 10.1074/jbc.270.15.8667. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Iijima M, Tang M, Landree MA, Huang YE, et al. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;12:603–14. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Janetopoulos C, Huang YE, Iijima M, Borleis J, Devreotes PN. Two phases of actin polymerization display different dependencies on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol Biol Cell. 2003;14:5028–37. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- 18.Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11:3218–31. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung CY, Firtel RA. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J Cell Biol. 1999;147:559–76. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung CY, Lee S, Briscoe C, Ellsworth C, Firtel RA. Role of Rac in controlling the actin cytoskeleton and chemotaxis in motile cells. Proc Natl Acad Sci USA. 2000;97:5225–30. doi: 10.1073/pnas.97.10.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung CY, Potikyan G, Firtel RA. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol Cell. 2001;7:937–47. doi: 10.1016/s1097-2765(01)00247-7. [DOI] [PubMed] [Google Scholar]
- 22.Clarke M, Muller-Taubenberger A, Anderson KI, Engel U, Gerisch G. Mechanically induced actin-mediated rocketing of phagosomes. Mol Biol Cell. 2006;17:4866–75. doi: 10.1091/mbc.E06-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comer FI, Lippincott CK, Masbad JJ, Parent CA. The PI3K-mediated activation of CRAC independently regulates adenylyl cyclase activation and chemotaxis. Curr Biol. 2005;15:134–39. doi: 10.1016/j.cub.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Comer FI, Parent CA. Phosphoinositide 3-kinase activity controls the chemoattractant-mediated activation and adaptation of adenylyl cyclase. Mol Biol Cell. 2006;17:357–66. doi: 10.1091/mbc.E05-08-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Condeelis J, Hall A, Bresnick A, Warren V, Hock R, et al. Actin polymerization and pseudopod extension during amoeboid chemotaxis. Cell Motil Cytoskelet. 1988;10:77–90. doi: 10.1002/cm.970100113. [DOI] [PubMed] [Google Scholar]
- 26.Coukell MB, Cameron AM. Characterization of revertants of stmF mutants of Dictyostelium discoideum: evidence that stmF is the structural gene of the cGMP-specific phosphodiesterase. Dev Genet. 1986;6:163–77. doi: 10.1002/dvg.1020060303. [DOI] [PubMed] [Google Scholar]
- 27.Dayel MJ, Akin O, Landeryou M, Risca V, Mogilner A, Mullins RD. In silico reconstitution of actin-based symmetry breaking and motility. PLoS Biol. 2009;7:e1000201. doi: 10.1371/journal.pbio.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–48. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 29.Dormann D, Weijer G, Dowler S, Weijer CJ. In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J Cell Sci. 2004;117:6497–509. doi: 10.1242/jcs.01579. [DOI] [PubMed] [Google Scholar]
- 30.Dumstrei K, Mennecke R, Raz E. Signaling pathways controlling primordial germ cell migration in zebrafish. J Cell Sci. 2004;117:4787–95. doi: 10.1242/jcs.01362. [DOI] [PubMed] [Google Scholar]
- 31.Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–71. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 32.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, et al. PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat Cell Biol. 2007;9:86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- 34.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–23. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh P, Garcia-Marcos M, Bornheimer SJ, Farquhar MG. Activation of Galphai3 triggers cell migration via regulation of GIV. J Cell Biol. 2008;182:381–93. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gundersen RE, Devreotes PN. In vivo receptor-mediated phosphorylation of a G protein in Dictyostelium. Science. 1990;248:591–93. doi: 10.1126/science.2110382. [DOI] [PubMed] [Google Scholar]
- 37.Hadwiger JA. Developmental morphology and chemotactic responses are dependent on G alpha subunit specificity in Dictyostelium. Dev Biol. 2007;312:1–12. doi: 10.1016/j.ydbio.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han JW, Leeper L, Rivero F, Chung CY. Role of RacC for the regulation of WASP and phosphatidylinositol 3-kinase during chemotaxis of Dictyostelium. J Biol Chem. 2006;281:35224–34. doi: 10.1074/jbc.M605997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hereld D, Vaughan R, Kim JY, Borleis J, Devreotes P. Localization of ligand-induced phosphorylation sites to serine clusters in the C-terminal domain of the Dictyostelium cAMP receptor, cAR1. J Biol Chem. 1994;269:7036–44. [PubMed] [Google Scholar]
- 40.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–17. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Huang YE. PhD diss. Baltimore, MD: Johns Hopkins Univ. Press; 2004. Phosphatidylinositol signaling in chemotaxis; p. 90. [Google Scholar]
- 42.Huang YE, Iijima M, Parent CA, Funamoto S, Firtel RA, Devreotes P. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol Biol Cell. 2003;14:1913–22. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hynes TR, Tang L, Mervine SM, Sabo JL, Yost EA, et al. Visualization of G protein betagamma dimers using bimolecular fluorescence complementation demonstrates roles for both beta and gamma in subcellular targeting. J Biol Chem. 2004;279:30279–86. doi: 10.1074/jbc.M401432200. [DOI] [PubMed] [Google Scholar]
- 44.Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr Opin Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 46.Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3:469–78. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 47.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–13. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 48.Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS ONE. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–29. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- 50.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–22. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Insall RH, Soede RD, Schaap P, Devreotes PN. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol Biol Cell. 1994;5:703–11. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isik N, Brzostowski JA, Jin T. An Elmo-like protein associated with myosin II restricts spurious F-actin events to coordinate phagocytosis and chemotaxis. Dev Cell. 2008;15:590–602. doi: 10.1016/j.devcel.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–77. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 54.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–11. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 55.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101:8951–56. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeon TJ, Lee DJ, Lee S, Weeks G, Firtel RA. Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J Cell Biol. 2007;179:833–43. doi: 10.1083/jcb.200705068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeon TJ, Lee DJ, Merlot S, Weeks G, Firtel RA. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J Cell Biol. 2007;176:1021–33. doi: 10.1083/jcb.200607072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Localization of the G protein betagamma complex in living cells during chemotaxis. Science. 2000;287:1034–36. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 59.Kabacoff C, Xiong Y, Musib R, Reichl EM, Kim J, et al. Dynacortin facilitates polarization of chemotaxing cells. BMC Biol. 2007;5:53. doi: 10.1186/1741-7007-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kae H, Kortholt A, Rehmann H, Insall RH, Van Haastert PJ, et al. Cyclic AMP signaling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 2007;8:477–82. doi: 10.1038/sj.embor.7400936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–6. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. PIP3-independent activation of TorC2 and PKB at the cell’s leading edge mediates chemotaxis. Curr Biol. 2008;18:1034–43. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–63. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- 64.Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–52. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- 65.Keizer-Gunnink I, Kortholt A, Van Haastert PJ. Chemoattractants and chemorepellents act by inducing opposite polarity in phospholipase C and PI3-kinase signaling. J Cell Biol. 2007;177:579–85. doi: 10.1083/jcb.200611046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khosla M, Spiegelman GB, Insall R, Weeks G. Functional overlap of the Dictyostelium RasG, RasD and RasB proteins. J Cell Sci. 2000;113(Pt. 8):1427–34. doi: 10.1242/jcs.113.8.1427. [DOI] [PubMed] [Google Scholar]
- 67.Kim JY, Borleis JA, Devreotes PN. Switching of chemoattractant receptors programs development and morphogenesis in Dictyostelium: receptor subtypes activate common responses at different agonist concentrations. Dev Biol. 1998;197:117–28. doi: 10.1006/dbio.1998.8882. [DOI] [PubMed] [Google Scholar]
- 68.Kim JY, Caterina MJ, Milne JL, Lin KC, Borleis JA, Devreotes PN. Random mutagenesis of the cAMP chemoattractant receptor, cAR1, of Dictyostelium. Mutant classes that cause discrete shifts in agonist affinity and lock the receptor in a novel activational intermediate. J Biol Chem. 1997;272:2060–68. doi: 10.1074/jbc.272.4.2060. [DOI] [PubMed] [Google Scholar]
- 69.Kim JY, Devreotes PN. Random chimeragenesis of G-protein-coupled receptors. Mapping the affinity of the cAMP chemoattractant receptors in Dictyostelium. J Biol Chem. 1994;269:28724–31. [PubMed] [Google Scholar]
- 70.Kim JY, Soede RD, Schaap P, Valkema R, Borleis JA, et al. Phosphorylation of chemoattractant receptors is not essential for chemotaxis or termination of G-protein-mediated responses. J Biol Chem. 1997;272:27313–18. doi: 10.1074/jbc.272.43.27313. [DOI] [PubMed] [Google Scholar]
- 71.Kortholt A, King JS, Keizer-Gunnink I, Harwood AJ, Van Haastert PJ. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol Biol Cell. 2007;18:4772–79. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kortholt A, Rehmann H, Kae H, Bosgraaf L, Keizer-Gunnink I, et al. Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J Biol Chem. 2006;281:23367–76. doi: 10.1074/jbc.M600804200. [DOI] [PubMed] [Google Scholar]
- 73.Kriebel PW, Barr VA, Parent CA. Adenylyl cyclase localization regulates streaming during chemotaxis. Cell. 2003;112:549–60. doi: 10.1016/s0092-8674(03)00081-3. [DOI] [PubMed] [Google Scholar]
- 74.Kumagai A, Hadwiger JA, Pupillo M, Firtel RA. Molecular genetic analysis of two G alpha protein subunits in Dictyostelium. J Biol Chem. 1991;266:1220–28. [PubMed] [Google Scholar]
- 75.Lee S, Comer FI, Sasaki A, McLeod IX, Duong Y, et al. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell. 2005;16:4572–83. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S, Rivero F, Park KC, Huang E, Funamoto S, Firtel RA. Dictyostelium PAKc is required for proper chemotaxis. Mol Biol Cell. 2004;15:5456–69. doi: 10.1091/mbc.E04-04-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leslie NR, Batty IH, Maccario H, Davidson L, Downes CP. Understanding PTEN regulation: PIP2, polarity and protein stability. Oncogene. 2008;27:5464–76. doi: 10.1038/onc.2008.243. [DOI] [PubMed] [Google Scholar]
- 78.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci USA. 2006;103:9761–66. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang W, Licate L, Warrick H, Spudich J, Egelhoff T. Differential localization in cells of myosin II heavy chain kinases during cytokinesis and polarized migration. BMC Cell Biol. 2002;3:19. doi: 10.1186/1471-2121-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maeda M, Lu S, Shaulsky G, Miyazaki Y, Kuwayama H, et al. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304:875–78. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- 81.Marin I, van Egmond WN, van Haastert PJ. The Roco protein family: a functional perspective. FASEB J. 2008;22:3103–10. doi: 10.1096/fj.08-111310. [DOI] [PubMed] [Google Scholar]
- 82.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112(Pt. 17):2867–74. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 83.Merlot S, Firtel RA. Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J Cell Sci. 2003;116:3471–78. doi: 10.1242/jcs.00703. [DOI] [PubMed] [Google Scholar]
- 84.Milne JL, Devreotes PN. The surface cyclic AMP receptors, cAR1, cAR2, and cAR3, promote Ca2+ influx in Dictyostelium discoideum by a G alpha 2-independent mechanism. Mol Biol Cell. 1993;4:283–92. doi: 10.1091/mbc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Milne JL, Kim JY, Devreotes PN. Chemoattractant receptor signaling: G protein-dependent and -independent pathways. Adv Second Messenger Phosphoprotein Res. 1997;31:83–104. doi: 10.1016/s1040-7952(97)80011-0. [DOI] [PubMed] [Google Scholar]
- 86.Milne JL, Wu L, Caterina MJ, Devreotes PN. Seven helix cAMP receptors stimulate Ca2+ entry in the absence of functional G proteins in Dictyostelium. J Biol Chem. 1995;270:5926–31. doi: 10.1074/jbc.270.11.5926. [DOI] [PubMed] [Google Scholar]
- 87.Mondal S, Bakthavatsalam D, Steimle P, Gassen B, Rivero F, Noegel AA. Linking Ras to myosin function: RasGEF Q, a Dictyostelium exchange factor for RasB, affects myosin II functions. J Cell Biol. 2008;181:747–60. doi: 10.1083/jcb.200710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Newell PC, Liu G. Streamer F mutants and chemotaxis of Dictyostelium. Bioessays. 1992;14:473–79. doi: 10.1002/bies.950140708. [DOI] [PubMed] [Google Scholar]
- 89.Para A, Krischke M, Merlot S, Shen Z, Oberholzer M, et al. Dictyostelium Dock180-related RacGEFs regulate the actin cytoskeleton during cell motility. Mol Biol Cell. 2009;20:699–707. doi: 10.1091/mbc.E08-09-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 91.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–70. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 92.Park KC, Rivero F, Meili R, Lee S, Apone F, Firtel RA. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004;23:4177–89. doi: 10.1038/sj.emboj.7600368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pitt GS, Milona N, Borleis J, Lin KC, Reed RR, Devreotes PN. Structurally distinct and stagespecific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–15. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- 94.Postma M, Van Haastert PJ. A diffusion-translocation model for gradient sensing by chemotactic cells. Biophys J. 2001;81:1314–23. doi: 10.1016/S0006-3495(01)75788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pramanik MK, Iijima M, Iwadate Y, Yumura S. PTEN is a mechanosensing signal transducer for myosin II localization in Dictyostelium cells. Genes Cells. 2009;14:821–34. doi: 10.1111/j.1365-2443.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 96.Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA. 2009;106:480–85. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rehberg M, Kleylein-Sohn J, Faix J, Ho TH, Schulz I, Graf R. Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol Biol Cell. 2005;16:2759–71. doi: 10.1091/mbc.E05-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roelofs J, Van Haastert PJ. Characterization of two unusual guanylyl cyclases from Dictyostelium. J Biol Chem. 2002;277:9167–74. doi: 10.1074/jbc.M111437200. [DOI] [PubMed] [Google Scholar]
- 99.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–18. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schneider IC, Haugh JM. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 2006;5:1130–34. doi: 10.4161/cc.5.11.2770. [DOI] [PubMed] [Google Scholar]
- 101.Senda S, Lee SF, Cote GP, Titus MA. Recruitment of a specific amoeboid myosin I isoform to the plasma membrane in chemotactic Dictyostelium cells. J Biol Chem. 2001;276:2898–904. doi: 10.1074/jbc.M008059200. [DOI] [PubMed] [Google Scholar]
- 102.Soede RD, Insall RH, Devreotes PN, Schaap P. Extracellular cAMP can restore development in Dictyostelium cells lacking one, but not two subtypes of early cAMP receptors (cARs). Evidence for involvement of cAR1 in aggregative gene expression. Development. 1994;120:1997–2002. doi: 10.1242/dev.120.7.1997. [DOI] [PubMed] [Google Scholar]
- 103.Somesh BP, Vlahou G, Iijima M, Insall RH, Devreotes P, Rivero F. RacG regulates morphology, phagocytosis, and chemotaxis. Eukaryot Cell. 2006;5:1648–63. doi: 10.1128/EC.00221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steimle PA, Yumura S, Cote GP, Medley QG, Polyakov MV, et al. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr Biol. 2001;11:708–13. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 105.Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Curr Biol. 2008;18:R485–94. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 106.Swanson JA, Taylor DL. Local and spatially coordinated movements in Dictyostelium discoideum amoebae during chemotaxis. Cell. 1982;28:225–32. doi: 10.1016/0092-8674(82)90340-3. [DOI] [PubMed] [Google Scholar]
- 107.Tang L, Franca-Koh J, Xiong Y, Chen MY, Long Y, et al. tsunami, the Dictyostelium homolog of the Fused kinase, is required for polarization and chemotaxis. Genes Dev. 2008;22:2278–90. doi: 10.1101/gad.1694508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T. Single-molecule analysis of chemotactic signaling in Dictyostelium cells. Science. 2001;294:864–67. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 109.van Egmond WN, Kortholt A, Plak K, Bosgraaf L, Bosgraaf S, et al. Intramolecular activation mechanism of the Dictyostelium LRRK2 homolog Roco protein GbpC. J Biol Chem. 2008;283:30412–20. doi: 10.1074/jbc.M804265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Es S, Weening KE, Devreotes PN. The protein kinase YakA regulates G-protein-linked signaling responses during growth and development of Dictyostelium. J Biol Chem. 2001;276:30761–65. doi: 10.1074/jbc.M103365200. [DOI] [PubMed] [Google Scholar]
- 111.Van Haastert PJ. Guanine nucleotides modulate cell surface cAMP-binding sites in membranes from Dictyostelium discoideum. Biochem Biophys Res Commun. 1984;124:597–604. doi: 10.1016/0006-291x(84)91596-1. [DOI] [PubMed] [Google Scholar]
- 112.van Haastert PJ, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J Cell Biol. 2007;177:809–16. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Haastert PJ, Kuwayama H. cGMP as second messenger during Dictyostelium chemotaxis. FEBS Lett. 1997;410:25–28. doi: 10.1016/s0014-5793(97)00416-x. [DOI] [PubMed] [Google Scholar]
- 114.van Rheenen J, Song X, van Roosmalen W, Cammer M, Chen X, et al. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J Cell Biol. 2007;179:1247–59. doi: 10.1083/jcb.200706206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vaughan RA, Devreotes PN. Ligand-induced phosphorylation of the cAMP receptor from Dictyostelium discoideum. J Biol Chem. 1988;263:14538–43. [PubMed] [Google Scholar]
- 116.Vazquez F, Matsuoka S, Sellers WR, Yanagida T, Ueda M, Devreotes PN. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc Natl Acad Sci USA. 2006;103:3633–38. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Veeranki S, Kim B, Kim L. The GPI-anchored superoxide dismutase SodC is essential for regulating basal Ras activity and for chemotaxis of Dictyostelium discoideum. J Cell Sci. 2008;121:3099–108. doi: 10.1242/jcs.030056. [DOI] [PubMed] [Google Scholar]
- 118.Vicker MG. Reaction-diffusion waves of actin filament polymerization/depolymerization in Dictyostelium pseudopodium extension and cell locomotion. Biophys Chem. 2000;84:87–98. doi: 10.1016/s0301-4622(99)00146-5. [DOI] [PubMed] [Google Scholar]
- 119.von Philipsborn A, Bastmeyer M. Mechanisms of gradient detection: a comparison of axon pathfinding with eukaryotic cell migration. Int Rev Cytol. 2007;263:1–62. doi: 10.1016/S0074-7696(07)63001-0. [DOI] [PubMed] [Google Scholar]
- 120.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–37. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 121.Wang B, Kuspa A. Dictyostelium development in the absence of cAMP. Science. 1997;277:251–54. doi: 10.1126/science.277.5323.251. [DOI] [PubMed] [Google Scholar]
- 122.Weeks G, Spiegelman GB. Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell Signal. 2003;15:901–9. doi: 10.1016/s0898-6568(03)00073-1. [DOI] [PubMed] [Google Scholar]
- 123.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR. A PtdInsP(3)-and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat Cell Biol. 2002;4:509–13. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wessels D, Lusche DF, Kuhl S, Heid P, Soll DR. PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J Cell Sci. 2007;120:2517–31. doi: 10.1242/jcs.010876. [DOI] [PubMed] [Google Scholar]
- 126.Wessels D, Soll DR, Knecht D, Loomis WF, De Lozanne A, Spudich J. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev Biol. 1988;128:164–77. doi: 10.1016/0012-1606(88)90279-5. [DOI] [PubMed] [Google Scholar]
- 127.Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–75. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xiao Z, Zhang N, Murphy DB, Devreotes PN. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J Cell Biol. 1997;139:365–74. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol Biol Cell. 2005;16:676–88. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang B, Ma Y, Guo H, Sun B, Niu R, et al. Akt2 is required for macrophage chemotaxis. Eur J Immunol. 2009;39:894–901. doi: 10.1002/eji.200838809. [DOI] [PubMed] [Google Scholar]
- 131.Zhang H, Wessels D, Fey P, Daniels K, Chisholm RL, Soll DR. Phosphorylation of the myosin regulatory light chain plays a role in motility and polarity during Dictyostelium chemotaxis. J Cell Sci. 2002;115:1733–47. doi: 10.1242/jcs.115.8.1733. [DOI] [PubMed] [Google Scholar]
- 132.Zhang M, Goswami M, Hereld D. Constitutively active G protein-coupled receptor mutants block Dictyostelium development. Mol Biol Cell. 2005;16:562–72. doi: 10.1091/mbc.E04-06-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang N, Long Y, Devreotes PN. Ggamma in Dictyostelium: Its role in localization of gbetagamma to the membrane is required for chemotaxis in shallow gradients. Mol Biol Cell. 2001;12:3204–13. doi: 10.1091/mbc.12.10.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18:1587–93. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zigmond SH, Joyce M, Borleis J, Bokoch GM, Devreotes PN. Regulation of actin polymerization in cell-free systems by GTPgammaS and Cdc42. J Cell Biol. 1997;138:363–74. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
RELATED RESOURCES
- Bagorda A, Parent CA. Eukaryotic chemotaxis at a glance. J Cell Sci. 2008;121:2621–24. doi: 10.1242/jcs.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca-Koh J, Kamimura Y, Devreotes P. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr Opin Genet Dev. 2006;16:333–38. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–85. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–63. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJ, Veltman DM. Chemotaxis: navigating by multiple signaling pathways. Sci STKE. 2007:pe40. doi: 10.1126/stke.3962007pe40. [DOI] [PubMed] [Google Scholar]


