Abstract
White-nose syndrome (WNS) caused by the pathogenic fungus Pseudogymnoascus destructans is decimating the populations of several hibernating North American bat species. Little is known about the molecular interplay between pathogen and host in this disease. Fluorescence microscopy ambient ionization mass spectrometry was used to generate metabolic profiles from the wings of both healthy and diseased bats of the genus Myotis. Fungal siderophores, molecules that scavenge iron from the environment, were detected on the wings of bats with WNS, but not on healthy bats. This work is among the first examples in which microbial molecules are directly detected from an infected host and highlights the ability of atmospheric ionization methodologies to provide direct molecular insight into infection.
Introduction
Fungal diseases of animals and plants have long been a feature of natural ecosystems, but evidence suggests that incidences of mycoses are increasing in frequency and severity and that these emerging infectious diseases pose a threat in terms of loss of biodiversity and food security [1]. Mycoses have garnered a large amount of attention recently due to their detrimental impacts on populations of organisms as varied as sea fans, turtles, bees, corals, frogs, crayfish, and bats [1–7]. The fungal infection affecting hibernating North American bats, white-nose syndrome (WNS), has killed an estimated 5.7–6.7 million bats since it was first described in 2006 and has spread to 25 states and five Canadian provinces [8–11]. The psychrophilic fungus identified as the causative agent of WNS, Pseudogymnoascus destructans, is believed to be an exotic pathogen of European origin that was introduced to the U.S. through human activity [12–17].
The mechanism through which WNS induces mortality remains to be fully elucidated. It is hypothesized that cutaneous infection of bat wing skin with P. destructans disrupts electrolyte balance [18–19]. This imbalance or some other mechanism causes increases in the frequency with which infected bats arouse from torpor during winter [19]. Frequent arousals may in turn deplete fat stores and lead to death by starvation [20].
Identifying molecules that fungal pathogens use to interact with a host provides insight into the mechanism of their pathogenicity. Better understanding these mechanisms may lead to improved management strategies that account for the biology of the pathogen. We therefore set out to directly profile the metabolic milieu on the wings of bats with WNS with a focus on fungal molecules. We used fluorescent microscopy-guided ambient mass spectrometry to metabolically profile the wings of bats of the genus Myotis that were healthy (n = 5) or showed signs of WNS infection (n = 11). This will enable us to begin to establish a ‘molecular signature’ for WNS and to determine if there are molecules that the fungus may use to facilitate infection.
Materials and Methods
Animal Collection & Tissue Sample Preparation
Five healthy Myotis lucifugus, 10 M. lucifugus with WNS, and one Myotis septentrionalis with WNS were included in the analysis (Table 1). All bats were found deceased during routine monitoring of maternity or hibernation roosts being conducted by state agency biologists. In Pennsylvania, personnel of the Pennsylvania Game Commission collected the specimens in compliance with Pennsylvania Statute Title 34, Section 322. Deceased bats collected in West Virginia were collected by personnel of the West Virginia Department of Natural Resources and no permits were required. Healthy bats were collected from 2004 to 2008 in Pennsylvania. All but one of these bats were collected prior to the emergence of WNS in North America and the bat collected after WNS emergence had no obvious signs of P. destructans infection (e.g. cupping erosions) when evaluated microscopically. WNS bats were collected in 2011 in Pennsylvania and in West Virginia during a WNS-associated mass mortality event. No bats were euthanized for this study. Two 6 mm tissue punches were collected from the wings of each of the deceased bats, affixed to 1x3 inch microscope glass slides, and stored at -80°C prior to mass spectrometric analysis.
Table 1. Specimen information for the Myotis used in the study.
| WNS Status | Species | Sex | Collection Date | Collection Location |
|---|---|---|---|---|
| Pre-WNS | Myotis lucifugus | unknown | 11/12/08 | Barton Cave, Fayette County, PA |
| Pre-WNS | Myotis lucifugus | female | 8/15/05 | Shaver’s Creek, Huntington County, PA |
| Pre-WNS | Myotis lucifugus | male | 9/13/04 | Harlansburg Cave Gate, Lawrence County, PA |
| Pre-WNS | Myotis lucifugus | female | 4/16/05 | Canoe Creek Mine, Blair County, PA |
| Pre-WNS | Myotis lucifugus | female | 4/16/05 | Canoe Creek Mine, Blair County, PA |
| WNS | Myotis lucifugus | female | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | female | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | female | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | female | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis lucifugus | male | 2/26/11 | Hellhole Cave, Pendelton County, WV |
| WNS | Myotis septentrionalis | unknown | 1/7/11 | Lincoln Cavern, Huntington County, PA |
Microscopy Ambient Ionization Mass Spectrometry of Bat Wings
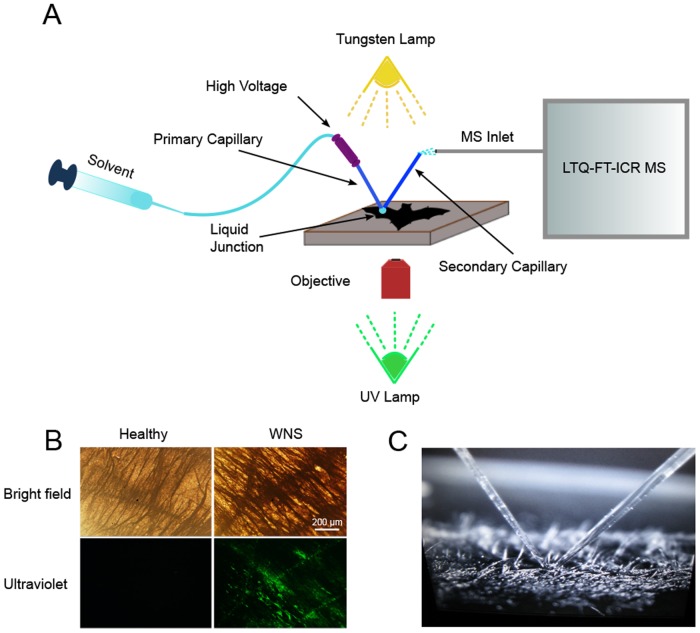
The mass spectrometric interrogation of bat wing skin was performed using a hybrid microscopy/ionization technique which combines an ambient nanospray desorption electrospray ionization (nanoDESI) source and an inverted microscope as described [21] (Fig. 1A). Unlike previous usage of these tools, our analysis incorporated the use of fluorescence. This allowed us to illuminate the wings with UV light and visualize areas of fluorescence that correlate with the development of cupping erosions due to infection with P. destructans [22] (Fig. 1B). We could then directly target these areas with the nanoDESI probe for MS analysis (Fig. 1C). The sample slide with wing punches was placed on the stage of a Nikon DIAPHOT 300 microscope and bright field and fluorescent images of the tissue were captured using a CCD camera (Nikon D40 DSLR) to confirm the presence or absence of fungal infection. The stage was then manipulated to move the tissue sample to the desired position under the micrometer-sized liquid junction formed by the two flame-pulled fused silica capillaries of the nanoDESI. The capillary tubes were flamed-pulled from original 150/50 μm (O.D./I.D.) to ~60 μm O.D. and a voltage of 2.2 kV was applied to them throughout the experiment. The capillaries were aligned in a “V” configuration so that they abutted one another at the bottom of the “V” and then angled 45° away from the point of contact in either direction. A syringe pump was used to continuously deliver the solvent (either acetonitrile in water (65/35, vol/vol) with 0.2% formic acid, methanol in water (50/50, vol/vol) with 0.2% formic acid, or methanol, acetonitrile, and toluene (50/35/15, vol/vol/vol) with 0.2% formic acid) at a rate of ~1.0 μL/min through 300/100 μm (O.D./I.D.) fused silica capillary tubing to the primary flame-pulled capillary. This solvent was then aspirated by the secondary capillary resulting in the formation of a dynamic liquid droplet of approximately 100 μm in diameter at the junction of the two capillaries. The sample stage was raised until the tissue contacted the liquid droplet and a continuous stream of solvent containing analytes desorbed from the tissue was delivered to the inlet of the mass spectrometer (hybrid 6.4-T LTQ-FT; Thermo Electron, North America) via electrospray ionization generated at the terminal end of the secondary capillary. Multiple spots were sampled along each tissue slide.
Fig 1. Microscopy ambient ionization mass spectrometry of bat wings.
Tissue samples from the wings of healthy bats and individuals with white-nose syndrome were subjected to microscopy ambient ionization mass spectrometry (A). Pseudogymnoascus destructans infection was confirmed by the presence of fluorescent lesions when the tissue was excited with UV light (B). A nanoDESI source was used to desorb analytes from fluorescent tissue regions for MS analysis (capillary junction making contact with the tissue surface, C).
Fungal Extraction for MS Analysis
Replicates of P. destructans were grown on ISP2 agar plates (4 g/liter yeast extract, 10 g/liter malt extract, 4 g/liter dextrose, and 1.5 to 2% agar). Small plugs of agar and fungal biomass were collected and combined with 100 μl of n-butanol in microcentrifuge tubes. Extractions were allowed to proceed at room temperature for one hour. The organic layers were centrifuged and the supernatant was subsequently analyzed. A Triversa nanomate-electrospray ionization source was used to introduce the extracts (diluted in methanol in water (50:50) with 1% formic acid) directly into the MS inlet. This was accomplished using a spray voltage setting of 1.3–1.45 kV and a pressure of 0.35–0.5 psi as set with Chipsoft software version 7.2.0. Data were collected using Xcalibur software version 1.4 SR1 running the data-dependent method described below.
Mass Spectrometric Analysis
A 6.4 T Finnigan LTQ-FT-ICR MS (Thermo Electron, North America) was used to analyze tissue samples and fungal extracts. The instrument was tuned to 816 m/z using bovine cytochrome c (charge state 15, Sigma Aldrich) in a 65/35 (vol/vol) acetonitrile/0.2% formic acid aqueous solution with Tune Plus software version 1.0. Ion spectra from 150–2,000 m/z were collected in positive ion mode. In cases where there was a high background, the ion spectra observation range was reduced. The automated instrument scan cycle consisted of two segments. The first segment had two profile mode MS1 scans: one full scan in the IT mode with 200 ms max fill time; one full scan in the FT cell (50,000 resolution) with 8 sec max inject time. This was followed by an MS1-dependent tandem mass (MS/MS) acquisition which consisted of 9 scans in a cycle with a Δm/z = 3 isolation window. The top 9 most abundant peaks in the MS1 scan were sequentially isolated and fragmented by applying collision induced dissociation (CID) energy using nitrogen as the collision gas. Ion peaks that had been selected twice were excluded and not fragmented again in subsequent cycles in order to cover as many individual ions as possible. These data-dependent MS/MS scans (IT mode, profile spectra) consisted of a maximum 500 ms fill time, 35% normalized collision energy, 0.25 activation Q, and 0.05 s activation time. Each scan contained 4 microscans and was recorded in average. Data were acquired for 10 minutes.
Molecular Networking
The structural relationships between different masses in MS1 scans (precursor ions) were mapped in Cytoscape version 2.8.3 based on the similarity of their tandem MS fragmentation patterns as assessed through molecular networking using the web-based Global Natural Products Social Molecular Networking tool (GNPS; http://gnps.ucsd.edu/)[23–25]. A cosine cutoff of 0.6 was selected for generation of the network. Background signals arising from solvents or agar were subtracted during the network processing stage. The network was then imported into Cytoscape for visualization. Precursor ions were represented by nodes and the similarity between two precursor ions as determined by comparison of their fragments/cosine score was represented as an edge between two nodes. The magnitude of the cosine score was represented by edge thickness with pairs of compounds with high scores having thicker lines. A search of the publicly curated GNPS standards library was also performed to determine if any of the masses represented in the network matched to known compounds that we had not considered in our analysis. All data files used in the generation of the network were deposited in the GNPS MassIVE data repository and are publicly available (MassIVE ID MSV000078620).
Seeding the Network with Desferrichrome, Ferrichrome, and Triacetylfusarinine C Standards
Iron-free ferrichrome (Santa Cruz Biotechnology), ferrichrome (Iron-free ferrichrome with the addition of iron (III) chloride hexahydrate), and triacetylfusarinine C (EMC microcollections GmbH) were used as standards to verify the molecular assignment of the siderophores in the tissue samples through a process known as dereplication [25]. The solubilized standards were introduced directly into the MS inlet with a Triversa nanomate-electrospray ionization source and analyzed using the data-dependent method described above. A molecular network was then generated using the data files from the standards, fungal extracts, and tissue samples. The nodes correlated with the standard data files were examined to establish if they (1) clustered with experimental nodes or (2) were incorporated into consensus nodes that also included experimental files. Following this neighborhood analysis, the spectra were manually examined and the fragmentation of the raw spectra evaluated for similarity. Through this analysis it was possible to definitively determine whether or not desferrichrome, ferrichrome and triacetylfusarinine C were detected from the wing surfaces and/or from fungal cultures.
Results and Discussion
Detection of fungal metabolites on bat wings by fluorescence microscopy ambient ionization mass spectrometry and molecular networking
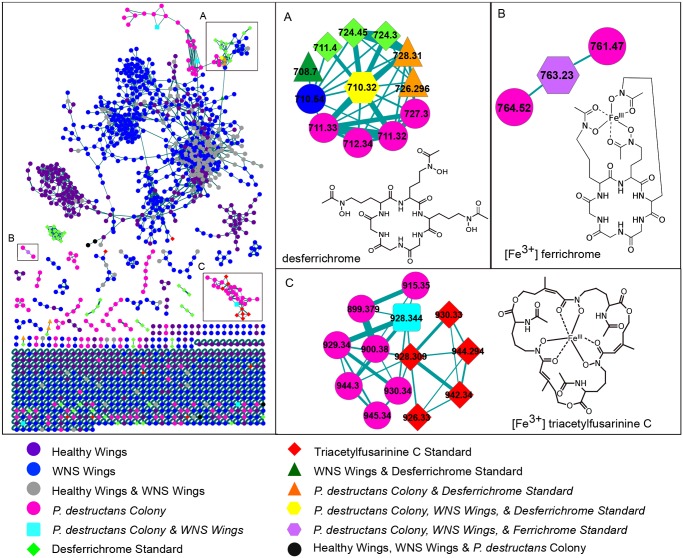
Fluorescent microscopy ambient mass spectrometry of bat wings, extracts from cultured P. destructans, and commercial standards generated a large number of MS/MS spectra. These spectra were subjected to molecular networking [23,25] giving rise to a network comprised of 1,503 nodes representing precursor ions derived from intact chemical compounds and 2,463 edges representing their structural relationships (Fig. 2A; S1 Fig.). Each node was colored with respect to the sample type(s) from which it derived. The resulting molecular network provides a visual picture of the diversity and origin of MS/MS spectra that were collected.
Fig 2. Detection of siderophores on the wings of bats with white-nose syndrome via molecular networking.
Molecular networking of the MS/MS data was used to determine the structural relationships between the metabolites detected from wing surfaces, cultured Pseudogymnoascus destructans, and commercial siderophore standards. The siderophores desferrichrome (m/z 710.324) and ferrichrome (m/z 763.230) were observed from the wings of bats with white-nose syndrome and from cultured P. destructans and formed consensus nodes with commercial standards (A,B). Triacetylfusarinine C (m/z 928.344) was also present on WNS wings and in the P. destructans colony (C). None of the three siderophores were detected on the wings of healthy bats. In A and C insets only the node with the m/z of interest and its first and second neighbors have been displayed from the overall cluster for simplicity.
The network highlighted a number of molecules that were present on the wings of bats with WNS that were not detected from healthy bats and vice versa. These distinct chemical signatures were found to be due to the presence of fungal metabolites on the WNS wings. Individual variation and a host response to infection likely also contribute the observed difference. Several of the MS/MS spectra from the experimental samples matched those of known fungal metabolites and these nodes were inspected in further detail.
In the first node we inspected, spectra from fungal extracts matched to a compound in the GNPS standards library, the mycotoxin citrinin, indicating that P. destructans may have the capacity to produce citrinin or a citrinin-like molecule (S2A, S2B Fig.). This skin-permeable nephrotoxin is known to be produced by fungi of the genera Penicillium, Monascus, and Aspergillus [26–27]. A protein blast of the P. destructans genome using the citrinin polyketide synthase from Monascus purpureus as the query revealed a homologous sequence with 44% identity. (S2C Fig.). It may be interesting to evaluate the overall ability of the fungus to produce mycotoxins in the future and to evaluate the contributions of any such molecules to pathogenicity.
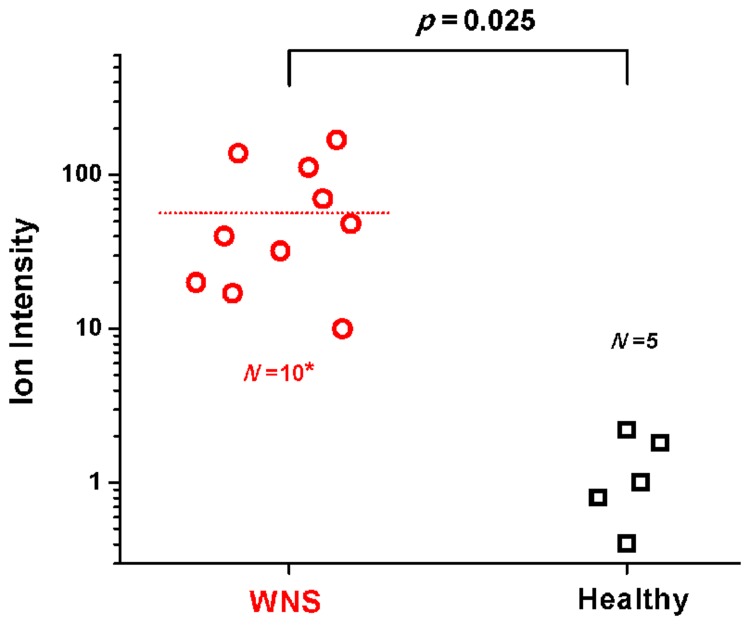
Another node was a match for the fungal siderophore desferrichrome (m/z 710.324, Fig. 2A). Siderophores are microbial iron-chelating molecules that are either retained intracellularly as a means of iron storage or are of a diffusible type that gather ferric iron from the extracellular environment and are then retrieved by the organism [28]. The node incorporated scans from WNS wings, the cultured P. destructans colony, and the desferrichrome standard; scans from the wings of healthy bats were not included (Fig. 2A). Manual inspection of the MS/MS fragmentation data for these precursors confirmed that the pattern observed from the WNS wings and the fungal colony matched that of the commercial desferrichrome standard (S3 Fig.). This indicates that the masses observed from the cultured fungus and on the wing skin of infected bats derive from desferrichrome and that, presumably, P. destructans is producing this siderophore during its colonization of bats. When the signal for the siderophore was analyzed in a targeted fashion, by performing MS/MS directly on the isolated parent ion during microscopy nanoDESI analysis, desferrichrome was detected on 10 of the 11 WNS bats and was virtually undetectable in all of the samples from healthy bat wings (ANOVA, p = 0.025; Fig. 3; S4 Fig.). In addition, the iron-chelated form of the molecule, ferrichrome, was detected (m/z 763.230, Fig. 2B; S5 Fig.). This indicates that there is sufficient iron present on bat wings to be chelated by the siderophore.
Fig 3. Statistical analysis of desferrichrome daughter ion intensity among healthy and diseased bat wings.
Analysis of variance (ANOVA) of the absolute intensities of the desferrichrome daughter ion, m/z 650 (neutral loss of C2H4O2), observed on WNS and healthy bat wings (See also S4 Fig.). A ± 3 m/z ion window was allowed when selecting the precursor, m/z 710. Differences in the absolute intensities of the daughter ions among 10 of the 11 WNS wings and 5 healthy wings were statistically significant (p = 0.025). On the eleventh WNS wing, no ions within the m/z 710 ± 3 m/z range was selected for fragmentation by the automatic data-dependent method due to low ion intensity and the sample could therefore not be included in the plot.
A second siderophore, triacetylfusarinine C, was also detected by molecular networking (Fig. 2C). A node with m/z 928.344 incorporated scans from a WNS wing and the cultured P. destructans colony and clustered with triacetylfusarinine C standard nodes. Manual inspection of the tandem mass spectra confirmed that the node derived from the fragmentation of triacetylfusarinine C, although it failed to form a consensus node with the standard under the molecular networking parameters selected (S6 Fig.). This indicates that this siderophore is also produced by P. destructans during infection.
Interrogation of the P. destructans genome for secondary metabolite enzymes involved in siderophore biosynthesis and imaging mass spectrometry of the cultured P. destructans colony further support the production of ferrichrome and triacetylfuarinine C by the fungus (S1 Supporting Information; S7 Fig.; S1 Table). In the absence of a controlled laboratory experiment in which bats are analyzed before and after infection with P. destructans, it cannot be unequivocally stated that the observed metabolites are produced by P. destructans since other fungi also have the ability to form these siderophores. However, this seems unlikely given the marked difference we observe between WNS and healthy wings and we hypothesize that P. destructans requires iron in order to grow and mount infection and therefore produces siderophores during its colonization of bats.
Microbes require iron as a cofactor in redox reactions involved in a variety of cellular processes. Consequently, a large part of the host response to infection centers on limiting the amount of iron available to invading microbes [29–32]. Because of this, the amount of available iron is, in general, sub-optimal for microbial growth [30,33]. Whether this is the case for hibernating bats is unknown.
High-affinity siderophores are important for satisfying microbial iron demand in the face of host defenses and a number of investigations have demonstrated that they are essential for growth and virulence [34–38]. In addition, production of hydroxamate siderophores is a feature of the dermatophytic pathogenic fungi Microsporum and Trichophyton and ferrichrome-type siderophores, specifically, have been demonstrated to be necessary for epithelial invasion by C. albicans in an in vitro model of oral candidiasis [39–40]. Our detection of fungal siderophores on wings of bats with WNS suggests that these molecules may have a role in infection and/or tissue invasion.
Because of the importance of iron in microbial infections, a number of antimicrobial strategies that target siderophores have recently emerged. These include efforts that link antibiotics or antifungals to siderophores to mediate their uptake, inundation with irrelevant siderophores that the organism cannot use or analogues that cannot bind iron, addition of iron chelators such as EDTA, and disruption of endogenous siderophore pathways through the blocking of biosynthetic enzymes or transporters [41–44]. We contend that such methods of iron deprivation should be investigated for their effectiveness against P. destructans infection.
Conclusion
Here we used fluorescence microscopy ambient mass spectrometry in combination with molecular networking to detect metabolites directly from the wing skin of bats with WNS. The approach established the presence of fungal siderophores on infected bat wings. Siderophores are important for fungal growth and virulence and methods that interfere with their biosynthesis or activity may prove effective in limiting P. destructans infection.
Supporting Information
(XLSX)
The molecular network was composed of nodes that incorporated MS/MS scans from only a single sample type as well as consensus nodes that incorporated spectra from different experimental sample types. The number of nodes in each category and their relationships to each other are conveyed in a Venn diagram.
(TIF)
A comparison of the data to a library of standards from the Global Natural Products Social Molecular Networking database resulted in a putative match, citrinin, to the P. destructans culture extract (A,B). A protein blast of the P. destructans genome using the polyketide synthase responsible for citrinin biosynthesis as the query returned a homologous amino acid sequence (C).
(TIF)
MS/MS of the desferrichrome standard m/z 710 (A). The MS/MS fragmentation patterns from the P. destructans colony (B) and WNS wing (C) matched the fragmentation of the standard precursor.
(TIF)
In cases where desferrichrome precursor ions display high intensities relative to other metabolites, their fragmentation patterns will be less complicated and they will have a greater cosine correlation with the standard (A). When the intensities of the desferrichrome ions are less intense, background noise or peaks from other compounds of similar mass may be fragmented along with them resulting in more complicated MS/MS spectra and a lower cosine correlation with the standard even though the molecule is present (B).
(TIF)
FT-MS/MS of ferrichrome Fe3+ complex standard m/z 763 (A). The IT-MS/MS fragmentation patterns for m/z 763 from the P. destructans colony (B) and WNS wings (C) matched the fragmentation of the standard precursor.
(TIF)
FT-MS/MS of triacetylfusarinine C Fe3+ complex standard m/z 928 (A). The IT-MS/MS fragmentation patterns for m/z 928 from the P. destructans colony (C) and WNS wings (D) matched the fragmentation of the standard precursor.
(TIF)
P. destructans was grown directly on ISP2 agar (Ai) or on top of a permeable membrane overlaid on agar (Aii) which was later removed for IMS analysis. Colonies and agar were excised and placed on a MALDI target plate along with a portion of plain agar which served as a negative control (B). Aerial hyphae were removed from the P. destructans colony grown directly on the agar, and the membrane and P. destructans colony were removed from the second sample to reveal the agar underneath (C). Several metabolites were observed to be associated with the fungal colony (Di) or secreted into the agar underneath it (Dii). Among the metabolites for which ions were observed were the siderophores desferrichrome and triacetylfusarinine C.
(TIF)
(PDF)
Acknowledgments
The authors would like to thank Dr. Chris Hoefler and the Dr. Paul Straight Laboratory for the gift of cellophane membranes, Dr. Craig Stihler for collection of specimens from West Virginia, Dr. Laura Sanchez and Paul D. Boudreau for clip art and assistance with figures, and Dr. Laura Sanchez, Don Nguyen, and Dr. Vanessa V. Phelan for training and assistance with mass spectrometry and molecular networking. We also thank Dr. Kevin Tidgewell for reviewing the manuscript and providing helpful insights. The use of trade, product or firm names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Research in the laboratory of PCD for the development of real-time MS was supported by National Institutes of Health Grant GM094802 (http://www.nih.gov/). Research in the laboratory of AMK was supported by National Science Foundation grant DEB-1115895 (http://www.nsf.gov/). Work by SJM was partially supported by a P.E.O. fellowship (http://www.peointernational.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim K, Harvell CD. The rise and fall of a six-year coral-fungal epizootic. Am Nat. 2004;164: S52–S63. [DOI] [PubMed] [Google Scholar]
- 3. Sarmiento-Ramirez JM, Abella E, Martin MP, Telleria MT, Lopez-Jurado LF, Marco A, et al. Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. Fems Microbiol Lett. 2010;312: 192–200. 10.1111/j.1574-6968.2010.02116.x [DOI] [PubMed] [Google Scholar]
- 4. Cameron SA, Lozier JD, Strange JP, Koch JB, Cordes N, Solter LF, et al. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci USA. (2011);108: 662–667. 10.1073/pnas.1014743108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ, et al. Emerging marine diseases—Climate links and anthropogenic factors. Science. 1999;285: 1505–1510. [DOI] [PubMed] [Google Scholar]
- 6. Holdich DM, Reynolds JD, Souty-Grosset C, Sibley PJ. A review of the ever increasing threat to European crayfish from non-indigenous crayfish species. Knowl Manag Aquat Ec. 2009;394–395: 11. [Google Scholar]
- 7. Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, et al. Bat White-Nose Syndrome: An Emerging Fungal Pathogen? Science. 2009;323: 227–227. 10.1126/science.1163874 [DOI] [PubMed] [Google Scholar]
- 8. Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, et al. An Emerging Disease Causes Regional Population Collapse of a Common North American Bat Species. Science. 2010;329: 679–682. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- 9. Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Kilpatrick AM. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecology Letters. 2012;15: 1050–1057. 10.1111/j.1461-0248.2012.01829.x [DOI] [PubMed] [Google Scholar]
- 10.News Release: North American bat death toll exceeds 5.5 million from white-nose syndrome. Top of FormUnited States Fish and Wildlife Service. 2012. Available: http://www.fws.gov/northeast/feature_archive/Feature.cfm?id=794592078.
- 11.News Release: Where is it now? Whitenosesyndrome.org. 2014. Available: http://www.whitenosesyndrome.org/about/where-is-it-now.
- 12. Gargas A, Trest MT, Christensen M, Volk TJ, Blehert D. Geomyces destructans sp. nov., associated with Bat White-Nose Syndrome. Mycotaxon. 2009;108: 147–154. [Google Scholar]
- 13. Minnis AM, Lindner DL. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 2013;117: 638–649. 10.1016/j.funbio.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 14. Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480: 376–378. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- 15. Puechmaille SJ, Frick WF, Kunz TH, Racey PA, Voigt CC, Wibbelt G, et al. White-nose syndrome: is this emerging disease a threat to European bats? Trends Ecol Evol. 2011;26: 570–576. 10.1016/j.tree.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 16. Puechmaille SJ, Wibbelt G, Korn V, Fuller H, Forget F, Muhldorfer K, et al. Pan-European Distribution of White-Nose Syndrome Fungus (Geomyces destructans) Not Associated with Mass Mortality. PLoS ONE. 2011;6(4): e19167 10.1371/journal.pone.0019167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warnecke L, Turner JM, Bollinger TK, Lorch JM, Misra V, Cryan PM, et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc Natl Acad Sci USA. 2012;109: 6999–7003. 10.1073/pnas.1200374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meteyer CU, Buckles EL, Blehert DS, Hicks AC, Green DE, Shearn-Bochsler V, et al. Histopathologic criteria to confirm white-nose syndrome in bats. J Vet Diagn Inves. 2009;21: 411–414. [DOI] [PubMed] [Google Scholar]
- 19. Cryan PM, Meteyer CU, Blehert DS, Lorch JM, Reeder DM, Turner GG, et al. Electrolyte depletion in white-nose syndrome bats. J Wildl Dis. 2013;49: 398–402. 10.7589/2012-04-121 [DOI] [PubMed] [Google Scholar]
- 20. Reeder DM, Frank CL, Turner GG, Meteyer CU, Kurta A, Britzke ER, et al. Frequent Arousal from Hibernation Linked to Severity of Infection and Mortality in Bats with White-Nose Syndrome. PLoS ONE. 2012;7(6): e38920 10.1371/journal.pone.0038920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu CC, White NM, Hayashi M, Lin EC, Poon T, Banerjee I, et al. Microscopy ambient ionization top-down mass spectrometry reveals developmental patterning. Proc Natl Acad Sci USA. 2013;110: 14855–14860. 10.1073/pnas.1310618110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turner GG, Meteyer CU, Barton H, Gumbs JF, Reeder DM. Non-Lethal Screening of bat wing skin using UV fluorescence to detect lesions indicative of white-nose syndrome. J Wildl Dis. 2014;50(3): 000–000. [DOI] [PubMed] [Google Scholar]
- 23. Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109: E1743–E1752. 10.1073/pnas.1203689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3): 431–432. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76(9): 1686–1699. 10.1021/np400413s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flajs D, Peraica M. Toxicological properties of citrinin. Arh Hig Rada Toksikol. 2010;60(4) 457–464. 10.2478/10004-1254-60-2009-1992 [DOI] [PubMed] [Google Scholar]
- 27. Boonen J, Malysheva SV, Taevernier L, di Mavungu JD, de Saeger S, De Speigeleer B. Human skin penetration of selected model mycotoxins. Toxicol. 2012;301(1–3): 21–32. [DOI] [PubMed] [Google Scholar]
- 28. Winkelmann G. Ecology of siderophores with special reference to the fungi. Biometals. 2007;20: 379–392. [DOI] [PubMed] [Google Scholar]
- 29. Hood HM, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10: 525–537. 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parrow NL, Fleming RE, Minnick MF. Sequestration and scavenging of iron in infection. Infect Immun. 2013;81(10): 3503–3514. 10.1128/IAI.00602-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Theurl I, Fritsche G, Ludwiczek S, Garimorth K, Bellmann-Weiler R, Weis G. The macrophage: a cellular factory at the interphase between iron and immunity for the control of infections. Biometals. 2005;18: 359–367. [DOI] [PubMed] [Google Scholar]
- 32. Fluckinger M, Haas H, Merschak P, Glasgow BJ, Redl B. Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob Agents and Chemother. 2004;48: 3367–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71: 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hissen AHT, Wan ANC, Warwas ML, Pinto LJ, Moore MM. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N-5-oxygenase, is required for virulence. Infect Immun. 2005;73: 5493–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigates virulence. J Exp Med. 2004;200: 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, et al. Distinct Roles for Intra- and Extracellular Siderophores during Aspergillus fumigatus Infection. PLoS Pathog. 2007;3(9): e128 10.1371/journal.ppat.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson L. Iron and siderophores in fungal-host interactions. Mycol Res. 2008;112: 170–183. 10.1016/j.mycres.2007.11.012 [DOI] [PubMed] [Google Scholar]
- 38. Eisendle M, Schrettl M, Kragl C, Mueller D, Illmer P, Haas H. The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination, and sexual development in Aspergillus nidulans . Eukaryotic Cell. 2006;5: 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mor H, Kashman Y, Winkelmann G, Barash I. Characterization of siderophores produced by different species of the dermatophytic fungi Microsporum and Trichophyton . Biometals. 1992;5: 213–216. [Google Scholar]
- 40. Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70: 5246–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46: 149–187. 10.1146/annurev.phyto.45.062806.094338 [DOI] [PubMed] [Google Scholar]
- 42. Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71: 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schrettl M, Haas H. Iron homeostasis-Achilles’ heel of Aspergillus fumigatus? Curr Opin Microbiol. 2011;14: 400–405. 10.1016/j.mib.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller MJ, Zhu H, Xu Y, Wu C, Walz AJ, Vergne A, et al. Utilization of microbial iron assimilation processes for the development of new antibiotics and inspiration for the design of new anticancer agents. Biometals. 2009;22: 61–75. 10.1007/s10534-008-9185-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
The molecular network was composed of nodes that incorporated MS/MS scans from only a single sample type as well as consensus nodes that incorporated spectra from different experimental sample types. The number of nodes in each category and their relationships to each other are conveyed in a Venn diagram.
(TIF)
A comparison of the data to a library of standards from the Global Natural Products Social Molecular Networking database resulted in a putative match, citrinin, to the P. destructans culture extract (A,B). A protein blast of the P. destructans genome using the polyketide synthase responsible for citrinin biosynthesis as the query returned a homologous amino acid sequence (C).
(TIF)
MS/MS of the desferrichrome standard m/z 710 (A). The MS/MS fragmentation patterns from the P. destructans colony (B) and WNS wing (C) matched the fragmentation of the standard precursor.
(TIF)
In cases where desferrichrome precursor ions display high intensities relative to other metabolites, their fragmentation patterns will be less complicated and they will have a greater cosine correlation with the standard (A). When the intensities of the desferrichrome ions are less intense, background noise or peaks from other compounds of similar mass may be fragmented along with them resulting in more complicated MS/MS spectra and a lower cosine correlation with the standard even though the molecule is present (B).
(TIF)
FT-MS/MS of ferrichrome Fe3+ complex standard m/z 763 (A). The IT-MS/MS fragmentation patterns for m/z 763 from the P. destructans colony (B) and WNS wings (C) matched the fragmentation of the standard precursor.
(TIF)
FT-MS/MS of triacetylfusarinine C Fe3+ complex standard m/z 928 (A). The IT-MS/MS fragmentation patterns for m/z 928 from the P. destructans colony (C) and WNS wings (D) matched the fragmentation of the standard precursor.
(TIF)
P. destructans was grown directly on ISP2 agar (Ai) or on top of a permeable membrane overlaid on agar (Aii) which was later removed for IMS analysis. Colonies and agar were excised and placed on a MALDI target plate along with a portion of plain agar which served as a negative control (B). Aerial hyphae were removed from the P. destructans colony grown directly on the agar, and the membrane and P. destructans colony were removed from the second sample to reveal the agar underneath (C). Several metabolites were observed to be associated with the fungal colony (Di) or secreted into the agar underneath it (Dii). Among the metabolites for which ions were observed were the siderophores desferrichrome and triacetylfusarinine C.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.