Abstract
Purpose of review
To provide both an overview and evidence of the potential etiology of oxidative DNA base damage and repair-signaling in chronic inflammation and histological changes associated with asthma.
Recent findings
Asthma is initiated/maintained by immunological, genetic/epigenetic and environmental factors. It is a world-wide health problem, as current therapies suppress symptoms rather than prevent/reverse the disease, largely due to gaps in understanding its molecular mechanisms. Inflammation, oxidative stress and DNA damage are inseparable phenomena, but their molecular roles in asthma pathogenesis are unclear. It was found that among oxidatively modified DNA bases, 8-oxoguanine (8-oxoG) is one of the most abundant, and its levels in DNA and body fluids are considered a biomarker of ongoing asthmatic processes. Free 8-oxoG forms a complex with 8-oxoguanine DNA glycosylase-1 (OGG1) and activates RAS-family GTPases that induce gene expression to mobilize innate and adaptive immune systems, along with genes regulating airway hyperplasia, hyper-responsiveness and lung remodeling in atopic and non-atopic asthma.
Summary
DNA’s integrity must be maintained to prevent mutation, so its continuous repair and downstream signaling “fuels” chronic inflammatory processes in asthma, and forms the basic mechanism whose elucidation will allow the development of new drug targets for the prevention/reversal of lung diseases.
Keywords: DNA repair, OGG1, 8-oxoG, inflammation, asthma
INTRODUCTION
Asthma is a complex chronic inflammatory lung disease, thought to result from the interaction between individuals’ genetic susceptibility and epigenetic changes induced by environmental factors. It is characterized by heterogeneity in pathogenesis, ventilatory/gas exchange impairment, mediated by multiple inflammatory mediators and oxidative stress induced by reactive oxygen species (ROS) [1,2,3,4].
Environmental agents acting singly or in combination primarily impact the surface of the airway, and when in excess, their interactions with airway epithelium and resident immune cells lead to increased ROS generation and both the onset and worsening of lung diseases, including asthma [1,2]. ROS are signaling molecules, and due to their reactivity, modify proteins, lipids, and DNA [5■]. In the DNA, one of the most common oxidation products is 8-oxo-7,8-dihydroguanine (8-oxoG) [6,7], which is preferentially repaired by the DNA base excision repair (BER) pathway [8], that utilizes glycosylases to excise the lesion by cleaving its N-glycosidic bond, followed by endonucleolytic cleavage and subsequent gap filling [8,9]. Although increases in oxidatively modified DNA bases are common, accumulation of 8-oxoG in the DNA has been linked to various inflammatory disease processes [9,10].
The genetics, development, imunopathogeneses, and pathophysiology of asthma have been elegantly reviewed by leading experts in the field [2,4,11,12,13,14,15,16,17,18]. It mostly agreed that asthma is a highly complex interplay among dysregulated airway epithelial, mast cells, basophils, dendritic cells, B and T cells, neutrophils and eosinophils, and is clinically manifested via a multitude of mediators leading to pathological pulmonary physiology. This review discusses present views the on role of OGG1-initiated repair of oxidatively damaged DNA and provides data to support a novel concept – the association of OGG1-BER with the expression of genes that are implicated in deregulation of the immune system and fundamental to histopathological changes in asthma.
GUANINE OXIDATION IS A MARKER OF ASTHMA
In DNA or RNA, the primary target of ROS is guanine, due to its lowest redox potential among nucleic acid bases [7]. Thus, 8-oxoG is one of the most abundant oxidatively modified DNA base lesions; it is estimated that up to 100,000 8-oxoG DNA lesions could be formed daily per cell [19]. The level of genomic 8-oxoG also correlates well with the dose and length of exposure, chemical composition, and physical nature of inhaled environmental agents [20,21,22,23,24,25,26]. For example, exposure to environmental pollutants and ROS generated by inflammatory cells significantly increased genomic 8-oxoG levels in the lung epithelium, resident macrophages, and peripheral blood monocytes, or of 8-oxoG base or its nucleoside levels in such body fluids as serum, urine, sputum and bronchoalveolar lavage fluid (BALF) [24,25,26,27,28,29,30,31,32].
In asthmatic patients and experimental animal models of asthma, one of the most referenced form of DNA base damage is 8-oxoG and its open-ringed form FapyG (2,6-diamino-4-hydroxy-5-formamidopyrimidine) [33,34,35,36,37,38,39]. Comprehensive studies have also shown that asthmatic patients have elevated levels of 8-oxoG in their sputum, serum, urine and BALF compared to controls [36,40,41]. Increased 8-oxoG levels in the genome and body fluids are traditionally considered markers of inflammation; however, it remains unknown as to whether these lesions contribute to the development, maintenance and/or progression of inflammatory processes underlying asthma.
PARADOXICAL ROLE OF GENOMIC 8-OXOGUANINE IN INNATE INFLAMMATION
To elucidate the role of genomic 8-oxoG and OGG1 in pathophysiological processes, Ogg1null mice were developed [42,43]. Intriguingly, the lack of OGG1 activity and consequent supraphysiological 8-oxoG levels did not affect embryonic development or life span. Despite high levels of 8-oxoG in the mitochondrial DNA (> 20-fold increase vs. wild-type), the mitochondria were functionally normal, with no detectable changes in maximal respiration rates or mitochondrial ROS generation [44]. Under chronic oxidative stress (such as that occurring in asthma), 8-oxoG levels increased by 250-fold in Ogg1null mice without apparent consequences, and there was no increased incidence of precancerous lesions or tumors including lungs [45].
Unexpectedly, Ogg1null mice show decreased inflammatory responses to bacterial infection [46], and increased resistance to lipopolysaccharide (LPS)-induced inflammation and organ dysfunction [47]. The decreased immune response was associated with significantly lower serum chemokine/cytokine levels and prolonged survival after LPS exposure, despite a marked increase in LPS-induced oxidative stress in the lungs, heart, kidneys, and liver. Moreover, sensitized Ogg1null showed significantly decreased contact hypersensitivity to oxazolone when compared to wild-type ones as shown by attenuation of chemokine/cytokine responses including interleukin (IL)-1β, tumor necrosis factor-alpha (TNFα), macrophage inflammatory protein-1 (MIP1-a), IL-4 and lower inflammatory cell accumulation [47]. These data raise the possibility that OGG1 itself and/or OGG1-BER could be the link to immune processes and associated diseases.
DECREASED ALLERGIC IMMUNE RESPONSES IN THE ABSENCE OF 8-OXOG REPAIR BY OGG1
Li and colleagues [48] documented that, compared to wild-type, ovalbumin (OVA) challenge of sensitized Ogg1null mice resulted in significantly lower levels of Th1 TNF-α, interferon-gamma (IFNγ), IL-2, IL-12, and T-helper-2 (Th2) cytokines (IL-4, IL-13), and IL-6, IL-17 levels and inflammatory cell infiltration in lung tissues due to decreased NF-κB activation after OVA challenge. In lung epithelial cells, OGG1 downregulation led to both decreased ROS generation and higher IFN-γ production. The authors concluded that OGG1 may influence airway inflammation by regulating the cellular oxidative metabolism [48].
Environmental pollutants primarily affect the epithelium and its constituent cells [2,49,50]. Therefore, Bacsi and colleagues [51] investigated whether OGG1–initiated repair of genomic 8-oxoG in the airway epithelium impacted innate and allergic immune responses. OGG1 was therefore ablated from the airway epithelium of ragweed pollen grain extract (RWPE)-sensitized animals [51] before RWPE challenge, which-induced significantly lower allergic inflammatory responses as determined by the expression of Th2 cytokines, the number of eosinophils recruited to airways, epithelial metaplasia, and airway hyperresponsiveness (AHR). In contrast, challenging OGG1-proficient lungs with RWPE led to a robust innate and late allergic inflammation [52,53,54].
Besides protein allergens, pollens carry a myriad of molecules with a variety of biological functions, including an NADPH oxido-reductase that generates ROS upon exposure. Indeed, ragweed and 39 other pollens tested induce ROS in the airways, increase genomic 8-oxoG levels, and activate OGG1-BER before innate and allergic inflammation [54]. Ablation of NADPH oxidase activity prevented RWPE challenge-induced recruitment of neutrophils and eosinophils, airway hyperplasia, and AHR [54]. Taken together, these results suggest that oxidative stress, as well as oxidative DNA damage/repair via OGG1 generate activation signals relevant to a robust inflammatory response in sensitized subjects.
ACTIVATION OF SMALL GTPASES BY OGG1-INITIATED DNA REPAIR
The linkage between OGG1-BER and innate/allergic inflammation was not obvious until recent observations showing that OGG1 binds its repair product 8-oxoG base with high affinity, and the resulting complex (OGG1•8-oxoG) physically interacts with small GTPases [55■■, 56]. Importantly, the OGG1•8-oxoG complex caused GDP→GTP exchange in canonical RAS family proteins, and so functions as a guanine nucleotide exchange factor (GEF) in a manner similar to other GEFs [57].
The high sequence homology among the RAS and RHO GTPases [58], provided the rationale to examine whether OGG1-BER and the consequent formation of a OGG1•8-oxoG complex activate the RHO family member RAC1. Hajas and colleagues reported that OGG1•8-oxoG physically interacts with guanine nucleotide-free and GDP-bound RAC1. This interaction resulted in rapid GDP→GTP exchange, indicating that OGG1•8-oxoG functions as a prototypic GEF [55■■,59■]. Luo et al., 2014 provided further insights into the biological consequences of OGG1-initiated release of 8-oxoG from DNA [60■]. These authors demonstrated that only OGG1-expressing cells display increased activation of RHOA-GTPase in oxidatively stressed cells. These observations were intriguing, as many small GTPases are redox-sensitive, and ROS have an effect similar to GEFs in that they modulate guanine nucleotide binding of GTPases [58], which could be observed in OGG1-expressing but not OGG1-deficient cells.
The biological significance of the above observation became evident from studies showing that increasing the cellular 8-oxoG level by adding it to cells, into airways, or activating OGG1-BER in cellulo, rapidly increased the GTP-bound levels of RAS, RAC1 and RHOA GTPases. It has been shown that RAS-GTPases activate downstream targets, including mitogen-activated protein kinase kinase, and extracellular signal-regulated kinase, and the latter’s nuclear translocation [56,61■]. Activated RAC1 facilitated a spatially controlled increase in cellular ROS levels via a nuclear membrane-associated type 4 NADPH oxidase [59■]. Moreover, RHOA-GTP induced smooth muscle alpha-actin synthesis and its polymerization into stress fibers in cultured cells and lungs [60■]. It was also shown that in the airway epithelium OGG1-BER is a prerequisite for GDP→GTP exchange, KRAS-GTP-driven signaling via mitogen-activated-, phosphoinositide-3-, and mitogen- and stress-activated protein kinases, leading to activation of the NF-κB pathway by inducing RelA phosphorylation at Serine276 and nuclear translocation [62■■,63■■]. These events are essential for NF-κB-orchestrated activation of the pro-inflammatory innate and adaptive networks, including C-C and C-X-C chemokines and ILs expression leading to mucosal airway inflammation [64,65].
ASTHMA SIGNATURE GENES INDUCED BY 8-OXOG BASE CHALLENGE OF AIRWAYS
The perturbed, primarily NF-κB-driven sub-networks change the sensitivity and response to stimuli of airway epithelial, mast, and dendritic cells, as well as basophils, lymphocytes, neutrophils and eosinophils [64,65]. The multitude of mediators they generate impacts not only the immune system but also airway smooth muscle, vascular endothelium, nerve and other cell types leading to deregulation of cellular interactions and clinical manifestations of asthma [2,13,14,15,16,17,18,66].
In examining the relevance of OGG1-BER to immune deregulation and pulmonary pathophysiology, airways were challenged with the OGG1-BER product 8-oxoG base, and resulting gene expression was determined via RNA sequencing (RNA-Seq). 8-oxoG challenge mimics the impact of OGG1-BER [56,67] and excludes ROS signaling that has been associated with the pathogenesis of asthma [5■,68]. In order to define the primary effect of 8-oxoG challenge, experiments were restricted to early time-points (30, 60, 120 min). From 2 h on, high levels of TNFα, C-C and C-X-C chemokines were present in the lungs. RNA-Seq analyses showed changes in the expression of an unexpectedly large number of transcripts (mRNAs, miRNAs and non-coding RNAs – 18,874). When threshold values of mRNA levels were set to ≥3-fold, a total of 2,381 genes were modulated (983 up- and 1398 downregulated). Gene ontology analysis (PANTHER database) showed that coding transcripts were related to various biological processes, among which immune system processes and inflammation were overrepresented (data not shown).
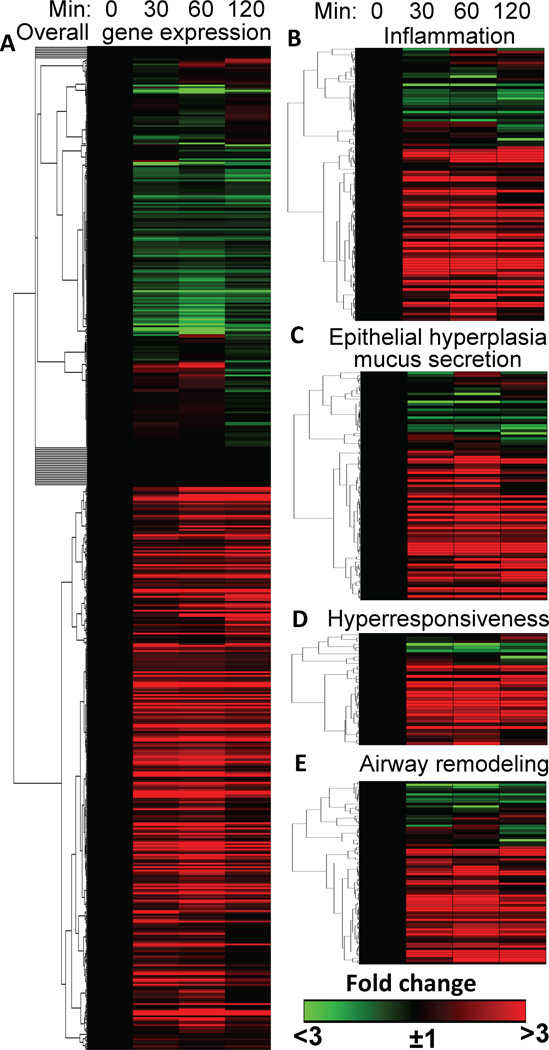
To elucidate the relevance of 8-oxoG challenge-induced gene expression to immune deregulation and asthma pathophysiology, we inserted our 2,381 genes into a set of 572 genes upregulated (http://www.jci.org/cgi/content/full/111/12/1863/DC1) in A. fumigatus- and OVA-induced experimental mouse models of asthma [69,70] and hierarchically clustered them. Fig. 1A shows that 8-oxoG challenge upregulated 344 genes essential in experimental asthma (119 were downregulated, and 109 unchanged; Fig. 1A). These unexpected data were further examined for the relevance of 8-oxoG–induced genes to immune and histopathological changes induced by A. fumigatus and OVA. 8-oxoG challenge up-regulated (>3-fold) 85 out of 101 genes associated with immune deregulation by A. fumigatus and OVA challenge. These data are visually depicted in Fig. 1B. Examples include C-C motif chemokines [e.g., MIP-1-α,-β, MIP-1-related-protein 6, MIP-1-γ, Th2-attracting C-C motif chemokine ligand-17 and -22, C-X-C motif chemokine ligand-1,-2,-5,-9, their receptors-1,4, and 5, IL-1-α,-1-β, IL-17, IL-6, and IL receptor-2,-3, -ra2]. Moreover, 8-oxoG challenge upregulated (by 3- to >50-fold) the expression of genes encoding proteins important in inflammatory cell attachment, migration (e.g. C-X-C motif chemokine ligand-1,-2; transcription factor activating protein 3, chemokine ligand 22, integrin alpha-M, integrin beta chain beta 2, mannose receptor gene-1, serum soluble E-selectin, thrombospondin-1 protein), T cell development and functions (e.g., plasminogen activator inhibitor-2, TNF-α-induced protein 6 and 9), B-cell responses (e.g. membrane-spanning 4-domain protein), allergen-induced cytokine release (e.g. T-cell surface glycoprotein CD28, granzyme B, monocyte differentiation antigen CD14, secretory leukocyte protease inhibitor protein, Tenascin C, prostaglandin I receptor family) (Fig. 1B).
FIGURE 1.
Visual depiction of 8-oxoG-challenge-induced gene expression documented to be signatures of A. fumigatus-/OVA-induced experimental asthma. A, 8-oxoG challenge alters the expression of genes that were also up-regulated in A. fumigatus-/OVA-induced experimental asthma. B, 8-oxoG challenge modulates expression of the inflammatory genes induced during A. fumigatus-/OVA-induced allergic inflammation and those implicated in C, airway epithelial hyperplasia and mucus production; D, airway hyper-responsiveness and E, airway remodeling. Lungs of naïve mice were challenged with 8-oxoG base, RNA was extracted, pooled from five mice for each time point, and subjected to RNA-sequence analysis (Illumina HiSeq 1000 sequencing system, UTMB Next-Generation Sequencing Core Facility). For data specificity and validity, three time points were utilized. Genes were hierarchically clustered and heat maps generated using the matrix visualization and analysis platform GENE-E (Broad Institute; Cambridge, MA). The threshold values were set to ±3-fold change in RNA levels to identify genes involved in immuno and pulmonary pathophysiology. Animal experiments were performed according to the NIH Guide for the Care and Use of Experimental Animals and approved by the UTMB IACUC (no. A0807044).
Given that OGG1•8-oxoG activates RAS, RAC1 and RHOA [55■■,56,59■,60■], it was not entirely unexpected that challenging lungs with 8-oxoG increased the expression of 78 of 92 genes involved in epithelial hyperplasia and mucus secretion (Fig. 1C); 28 of 35 genes linked to airway AHR (Fig. 1D) and 58 of 76 genes important in airway remodeling processes were up-regulated (Fig. 1E). These analyses suggest that OGG1•8-oxoG-induced gene expression is similar to that responsible for immune- and pulmonary pathophysiology in experimental models of allergic asthma induced by A. fumigatus or OVA. A detailed gene list is shown in Supplementary Materials (Table 1).
OGG1-BER-DRIVEN GENE EXPRESSION IN HUMAN ASTHMA
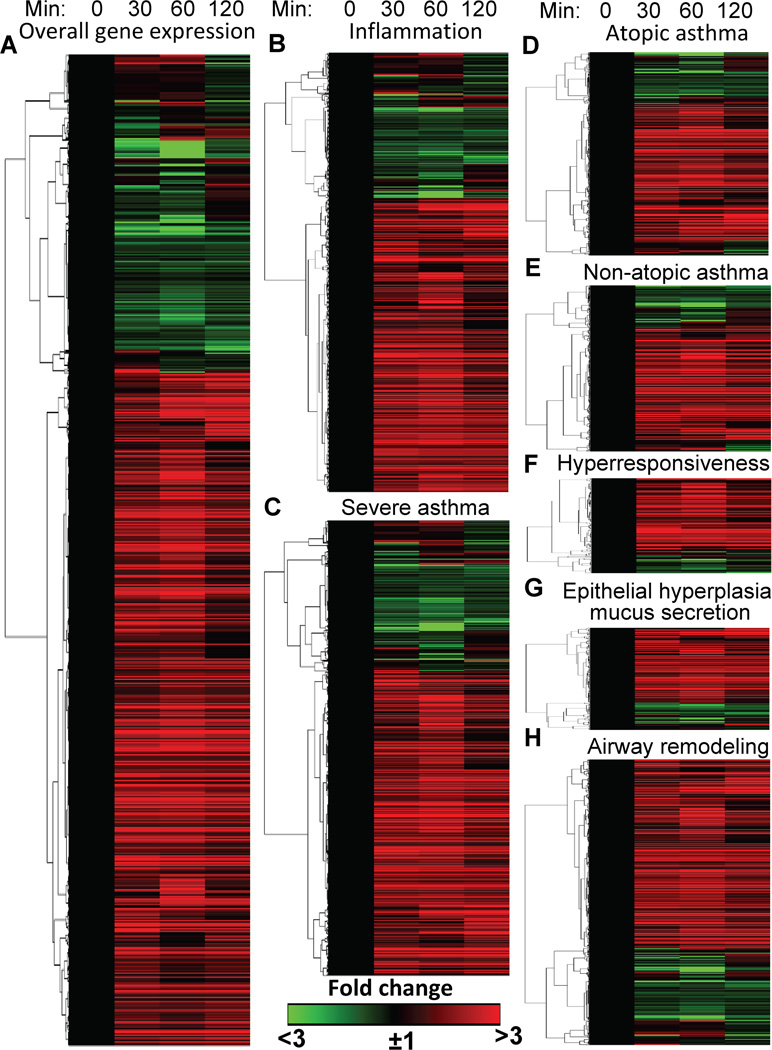
Next, it was examined whether 8-oxoG challenge-induced changes in gene expression are similar to those previously associated with immune deregulation and pulmonary pathology in human asthma. To address this, the human equivalent of the mouse gene list was created and compared to a list of human asthma-related genes identified and documented in the GeneCards database (www.genecards.org). GeneCards’ database is integrated from the Human Genome Organization, Gene Nomenclature Committee, European Bioinformatics Institute, and National Center for Biotechnology Information and Database of Allergy and Asthma Biomarkers and others. Strikingly, of the 2,381 8-oxoG challenge-regulated genes 1,051 were previously linked to human asthma (731 were upregulated, 169 downregulated and 151 unchanged; Fig. 2A). To better define gene expression resulting from pulmonary challenge with 8-oxoG, genes associated with inflammation, atopic, non-atopic and severe asthma, as well as AHR, epithelial hyperplasia, and remodeling were further analyzed. Six-hundred and fifty-nine genes were identified as immune response-related, of which 454 were up- and 100 downregulated (95 genes were unaltered) by 8-oxoG challenge (Fig. 2B). 758 genes shown to be involved in severe asthma of which 519 were up-, 122 down-regulated and 117 were unaltered (Fig. 2C). Ninety percent of the 320 atopic and 93% of 281 genes up-regulated in non-atopic asthma by 8-oxoG exposure (Fig. 2D,E). Further analysis revealed that >82% of genes previously associated with AHR (158; Fig. 2F), and mucus production/secretion (172 genes; Fig. 2G) were up-regulated by 8-oxoG challenge of airways. GeneCards’ database contains 540 genes linked to changes in molecular and cellular (airway smooth muscle, epithelium) composition, and extracellular matrix during airway remodeling [71]. 8-oxoG challenge up- (392 genes), down- (73 genes) regulated and did not change 85 of them (Fig. 2H). These results suggest that OGG1-BER-associated gene expression regulates pulmonary inflammation and cellular/tissue pathology in human asthma. A detailed gene list is in Supplementary Materials (Table 2).
FIGURE 2.
Visual depiction of 8-oxoG challenge-induced alterations in gene expression is similar to the signatures of human asthma. RNA-Seq analysis was carried out as in the legend to Figure 1. Gene sets associated with deregulation of innate/adaptive immune system in asthma (atopic, non-atopic, severe asthma) as well as airway hyperresponsiveness, hyperlasia-mucus secretion and remodeling were defined by GeneCards’ database. Genes were hierarchically clustered using the GENE-E analysis platform.
CONCLUSION
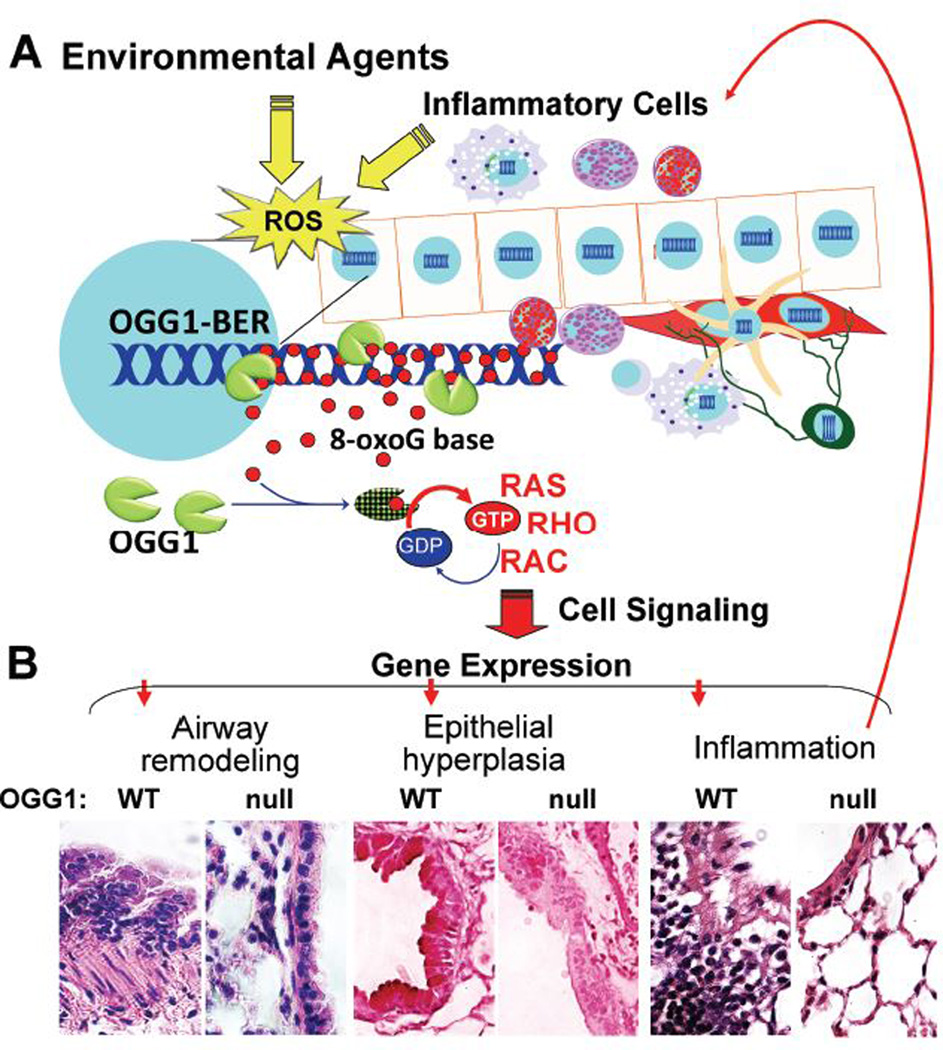
A close association between intrahelical 8-oxoG and 8-oxoG base levels in the body fluids of asthmatic subjects has been extensively documented, implying that genomic integrity is continually maintained via OGG1-BER. A review of the literature and data introduced here strongly suggest that OGG1-BER and the consequent OGG1•8-oxoG-driven signaling induces gene expression implicated in innate immune and adaptive immune regulation, AHR, epithelial hyperplasia, mucin production and remodeling (Fig. 3). Together these data imply a role of OGG1-BER signaling as a basic molecular mechanism underlying gene expression in atopic, non-atopic and severe asthma. In support of this, the resistance of OGG1-null mice to deregulation of the innate and adaptive immune systems and decreased pulmonary pathology further underlines the importance of OGG1-BER. Importantly, data provided here point to a novel mechanism – a role of OGG1-BER in the initiation and maintenance of chronic inflammatory disease conditions, not only in the lungs, but other organs. A greater understanding of the molecular mechanisms of OGG1-BER signaling will be essential to the development of better therapeutic modalities to prevent/reverse related disease processes.
FIGURE 3.
A proposed role of OGG1-initiated DNA BER in the maintenance of chronic inflammatory processes in asthma. Environmental exposures and inflammatory cells generate ROS and intrahelical 8-oxoG. 8-OxoG is repaired via OGG1-BER, then bound by OGG1 to form OGG1•8-oxoG, which activates small GTPases. Signaling downstream from RAS, RHO and RAC induces gene expression implicated in deregulation of the innate and adaptive immune systems. Recruited inflammatory cells generate ROS, and increase DNA damage/repair, leading to a vicious cycle of chronic inflammation. B, Patholohistological changes resulting from OGG1-initiated DNA repair. Sensitized OGG1 null and wild-type mice were RWPE-challenged, their lungs excised, sectioned and stained to demonstrate histological features of experimental asthma. Panel right to left: Inflammatory cells in sub-epithelium, airway epithelial hyperplasia, and increased smooth muscle mass in wild-type (left panels) vs. Ogg1null (right panels) mice. WT, wild-type, null, Ogg1null
Supplementary Material
KEY POINTS.
Asthma is a complex interplay among aberrant pulmonary responses to environmental challenge and intrinsic determinants
Inflammation in asthma and oxidative damage to DNA are inseparable twins
Continuous OGG1-driven DNA repair-signaling “fuels” unscheduled gene expression via small GTPases for inflammation and histological changes in asthma
Elucidating the involved molecular pathways will aid to identify novel targets for prevention of pulmonary inflammation and histological changes
Acknowledgements
We thank Dr. David Konkel (Institute for Translational Sciences) and Mardelle Susman for their scientific input and critically editing the manuscript.
Funding: This work was supported by grants NIEHS RO1 ES018948 (IB), NIAID/AI062885 (IB), P30 ES06676 (IB, Director: Dr. Elferink), N01HV00245 (IB, Director: Dr. A. Kurosky) and NIEHS T32 ES007254 (LA, Director: Dr. B. Ameredes).
Footnotes
Conflict of interest
The authors declare that no conflict of interest exists
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual review period, have been highlighted as
■ of special interest
■■ of outstanding interest
- 1.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 2.Holgate ST. The sentinel role of the airway epithelium in asthma pathogenesis. Immunol Rev. 2011;242:205–219. doi: 10.1111/j.1600-065X.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 3.Lotvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127:355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. Reactive oxygen species function as cellular signaling molecules, a concept which has been well described but is not fully accepted. This uncertainty is associated with the apparent paradox between the specificity that is needed for signaling and their indiscriminate reactivity with cellular molecules
- 6.Margolin Y, Cloutier JF, Shafirovich V, Geacintov NE, Dedon PC. Paradoxical hotspots for guanine oxidation by a chemical mediator of inflammation. Nat Chem Biol. 2006;2:365–366. doi: 10.1038/nchembio796. [DOI] [PubMed] [Google Scholar]
- 7.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. Sequence and stacking dependence of 8-oxoguanine oxidation: Comparison of one-electron vs. singlet oxygen mechanisms. Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 8.Mitra S. DNA glycosylases: specificity and mechanisms. Prog Nucleic Acid Res Mol Biol. 2001;68:189–192. doi: 10.1016/s0079-6603(01)68099-1. [DOI] [PubMed] [Google Scholar]
- 9.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587–596. doi: 10.1016/j.freeradbiomed.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busse WW. The role of allergy in disease. Immunol Rev. 2011;242:5–9. doi: 10.1111/j.1600-065X.2011.01036.x. [DOI] [PubMed] [Google Scholar]
- 12.Barnes KC. Genetic studies of the etiology of asthma. Proc Am Thorac Soc. 2011;8:143–148. doi: 10.1513/pats.201103-030MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev. 2011;242:31–50. doi: 10.1111/j.1600-065X.2011.01020.x. [DOI] [PubMed] [Google Scholar]
- 14.Peden DB. The role of oxidative stress and innate immunity in O(3) and endotoxin-induced human allergic airway disease. Immunol Rev. 2011;242:91–105. doi: 10.1111/j.1600-065X.2011.01035.x. [DOI] [PubMed] [Google Scholar]
- 15.Minnicozzi M, Sawyer RT, Fenton MJ. Innate immunity in allergic disease. Immunol Rev. 2011;242:106–127. doi: 10.1111/j.1600-065X.2011.01025.x. [DOI] [PubMed] [Google Scholar]
- 16.Koziol-White CJ, Panettieri RA., Jr Airway smooth muscle and immunomodulation in acute exacerbations of airway disease. Immunol Rev. 2011;242:178–185. doi: 10.1111/j.1600-065X.2011.01022.x. [DOI] [PubMed] [Google Scholar]
- 17.Proud D, Leigh R. Epithelial cells and airway diseases. Immunol Rev. 2011;242:186–204. doi: 10.1111/j.1600-065X.2011.01033.x. [DOI] [PubMed] [Google Scholar]
- 18.Bhakta NR, Woodruff PG. Human asthma phenotypes: from the clinic, to cytokines, and back again. Immunol Rev. 2011;242:220–232. doi: 10.1111/j.1600-065X.2011.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 20.Kirkham P, Rahman I. Oxidative stress in asthma and COPD: antioxidants as a therapeutic strategy. Pharmacol Ther. 2006;111:476–494. doi: 10.1016/j.pharmthera.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Nehls P, Seiler F, Rehn B, Greferath R, Bruch J. Formation and persistence of 8-oxoguanine in rat lung cells as an important determinant for tumor formation following particle exposure. Environ Health Perspect. 1997;105(Suppl 5):1291–1296. doi: 10.1289/ehp.97105s51291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunec J. ESCODD: European Standards Committee on Oxidative DNA Damage. Free Radic Res. 1998;29:601–608. doi: 10.1080/10715769800300651. [DOI] [PubMed] [Google Scholar]
- 23.Seiler JP. Chirality--from molecules to organisms. Arch Toxicol Suppl. 1995;17:491–498. doi: 10.1007/978-3-642-79451-3_42. [DOI] [PubMed] [Google Scholar]
- 24.Loft S, Poulsen HE, Vistisen K, Knudsen LE. Increased urinary excretion of 8-oxo-2'-deoxyguanosine, a biomarker of oxidative DNA damage, in urban bus drivers. Mutat Res. 1999;441:11–19. doi: 10.1016/s1383-5718(99)00034-0. [DOI] [PubMed] [Google Scholar]
- 25.Son J, Pang B, McFaline JL, Taghizadeh K, Dedon PC. Surveying the damage: the challenges of developing nucleic acid biomarkers of inflammation. Mol Biosyst. 2008;4:902–908. doi: 10.1039/b719411k. [DOI] [PubMed] [Google Scholar]
- 26.Barbato DL, Tomei G, Tomei F, Sancini A. Traffic air pollution and oxidatively generated DNA damage: can urinary 8-oxo-7,8-dihydro-2-deoxiguanosine be considered a good biomarker? A meta-analysis. Biomarkers. 2010;15:538–545. doi: 10.3109/1354750X.2010.493974. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki J, Inoue Y, Suzuki S. Changes in the urinary excretion level of 8-hydroxyguanine by exposure to reactive oxygen-generating substances. Free Radic Biol Med. 1995;18:431–436. doi: 10.1016/0891-5849(94)00152-a. [DOI] [PubMed] [Google Scholar]
- 28.Lee MW, Chen ML, Lung SC, Tsai CJ, Yin XJ, et al. Exposure assessment of PM2.5 and urinary 8-OHdG for diesel exhaust emission inspector. Sci Total Environ. 2010;408:505–510. doi: 10.1016/j.scitotenv.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Malayappan B, Garrett TJ, Segal M, Leeuwenburgh C. Urinary analysis of 8-oxoguanine, 8-oxoguanosine, fapy-guanine and 8-oxo-2'-deoxyguanosine by high-performance liquid chromatography-electrospray tandem mass spectrometry as a measure of oxidative stress. J Chromatogr A. 2007;1167:54–62. doi: 10.1016/j.chroma.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Andreoli R, Protano C, Manini P, De Palma G, Goldoni M, et al. Association between environmental exposure to benzene and oxidative damage to nucleic acids in children. Med Lav. 2012;103:324–337. [PubMed] [Google Scholar]
- 31.Tsurudome Y, Hirano T, Yamato H, Tanaka I, Sagai M, et al. Changes in levels of 8-hydroxyguanine in DNA, its repair and OGG1 mRNA in rat lungs after intratracheal administration of diesel exhaust particles. Carcinogenesis. 1999;20:1573–1576. doi: 10.1093/carcin/20.8.1573. [DOI] [PubMed] [Google Scholar]
- 32.Moller P, Danielsen PH, Jantzen K, Roursgaard M, Loft S. Oxidatively damaged DNA in animals exposed to particles. Crit Rev Toxicol. 2013;43:96–118. doi: 10.3109/10408444.2012.756456. [DOI] [PubMed] [Google Scholar]
- 33.Zeyrek D, Cakmak A, Atas A, Kocyigit A, Erel O. DNA damage in children with asthma bronchiale and its association with oxidative and antioxidative measurements. Pediatr Allergy Immunol. 2009;20:370–376. doi: 10.1111/j.1399-3038.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 34.Hasbal C, Aksu BY, Himmetoglu S, Dincer Y, Koc EE, et al. DNA damage and glutathione level in children with asthma bronchiale: effect of antiasthmatic therapy. Pediatr Allergy Immunol. 2010;21:e674–678. doi: 10.1111/j.1399-3038.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- 35.Al-Afaleg NO, Al-Senaidy A, El-Ansary A. Oxidative stress and antioxidant status in Saudi asthmatic patients. Clin Biochem. 2011;44:612–617. doi: 10.1016/j.clinbiochem.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Proklou A, Soulitzis N, Neofytou E, Rovina N, Zervas E, et al. Granule cytotoxic activity and oxidative DNA damage in smoking and nonsmoking patients with asthma. Chest. 2013;144:1230–1237. doi: 10.1378/chest.13-0367. [DOI] [PubMed] [Google Scholar]
- 37.Deslee G, Woods JC, Moore C, Conradi SH, Gierada DS, et al. Oxidative damage to nucleic acids in severe emphysema. Chest. 2009;135:965–974. doi: 10.1378/chest.08-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igishi T, Hitsuda Y, Kato K, Sako T, Burioka N, et al. Elevated urinary 8-hydroxydeoxyguanosine, a biomarker of oxidative stress, and lack of association with antioxidant vitamins in chronic obstructive pulmonary disease. Respirology. 2003;8:455–460. doi: 10.1046/j.1440-1843.2003.00490.x. [DOI] [PubMed] [Google Scholar]
- 39.Caramori G, Adcock IM, Casolari P, Ito K, Jazrawi E, et al. Unbalanced oxidant-induced DNA damage and repair in COPD: a link towards lung cancer. Thorax. 2011;66:521–527. doi: 10.1136/thx.2010.156448. [DOI] [PubMed] [Google Scholar]
- 40.Fitzpatrick AM, Teague WG, Burwell L, Brown MS, Brown LA. Glutathione oxidation is associated with airway macrophage functional impairment in children with severe asthma. Pediatr Res. 2011;69:154–159. doi: 10.1203/PDR.0b013e3182026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortoul TI, Valverde M, Lopez Mdel C, Bizarro P, Lopez I, et al. Single-cell gel electrophoresis assay of nasal epithelium and leukocytes from asthmatic and nonasthmatic subjects in Mexico City. Arch Environ Health. 2003;58:348–352. [PubMed] [Google Scholar]
- 42.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, et al. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 45.Arai T, Kelly VP, Minowa O, Noda T, Nishimura S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis. 2002;23:2005–2010. doi: 10.1093/carcin/23.12.2005. [DOI] [PubMed] [Google Scholar]
- 46.Touati E, Michel V, Thiberge JM, Ave P, Huerre M, et al. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter. 2006;11:494–505. doi: 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 47.Mabley JG, Pacher P, Deb A, Wallace R, Elder RH, et al. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. Faseb J. 2005;19:290–292. doi: 10.1096/fj.04-2278fje. [DOI] [PubMed] [Google Scholar]
- 48.Li G, Yuan K, Yan C, Fox J, 3rd, Gaid M, et al. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic Biol Med. 2012;52:392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swamy M, Jamora C, Havran W, Hayday A. Epithelial decision makers: in search of the 'epimmunome'. Nat Immunol. 2010;11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Locksley RM. Asthma and allergic inflammation. Cell. 2010;140:777–783. doi: 10.1016/j.cell.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bacsi A, Aguilera-Aguirre L, Szczesny B, Radak Z, Hazra TK, et al. Down-regulation of 8-oxoguanine DNA glycosylase 1 expression in the airway epithelium ameliorates allergic lung inflammation. DNA Repair (Amst) 2013;12:18–26. doi: 10.1016/j.dnarep.2012.10.002. This study documents that silencing Ogg1 expression and thereby 8-oxoG repair in the airway epithelium nearly prevented inflammatory response after ragweed pollen extract challenge of sensitized mice, as determined by expression of Th2 cytokines, eosinophilia, epithelial methaplasia, and airway hyperresponsiveness
- 52.Bacsi A, Choudhury BK, Dharajiya N, Sur S, Boldogh I. Subpollen particles: carriers of allergenic proteins and oxidases. J Allergy Clin Immunol. 2006;118:844–850. doi: 10.1016/j.jaci.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bacsi A, Dharajiya N, Choudhury BK, Sur S, Boldogh I. Effect of pollen-mediated oxidative stress on immediate hypersensitivity reactions and late-phase inflammation in allergic conjunctivitis. J Allergy Clin Immunol. 2005;116:836–843. doi: 10.1016/j.jaci.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, et al. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. J Clin Invest. 2005;115:2169–2179. doi: 10.1172/JCI24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pandita TK. Unraveling the novel function of the DNA repair enzyme 8-oxoguanine-DNA glycosylase in activating key signaling pathways. Free Radic Biol Med. 2014;73:439–440. doi: 10.1016/j.freeradbiomed.2014.05.013. A commentary on novel discoveries —activation by 8-oxoguanine DNA glycosylase-1:8-oxoguanine base complex of small GTPases RAS, RHO and RAC1, master regulators of gene expression driving inflammatory processes, and pathophysiological changes observed in asthma and airway remodeling
- 56.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, et al. Activation of Ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J Biol Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ksionda O, Limnander A, Roose JP. RasGRP Ras guanine nucleotide exchange factors in cancer. Front Biol (Beijing) 2013;8:508–532. doi: 10.1007/s11515-013-1276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heo J. Redox control of GTPases: from molecular mechanisms to functional significance in health and disease. Antioxid Redox Signal. 2011;14:689–724. doi: 10.1089/ars.2009.2984. [DOI] [PubMed] [Google Scholar]
- 59. Hajas G, Bacsi A, Aguilera-Aguirre L, Hegde ML, Tapas KH, et al. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free Radic Biol Med. 2013;61:384–394. doi: 10.1016/j.freeradbiomed.2013.04.011. This report documents that, in the presence of the 8-oxoG base, 8-oxoguanine DNA glycosylase-1 (OGG1) physically interacts with guanine nucleotide-free and GDP-bound RAC1 protein. This interaction results in rapid GDP→GTP, but not GTP→GDP, exchange in vitro and intracellularly. In turn, RAC1-GTP mediates an increase in ROS levels via nuclear membrane-associated NADPH oxidase type 4
- 60. Luo J, Hosoki K, Bacsi A, Radak Z, Hegde ML, et al. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and alpha-smooth muscle actin polymerization. Free Radic Biol Med. 2014;73:430–438. doi: 10.1016/j.freeradbiomed.2014.03.030. This study show for the first time that 8-oxoguanine DNA glycosylase-1 (OGG1) physically interacts with RHOA GTPase and, in the presence of 8-oxoG base, increases Rho-GTP levels in cultured cells and lungs, which mediates α-smooth muscle actin (α-SMA) polymerization into stress fibers and increases the level of α-SMA in insoluble cellular and lung fractions.
- 61.German P, Szaniszlo P, Hajas G, Radak Z, Bacsi A, et al. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1-initiated DNA base excision repair. DNA Repair (Amst) 2013;12:856–863. doi: 10.1016/j.dnarep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ba X, Bacsi A, Luo J, Aguilera-Aguirre L, Zeng X, et al. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J Immunol. 2014;192:2384–2394. doi: 10.4049/jimmunol.1302472. For the first time this paper documents that exposure of cells to TNF-α altered cellular redox increased the 8-oxoG level in DNA, and recruited 8-oxoguanine DNA glycosylase-1 (OGG1) to promoter sequences. Promoter-associated OGG1 then enhanced NF-κB/RelA binding to cis-elements and facilitated the recruitment of specificity protein 1 (SP1), transcription initiation factor II-D (TFIID), and phospho-RNA polymerase II, resulting in the rapid expression of chemokines and cytokines and of inflammatory cell accumulation in mouse airways.
- 63. Aguilera-Aguirre L, Bacsi A, Radak Z, Hazra TK, Mitra S, Sur S, Brasier AR, Ba X, Boldogh I. Innate Inflammation Induced by 8-Oxoguanine DNA Glycosylase-1-KRAS-NF-κB Pathway Journal of Immunology # 14-01625-FL. 2014 doi: 10.4049/jimmunol.1401625. In Press. It is reported that the 8-oxoguanine DNA glycosylase-1-intiated repair of oxidatively damaged DNA is a prerequisite event for GDP→GTP exchange, KRAS-GTP-driven signaling via MAP-, PI3-, and MS kinases for NF-κB activation, pro-inflammatory chemokine/cytokine expression, and inflammatory cell recruitment to the airways.
- 64.Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 65.Kalita M, Tian B, Gao B, Choudhary S, Wood TG, et al. Systems approaches to modeling chronic mucosal inflammation. Biomed Res Int. 2014;2013:505864. doi: 10.1155/2013/505864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis MJ, Short AL, Lewis KE. Autonomic nervous system control of the cardiovascular and respiratory systems in asthma. Respir Med. 2006;100:1688–1705. doi: 10.1016/j.rmed.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 67.Hajas G, Bacsi A, Aguilerra-Aguirre L, German P, Radak Z, et al. Biochemical identification of a hydroperoxide derivative of the free 8-oxo-7,8-dihydroguanine base. Free Radic Biol Med. 2012;52:749–756. doi: 10.1016/j.freeradbiomed.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang L, Diaz PT, Best TM, Stimpfl JN, He F, et al. Molecular characterization of redox mechanisms in allergic asthma. Ann Allergy Asthma Immunol. 2014;113:137–142. doi: 10.1016/j.anai.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 69.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, et al. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–1824. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 71.Hirota N, Martin JG. Mechanisms of airway remodeling. Chest. 2013;144:1026–1032. doi: 10.1378/chest.12-3073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.