Abstract
Treatment with recombinant human growth hormone (rhGH) has been consistently reported to induce transcriptional changes in various human tissues including peripheral blood. For other hormones it has been shown that the induction of such transcriptional effects is conferred or at least accompanied by DNA-methylation changes. To analyse effects of short term rhGH treatment on the DNA-methylome we investigated a total of 24 patients at baseline and after 4-day rhGH stimulation. We performed array-based DNA-methylation profiling of paired peripheral blood mononuclear cell samples followed by targeted validation using bisulfite pyrosequencing. Unsupervised analysis of DNA-methylation in this short-term treated cohort revealed clustering according to individuals rather than treatment. Supervised analysis identified 239 CpGs as significantly differentially methylated between baseline and rhGH-stimulated samples (p<0.0001, unadjusted paired t-test), which nevertheless did not retain significance after adjustment for multiple testing. An individualized evaluation strategy led to the identification of 2350 CpG and 3 CpH sites showing methylation differences of at least 10% in more than 2 of the 24 analyzed sample pairs. To investigate the long term effects of rhGH treatment on the DNA-methylome, we analyzed peripheral blood cells from an independent cohort of 36 rhGH treated children born small for gestational age (SGA) as compared to 18 untreated controls. Median treatment interval was 33 months. In line with the groupwise comparison in the short-term treated cohort no differentially methylated targets reached the level of significance in the long-term treated cohort. We identified marked intra-individual responses of DNA-methylation to short-term rhGH treatment. These responses seem to be predominately associated with immunologic functions and show considerable inter-individual heterogeneity. The latter is likely the cause for the lack of a rhGH induced homogeneous DNA-methylation signature after short- and long-term treatment, which nevertheless is well in line with generally assumed safety of rhGH treatment.
Introduction
Recombinant human growth hormone (rhGH) has been introduced in 1985 for the treatment of children with short stature [1]. Current indications include among others growth hormone (GH) deficiency, children born small for gestational age (SGA), Turner syndrome, Prader–Willi syndrome, chronic renal insufficiency and short stature homeobox (SHOX) gene deficiency [2]. While some authors reported rhGH treatment to be associated with adverse side effects like increased risk for leukemia or secondary neoplasms [3,4,5,6,7], this has not been confirmed by others [8,9,10].
Physiologically, GH is secreted from pituitary somatotrophic cells. It regulates postnatal growth through a complex network of direct and indirect effects involving amongst others the stimulation of insulin-like growth factor 1 (IGF1) synthesis and release in the liver [11]. Analogous to the physiologic GH effect, treatment with rhGH enhances circulating IGF1 levels. The rise of IGF1 concentration in the peripheral blood following rhGH treatment (so called IGF1 generation test, IGFGT) serves, thus, in the clinical setting as basic laboratory parameter to estimate the response to rhGH treatment in vivo [12].
In order to identify in vivo targets of the GH-axis and eventually develop better prediction models to monitor response to rhGH therapy a series of microarray-based gene expression studies has been conducted [13,14,15]. Indeed, albeit variable in design and of restricted power due to small sample sizes, transcriptional changes following rhGH treatment were consistently observed in peripheral blood mononuclear cells (PBMCs).
Determination of gene expression patterns involves epigenetic modifications, including DNA-methylation [16,17]. For hormones other than GH it has been shown that transcriptional effects induced by these hormones are conferred or at least accompanied by DNA-methylation changes [18,19,20]. Altered DNA-methylation is also a feature of many diseases, including diabetes and cancer [21,22]. Thus, the characterisation of epigenetic modifications induced by rhGH treatment might not only contribute to the understanding of the GH response but could also provide clues for potential adverse effects of rhGH treatment.
To analyse effects of rhGH treatment on the DNA-methylome, we performed an array-based analysis of DNA-methylation in two independent cohorts of 24 and 36 children treated short-term and long-term with rhGH, respectively.
Material and Methods
Ethics Statement
The study has been approved by the Ethics Committee of the Medical Faculty of the Christian-Albrechts-University Kiel (AZ B305/08, D401/08, Amendment 06/03/2012 and AZ D422/07). All patients and in minors their caretakers gave written informed consent upon inclusion into the study.
Study population for short-term effects of rhGH on DNA-methylation
PBMC samples from 24 patients were included upon initialisation of rhGH treatment. Diagnoses leading to rhGH treatment in this cohort were classical GH deficiency (n = 7, one with panhypopituitarism, one also born SGA), neurosecretory dysfunction leading to GH deficiency (n = 6), SGA with lack of catch-up growth (n = 7), qualitative GH deficiency (Kowarski syndrome (n = 2)), Turner syndrome (n = 1), primary IGF1 deficiency (n = 1). Median age of the patients in this cohort was 9.17 years (range: 5.17–15.42 years). For details on patients’ characteristics see Table 1. As part of routine clinical work-up all patients underwent an IGFGT. The first blood sample (baseline) was drawn prior to the first dose of rhGH and the second blood sample (stimulated) in the first morning between 8 and 10 a.m. after 4 days of rhGH treatment (30 μg/kg body weight/day). The actual lower leg length was recorded with a knemometer according to the method extensively described by Hermanussen et al. in [23] upon baseline and stimulated blood sampling and the knemometry rate over the lower limbs was calculated from both measurements (Table 1). IGF1 and IGFBP3 concentrations (RIA, Mediagnost, Reutlingen, Germany) were determined upon baseline and stimulated blood sampling (Table 1, Fig. 1).
Table 1. Clinical characterisation of the 24 children investigated for short-term effects of rhGH on DNA-methylation.
| Sample ID | Age (years) | Diagnosis | Sex | IGF1 | IGFBP3 baseline in μg/ml (centile) | IGF1 stimulated in ng/ml | IGFBP3 stimulated in μg/ml | Knemometry rate (mm) |
|---|---|---|---|---|---|---|---|---|
| baseline in ng/ml (centile) | ||||||||
| P1 | 13.00 | NSD | m | 126 (2) | 2.23 (6) | 200 | 3.49 | 0.1 |
| P2 | 11.58 | SGA | f | 232 (24) | 3.29 (45) | 243 | 3.4 | 0.75 |
| P3 | 13.33 | NSD | m | 88 (<0.1) | 1.97 (1) | 144 | 2.98 | 0.4 |
| P4 | 8.92 | STH-D~ | f | 82 (2) | 2.37 (27) | 170 | 2.74 | -0.1 |
| P7 | 7.92 | Q-STH-D | m | 77 (4) | 1.22 (<0.1) | 170 | 1.38 | 0.5 |
| P8 | 7.67 | SGA | m | 229 (86) | 3.5 (93) | 332 | 2.1 | 4.1 |
| P9 | 12.33 | Q-STH-D | m | 180 (18) | 2.43 (12) | 287 | 3.53 | 0.9 |
| P10 | 10.67 | NSD | m | 118 (5) | 1.8 (2) | 227 | 2.47 | 1 |
| P11 | 7.33 | STH-D | m | 46 (0.4) | 1.58 (2.6) | 72 | 2.53 | 0.55 |
| P12 | 9.42 | SGA | m | 212 (76) | 3.95 (94) | 442 | 4.39 | 1.3 |
| P13 | 8.67 | NSD | f | 133 (18) | 2.34 (25) | 222 | 2.78 | 0.2 |
| P14 | 15.42 | STH-D | m | 160 (0.2) | 3.27 (42.1) | 283 | 3.87 | 1.1 |
| P15 | 12.92 | NSD | f | 172 (2.2) | 3.14 (37.9) | 156 | 3.71 | 0 |
| P16 | 6.00 | SGA | m | 69 (16.5) | 2.66 (64.1) | 139 | 2.85 | 1.2 |
| P17 | 11.58 | NSD | f | 171 (9.6) | 3.35 (53.3) | 338 | 3.5 | 0.75 |
| P18 | 6.42 | IGF1-D | f | 82 (22.3) | 1.75 (7.7) | 71 | 1.67 | 0.3 |
| P19 | 6.17 | SGA | m | 58 (9) | 1.14 (0.1) | 64 | 2.82 | -0.5 |
| P20 | 13.33 | STH-D, PAN | m | 67 (0) | 1.6 (0.1) | 286 | 4.39 | 0.6 |
| P21 | 11.75 | STH-D | f | 202 (16.5) | 3.03 (34.4) | 321 | 3.14 | 0 |
| P22 | 5.17 | SGA | m | 37 (1.1) | 1.69 (9.8) | 62 | 2.44 | 0.3 |
| P23 | 6.92 | SGA, TS | f | 192 (85.5) | 1.85 (11.6) | 343 | 4.66 | 1.1 |
| P24 | 13.17 | STH-D | m | 198 (29.4) | 2.77 (27.2) | 288 | 1.89 | 0.5 |
| P25 | 8.00 | STH-D, SGA | f | 100 (15.2) | 2.76 (52.5) | 105 | 2.44 | 0.2 |
| P26 | 5.83 | UTS | f | 121 (59.3) | 3.02 (83) | 233 | 4.04 | 0.1 |
NSD: GH deficiency due to neurosecretory dysfunction; SGA: small for gestational age; STH-D: classical GH deficiency; Q-STH-D: qualitative GH deficiency (Kowarski syndrome); IGF1-D: IGF1 deficiency; PAN: panhypopituitarism; TS: Temple-Syndrome (UPD(14)mat syndrome); UTS: Turner-Syndrome. ~: STH-D following pontine tumor. IGF1 and IGFBP3 concentration measured at collection of baseline blood sample. Knemometry rate measured according to [23]. m: male; f: female.
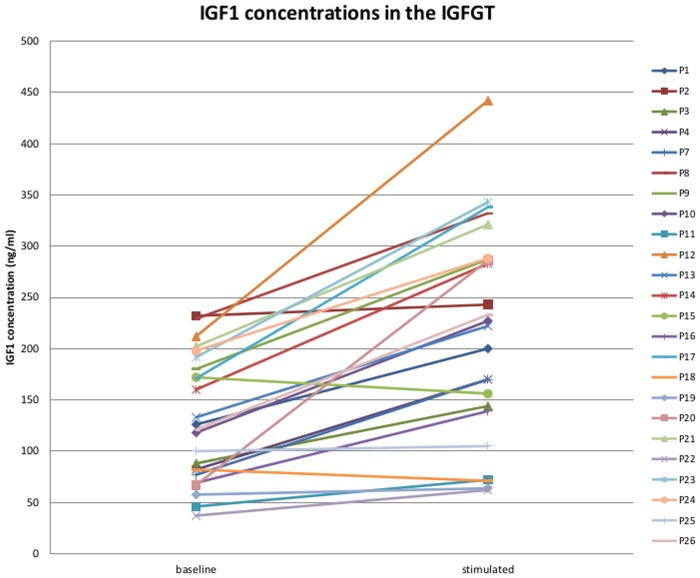
Fig 1. Increase of IGF1 concentration upon rhGH treatment in the IGFGT.
Scatter plot of the IGF1 concentrations (ng/ml) at baseline and rhGH stimulated blood sampling among the short-term rhGH treatment study cohort. The two measurements per sample are connected. Sample numbering corresponds to the sample identifier in Table 1. A significant increase in IGF1 concentrations after rhGH treatment is observed (paired t-test, p = 1.93 x 10–6).
Study population for long-term effects of rhGH on DNA-methylation
Peripheral blood (PB) samples from 54 children born SGA, defined as birth length and/or birth weight -2SD below the mean for gestational age according to the reference data published by Niklasson et al. [24], were included. The cohort has been recruited in the framework of the consortium “Diseases caused by imprinting defects: clinical spectrum and pathogenetic mechanisms” and partially overlaps with the previously described cohort of SGA born children studied for their DNA-methylation pattern at 10 imprinted loci by bisulfite pyrosequencing [25]. Of note, the children with evidence for DNA-methylation defects at the loci IGF2, H19, GRB10, MEG3 and NDN in the latter study were excluded from the present study. Out of 54 children, 36 received treatment with rhGH (median therapy interval at inclusion: 33 months, interquartile range: 15.5–62.25 months) and 18 not. Clinical parameters were obtained by standardised questionnaires and are summarized in Table 2.
Table 2. Clinical characterisation of the 54 children born SGA investigated for long-term effects of rhGH treatment on DNA-methylation.
| Criteria | Children treated with rhGH | No data | Children without rhGH treatment | No data |
|---|---|---|---|---|
| number of included children | 36 | 0 | 18 | 0 |
| female | 8 | 0 | 5 | 0 |
| male | 28 | 0 | 13 | 0 |
| rhGH treatment in months (median and IQR) | 33 [15.5–62.25] | 0 | 0 | 0 |
| developmental delay | 6 | 3 | 4 | 0 |
| Prenatal factors | ||||
| maternal nicotin consumption | 8 | 5 | 4 | 1 |
| EPH gestosis | 2 | 8 | 2 | 3 |
| placenta insufficiency | 9 | 14 | 5 | 4 |
| Parametric data (median and IQR) | ||||
| age at investigation (in years) | 10.63 [7.75–12.65] | 0 | 5.67 [4.33–9.23] | 0 |
| birth weight (SDS) | -3.29 [-2.59–-4.25] | 0 | -2.88 [-2.43–-3.76] | 0 |
| birth length (SDS) | -2.54 [-2.08–-3.42] | 0 | -2.5 [-2.22–-3.37] | 0 |
rhGH: recombinant human growth hormone; EPH gestosis: edema-proteinuria-hypertension gestosis; IQR: interquartile range; SDS: standard deviation score.
PBMC isolation, DNA extraction and bisulfite conversion
For the short-term rhGH treatment cohort PBMC isolation from freshly drawn blood samples was performed according to [15]. Briefly, PBMC were isolated by Ficoll (Biochrom AG, Berlin, Germany) gradient centrifugation, resuspended in RPMI 1640 medium (Gibco, Karlsruhe, Germany) with 10% DMSO, 10% heat-inactivated FCS, penicillin (100 U/mL), streptomycin (100 μg/mL), and L-glutamine (2 mM) and frozen in liquid nitrogen.
DNA was extracted from PBMC (short-term cohort) or PB (long-term cohort) using standard methods and 1 μg DNA was bisulfite converted using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions.
Array-based DNA-methylation analysis
For DNA-methylation analysis the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA) was used according to the manufacturer’s instruction. This platform allows the interrogation of 485,577 assays (482,421 CpG sites, 3091 non-CpG sites, 65 random SNPs) in parallel at a single-nucleotide resolution per sample [26]. Arrays were scanned using the Illumina iScan. Raw hybridisation signals were processed using the GenomeStudio software (version 2011.1; Methylation Analysis Module version 1.9.0, Illumina) applying the default settings and internal controls for normalisation. Quality criteria referring to the technical results of hybridisation were set as a gene call rate above 98% per sample and a detection p-value <0.01 per CpG site. After quality filtering a total of 48 hybridisations and 482,000 targets entered the final analyses in the short-term rhGH treatment cohort. In the long-term rhGH treatment cohort CpGs were additionally filtered for those targets corresponding to autosomes and those labeled as SNPs. This led to a total of 54 samples and 471,808 targets entering final analyses. Results are available in a MIAMI compliant format from Gene Expression Omnibus (GSE57205).
Analyses of DNA-methylation data
For unsupervised and groupwise comparisons the Omics Explorer (ver.3.0 (25); Qlucore, Lund, Sweden) was used. For unsupervised analysis false discovery rate (FDR) and/or variance (σ/σmax) filters applied for the different analyses are given in the respective descriptions of the results. Groupwise comparisons were performed as supervised analysis applying t-test statistics.
In the short-term rhGH treatment group in addition a paired t-testing of avg.beta values was performed, both unadjusted and adjusted for multiple testing. For targets with a p-value <0.0001 in the paired t-test avg.beta values of baseline and stimulated samples were subjected to a cluster analysis (R version 3.0.1 and package ‘gplots’ function heatmap.2).
Moreover, in the short-term cohort delta beta values (avg.beta value stimulated sample subtracted from avg.beta value baseline sample) were calculated for all 482,000 targets entering final analysis. Delta beta values >0.1 or <-0.1 (corresponding roughly to a difference of DNA-methylation between both samples of a pair of +/-10%) were classified as differentially methylated. The cut-off of 10% was arbitrarily chosen to enrich for differentially methylated CpGs with likely biological significance [17]. Those targets in which at least 3 of the 24 sample pairs (>10%) show delta beta values of >0.1 or <-0.1 were subsequently subjected to cluster analysis as described above. Clustering was performed using the delta beta values as continuous variables as well as by classifying the targets as gained methylation (labeled 1 for delta beta values >0.1), lost methylation (labeled -1 for delta beta values <-0.1) and unchanged (labeled 0 for delta beta values <0.1->-0.1) in response to rhGH treatment.
Information about the location of differentially methylated CpG loci in regulatory regions in the lymphoblastoid cell line GM12878 given in S2 Table and S5 Table were available from Ensembl (ftp://ftp.ensembl.org/pub/release-78/regulation/homo_sapiens/RegulatoryFeatures_GM12878.gff.gz).
Gene ontology analysis
Gene ontology analysis for enrichment of biological processes, function and protein binding among differentially methylated genes was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
Validation of DNA-methylation differences by bisulfite pyrosequencing
Based on delta beta values in the rhGH short-term cohort 5 loci (GNLY, TRIM39, SLC15A4, IGF1R, SLC6A16) were selected for validation. Selection criteria comprised at least three differentially methylated CpGs per locus in at least one, preferably multiple patient samples. Bisulfite pyrosequencing assays were designed to cover two of these differentially methylated CpGs per locus. Bisulfite pyrosequencing assays were designed using the PyroMark Assay Design Software (Version 2.0, Qiagen, Hilden Germany). For PCR and primer conditions as well as analysed sample pairs from the short term cohort see S1 Table. We used 1 μl of bisulfite converted DNA from PBMC as template for each PCR in a final volume of 25μl. The correct size of the PCR product was verified by gel electrophoresis. For purification of PCR products and single strand preparation a mastermix of 5μl streptavidin sepharose beads and 40μl binding buffer was prepared. 43μl of this mastermix was pipetted to the PCR products. Purification and single strand preparation of the PCR product were performed with the Vacuum Prep Tool (Biotage), followed by denaturation at 85°C for 2 minutes and sequencing primer hybridisation. Pyrosequencing was performed using the PyroMark ID and evaluated using the PyroMark CpG Software 1.0.11 (Biotage). Assays were validated using in vitro methylated DNA as positive control (Millipore, Hilden, Germany) and whole genome amplified DNA as unmethylated control.
Results
In order to investigate the short-term and long-term influence of rhGH on DNA-methylation we analysed DNA-methylation patterns in two independent cohorts using the HumanMethylation450 BeadChip [26]. We will first separately report the results obtained in the short-term and long-term cohorts, respectively, and then provide data on the intersection of the results between both cohorts.
Analyses of the short-term study cohort
Evaluation of biological response to rhGH treatment
The short-term study cohort comprised paired PBMC samples from 24 patients obtained during the initialisation of rhGH treatment (Table 1). There was a significant increase of the IGF1 concentrations after 4 days of rhGH treatment in this cohort (paired t-test p-value = 1.93 x 10-6) pointing to a significant biological response. On an individual level, an increase of IGF1 concentration was observed in 22/24 patients (Fig. 1). In one of the two cases without IGF1 rise the knemometry rate points to biological response.
Comparison of DNA-methylation of baseline and rhGH stimulated PBMC samples
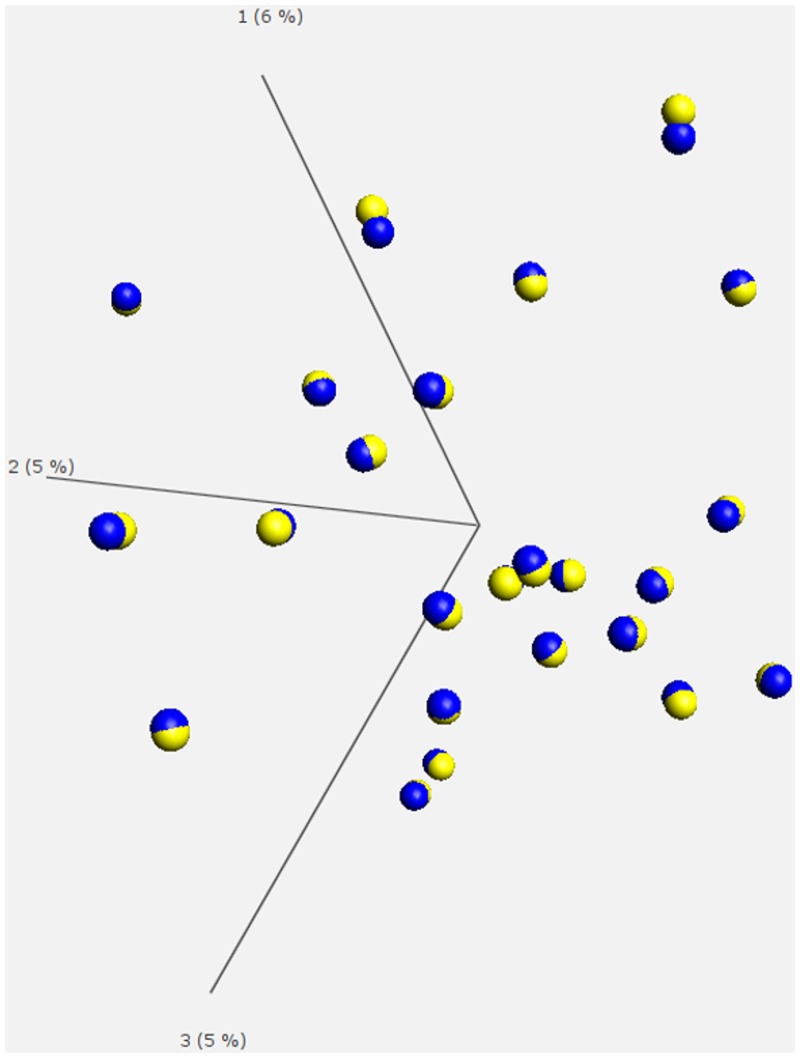
To investigate the short-term effects of rhGH on in vivo DNA-methylation we first globally compared the DNA-methylation values of all baseline and stimulated PBMC samples in the short-term cohort. A principal component analysis (PCA) of the complete short-term data set revealed no significant separation in different subgroups. PCA analysis using the 4262 loci with the highest variance (filter of variance σ/ σmax>0.275) grouped the sample pairs of each patient but failed to segregate the cohort into treatment groups thereby proving that the individual origin of the samples has much stronger influence on DNA-methylation than rhGH treatment (Fig. 2). An unpaired supervised comparison according to rhGH treatment revealed none of the evaluated CpG loci to be significantly (FDR<0.05) differentially methylated between baseline and stimulated samples. Nevertheless, a paired t-test of the 482,000 targets passing quality filtering identified 239 CpGs as significantly differentially methylated (p < 0.0001) among baseline and stimulated samples (S2 Table). The absolute differences of the mean DNA-methylation between baseline and stimulated samples for the significantly differentially methylated loci reached a maximum of 0.041 (corresponding roughly to a difference of DNA-Methylation of 4.1%). This maximum was determined for the CpG site cg11283402 associated with the gene ARID5B. Gene ontology analysis of the 162 genes associated with the 239 CpG loci revealed significant enrichment among the annotation category “protein binding” for e.g. BCL6 (B-cell CLL/lymphoma 6), HDAC4 (histone deacetylase 4) and HDAC7A (histone deacetylase 7A) (S3 Table and S4 Table). However, in line with the results of the unsupervised and unpaired supervised analysis, none of the 239 CpG loci remained significantly differentially methylated any more after adjustment for multiple comparisons at a FDR of 0.05.
Fig 2. Principal component analysis (PCA) in the overall short-term rhGH treatment study cohort.
Results of the PCA analysis performed by using a set of 4262 loci with the highest variance (filter of variance of σ/σmax>0.275) does not segregate the cohort into treatment groups. Instead, each sample pair showed close relationship to one another thereby proving that the individual origin of the samples has much stronger influence on DNA-methylation than rhGH treatment. rhGH stimulated samples are depicted in blue, baseline samples are depicted in yellow.
Because both, the diagnosis leading to rhGH treatment and the response to rhGH—measured by IGF1 and IGFBP3 increases in plasma as well as by increases of knemometry rates—showed marked inter-individual heterogeneity we additionally aimed at a more individualised evaluation of the DNA-methylation changes. Thus, the difference of avg.beta values between stimulated and baseline samples (delta beta) of each pair was calculated. A total of 2350 CpG and 3 CpH loci showed absolute differences between baseline and stimulation of at least 0.1 in at least 3 of the 24 pairs (Fig. 3, S5 Table). Gene ontology analysis of the 1426 genes associated with these 2353 loci revealed among others significant enrichment for the processes “cell communication”, “immune response” and”signal transduction” (S6 Table and S7 Table).
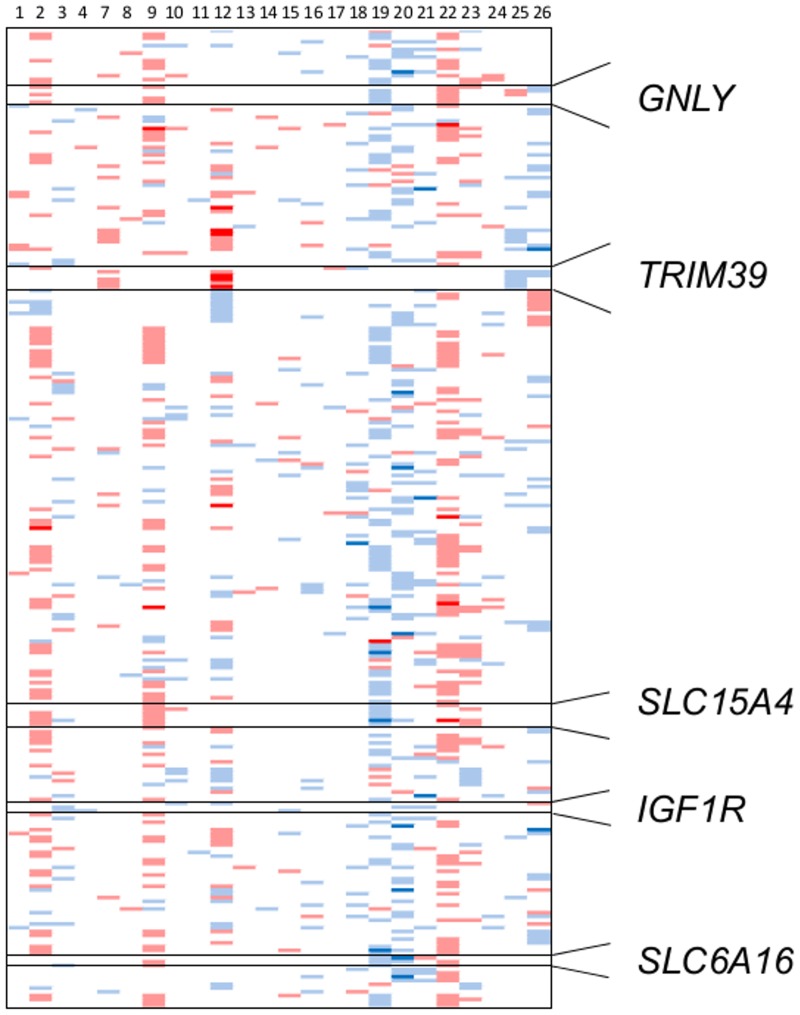
Fig 3. DNA-methylation results in the short-term rhGH treatment study cohort using an individualised evaluation approach.
Differences of avg.beta values between stimulated and baseline samples (delta beta) of each pair were calculated. Depicted are those loci, in which at least 3/24 analysed sample pairs showed absolute delta beta values below -0.1 or above 0.1 (roughly corresponding to a difference of DNA-methylation of +/- 10% between each analysed sample pair) and in which at least 3 CpGs were affected per gene (n = 259). Red: delta beta values above 0.2, pale red: delta beta values between 0.1 and 0.2. Blue: delta beta values below -0.2, pale blue: delta beta values between -0.1 and -0.2. Samples are sorted according to Table 1 with P1 at the left margin and P26 at the right margin. Those loci used for bisulfite pyrosequencing validation are zoomed out and shown at the right side of the figure.
Comparison of the 162 and 1426 genes identified with the unadjusted paired t-test and the individualised approach revealed 23 overlapping genes (S8 Table). Those 23 genes showed significant enrichment for the processes “imprinting”, “DNA methylation” and “DNA alkylation” as well as protein binding to “BCL6” and “HDAC1, HDAC4, HDAC6” (S9 Table and S10 Table). Neither cluster analysis of the 239 CpGs detected by the unadjusted paired t-test nor of the 2353 loci detected with the individualised approach as differentially methylated between baseline and stimulated samples showed grouping according to the underlying diagnosis.
Validation of DNA-methylation differences by bisulfite pyrosequencing
Differences observed by the array-based analyses were validated by bisulfite pyrosequencing for 5 loci selected based on delta beta values (GNLY, TRIM39, SLC15A4, IGF1R, SLC6A16) in the short-term cohort. The bisulfite pyrosequencing assays interrogated for each selected locus two CpGs previously investigated on the array. For each of the 10 investigated CpGs 1 to 6 sample pairs of the short-term cohort were analysed, resulting in a total of 34 paired measurements (see S1 Table for details). A comparison of the results obtained by both techniques at the investigated CpGs showed high correlation (Pearson r = 0.78, p-value = 4.015 x 10-8; S1 Fig).
Analyses of the long-term study cohort
Comparison of DNA-methylation between rhGH treated and untreated peripheral blood samples
Complementary to the investigations in the short-term cohort we analysed an independent cohort of children born SGA for long-term effects of rhGH treatment. This cohort consisted of peripheral blood (PB) samples from 54 children born SGA, among which 36 received rhGH treatment for a median of 33 months (interquartile range 15.5–62.25 months) and 18 received no rhGH treatment (Table 2). Importantly, no paired pre-treatment and follow-up samples were available from the rhGH treated patients in this cohort. Thus, and because the long-term comparison in a longitudinal study could be confounded by age-related DNA methylation changes, the statistical approach differed from the short-term cohort and we compared the rhGH treated patients to an independent cohort of untreated patients matched with regard to SGA (see methods section and Table 2). All autosomal target loci fulfilling quality filtering (n = 471,808) were subjected to the analysis. Nevertheless, neither unsupervised nor supervised analyses (FDR<0.05) revealed a separation of the overall cohort into distinct subgroups or lead to the identification of significantly differentially methylated CpGs between rhGH treated and untreated individuals. A linear regression analysis of DNA-methylation results and period of rhGH treatment revealed no correlation for any of the CpG sites analysed. Thus, in this cohort we could not identify a homogeneous effect of long-term rhGH treatment on DNA-methylation in the peripheral blood.
Analysis of the long-term cohort for loci identified as differentially methylated in the short term cohort
Finally, we investigated the long-term cohort only for those targets detected as differentially methylated in the short-term rhGH treatment cohort. A total of 235 of the 239 CpGs and 2280 of the 2353 targets, respectively, identified in the short-term cohort matched the (quality) filter criteria in the long-term cohort. Of those none was differentially methylated between the treatment groups in the long-term cohort (supervised analysis, FDR<0.05). Similarly, there was no significant correlation of the methylation levels of those targets and the treatment interval.
Discussion
In this study we aimed at characterising short- and long-term effects of rhGH treatment on the DNA-methylome. We identified marked intra-individual responses of the DNA-methylation to rhGH treatment, primarily when comparing paired samples in the short-term treatment setting. These, however, seem to be inter-individually heterogeneous.
Groupwise DNA-methylation changes associated with rhGH treatment
During the initial phase of rhGH treatment 162 genes were detected as significantly differentially methylated by paired unadjusted t-testing in the short-term treatment cohort. Despite none of these genes retained significance after correction for multiple testing, we investigated whether these genes represent putative candidate genes to eventually integrate in a (epi)genetic prediction model for rhGH response and whether these targets are persistently affected during long-term rhGH treatment. Nevertheless, by analysing paired samples in a longitudinal approach over a long period of time (i.e. several years) and during a critical phase in development (i.e. childhood and adolescence) it needs to be taken into account that the effects of rhGH treatment on the IGF1 signaling pathway are inevitably influenced by the effects of ageing [28,29,30]. Indeed, it is well known that ageing affects DNA methylation in peripheral blood [29,30]. We therefore had to adapt the study design in the long-term treatment cohort. Though our approach takes aging into account, it lacks power as compared to a paired design and is less controlled for diagnosis heterogeneity. We therefore chose for the long-term analysis a homogeneous cohort of probands, which uniformly received rhGH treatment due to the condition SGA. Principal component analysis indeed indicated the long-term cohort to represent a homogeneous study batch. Neither unsupervised nor supervised analysis regarding rhGH treatment among all CpGs after quality filtering and in the subset of CpGs detected as differentially methylated by unadjusted paired t-testing in the short-term cohort revealed a separation among the long-term cohort. These findings uniformly point to a lack of a rhGH induced homogeneous DNA-methylation signature after rhGH treatment, which is well in line with the generally assumed safety of rhGH treatment.
Individual DNA-methylation changes associated with rhGH treatment
Indications to start rhGH treatment are numerous, pointing to high variability of the individual environment in which the rhGH exerts its effect. To address this point we performed unsupervised analyses with the various data sets. Whereas these analyses did not identify considerable subgroups of patients they clearly showed the closest relation of DNA-methylation patterns between both samples of the same individual in the short-term cohort (Fig. 2). These findings prompted us to take advantage of the paired study design in the short-term cohort and compare the sample pairs in an individualised approach leading to the detection of 1426 recurrently differentially methylated genes in at least 3/24 pairs. Exemplary validation for the targets detected as recurrently differentially methylated in the individualised approach showed good correlation suggesting that these differences are technically—and likely also biologically—robust. The direction of identified and validated DNA-methylation changes in response to rhGH stimulation is frequently heterogeneous, which is in concordance to the observed effects of androgens on the DNA methylome [18,19].
rhGH and immune system
We cannot reliably distinguish whether the DNA-methylation changes observed in the individualised approach are attributable to selective de novo DNA-(de)methylation, to more general effects like individual host factors or to expansion of a distinct cell population. Nevertheless, gene ontology analyses of these genes indicate that a subset of patients shows a response of immune cells in the initial phase of rhGH treatment. Of note previous studies showed no changes in the number of T- and B-lymphocytes upon short-term in vitro incubation with growth hormone releasing hormone [31], while the number of B-lymphocytes in long-term rhGH treated children was reported to decrease [32,33,34]. Approximately 90% of B-lymphocytes and monocytes express GH receptors on their cell surfaces, while they were variably detected on T-lymphocytes and natural killer cells [35,36]. Thus, rhGH has the potential to regulate all major immune cell types and influence immune function [37]. The association of rhGH treatment and the immune system we observed in our study corroborates several previous studies on gene expression [13,14,15]. Welzel et al. performed gene expression studies in PBMC in the frame of the IGFGT and detected inter-individual variability in rhGH-related response. Nevertheless, gene ontology analyses revealed significant over-representation of T-cell and B-cell activation, Toll receptor and JAK/STAT signaling pathways among the 313 transcripts associated with GH treatment [15]. Of those 20 transcripts overlap with the differentially methylated loci identified herein. Among them is BCL6 encoding a master regulator of B- and T-cell development [38]. The implication of the JAK/STAT signaling pathway in GH treatment has been further proven by Trovato et al. [39]. By analysing GH gene expression changes in PBMC of 10 adult GH deficient women after 4 weeks of GH treatment Fernández-Pérez et al. proposed 24 candidate genes to discriminate between samples before and after GH treatment, which also include genes involved in immune response and signal transduction [14]. Whatmore and colleagues studied gene expression changes in PBMC after 3 months of rhGH treatment [13]. Gene ontology analysis of one of the cluster patterns associated with GH treatment, referring to 179 probes, again included immune function genes [13]. Thus, aside from a recurrent theme also in the gene expression studies pointing to considerable individual differences there is compelling evidence that (rh)GH treatment affects expression of immune-related genes either directly or through regulating growth of certain immune cell populations.
Conclusions
In summary we observed strong inter-individual differences of DNA-methylation upon rhGH treatment in the short-term treatment cohort, primarily related to a response of immune cells. These might be caused by a direct effect of rhGH on DNA-methylation or by growth differences in cell populations with differential rhGH sensitivity in the peripheral blood. The observed heterogeneity of DNA-methylation changes might be influenced by the host variability, e.g. individual differences in GH receptor and IGF1 receptor expression or the complex polygenic background on which GH effects are exerted. We did not observe the establishment of a rhGH induced homogeneous DNA-methylation signature after short- and long-term treatment, thereby corroborating the generally assumed safety of rhGH treatment. However, these results cannot preclude uniform DNA-methylation changes in rhGH target tissues, like liver and cartilage, tissues hardly available in a clinical setting based on ethical considerations. Moreover, DNA methylation changes are generally considered to represent rather long-term epigenetic modifications that may not be affected by a short-term rhGH treatment. Therefore, in a future approach, it would be conceivable to investigate if short-term treatment has an impact on more dynamic (short-term) epigenetic modifications such as histone methylation and acetylation by analyzing genome-wide changes in histone modifications by ChIP-Seq in the studied short-term cohort.
Supporting Information
Bisulfite pyrosequencing assays were designed for 5 loci (GNLY, TRIM39, SLC15A4, IGF1R, SLC6A16). Within each locus 2 CpGs covered by the array-based approach were evaluated. For each of the 10 investigated CpGs 1 to 6 sample pairs were analysed, resulting in a total of 34 paired measurements. A: Correlation analysis. Depicted are the delta beta values (value of the stimulated samples subtracted from the value of the baseline sample) for the 34 paired measurements. A comparison of the results obtained by both techniques at the investigated CpGs showed high correlation (Pearson r = 0.78, p-value = 4.015 x 10-8). B: Bland-Altman plot. Bland-Altman plot of DNA-methylation results from all 68 single measurements obtained by bisulfite pyrosequencing and the corresponding avg.beta values obtained from the array-based measurements. For this figure avg.beta-values as well as methylation levels determined by bisulfite pyrosequencing are displayed as percent values in order to obtain data sets of the same size range. The difference between both methods for every individual measurement is plotted against the average of both methods. The average of the differences +/− two times the standard deviation denotes the 95% range for the limits of agreement (marked by the dotted horizontal lines).
(TIF)
(XLSX)
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Delta beta values for each sample pair as well as classifiers (“1” for delta beta values >0.1, “0” for delta beta values between 0.1 and -0.1 and “-1” for delta beta values <-0.1) are given.
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Acknowledgments
The authors thank the technical staff of the epigenetic laboratory of the Institute of Human Genetics for expert assistance, the technical staff of the pediatric endocrine laboratory in the Department of Pediatrics as well as the clinical colleagues for support.
Data Availability
Results are available in a MIAMI compliant format from Gene Expression Omnibus (GSE57205).
Funding Statement
The research leading to these results received funding from the German national BMBF (Ministry of Education and Science)-funded consortium ‘Diseases caused by imprinting defects: clinical spectrum and pathogenetic mechanisms’ (FKZ: 01GM0886 and 01GM1114) and the Medical Faculty of the Christian-Albrechts-University Kiel. The study used for the short-term rhGH treatment cohort left-over samples from a different study supported by Novo Nordisk. The authors confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Glasbrenner K. Technology spurt resolves growth hormone problem, ends shortage (Medical News). JAMA. 1986;255: 443–447. [DOI] [PubMed] [Google Scholar]
- 2. Takeda A, Cooper K, Bird A, Baxter L, Frampton GK, Gospodarevskaya E, et al. Recombinant human growth hormone for the treatment of growth disorders in children: a systematic review and economic evaluation. Health Technol Assess. 2010;14: 1–209. [DOI] [PubMed] [Google Scholar]
- 3. Swerdlow AJ, Higgins CD, Adlard P, Preece MA. Risk of cancer in patients treated with human pituitary growth hormone in the UK, 1959–85: a cohort study. Lancet. 2002;360: 273–277. [DOI] [PubMed] [Google Scholar]
- 4. Mills JL, Schonberger LB, Wysowski DK, Brown P, Durako SJ, Cox C, et al. Long-term mortality in the United States cohort of pituitary-derived growth hormone recipients. J Pediatr. 2004;144: 430–436. [DOI] [PubMed] [Google Scholar]
- 5. Ergun-Longmire B, Mertens AC, Mitby P, Qin J, Heller G, Shi W, et al. Growth hormone treatment and risk of second neoplasms in the childhood cancer survivor. J Clin Endocrinol Metab. 2006;91: 3494–3498. [DOI] [PubMed] [Google Scholar]
- 6. Carel JC, Ecosse E, Landier F, Meguellati-Hakkas D, Kaguelidou F, Rey G, et al. Long-term mortality after recombinant growth hormone treatment for isolated growth hormone deficiency or childhood short stature: preliminary report of the French SAGhE study. J Clin Endocrinol Metab. 2012;97: 416–425. 10.1210/jc.2011-1995 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe S, Tsunematsu Y, Fujimoto J, Komiyama A. Leukaemia in patients treated with growth hormone. Lancet. 1988;1: 1159–1160. [PubMed] [Google Scholar]
- 8. Bell J, Parker KL, Swinford RD, Hoffman AR, Maneatis T, Lippe B. Long-term safety of recombinant human growth hormone in children. J Clin Endocrinol Metab. 2010;95: 167–177. 10.1210/jc.2009-0178 [DOI] [PubMed] [Google Scholar]
- 9. Wilton P, Mattsson AF, Darendeliler F. Growth hormone treatment in children is not associated with an increase in the incidence of cancer: experience from KIGS (Pfizer International Growth Database). J Pediatr.2010;157: 265–270. 10.1016/j.jpeds.2010.02.028 [DOI] [PubMed] [Google Scholar]
- 10. Sävendahl L, Maes M, Albertsson-Wikland K, Borgström B, Carel JC, Henrard S, et al. Long-term mortality and causes of death in isolated GHD, ISS, and SGA patients treated with recombinant growth hormone during childhood in Belgium, The Netherlands, and Sweden: preliminary report of 3 countries participating in the EU SAGhE study. J Clin Endocrinol Metab. 2012;97: E213–217. 10.1210/jc.2011-2882 [DOI] [PubMed] [Google Scholar]
- 11. Le Roith D, Bondy C, Yakar S, Liu JL, Butler A. The somatomedin hypothesis: 2001. Endocr Rev. 2001;22: 53–74. [DOI] [PubMed] [Google Scholar]
- 12. Buckway CK, Guevara-Aquirre J, Pratt KL, Burren CP, Rosenfeld RG. The IGF-I generation test revisited: a marker of GH sensitivity. J Clin Endocrinol Metab. 2001;86: 5176–5183. [DOI] [PubMed] [Google Scholar]
- 13. Whatmore AJ, Patel L, Clayton PE. A pilot study to evaluate gene expression profiles in peripheral blood mononuclear cells (PBMCs) from children with GH deficiency and Turner syndrome in response to GH treatment. Clin Endocrinol (Oxf). 2009;70: 429–434. 10.1111/j.1365-2265.2008.03477.x [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Pérez L, Nóvoa J, Stahlberg N, Santana-Farré R, Boronat M, Marrero D, et al. The effect of in vivo growth hormone treatment on blood gene expression in adults with growth hormone deficiency reveals potential biomarkers to monitor growth hormone therapy. Clin Endocrinol (Oxf). 2010;72: 800–806. 10.1111/j.1365-2265.2009.03732.x [DOI] [PubMed] [Google Scholar]
- 15. Welzel M, Appari M, Brämswig N, Riepe FG, Holterhus PM. Transcriptional response of peripheral blood mononuclear cells to recombinant human growth hormone in a routine four-days IGF-1 generation test. Growth Horm IGF Res. 2011;21: 336–342. 10.1016/j.ghir.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 16. Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502: 489–498. 10.1038/nature12752 [DOI] [PubMed] [Google Scholar]
- 17. Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013; 18: 296–302. 10.1016/j.cmet.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 18. Bens S, Ammerpohl O, Martin-Subero JI, Appari M, Richter J, Hiort O, et al. Androgen receptor mutations are associated with altered epigenomic programming as evidenced by HOXA5 methylation. Sex Dev. 2011; 5: 70–76. 10.1159/000323807 [DOI] [PubMed] [Google Scholar]
- 19. Ammerpohl O, Bens S, Appari M, Werner R, Korn B, Drop SL, et al. Androgen receptor function links human sexual dimorphism to DNA methylation. PLoS ONE. 2013;8: e73288 10.1371/journal.pone.0073288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frick KM, Zhao Z, Fan L. The epigenetics of estrogen: epigenetic regulation of hormone-induced memory enhancement. Epigenetics. 2011;6: 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin-Subero JI, Kreuz M, Bibikova M, Bentink S, Ammerpohl O, Wickham-Garcia E, et al. New insights into the biology and origin of mature aggressive B-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113: 2488–2497. 10.1182/blood-2008-04-152900 [DOI] [PubMed] [Google Scholar]
- 22. Sookoian S, Pirola CJ. Epigenetics of insulin resistance: an emerging field in translational medicine. Curr Diab Rep. 2013;13: 229–237. 10.1007/s11892-012-0361-9 [DOI] [PubMed] [Google Scholar]
- 23. Hermanussen M, Geiger-Benoit K, Burmeister J, Sippell WG. Knemometry in childhood: accuracy and standardization of a new technique of lower leg length measurement. Ann Hum Biol. 1988;15: 1–15. [DOI] [PubMed] [Google Scholar]
- 24. Niklasson A, Albertsson-Wikland K. Continous growth reference from 24th week of gestation to 24 months by gender. BMC Pediatr. 2008;29: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bens S, Haake A, Richter J, Leohold J, Kolarova J, Vater I, et al. Frequency and characterization of DNA methylation defects in children born SGA. Eur J Hum Genet. 2013; 21: 838–43. 10.1038/ejhg.2012.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98: 288–295. 10.1016/j.ygeno.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 27. Chang GT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22: 2926–2933. [DOI] [PubMed] [Google Scholar]
- 28. Campisi J, Yaswen P. Aging and cancer cell biology. Aging Cell. 2009; 8: 221–225. 10.1111/j.1474-9726.2009.00475.x [DOI] [PubMed] [Google Scholar]
- 29. Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20: 440–446. 10.1101/gr.103606.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22: 623–632. 10.1101/gr.125187.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7: e41361 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiess W, Malozowski S, Gelato M, Butenand O, Doerr H, Crisp B, et al. Lymphocyte subset distribution and natural killer activity in growth hormone deficiency before and during short-term treatment with growth hormone releasing hormone. Clin Immunol Immunopathol. 1988; 48: 85–94. [DOI] [PubMed] [Google Scholar]
- 33. Rapaport R, Oleske J, Ahdieh H, Skuza K, Holland BK, Passannante MR, et al. Effects of human growth hormone on immune functions: in vitro studies on cells of normal and growth hormone-deficient children. Life Sci. 1987;41: 2319–2324. [DOI] [PubMed] [Google Scholar]
- 34. Petersen BH, Rapaport R, Henry DP, Huseman C, Moore WV. Effect of treatment with biosynthetic human growth hormone (GH) on peripheral blood lymphocyte populations and function in growth hormone-deficient children. J Clin Endocrinol Metab. 1990;70: 1756–1760. [DOI] [PubMed] [Google Scholar]
- 35. Rapaport R, Sills IN, Green L, Barrett P, Labus J, Skuza KA, et al. Detection of human growth hormone receptors on IM-9 cells and peripheral blood mononuclear cell subsets by flow cytometry: correlation with growth hormone-binding protein levels. J Clin Endocrinol Metab. 1995;80: 2612–2619. [DOI] [PubMed] [Google Scholar]
- 36. Badolato R, Bond HM, Valerio G, Petrella A, Morrone G, Waters MJ, et al. Differential expression of surface membrane growth hormone receptor on human peripheral blood lymphocytes detected by dual fluorochrome flow cytometry. J Clin Endocrinol Metab. 1994;79: 984–990. [DOI] [PubMed] [Google Scholar]
- 37. Kelley KW. The role of growth hormone in modulation of the immune response. Ann N Y Acad Sci. 1990;594: 95–103. [DOI] [PubMed] [Google Scholar]
- 38. Bunting KL, Melnick AM. New effector function and regulatory mechanisms of BCL6 in normal and malignant lymphocytes. Curr Opin Immunol. 2013;25: 339–346. 10.1016/j.coi.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trovato L, Riccomagno S, Prodam F, Genoni G, Walker GE, Moia S, et al. Isolated GHD: investigation and implication of JAK/STAT related genes before and after rhGH treatment. Pituitary. 2012;15: 482–489. 10.1007/s11102-011-0354-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bisulfite pyrosequencing assays were designed for 5 loci (GNLY, TRIM39, SLC15A4, IGF1R, SLC6A16). Within each locus 2 CpGs covered by the array-based approach were evaluated. For each of the 10 investigated CpGs 1 to 6 sample pairs were analysed, resulting in a total of 34 paired measurements. A: Correlation analysis. Depicted are the delta beta values (value of the stimulated samples subtracted from the value of the baseline sample) for the 34 paired measurements. A comparison of the results obtained by both techniques at the investigated CpGs showed high correlation (Pearson r = 0.78, p-value = 4.015 x 10-8). B: Bland-Altman plot. Bland-Altman plot of DNA-methylation results from all 68 single measurements obtained by bisulfite pyrosequencing and the corresponding avg.beta values obtained from the array-based measurements. For this figure avg.beta-values as well as methylation levels determined by bisulfite pyrosequencing are displayed as percent values in order to obtain data sets of the same size range. The difference between both methods for every individual measurement is plotted against the average of both methods. The average of the differences +/− two times the standard deviation denotes the 95% range for the limits of agreement (marked by the dotted horizontal lines).
(TIF)
(XLSX)
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Delta beta values for each sample pair as well as classifiers (“1” for delta beta values >0.1, “0” for delta beta values between 0.1 and -0.1 and “-1” for delta beta values <-0.1) are given.
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Analysis was performed with the Gene Annotation Tool to Help Explain Relationships GATHER, http://gather.genome.duke.edu/, accessed 05/06/2014 [27].
(XLSX)
Data Availability Statement
Results are available in a MIAMI compliant format from Gene Expression Omnibus (GSE57205).