Abstract
Background
The hormone auxin plays an important role not only in the growth and development of rice, but also in its defense responses. We’ve previously shown that the P450 gene CYP71Z2 enhances disease resistance to pathogens through regulation of phytoalexin biosynthesis in rice, though it remains unclear if auxin is involved in this process or not.
Methodology and Principal Findings
The expression of CYP71Z2 was induced by Xanthomonas oryzae pv. oryzae (Xoo) inoculation was analyzed by qRT-PCR, with GUS histochemical staining showing that CYP71Z2 expression was limited to roots, blades and nodes. Overexpression of CYP71Z2 in rice durably and stably increased resistance to Xoo, though no significant difference in disease resistance was detected between CYP71Z2-RNA interference (RNAi) rice and wild-type. Moreover, IAA concentration was determined using the HPLC/electrospray ionization/tandem mass spectrometry system. The accumulation of IAA was significantly reduced in CYP71Z2-overexpressing rice regardless of whether plants were inoculated or not, whereas it was unaffected in CYP71Z2-RNAi rice. Furthermore, the expression of genes related to IAA, expansin and SA/JA signaling pathways was suppressed in CYP71Z2-overexpressing rice with or without inoculation.
Conclusions and Significance
These results suggest that CYP71Z2-mediated resistance to Xoo may be via suppression of IAA signaling in rice. Our studies also provide comprehensive insight into molecular mechanism of resistance to Xoo mediated by IAA in rice. Moreover, an available approach for understanding the P450 gene functions in interaction between rice and pathogens has been provided.
Introduction
Bacterial blight is an important disease in rice caused by Xoo that results in severe loss of rice yield worldwide [1]. Rice has evolved to utilize a network of sophisticated signaling pathways against invasion by phytopathogens, for example pathogen-associated molecular patterns (PAMPs), systemic acquired resistance (SAR) and hypersensitive response [2–5]. Plant hormones such as SA, JA and IAA mediate broad-spectrum disease resistance in rice and have been widely studied; the mechanisms of resistance have been elucidated [6, 7].
IAA, the major form of auxin in rice, is generally believed to play an important role in plant growth and development [8, 9]. However, recent studies demonstrate that IAA acts as a negative regulator in the plant immune response [7, 10, 11], as exogenous application of IAA or auxin analogs in rice and Arabidopsis significantly promotes disease symptoms. Treatment with IAA and 2,4-dichlorophenoxyacetic acid (2, 4-D; an analog of IAA) in rice resistant to various types of bacterial blight significantly stimulates phytopathogenic Xoo proliferation, resulting in high susceptibility to these compounds [12]. Similarly, treatment of resistant rice plants with IAA enhances the infectivity of Xanthomonas oryzae pv. oryzicola (Xooc) and Magnaporthe oryzae to rice [13]. In addition, exogenous application of 1-naphthalacetic acid (NAA) or 2,4-D on Arabidopsis accelerates the development of disease symptoms during infection by Pseudomonas syringae pv. tomato (Pto) DC3000 or Pseudomonas syringae pv. maculicola [14, 15].
On the other hand, many phytopathogens are capable of inducing significant IAA accumulation that weakens the host’s native defense barrier, the cell wall [16–20]. This inhibits accumulation of endogenous auxin, leading to high disease resistance rates in rice. The mechanism for this is largely believed to be due to inhibition of expansin gene expression, which induces overexpression of GH3 family genes OsGH3.1, GH3-2 and GH3-8 that enhance broad-spectrum resistance to bacterial Xoo, Xooc and Magnaporthe grisea [12, 13, 21]. Based on these studies, it can be concluded that the suppression of the auxin signaling pathway partly contributes to disease resistance in rice.
The synthesis of IAA is dependent on whether the precursor tryptophan (Trp) is involved, with Trp-dependent and Trp-independent pathways found in both the monocotyledonous model rice and the dicotyledonous model Arabidopsis [20, 22, 23]. In Trp-dependent pathways, indole 3-acetaldoxime (IAOx) is one of the key intermediate metabolites [22, 24]. IAOx is a common precursor of auxin, camalexin and indole glucosinolates biosynthesis, and is a crucial branching point from primary metabolism to secondary metabolism in Arabidopsis [25–28]. The cytochrome P450 monooxygenase CYP79B2 is responsible for catalyzing the conversion of tryptophan to IAOx in Arabidopsis [24, 25, 29, 30]. To date, many other cytochrome P450 monooxygenase genes involved in IAOx biosynthesis and metabolism have been cloned in Arabidopsis. The overexpression of cyp79B2 in Arabidopsis significantly increases IAA content [31], though less IAA is detected in the cyp79B2/cyp79B3 double mutant [31]. Another cytochrome P450 monooxygenase, CYP71A13, is capable of catalyzing IAOx to indole-3-acetonitrile (IAN) in the Trp-dependent IAA biosynthesis pathway [32]. P450 monooxygenases CYP83A1 and CYP83B1 have similar biochemical functions for IAA biosynthesis, which maintains the endogenous IAA balance in Arabidopsis [33–35]. Unfortunately, both IAOX and IAN have not been detected in rice, though indole-3-acetamide (IAM) is present [36, 37], thus we hypothesize that Arabidopsis and rice have different IAA biosynthesis pathways.
The pathway for IAA biosynthesis is very complex in rice, though a few genes involved in IAA signaling have been cloned, including the YUCCA family, the SMALL AUXIN-UP RNA (SAUR) family, the GH3 family and the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) family [38–42]. The cytochrome P450 family is the largest enzymatic protein family in rice and is largely responsible for both growth and development and the defense response [43–47]. In total, 356 P450 genes and 99 related pseudogenes have been identified in rice (indica and japonica) genomes using sequence information [48]. However, it remains unclear whether P450 genes involved in disease resistance to Xoo are responsible for regulating the IAA signaling pathway in rice.
A previous study by our group showed that the cytochrome P450 gene CYP71Z2 contributes to bacterial blight resistance by mediating diterpenoid phytoalexin accumulation in rice [47]. Here we present studies on the role of CYP71Z2 in auxin signaling pathway.
Materials and Methods
Constructs and transformation
To construct the CYP71Z2 promoter GUS reporter vector, the predicted 1098 bp DNA fragment upstream of the start codon was amplified from Nipponbare genomic DNA and then inserted into the binary expression vector pBI121. The primers used in this study are shown in S2 Table. The recombinant plasmids were transformed into Agrobacterium tumefaciens strain EHA105 using a freeze-thaw method. Subsequently, the T-DNA region with the predicted CYP71Z2 promoter was introduced into calli derived from mature Nipponbare embryos using the Agrobacterium-mediated method [47].
Plant materials and growth conditions
Three seedlings overexpressing CYP71Z2 and different RNAi-expressing rice were chosen to analyze the role of IAA in Xoo resistance. Transgenic seedlings (T5, T6 and T7) were grown in a growth chamber (12 h photoperiod; 28°C; 70% relative humidity; light strength, 30,000 lx), and a slow-release fertilizer was applied. All plants (wild-type and transgenic) were then transplanted into pots under normal growth conditions for Xoo inoculation, IAA quantification, RNA extraction and harvest.
Measurement of disease resistance
Resistance of rice to the bacterial blight pathogen Philippine Xoo strain PXO99A was evaluated by the leaf-clipping method at the booting stage. Level of resistance was classified into six groups and measured using the percentage of diseased area (lesion length/leaf length) at 2–3 weeks following inoculation. The six classifications are: 1) Leaves without obvious lesions have high resistance, 2) Leaves with diseased area less than 10% have resistance, 3) Leaves with diseased area ≥ 10% but < 20% have modest resistance, 4) Leaves with diseased area ≥ 20% and < 50%, have modest susceptibility, 5) Leaves with diseased area ≥ 50% and < 75% have susceptibility and 6) Leaves with diseased area ≥ 75% have high susceptibility [47]. The bacterial growth rate in rice leaves was determined by counting colony-forming units [47].
Bioinformatic analysis of CYP71Z2
The CYP71Z2 promoter region was predicted using the online Promoter Scan (http://www-bimas.cit.nih.gov/molbio/proscan/). The cis-acting regulatory DNA elements of the CYP71Z2 promoter were determined by searching in the PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) database. Phylogenetic analysis among eight species was performed by MEGA. Protein sequence alignment was performed by searching in the non-redundant protein sequences database of the NCBI.
Gene expression analyses
Leaf samples next to the sites of bacterial infection were collected for RNA extraction at different time points post-inoculation with Xoo PXO99A. Quantitative real-time PCR (qRT-PCR) was conducted on the Applied Biosystems 7500 real-time PCR system using SYBR Premix Ex TaqTM according to the manufacturer’s instructions. For qRT-PCR analysis, three independent biological samples were used, each with three technical replicates. The internal reference gene EF-1a (accession no. AK061464) was used to standardize RNA quantities. Primers used in this study for qRT-PCR are shown in S2 Table.
GUS histochemical staining and protein activity
Rice tissue, including leaf, root and stem, were put into GUS staining solution for ∼5 hours at 37°C. The staining solution was removed and 75% alcohol was added to wash off excess stain, as described previously [12]. After complete decolorization, photographs of the stained rice tissue were examined using electron microscopy.
IAA quantification
To determine the concentration of IAA in rice, at least 1 g of sample was cut from each leaf at different time points post-inoculation and kept frozen at −80°C until use. IAA measurement conditions are described in the methods of [12]. IAA concentration was determined using the HPLC/electrospray ionization/tandem mass spectrometry system and the peaks of the precursor ions 176.3 after purification with a C18-SepPak cartridge.
Results
Phylogenetic analysis of CYP71Z2
Our previous study showed that CYP71Z2, a cytochrome P450 gene involved in biosynthesis of diterpenoid phytoalexin, plays a role in resistance to bacterial blight in rice [47]. Other P450 genes, like cyp83B1, cyp79B2 and cyp79B3, are also known to play important roles for auxin metabolites in plants [31, 33]. Sequence alignment suggests that the amino acid sequence of CYP71Z2 shows similarity to the following proteins in various plants: AT3G26210 (46.4% identity, Arabidopsis thaliana, CYP71B23), LOC100824609 (62.1% identity, Brachypodium distachyon, CYP71D7-like), Sb01g020810 (61.7% identity, Sorghum bicolor), LOC100273457 (61.7% identity, Zea mays), 7467291 (50.3% identity, Populus trichocarpa, CYP71D26), LOC100794503 (50.2% identity, Glycine max), MTR-5g018990 (46.2% identity, Medicago truncatula) and LOC100263449 (46.0% identity, Vitis vinifera). However, no similarity was observed between amino acid sequences of CYP71Z2 and known P450s CYP83B1, CYP79B2 and CYP79B3. Further, a phylogenetic tree was constructed using MEGA (Molecular evolutional genetics analysis) and the eight homologous proteins having high identity with CYP71Z2 (Fig. 1). Considering the bioinformatic data, our results rationalize studying the function of the P450 gene CYP71Z2 in rice.
Fig 1. Phylogenetic relationship among the CYP71Z2 homologues in plants.
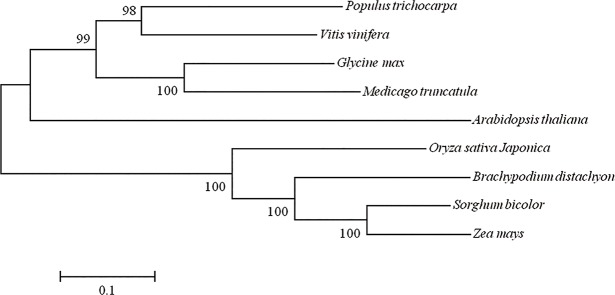
Amino acid sequences of the CYP71Z2 homologues were obtained from the NCBI Genbank. The scale bar indicates the number of amino acid substitutions per site. The numbers at the nodes indicate the level of bootstrap support.
The expression patterns of CYP71Z2
The predicted promoter region of CYP71Z2 was determined by using Promoter Scan (http://www-bimas.cit.nih.gov/molbio/proscan/) and PLACE (http://www.dna.affrc.go.jp/PLACE/). The TATA-box is located at 535 bp, and the DNA fragment of 301–551 bp is the predicted promoter region. The promoter sequence also contains cis-acting elements (CAAT-box, W-box, MYB, ASF-1, etc. binding motifs) involved in salicylic acid, auxin, photo-responsive and flavonoid biosynthesis processes.
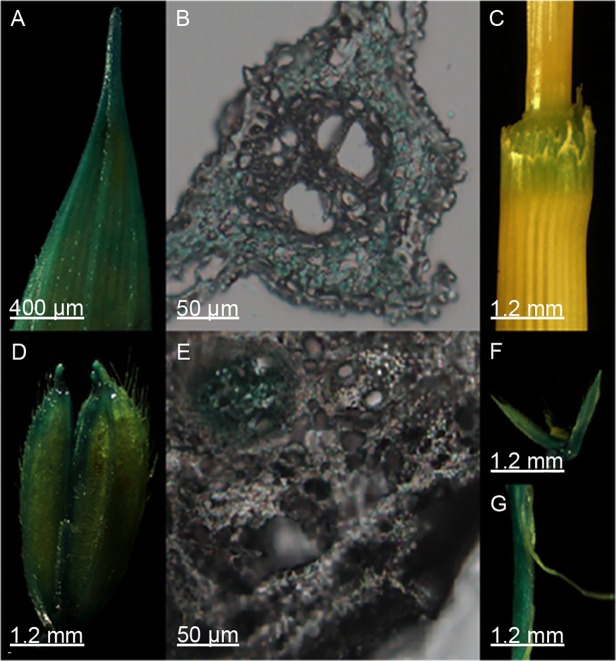
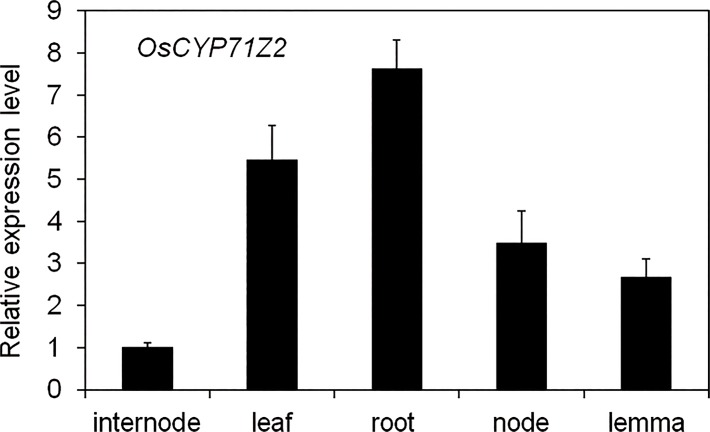
To assess the expression pattern of CYP71Z2, a CYP71Z2 promoter/GUS protein (β-glucuronidase) fusion expression vector was constructed (S1 Fig.). CYP71Z2 promoter/GUS transgenic plants were generated by transforming japonica cultivar Nipponbare. All seven confirmed transgenic lines showed a common pattern of GUS distribution, although differences were observed in GUS activity. GUS histochemical staining showed CYP71Z2 mainly expressed in the leaves, node, lemma, palea and primary roots, indicating tissue-specific expression patterns of CYP71Z2 in rice (Fig. 2). Gene expression patterns were analyzed by qRT-PCR, which showed high expression in the leaves, root node and internodes, consistent with results from GUS histochemical staining (Fig. 3).
Fig 2. Expression patterns of GUS driven by the CYP71Z2 promoter in various organs and tissues of transgenic rice plant.
Shown are leaf (A), node (C), lemma and palea (D, F), primary root (G) and transverse section of a leaf (B) and node (E). Scale bars are 400 μm (A), 50 μm (B, E) and 1.2 mm (C, D, F and G).
Fig 3. Expression levels of CYP71Z2 in leaf, root, node, internode and lemma.
Data represent means of three replicates ± standard deviation.
Expression patterns of CYP71Z2 during incompatible and compatible interactions between rice and bacterial blight were detected by qRT-PCR, with results showing that expression of CYP71Z2 in resistant rice NJH12 was higher than that in susceptible rice R109 (Fig. 4A). In addition, GUS activity in the leaves of CYP71Z2 promoter-driven transgenic rice plants significantly increased after inoculation with Xoo (Fig. 4B). These results suggest that CYP71Z2 is quickly activated in rice upon infection with the bacterial blight pathogen Xoo.
Fig 4. Expression of CYP71Z2 was induced in Xoo resistant rice cultivar upon Xoo infection.
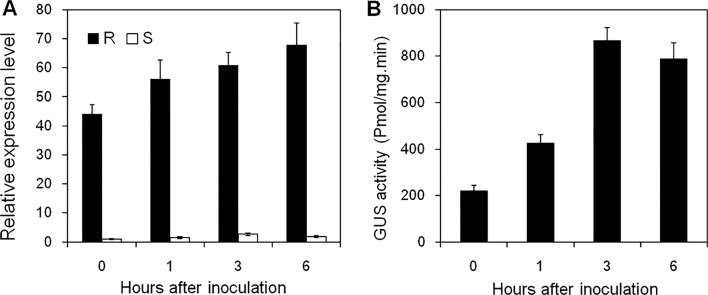
(A) Rice cultivar resistance to Xoo had a much higher basal expression level of CYP71Z2 than rice cultivar susceptible to Xoo, and showed an induced expression level of CYP71Z2 after inoculation with Xoo PXO99A. R, Xoo resistant rice cultivar NJH12. S, Xoo susceptible rice cultivar R109. (B) An increased GUS activity was observed in transgenic rice plants harbouring the PCYP71Z2::GUS construct after inoculation of Xoo.
Overexpressing CYP71Z2 increases resistance to Xoo
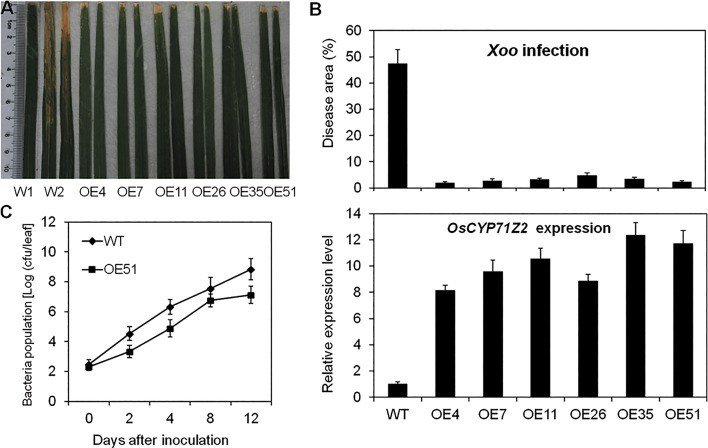
Previous studies have shown that the rice P450 gene CYP71Z2 is involved in diterpenoid phytoalexin biosynthesis, contributing to bacterial blight resistance [47]. In this study, we selected six CYP71Z2-overexpressing rice (Acceptor rice is the Nipponbare; T5, T6 and T7) to identify the role of auxin in CYP71Z2-mediated blight resistance at the booting stage. Six homozygous CYP71Z2-overexpressing lines showed resistance to Xoo strain PXO99A, with the average lesion area ranging from 1.86–4.75%, compared with an average of 47.37% in wild-type Nipponbare (Fig. 5A, B; S1 Table). The expression of CYP71Z2 in all six T7 CYP71Z2-overexpressing plants was higher than that in wild-type, showing increases of approximately 8.16- to 12.35-fold (Fig. 5B; S1 Table). Furthermore, bacterial growth analysis showed that the growth rate of PXO99A in T7 CYP71Z2-overexpressing line OE51 was lower than that in wild-type after inoculation (Fig. 5C). These results suggest that overexpression of CYP71Z2 confers rice with durable, stable resistance to Xoo.
Fig 5. Increased resistance to Xoo in CYP71Z2-overexpressing lines.
(A) CYP71Z2-overexpressing lines (T7) showed enhanced resistance to Xoo strain PXO99A. (B) Overexpression of CYP71Z2 was positively correlated with suppression of disease development. CYP71Z2 expression was analyzed by qRT-PCR. Data represent means of three replicates ± standard deviation. (C) Growth of Xoo strain PXO99A in leaves of rice plants overexpressing CYP71Z2 was suppressed. W1, Wild-type Nipponbare inoculated with water. W2, Wild-type Nipponbare inoculated with Xoo. cfu, colony-forming unit. Data represent means of three replicates ± standard deviation.
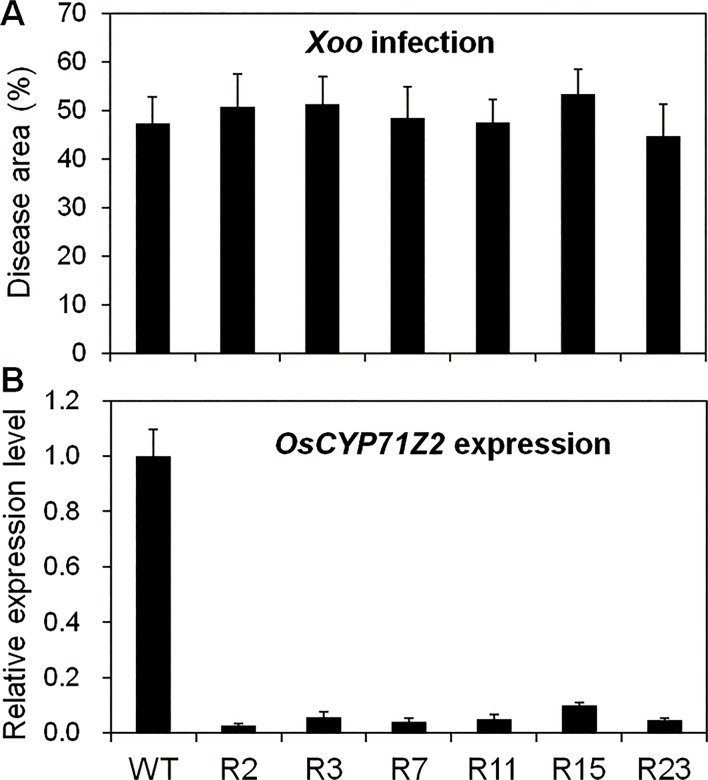
We also examined resistance to Xoo in CYP71Z2-RNAi lines (T5, T6 and T7) at the booting stage. Expression of CYP71Z2 in RNAi lines was significantly reduced, as shown in Fig. 6. Moreover, none of the RNAi lines showed a significant difference in response to PXO99A infection compared with wild-type (Fig. 6; S1 Table). These results show that suppression of CYP71Z2 expression does not significantly increase susceptibility to PXO99A in CYP71Z2-RNAi transgenic lines, suggesting that functional redundancy among the CYP71Z of family may mask the effect of CYP71Z2.
Fig 6. Knock-down CYP71Z2 had no impact on Xoo resistance.
(A) No significant difference in disease development was observed between CYP71Z2-RNAi lines and wild-type in response to inoculation of Xoo strain PXO99A. (B) Expression levels of CYP71Z2 in CYP71Z2-RNAi lines and wild-type. Data represent means of three replicates ± standard deviation.
CYP71Z2 negatively regulates IAA metabolism
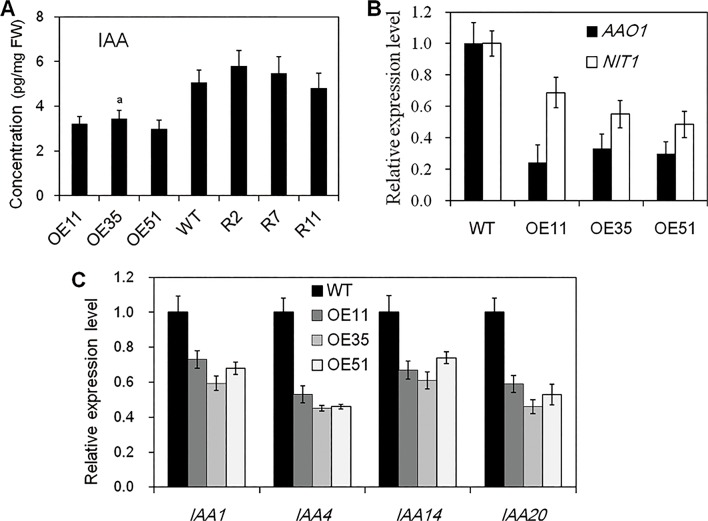
The endogenous phytohormone auxin is known to be involved in resistance of rice to bacterial blight [12], though it is unclear if auxin is involved in signaling pathways that contribute to CYP71Z2-mediated blight resistance. To examine this possibility, we measured free IAA concentration of CYP71Z2 transgenic lines at the booting stage. The concentration of endogenous free IAA in the CYP71Z2-overexpressing lines OE11, OE35 and OE51 was 3.21, 2.99 and 3.64 pg/mg fresh leaves, respectively. Compared with 5.07 pg/mg fresh leaves in wild-type, these results suggest that IAA expression is 1.47- to 1.7-fold lower than that of wild-type plants (Fig. 7A), which likely contributes to the resistance to Xoo of these transgenic plants. Moreover, the endogenous free IAA levels in the leaves of CYP71Z2-RNAi lines showed no significantly differences compared to those of wild-type (Fig. 7A), supporting the hypothesis of functional redundancy among CYP71Z family proteins in rice.
Fig 7. Overexpression of CYP71Z2 suppressed accumulation of endogenous IAA and the expression levels of genes related to auxin biosynthesis and signaling.
(A) Quantification of free IAA content in the leaves of CYP71Z2-overexpressing rice plants at the ripening stage. (B) Expression patterns of genes related to auxin biosynthesis in CYP71Z2-overexpressing rice plants. (C) Expression patterns of auxin-responsive genes in CYP71Z2-overexpressing rice plants. Data represent means of three replicates ± standard deviation. a indicates that a significant difference (P < 0.05) was detected between CYP71Z2-overexpressing rice and wild-type plants. WT, wild-type Nipponbare; FW, fresh weight.
To further analyze whether decreased IAA in CYP71Z2-overexpressing lines was caused by other genes of the IAA biosynthesis pathway, we quantified the expression of AAO1 (indole-3-acetaldehyde oxidase) and NIT1 (nitrilase) in rice using qRT-PCR. The sequence alignment showed 72% and 78% identity with homologous genes AAO1 and NIT1, respectively, in Arabidopsis [12]. Previous reports indicated that AAO1 and NIT1 function in two Trp-dependent IAA biosynthesis pathways (indole-3-pyruvic acid and indole-3-acetaldoxime) [12, 38]. Quantitative analysis showed that expression of AAO1 and NIT1 in OE11, OE35 and OE51 CYP71Z2–overexpressing plants was lower by 3.01 to 4.12-fold and 1.46 to 2.01-fold, respectively, than that in wild-type (Fig. 7B). These results support the notion that CYP71Z2 negatively regulates AAO1 and NIT1 expression to suppress IAA accumulation.
In addition, previous reports demonstrated that auxin signaling is also affected by changes in IAA concentration [12]. To evaluate this in our study, we analyzed the expression of auxin signaling-related genes (Aux/IAA families) in CYP71Z2–overexpressing lines by qRT-PCR. These results show that expression of IAA1, IAA4, IAA14 and IAA20 was lower in CYP71Z2–overexpressing lines (OE11, OE35 and OE51) than in wild-type, especially with respect to IAA4 and IAA20 (Fig. 7C).
IAA biosynthesis is suppressed in CYP71Z2–overexpressing rice after inoculation with Xoo
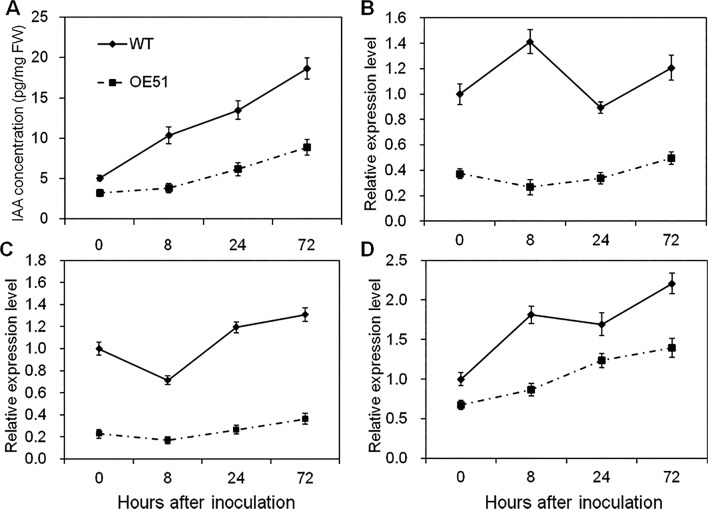
Previous reports demonstrated that auxin signaling is involved in rice-Xoo interactions, with auxin seeming to act as a negative regulator of resistance to Xoo in rice [10–12]. To further examine whether auxin signaling takes part in disease resistance to Xoo, we analyzed the IAA concentration in the CYP71Z2–overexpressing rice OE51 after inoculation with Xoo strain PXO99A. As shown in Fig. 8A, accumulation of IAA was induced at 8, 24 and 72 hours post-inoculation in both OE51 and wild-type. However, IAA accumulation in OE51 was found to be significantly lower than that in wild-type regardless of whether the plants were inoculated or not, with up to a 2.7-fold decrease in accumulation at 8 hours after innoculation (Fig. 8A). These results suggest that overexpression of CYP71Z2 in rice negatively regulates IAA biosynthesis in response to Xoo infection.
Fig 8. Overexpression of CYP71Z2 suppressed the IAA signaling pathway in rice after inoculation with PXO99A.
(A) Quantification of free IAA in the leaves of CYP71Z2-overexpressing rice after inoculation at the ripening stage. Transcript levels of genes AAO1 (B), IAA1 (C) and IAA20 (D) in CYP71Z2-overexpressing rice after inoculation were determined by qRT-PCR. Data represent means of three replicates ± standard deviation.
The expression of auxin signaling and biosynthetic genes was also analyzed by qRT-PCR in the resistant transgenic OE51 and the susceptible wild-type rice following Xoo inoculation. Expression of AAO1, IAA1 and IAA20 was induced in both OE51 and wild-type plants after infection, though expression of AAO1 and IAA20 was significantly decreased in wild-type at 24 h post-inoculation (Fig. 8B, C and D). However, we found that expression of AAO1, IAA1 and IAA20 in OE51 was significantly suppressed compared with wild-type after inoculation (Fig. 8B, C and D), suggesting that overexpression of CYP71Z2 in rice suppresses the expression of genes involved in auxin signaling.
Overexpressing CYP71Z2 inhibits expression of expansin genes
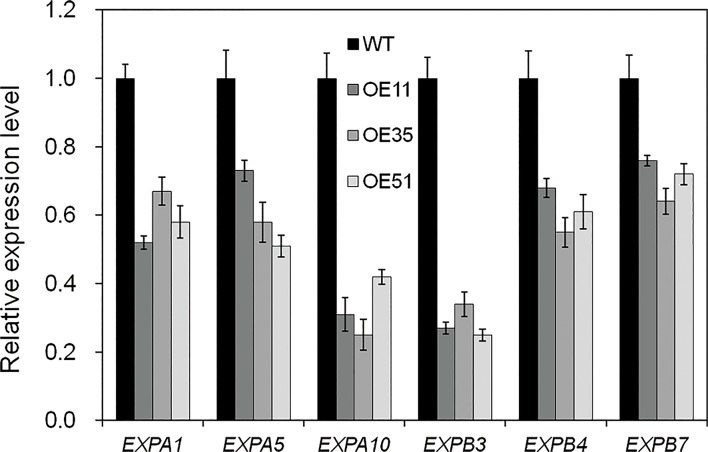
The plant cell wall is a natural protective barrier for phytopathogens. The loosening cell walls are easier to be infected by pathogenic bacteria. Suppression of expansion genes can prevent plant cell walls from loosening, resulting in enhanced physical protection of plants to phytopathogens [12]. To examine their role in the resistance to Xoo, we determined the expression of six expansin genes in CYP71Z2-overexpressing rice, including three rice α-expansin genes (EXPA1, EXPA5 and EXPA10) and three rice β-expansin genes (EXPB3, EXPB4 and EXPB7). qRT-PCR analysis showed that expression of all six expansin genes was decreased in CYP71Z2-overexpressing rice compared with wild-type under normal growth condition (Fig. 9). These results demonstrate that expansin genes may partly contribute to the resistance to Xoo in CYP71Z2-overexpressing rice.
Fig 9. Overexpression of CYP71Z2 had a negative impact on the expression of expansin genes.
Data represent means of three replicates ± standard deviation. WT, wild-type Nipponbare.
The SA/JA pathway is not involved in Xoo resistance of CYP71Z2-overexpressing rice
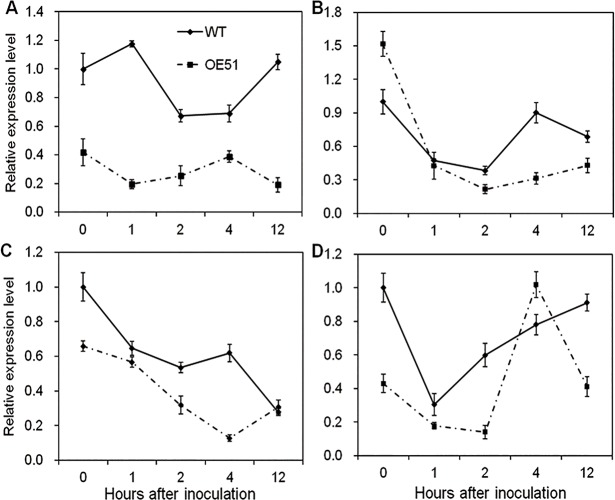
Previous studies suggested that SA/JA defense responses are independent of the resistance pathway mediated by auxin in rice [12]. To test whether the increased resistance to Xoo in CYP71Z2-overexpressing rice accompanied by the inactivation of SA- and JA-dependent defense pathways, we detected transcripts of four key genes (PR1a, PR1b, LOX and AOS2) that act in two distinct classes of defense signaling pathways. Relative expression levels analyzed by qRT-PCR showed that three genes had lower expression in OE51 than in wild-type without inoculation, with reductions in AOS2, LOX and PR1a being 1.52-, 2.33- and 2.39-fold, respectively, although PR1b was increased by 1.52-fold in OE51 compared with wild-type (Fig. 10). During Xoo infection, these four genes were largely suppressed in OE51 at most time points (Fig. 10). Gene expression analysis demonstrated that CYP71Z2 may function as a negative regulator of the SA/JA defenses signaling pathways during the incompatible interaction between rice and Xoo, which is consistent with results shown in a previous study [12].
Fig 10. Overexpression of CYP71Z2 inhibited the expression levels of genes involved in disease resistance pathway mediated by SA/JA.
The expression levels of four genes PR1a (A), PR1b (B), AOS2 (C) and LOX (D) functioning in the SA/JA-dependent disease resistance pathways in CYP71Z2-overexpressing rice plant. Data represent means of three replicates ± standard deviation.
Discussion
The mechanisms behind bacterial and fungal disease resistance in rice remain largely unknown, though some GH3 genes and auxin biosynthesis regulators have been implicated in the process. Our previous study showed that the P450 gene CYP71Z2 is involved in resistance to Xoo infection through activation of the phytoalexin biosynthesis pathway. In this study, we’ve found that overexpression of CYP71Z2 confers transgenic (T5, T6 and T7) rice with durable, stable resistance to bacterial blight, which is accompanied by up-regulation of genes related to IAA biosynthesis and IAA response pathways. Moreover, no significant differences were observed in resistance to Xoo and IAA accumulation between CYP71Z2-RNAi and wild-type lines, suggesting some functional redundancy to compensate for reduced CYP71Z2 expression. These results demonstrate that the cytochrome P450 gene CYP71Z2 is involved in disease resistance to Xoo, potentially through negative regulation of IAA/auxin biosynthesis.
The role of auxin in plant disease resistance has been widely studied, with recent evidence demonstrating IAA’s role as a negative regulator of plant disease resistance to bacterial and fungal pathogens [49, 50]. The role of GH3-like proteins in disease resistance mediated by IAA has also been gradually elucidated over recent years [12, 13, 21]. In this study, homozygous CYP71Z2-overexpressing rice showed durable resistance to Xoo accompanied by a reduction in IAA accumulation (Fig. 5, 7), consistent with the resistance phenotype of IAA-deficient plants in previous reports [12, 13, 21]. In addition, the putative indole-3-acetaldehyde oxidase (AAO1) and nitrilase (NIT1), two key genes related to auxin synthesis in rice [12, 38], were found to have reduced expression in CYP71Z2-overexpressing rice (Fig. 7). These results further confirm that suppression of auxin biosynthesis contributes to disease resistance of CYP71Z2-overexpressing rice, and overall importance of auxin regulation in response to pathogenic infection.
P450 genes have been reported to either act as either positive or negative regulators of auxin homeostasis in Arabidopsis. A plausible explanation for this may be that the substrates catalyzed by different P450 oxidases are different, resulting in changes in IAA production. For example, cyp83B1 mutants show significant overproduction of auxin, whereas CYP83B1-overexpressing lines display a loss of apical dominance that is typical of auxin deficiency [33–35]. These studies also showed that the CYP83B1 protein is responsible for converting IAOx to 1-aci-nitro-2-indolyl-ethane, which functions to maintain the dynamic balance between IAA and indole glucosinolate metabolism [34, 35]. In addition, the cytochrome P450 enzyme CYP79B2 catalyzes the transformation of tryptophan into IAOx, playing a positive role in IAA biosynthesis [29, 30]. In this study, we demonstrate that another P450 gene, CYP71Z2, shows similar results when overexpressed as the CYP83B1 mutants, suggesting that CYP71Z2 plays a negative regulatory role in IAA biosynthesis. Unfortunately, substrates for CYP71Z2 and the mechanism behind this role have not yet been identified.
IAOx and IAN are two key intermediates of camalexin metabolism and IAA biosynthesis in Arabidopsis, suggesting cross-talk between these two pathways [22, 24]. Overexpression of CYP79B2 in Arabidopsis has been shown to increase IAA content and lead to excessive auxin production, which was likely due to CYP79B2 catalyzing the transformation of tryptophan into IAOx. Interestingly, CYP79B2/CYP79B3 double mutants had reduced levels of both IAA and camalexin, suggesting some degree of similar regulation between the two pathways [24, 32]. Moreover, CYP71A13 was shown to catalyze the conversion of IAOx to IAN, which also led to reductions in IAN and camalexin upon cyp71A13 mutation [32]. Taken together, these data indicate that cross-talk likely exists between the auxin and camalexin biosynthetic pathways in Arabidopsis. Results from our study showing reduced IAA accumulation in CYP71Z2-overexpressing rice (Fig. 7A), in conjunction with previous reports showing that CYP71Z2 accelerates phytoalexin biosynthesis [47], lead us to speculate that cross regulation of IAA and phytoalexin biosynthesis also exists in rice, though this hypothesis requires further study.
Many phytopathogens produce IAA for survival and multiplication during the infection process [16–20]. Pathogen-produced IAA leads to induction of the expression of rice expansin genes, resulting in an increase in long-term cell wall flexibility [12, 51]. This process makes plant cell walls vulnerable and contributes to pathogen infection and multiplication in rice. In this study, the relative expression of expansin genes in Xoo-resistant, CYP71Z2-overexpressing rice was significantly decreased and correlated with suppression of IAA signaling (Fig. 7–9). These results suggest that the suppression of expansin genes may also contribute to disease resistance in CYP71Z2-overexpressing rice, though the mechanisms behind this remain unclear.
As has been shown, auxin biosynthesis is suppressed in resistant rice and is always accompanied by decreases in the expression of auxin-responsive genes. The expression of auxin signaling–related genes was found to be significantly decreased in auxin-deficient, GH3-8-overexpressing plants exhibiting resistance to Xoo [12, 52, 53]. Consistently, the accumulation of auxin signaling-related genes AAO1, IAA1 and IAA20 was inhibited in CYP71Z2-overexpressing rice (Fig. 8). This suggests that suppression of auxin response pathways results from reduced IAA accumulation in CYP71Z2-overexpressing rice.
JA and SA signaling pathways play an important role in broad-spectrum and durable disease resistance of rice. More studies are finding that immunity conferred by SA or JA is independent of IAA resistance signaling in plants, with no correlation reported between suppression of auxin signaling and the activation of SA/JA signaling pathways in resistant rice [12, 49]. Moreover, plant immunity mediated by SA is often accompanied by inhibition of auxin signaling, including down-regulation of auxin-response genes and IAA-amido synthase genes of the GH3 family [54]. In this study, qRT-PCR analysis showed that the expression of four key genes involved in SA/JA signaling was significantly decreased (Fig. 10), suggesting that SA and JA signaling pathways are inhibited by overexpression of CYP71Z2 in rice. These results demonstrate that activation of SA or JA signaling pathways is not required for disease resistance mediated by IAA in CYP71Z2-overexpressing rice.
The P450 family is the largest protein family in rice and plays an important role in the growth, development and defense responses of this plant. The function of some P450 genes in auxin biosynthesis has been studied in Arabidopsis, although so far, similar functionality has not been studied for P450 genes in rice. In this study, the overexpression of CYP71Z2 in rice increased resistance to Xoo PXO99A with suppression of IAA accumulation and IAA response genes, suggesting that the P450 gene CYP71Z2 takes part in IAA signaling in rice. Moreover, no significant differences in IAA accumulation were detected between CYP71Z2-RNAi rice and wild-type, which could be due to residual CYP71Z2 mRNA or functional redundancy among CYP71Z subfamily proteins. Regardless, these results show that a P450 gene plays a significant role in resistance to pathogen infection in transgenic rice by mediation of the auxin signaling pathway.
Supporting Information
(A) Promoter clone and vectors construction. 1, the amplification of CYP71Z2 promoter fragment; 3, the double enzyme digestion of P121/PRO plasmids; 2 and 5, DNA Marker DL 2000; 4, DNA Marker λ-HindIII. (B) Schematic representation of the transformation constructions for CYP71Z2 expression pattern. RB and LB indicate the right and left T-DNA borders, respectively; NOS indicates the nopaline synthase terminator; NOSP indicates the promoter of the gene encoding nopaline synthetase; NPTII indicates the bacterial kanamycin resistance gene (selection marker); Pro indicates the promoter of CYP71Z2; gus indicates the E. coli β-glucuronidase gene.
(TIF)
(DOC)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31301652), the Key Projects in the National Science & Technology Pillar Program (2011BAD16B03), the Natural Science Foundation of Jiangsu Province of China (BK20130723) and the Jiangsu Province Independent Innovation Project no. CX(14)5003.), the Special Fund for Agro-scientific Research in the Public Interest (201303102), the special fund of Jiangsu Province for the Transformation of Scientific and Technological Achievements (BAAA2014134). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhu YY, Chen HR, Fan JH, Wang YY, Li Y, Chen JB, et al. Genetic diversity and disease control in rice. Nature. 2000: 406: 718–722. [DOI] [PubMed] [Google Scholar]
- 2. Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004; 42: 185–209. [DOI] [PubMed] [Google Scholar]
- 3. Jones JD, Dangl JL. The plant immune system. Nature. 2006; 444: 323–329. [DOI] [PubMed] [Google Scholar]
- 4. Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009; 12: 414–420. 10.1016/j.pbi.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 5. Block A, Alfano JR. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol. 2011; 14: 39–46. 10.1016/j.mib.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, et al. Induction of systemic acquired disease resistance in plants by chemicals. Annu Rev Phytopathol. 1994; 32: 439–459. [DOI] [PubMed] [Google Scholar]
- 7. Spoel SH, Dong X. Making sense of hormone cross-talk during plant immune responses. Cell Host Microbe. 2008; 3: 348–351. 10.1016/j.chom.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 8. Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nature Reviews Mol Cell Biol. 2006; 7: 847–859. [DOI] [PubMed] [Google Scholar]
- 9. McSteen P. Auxin and monocot development. Cold Spring Harb Perspect Biol. 2010; 2(3): a001479 10.1101/cshperspect.a001479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang DL, Yang YN, He ZH. Roles of plant hormones and their interplay in rice immunity. Mol Plant. 2013; 6: 675–685. 10.1093/mp/sst056 [DOI] [PubMed] [Google Scholar]
- 11. Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006; 312: 436–439. [DOI] [PubMed] [Google Scholar]
- 12. Ding XH, Cao YL, Huang LL, Zhao J, Xu CG, Li XH, et al. Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. The Plant Cell. 2008; 20: 228–240. 10.1105/tpc.107.055657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu J, Liu HB, Li Y, Yu HH, Li XH, Xiao JH, et al. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 2011, 155: 589–602. 10.1104/pp.110.163774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, et al. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA. 2007; 104: 20131–20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007; 17: 1784–1790. [DOI] [PubMed] [Google Scholar]
- 16. Fett WF, Osman SF, Dunn MF. Auxin production by plant–pathogenic pseudomonads and xanthomonads. Appl Environ Microbiol. 1987; 53: 1839–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Canadian J Microbiol, 1996; 42: 207–220. [DOI] [PubMed] [Google Scholar]
- 18. Khalid A, Tahir S, Arshad M, Zahir ZA. Relative efficiency of rhizobacteria for auxin biosynthesis in rhizosphere and non-rhizosphere soils. Aus J Soil Res. 2004; 42: 921–926. [Google Scholar]
- 19. Kazan K, Manners JM. Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 2009; 14: 373–382. 10.1016/j.tplants.2009.04.005 [DOI] [PubMed] [Google Scholar]
- 20. Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol. 2011; 3: a001438 10.1101/cshperspect.a001438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact. 2009; 22: 201–210. 10.1094/MPMI-22-2-0201 [DOI] [PubMed] [Google Scholar]
- 22. Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis . Proc Natl Acad Sci USA. 2011; 108: 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012; 63: 2853–2872. 10.1093/jxb/ers091 [DOI] [PubMed] [Google Scholar]
- 24. Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis . Proc Natl Acad Sci USA. 2004; 101: 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000; 97: 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002; 7: 263–270. [DOI] [PubMed] [Google Scholar]
- 27. Schuhegger R, Rauhut T, Glawischnig E. Regulatory variability of camalexin biosynthesis. J Plant Physiol. 2007; 164: 636–644. [DOI] [PubMed] [Google Scholar]
- 28. Wang MY, Liu XT, Chen Y, Xu XJ, Yu B, Zhang SQ, et al. Arabidopsis acetyl-amido synthetase GH3.5 involvement in camalexin biosynthesis through conjugation of indole-3-carboxylic acid and cysteine and upregulation of camalexin biosynthesis genes. J Integr Plant Biol. 2012; 54: 471–485. 10.1111/j.1744-7909.2012.01131.x [DOI] [PubMed] [Google Scholar]
- 29. Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetal-doxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000; 275: 33712–33717. [DOI] [PubMed] [Google Scholar]
- 30. Mikkelsen MD, Naur P, Halkier BA. Arabidopsis mutants in the C-S lyase of glucosenolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004; 37: 770–777. [DOI] [PubMed] [Google Scholar]
- 31. Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, et al. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002; 16: 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, et al. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell. 2007; 19: 2039–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Malcolm Bennett, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA. 2000; 97: 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bak S, Feyereisen R. The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001; 127: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis . Plant Cell. 2001; 13: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Tomokazu Koshiba, et al. Biochemical analyses of indole-3-acetaldoxime dependent auxin biosynthesis in Arabidopsis . Proc Natl Acad Sci USA. 2009; 106: 5430–5435. 10.1073/pnas.0811226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012; 72: 523–536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- 38. Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot (Lond). 2005; 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics. 2006; 6: 47–59. [DOI] [PubMed] [Google Scholar]
- 40. Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin iosynthesis by the YUCCA genes in rice. Plant Physiol. 2007; 143: 1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA . Physiol Plant. 2012; 46: 487–499. [DOI] [PubMed] [Google Scholar]
- 42. Ljung K. Auxin metabolism and homeostasis during plant development. Dev. 2013; 140: 43–50. 10.1242/dev.085290 [DOI] [PubMed] [Google Scholar]
- 43. Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Annu Rev Plant Biol. 2003; 54: 629–667. [DOI] [PubMed] [Google Scholar]
- 44. Rupasinghe S, Schuler MA. Homology modeling of plant cytochrome P450s. Phytochem Rev. 2006; 5: 473. [Google Scholar]
- 45. Zhu YY, Nomura T, Xu YH, Zhang YY, Peng Y, Mao BZ, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006; 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, et al. Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell. 2010; 22: 173–190. 10.1105/tpc.109.070326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li WQ, Shao M, Yang J, Zhong WG, Okada K, Yamane H, et al. Oscyp71Z2 involves diterpenoid phytoalexin biosynthesis that contributes to bacterial blight resistance in rice. Plant Sci. 2013; 207: 98–107. 10.1016/j.plantsci.2013.02.005 [DOI] [PubMed] [Google Scholar]
- 48. Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 2004; 135: 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, et al. Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant, 2008; 1: 496–509. 10.1093/mp/ssn025 [DOI] [PubMed] [Google Scholar]
- 50. Eshraghi L, Anderson JP, Aryamanesh N, McComb JA, Shearer B, Hardy GS. Suppression of the auxin response pathway enhances susceptibility to Phytophthora cinnamomi while phosphite-mediated resistance stimulates the auxin signalling pathway. BMC Plant Biol. 2014; 14: 68 10.1186/1471-2229-14-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cosgrove DJ. Wall extensibility: its nature, measurement, and relationship to plant cell growth. New Phytol. 1993; 124: 1–23. [DOI] [PubMed] [Google Scholar]
- 52. Zhang J, Peng YL, Guo ZJ. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008; 18: 508–521. [DOI] [PubMed] [Google Scholar]
- 53. Song YL, You J, Xiong LZ. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol. 2009; 70: 297–309. 10.1007/s11103-009-9474-1 [DOI] [PubMed] [Google Scholar]
- 54. Fu J, Wang SP. Insights into auxin signaling in plant–pathogen interactions. Front Plant Sci. 2011; 2: 74 10.3389/fpls.2011.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Promoter clone and vectors construction. 1, the amplification of CYP71Z2 promoter fragment; 3, the double enzyme digestion of P121/PRO plasmids; 2 and 5, DNA Marker DL 2000; 4, DNA Marker λ-HindIII. (B) Schematic representation of the transformation constructions for CYP71Z2 expression pattern. RB and LB indicate the right and left T-DNA borders, respectively; NOS indicates the nopaline synthase terminator; NOSP indicates the promoter of the gene encoding nopaline synthetase; NPTII indicates the bacterial kanamycin resistance gene (selection marker); Pro indicates the promoter of CYP71Z2; gus indicates the E. coli β-glucuronidase gene.
(TIF)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.