Abstract
The dorsomedial hypothalamus (DMH) controls a number of essential physiological responses. We have demonstrated that the DMH plays an important role in the regulation of mammalian aging and longevity. To further dissect the molecular basis of the DMH function, we conducted microarray-based gene expression profiling with total RNA from laser-microdissected hypothalamic nuclei and tried to find the genes highly and selectively expressed in the DMH. We found neuropeptide VF precursor (Npvf),PR domain containing 13 (Prdm13), and SK1 family transcriptional corepressor (Skor1) as DMH-enriched genes. Particularly, Prdm13, a member of the Prdm family of transcription regulators, was specifically expressed in the compact region of the DMH (DMC), where Nk2 homeobox 1 (Nkx2-1) is predominantly expressed. The expression of Prdm13 in the hypothalamus increased under diet restriction, whereas it decreased during aging. Prdm13 expression also showed diurnal oscillation and was significantly upregulated in the DMH of long-lived BRASTO mice. The transcriptional activity of the Prdm13 promoter was upregulated by Nkx2-1, and knockdown of Nkx2-1 suppressed Prdm13 expression in primary hypothalamic neurons. Interestingly, DMH-specific Prdm13-knockdown mice showed significantly reduced wake time during the dark period and decreased sleep quality, which was defined by the quantity of electroencephalogram delta activity during NREM sleep. DMH-specific Prdm13-knockdown mice also exhibited progressive increases in body weight and adiposity. Our findings indicate that Prdm13/Nkx2-1-mediated signaling in the DMC declines with advanced age, leading to decreased sleep quality and increased adiposity, which mimic age-associated pathophysiology, and provides a potential link to DMH-mediated aging and longevity control in mammals.
Keywords: aging, age-associated pathophysiology, Dorsomedial hypothalamus, DMH-enriched gene, Nkx2-1, NREM delta power, Prdm13
Introduction
The hypothalamus is structurally divided into several functionally distinct areas called nuclei, such as the arcuate, ventromedial, dorsomedial, lateral, paraventricular, and suprachiasmatic nuclei of the hypothalamus (Arc, VMH, DMH, LH, PVN, and SCN, respectively). Each hypothalamic nucleus plays a crucial role in the regulation of various physiological responses, including feeding behavior, metabolism, endocrine regulation, physiological rhythm, emotion, and aging (Satoh & Imai, 2014). Recent findings demonstrate that the hypothalamus functions as a high-order ‘control center of aging,’ counteracting age-associated functional changes and thereby promoting longevity in mammals (Satoh et al., 2013; Zhang et al., 2013). Zhang et al. demonstrated that NF-κB signaling in the mediobasal hypothalamus (MBH) is significantly enhanced with advanced age, and suppressing NF-κB signaling in the MBH retards aging and extends lifespan in mice (Zhang et al., 2013). We also demonstrated that the mammalian NAD+-dependent protein deacetylase Sirt1 in the hypothalamus, particularly the DMH and LH, is critical to counteract age-associated declines in skeletal muscle mitochondrial function, physical activity, body temperature, oxygen consumption, and sleep quality, and promote longevity in mice (Satoh et al., 2013). Remarkably, increasing Sirt1 dosage only in the DMH is sufficient to regain physical activity in aged mice to a similar level as in young mice (Satoh et al., 2013). These results clearly demonstrate the importance of the hypothalamus, more specifically DMH function, in controlling mammalian aging.
Aging has a significant impact on DMH function. The DMH has been known to regulate body temperature (Zaretskaia et al., 2003; Morrison et al., 2008; Enriori et al., 2011), food-anticipating activity (Gooley et al., 2006; Acosta-Galvan et al., 2011), autonomic stress responses (Ulrich-Lai & Herman, 2009), food intake (Yang et al., 2009), and reproduction (Kirby et al., 2009; Soga et al., 2014). Studies illustrate several examples of the decline in DMH function with age. The capacity of thermoregulation declines with age (Reynolds et al., 1985). In humans, it has been reported that mean temperature declines with age, after controlling for sex, body mass index, and white blood cell count (Waalen & Buxbaum, 2011). Food-anticipating activity in response to timed-restricted feeding also declines in old rats (Shibata et al., 1994). The peripheral hormone ghrelin activates the DMH neurons, causing the enhancement of food intake, and this enhancement by ghrelin declines in old mice (Akimoto & Miyasaka, 2010). Finally, it has been reported that the ability of the reproductive system declines with advanced age, partly due to the reduction of DMH function (Soga et al., 2014). Given those findings, it is clear that DMH function declines over age. Thus, it is important to know the mechanisms by which the DMH controls these functions to treat and/or prevent age-associated pathophysiology.
To dissect the molecular basis of DMH function, we decided to search for the genes highly and selectively expressed in the DMH. Transcriptome studies have so far shown that particular subsets of neuronal genes exhibit clear spatial expression patterns across the adult mouse brain, suggesting that these spatial expression patterns are related to the unique physiological functions of distinct brain regions (Wang et al., 2010; Ko et al., 2013). Whereas other hypothalamic nuclei such as the Arc, VMH, LH, and PVN have been well characterized for their genetic and chemical identities (Elmquist et al., 1999; Leak & Moore, 2001; Gottsch et al., 2004; Segal et al., 2005), efforts to characterize the DMH have been very limited. Lee et al. (2012) reported a comprehensive gene profile for the ventral subdivision of the DMH (DMV) where the leptin receptor is exclusively expressed (Zhang et al., 2011). Although the DMH-enriched genes identified in this study are only moderately changed by dietary manipulations (Lee et al., 2012), subsequent studies have proven the importance of these genes in response to leptin (Bechtold et al., 2012). These findings indicate the potential for exploring the DMH-enriched genes to further dissect the physiological role of the DMH in neurobehavioral and metabolic regulation.
In this study, we identified several genes that display DMH-enriched expression patterns by performing gene expression profiling with RNA samples collected from laser-microdissected hypothalamic nuclei. We examined the distribution of these candidate genes in the subcompartments of the DMH and evaluated changes in gene expression under diet restriction (DR), through the diurnal cycle, and with age. Among those genes, we identified Prdm13, a member of the Prdm family (Fog et al., 2012; Hohenauer & Moore, 2012), as a highly DMH-enriched gene. The expression of Prdm13 increased under DR, whereas its expression decreased with advanced age. Moreover, the expression level of Prdm13 in the DMH of long-lived brain-specific Sirt1-overexpressing transgenic (BRASTO) mice is significantly higher than the level in wild-type control mice. Interestingly, we found that mice with DMH-specific knockdown of Prdm13 displayed a robust decline in sleep quality and a progressive increase in body weight. These findings provide a novel possibility that a specific subset of neurons, characterized by the expression of DMH-enriched genes, regulate sleep homeostasis and metabolism and that the functional deterioration of those specific DMH neurons could contribute to age-associated pathophysiology in mammals.
Results
Identification of genes enriched in the Arc, VMH, DMH, and LH
To identify genes selectively expressed in the DMH, we collected RNA samples from four hypothalamic nuclei, namely the Arc, VMH, DMH, and LH, using laser microdissection (Fig. S1) and conducted microarray analysis to compare their gene expression profiles. We determined genes selectively expressed greater than or equal to 10-fold in the Arc, VMH, DMH, and LH, respectively, compared to the other hypothalamic nuclei by calculating ratios of the signal intensities between each hypothalamic nucleus. Using this criterion, 21, 15, four, and eleven genes were found as genes enriched in the Arc, VMH, DMH, and LH, respectively (Fig.1 and Table S1). We identified neuropeptide VF precursor (Npvf) (NM_021892), gastrin-releasing peptide (Grp) (NM_175012), PR domain containing 13 (Prdm13) (NM_001080771), and SK1 family transcriptional corepressor (Skor1) (NM_172446) as the DMH-enriched genes (Table1). Our analysis also confirmed that agouti-related protein (Agrp) (NM_007427), neuropeptide Y (Npy) (NM_023456), and kisspeptin 1 (Kiss1) (NM_178260) are exclusively expressed in the Arc, and nuclear receptor subfamily 5, group A, member 1 (Nr5a1) (NM_139051) is exclusively expressed in the VMH (Table S1). These genes are well-known selective markers of the Arc and VMH (Elmquist et al., 1999; Gottsch et al., 2004; Segal et al., 2005), providing further validation of our results. Parvalbumin (Pvalb) (NM_013645) is among the top genes that show much higher expression levels in the LH compared to the other hypothalamic nuclei (Table S1), which was also confirmed by the Allen Brain Atlas (http://mouse.brain-map.org/), although Pvalb is also highly expressed in other brain regions such as the cerebellum and cortex.
Figure 1.
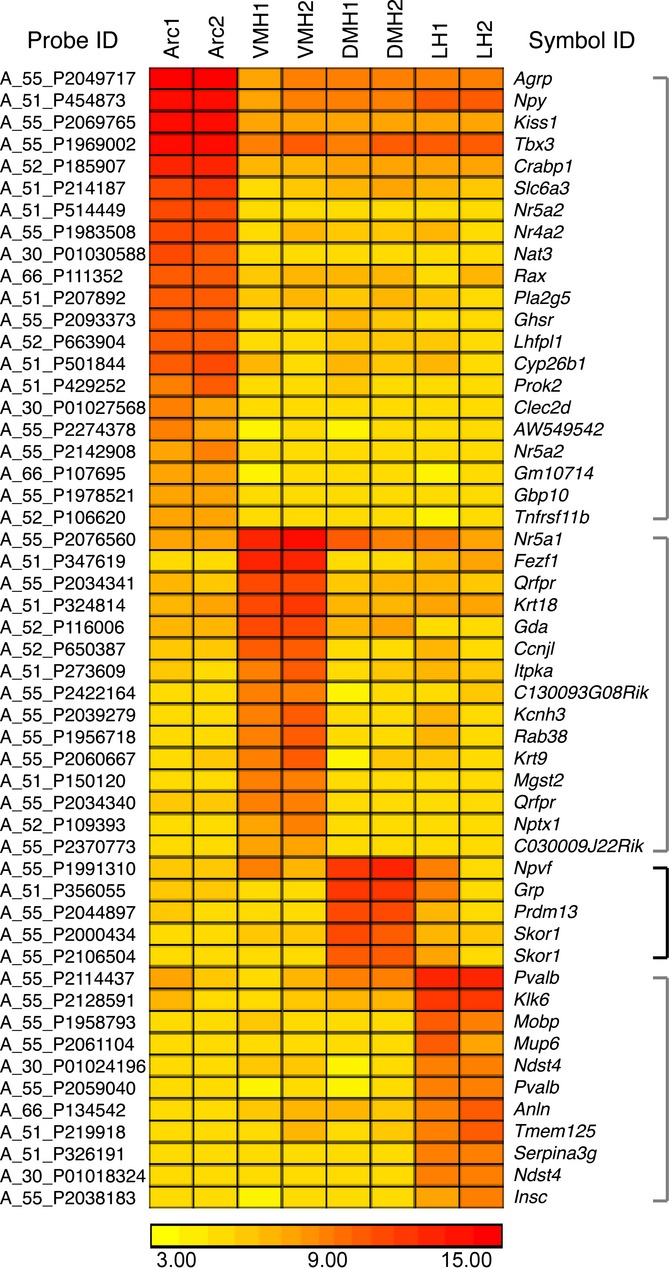
Heat map representation of genes enriched in the Arc, VMH, DMH, and LH. Genes expressed greater than or equal to 10-fold in each hypothalamic nucleus compared to other hypothalamic nuclei by calculating ratios of signal intensities between each hypothalamic nucleus are shown in the heat map. Microarray analysis was conducted using two individual RNA samples from the Arc (Arc1 and Arc2), VMH (VMH1 and VMH2), DMH (DMH1 and DMH2), and LH (LH1 and LH2). Expression levels are indicated by the colored log2 scale of signal intensities shown at the bottom of the figure. Probe ID from the Agilent Mouse_v1 8x60K array (left) and corresponding symbol ID (right) are shown for each gene.
Table 1.
Genes highly and selectively expressed in the DMH
| Probe ID | Acquisition number | Arc1 | Arc2 | VMH1 | VMH2 | DMH1 | DMH2 | LH1 | LH2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| A_55_P1991310 | NM_021892 | 75 | 82 | 369 | 92 | 4610 | 5816 | 427 | 25 | Npvf |
| A_51_P356055 | NM_175012 | 75 | 67 | 27 | 27 | 3242 | 5527 | 395 | 28 | Grp |
| A_55_P2044897 | NM_001080771 | 68 | 26 | 27 | 25 | 1942 | 2026 | 147 | 24 | Prdm13 |
| A_55_P2000434 | NM_172446 | 27 | 25 | 60 | 87 | 1655 | 1425 | 170 | 56 | Skor1 |
| A_55_P2106504 | NM_172446 | 25 | 23 | 26 | 24 | 1402 | 1363 | 193 | 25 | Skor1 |
Numbers indicate signal intensities from microarray analysis.
Expression patterns of Npvf,Prdm13, and Skor1 in the brain, in the hypothalamus, and within the DMH
We next determined the anatomical distribution of these DMH-enriched genes in the brain. Exploring DMH-enriched genes will have a potential to further address DMH function because the analysis of cell-type-specific gene expression patterns could elucidate brain function (Wang et al., 2010; Ko et al., 2013). We excluded Grp from the DMH-enriched genes we identified because it is also highly expressed in other peripheral tissues based on data available from the gene annotation portal BioGPS (http://biogps.org/#goto=welcome). We examined whether the expression of Npvf,Prdm13, and Skor1 was restricted to the hypothalamus. qRT-PCR was conducted with total RNAs from the hypothalamus, thalamus, hippocampus, cortex, and cerebellum. We found that Npvf and Prdm13 were selectively localized in the hypothalamus, whereas Skor1 was expressed in the hypothalamus, thalamus, hippocampus, cortex, and most extensively in the cerebellum (Fig.2A). We also examined the expression profiles of these three genes through the hypothalamic nuclei including the Arc, VMH, DMH, LH, PVN, and SCN. Consistent with the data from our microarray analysis, Npvf,Prdm13, and Skor1 were exclusively expressed in the DMH (Fig.2B). The DMH is further divided into three subdivisions: dorsal, compact, and ventral DMH (DMD, DMC, and DMV, respectively) (Fig. S1A). The highest expression of Npvf was observed in the DMV compared to the other two DMH subdivisions, whereas the expression levels of Prdm13 and Skor1 were highest in the DMC (Fig.2C).
Figure 2.
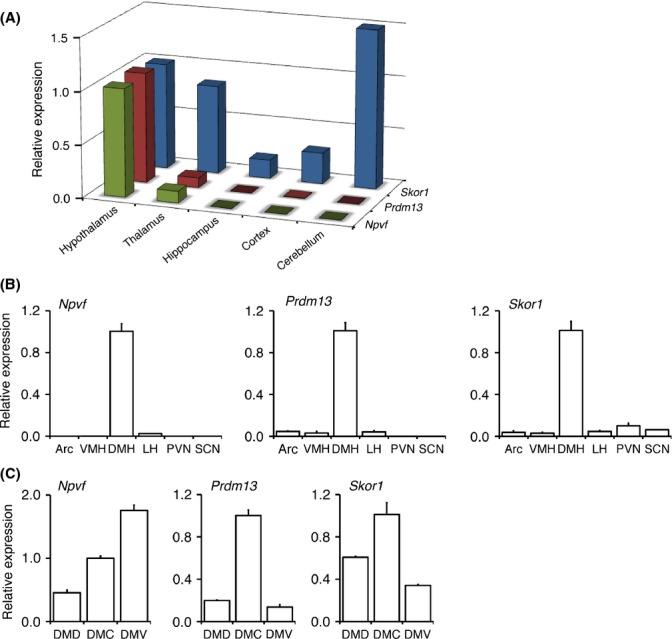
Distribution of Npvf,Prdm13, and Skor1 in the brain and specifically within the hypothalamus. (A, B, and C) RNA expression levels of Npvf,Prdm13, and Skor1 were examined in various brain regions including the hypothalamus, thalamus, hippocampus, cortex, and cerebellum (A), in the hypothalamic nuclei including the Arc, VMH, DMH, LH, PVN, and SCN (B) and in the DMH subdivisions including the DMD, DMC, and DMV (C). Expression levels were normalized to those of the hypothalamus (A), DMH (B), or DMC (C). Results are shown as mean ± S.E. (n = 2–3 mice for each group).
Expression changes in Npvf,Prdm13, and Skor1 in the hypothalamus under DR, through the 24-h diurnal cycle, and with age
As our recent findings indicate the importance of the DMH in the regulation of central adaptive responses to DR, physiological rhythms, and aging and longevity (Satoh et al., 2010, 2013), it is likely that these DMH-enriched genes show interesting expression changes in these conditions. To address this possibility, we first analyzed expression levels of Npvf,Prdm13, and Skor1 in the hypothalamus under DR, an experimentally proven dietary regimen that delays the aging process and extends lifespan in a wide variety of organisms (Guarente, 2013). The level of Npvf significantly decreased to about 40% under DR compared to that of ad libitum (AL)-fed controls (Fig.3A, left), whereas levels of Prdm13 and Skor1 significantly increased about 50% and 45%, respectively (Figs.3B and C, left). We next examined the diurnal expression patterns of Npvf,Prdm13, and Skor1. In the hypothalamus, both Npvf and Prdm13 displayed statistically significant 24-h diurnal cycles, with higher expression during the dark period and lower expression during the light period (Figs.3A and B, middle). The robust oscillation was not observed in the expression of Skor1 in the hypothalamus (Fig.3C, middle). Lastly, we compared the expression of Npvf,Prdm13, and Skor1 in the hypothalamus between mice at 4–6 months and 23–26 months of age. As the expression of Npvf and Prdm13 increases during the dark period, we collected hypothalamic samples from young and old mice during the dark period. The expression levels of Npvf in the hypothalamus significantly decreased in mice at 23–26 months of age, similar to its reduction under DR (Fig.3A, right). Intriguingly, the expression of Prdm13 significantly decreased with age, whereas its expression was elevated under DR (Fig.3B, right). The level of Skor1 was indistinguishable between young and old mice (Fig.3C, right). We also measured the expression levels of Npvf,Prdm13, and Skor1 in hypothalamic nuclei (Arc, VMH, DMH, and LH) of aged BRASTO mice, which showed significant delay in aging and lifespan extension (Satoh et al., 2013). Interestingly, the levels of Prdm13 and Skor1 in the DMH of aged BRASTO mice were significantly higher than the levels in wild-type control mice, whereas the levels of Npvf in the DMH were indistinguishable between genotypes (Fig.3D). These results suggest a possibility that Prdm13 and Skor1 contribute to the BRASTO phenotypes. Given that the data from DR, 24-h diurnal cycle, aging, and aged BRASTO mice, Prdm13 might be a key to counteract age-associated functional declines and to promote lifespan in mammals (Satoh et al., 2013).
Figure 3.
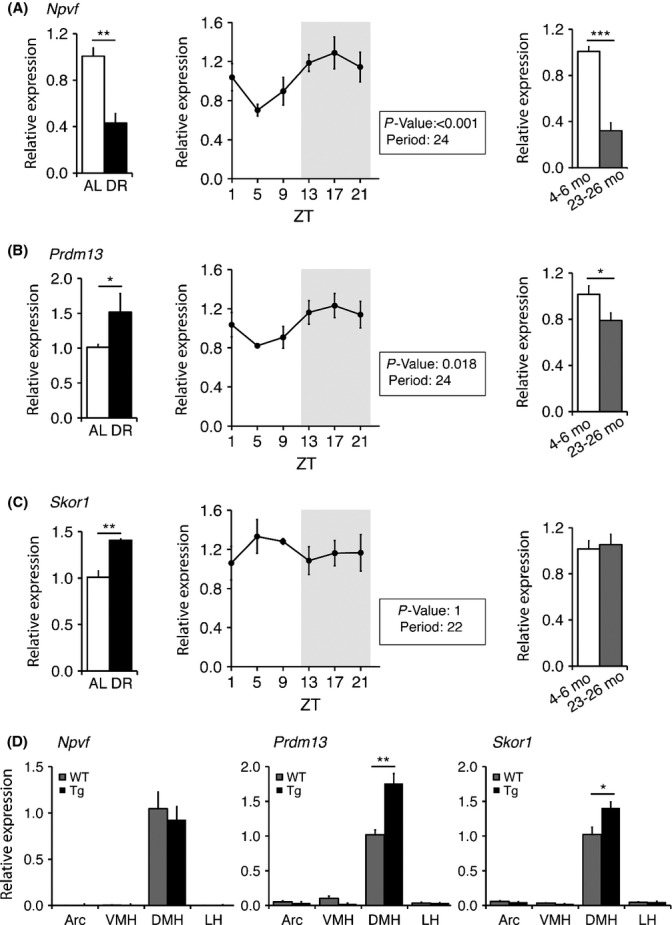
Alteration of Npvf,Prdm13, and Skor1 expression levels under DR, through the 24-h diurnal cycle, during the aging process, and in aged BRASTO mice. RNA expression levels of Npvf (A), Prdm13 (B), and Skor1 (C) in the hypothalamus were examined under ad libitum (AL)-fed versus diet restriction (DR) conditions at Zeitgeber time (ZT) 3 (left), every 4 h at different ZT points (middle), and in young (4–6 months of age) versus old (23–26 months of age) mice at ZT15 (right). Expression levels were normalized to those of AL (left) or young (right) groups. (D) RNA expression levels of Npvf,Prdm13, and Skor1 in the Arc, VMH, DMH, and LH of BRASTO mice (Tg) and wild-type mice (WT) at 20 months of age at ZT15. Results are shown as mean ± S.E. (*P < 0.05, **P < 0.01, ***P < 0.001, n = 4 mice each for AL and DR,n = 3–4 mice for each time point through 24 h, n = 7–8 mice for each group young vs. old, n = 4 mice each genotype). Shaded area represents the dark period.
The Nk2 homeobox 1 transcription factor Nkx2-1 shows DMC-predominant expression in the DMH and upregulates Prdm13 transcription
Interestingly, as well as Prdm13, Nkx2-1, a substrate and binding partner of Sirt1 in the DMH (Satoh et al., 2013), also displayed DMC-predominant expression. Immunohistochemistry using a specific antibody against Nkx2-1 revealed that Nkx2-1 is highly expressed in the DMC compared to other hypothalamic nuclei (Fig.4A). We also confirmed that Nkx2-1 is colocalized with NeuN in the DMC (Figs. S2A–D). qRT-PCR analysis clearly indicated that Nkx2-1 is most highly expressed in the DMC among the DMH subdivisions (Fig.4B). Given this anatomical overlap between Prdm13 and Nkx2-1 and the diurnal oscillation of Prdm13 expression, we hypothesized that Prdm13 could be a transcriptional target of Nkx2-1, whose activity is regulated by Sirt1 in a diurnal manner (Satoh et al., 2013). We found two predicted Nk2 binding sites in the Prdm13 promoter region (Fig.4C). To confirm whether the Prdm13 promoter indeed responds to Nkx2-1, we generated a luciferase reporter construct with the 360-bp genomic fragment of the Prdm13 promoter that contains these two Nk2 binding motif sequences. Increasing Nkx2-1 expression significantly enhanced the transcriptional activity of this Prdm13 promoter fragment in a dose-dependent manner in HEK293 cells (Fig.4D). Therefore, we next tested whether Nkx2-1 is sufficient to promote Prdm13 expression in primary hypothalamic neurons. Knocking down Nkx2-1 about 85–90% (data not shown) significantly but moderately reduced Prdm13 expression in the hypothalamic neurons (Fig.4E). These results suggest that Prdm13 transcription is regulated by Nkx2-1, although other factors or regulatory mechanisms are involved in the transcriptional regulation of Prdm13.
Figure 4.
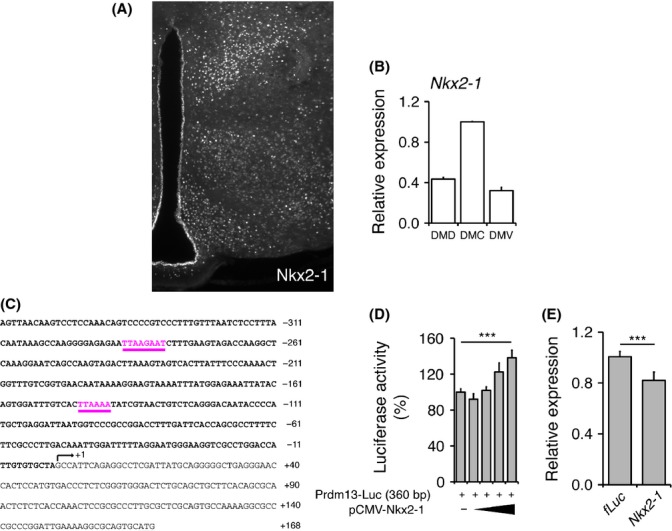
Prdm13 as a downstream gene regulated by Nkx2-1. (A) Immunohistochemistry staining of Nkx2-1 in the hypothalamus. White dots represent staining with an anti-Nkx2-1 antibody. We confirmed that these Nkx2-1-positive cells are NeuN-positive neurons (Fig. S2). (B) RNA expression levels of Nkx2-1 in the DMH subdivisions (DMD, DMC, and DMV). Expression levels were normalized to those of DMC. Results are shown as mean ± S.E. (n = 3 mice per subdivision). (C) Nucleotide sequence of the 360-bp upstream region of the mouse Prdm13 gene. The transcription initiation site is defined as +1, and the start codon (ATG) is underlined. Predicted Nk2 family binding sites are shown and underlined in pink color. (D) Luciferase activities driven by the 360-bp Prdm13 promoter fragment [Prdm13-Luc (360 bp)] in HEK293 cells cotransfected with Nkx2-1 (pCMV-Nkx2-1). Luciferase activities in cells cotransfected with a backbone vector were normalized to 100%. Results are shown as mean values ± S.E. (***P < 0.001 by one-way ANOVA with Tukey–Kramer post hoc test, n = 9). (E) RNA expression levels of Prdm13 after knocking down Nkx2-1 in primary hypothalamic neurons. Expression levels were normalized to those of firefly Luciferase (fLuc)-knockdown controls' cells. Results are shown as mean values ± S.E. (***P < 0.001 by Student's t-test, n = 7).
Prdm13 in the DMH affects sleep quality and body weight
To further elucidate the physiological significance of Prdm13 in the DMH, we generated DMH-specific Prdm13-knockdown mice by stereotactic injection of lentiviruses carrying Prdm13 shRNA. We then conducted electroencephalogram (EEG) recording on these mice. DMH-specific Prdm13-knockdown mice and control shRNA-injected mice showed indistinguishable sleep/wake architecture (Fig.5A–C), except that Prdm13-knockdown mice displayed a significantly lower amount of wakefulness during the dark period compared to controls on both Day 1 and Day 2 (Fig.5A). This reduction of wakefulness in Prdm13-knockdown mice was not accompanied by an alteration in the amount of NREM sleep (Fig.5B). We also found that the amount of REM sleep was significantly higher in Prdm13-knockdown mice compared to controls during the dark period only on Day 2 (Fig.5C). It should be noted that this statistically significant difference on Day 2 was dependent on a single data point showing an abnormal peak at ZT20 (shown as arrow in Fig.5C). Additionally, this difference was not observed on Day 1. These results suggest that Prdm13 in the DMH has a minimal effect on the normal diurnal oscillation of sleep. On the other hand, we found that the level of EEG delta power during NREM sleep, an indicator of the quality or depth of sleep, was significantly reduced in DMH-specific Prdm13-knockdown mice compared to control mice during both light and dark periods (Fig.5D), whereas these differences were not observed during the wake time (Fig.5E). This delta power defect caused by Prdm13 knockdown is very similar to that caused by Nkx2-1 knockdown (Satoh et al., 2013). Thus, these results indicate that Prdm13 in the DMH, as well as its upstream regulator Nkx2-1 (Satoh et al., 2013), significantly affects the level of sleep depth or quality in mice.
Figure 5.
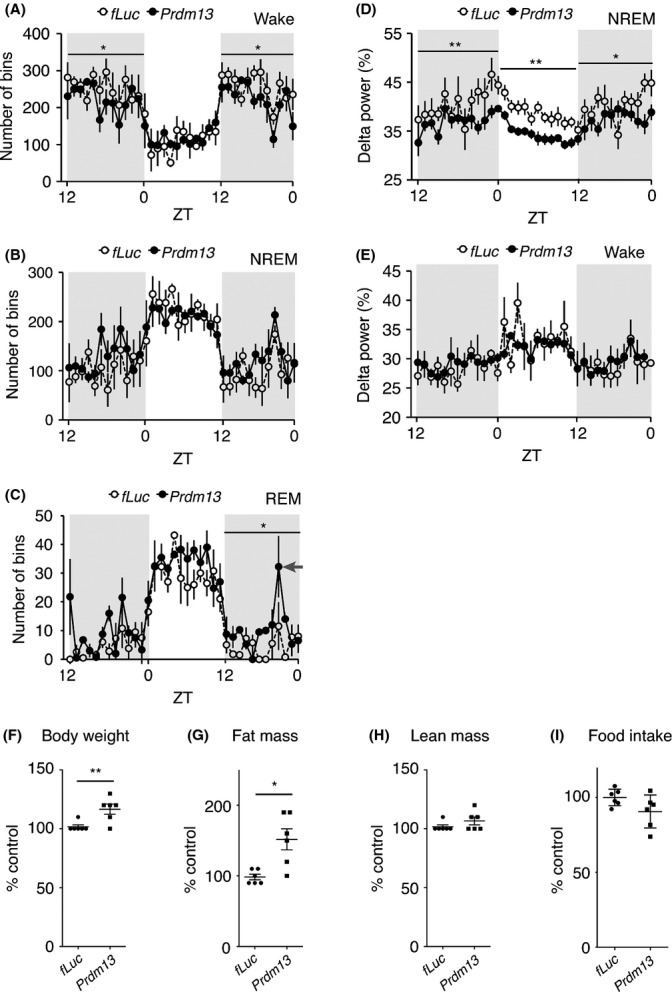
Sleep–wake patterns, NREM delta power, body weight, body composition, and daily food intake in DMH-specific Prdm13-knockdown mice. (A–C) The amount of wakefulness (wake) (A), NREM sleep (NREM) (B), and REM sleep (REM) (C) in DMH-specific firefly Luciferase (fLuc)-knockdown control mice and Prdm13-knockdown mice. Numbers of bins are shown as mean values ± S.E. (*P < 0.05 by Wilcoxon matched-pairs signed-rank test, n = 5–6 mice per group). Shaded areas represent the dark periods. A gray arrow in C indicates the ZT20 time point on Day 2 when an abnormal peak was observed in Prdm13-knockdown mice (see text). (D and E) Electroencephalogram delta power in NREM sleep (D) or in wake (E). Data are shown as mean values ± S.E. (*P < 0.05, **P < 0.01 by Wilcoxon matched-pairs signed-rank test, n = 5–6 mice per group). (F–H) Body weight (F), fat mass (G), lean mass (H), and daily food intake (I) for Prdm13-knockdown mice are normalized to the average values of fLuc-knockdown control mice within each cohort (3 cohorts). Fat mass and lean mass were measured by EchoMRI. Individual data are plotted, and mean values and S.E. are shown (*P < 0.05, **P < 0.01 by Student's t-test, n = 6 mice per group).
Poor sleep quality contributes to excessive weight gain, and better sleep quality confers protection against obesity (Mavanji et al., 2010). Therefore, we measured body weight in DMH-specific Prdm13-knockdown and control mice 2.5 months after stereotactic surgery. Because actual measured values of body weight, fat mass, and lean mass in control and Prdm13-knockdown mice fluctuated between cohorts, we calculated the percent changes compared to control mice in each individual cohort. The relative values of body weight and fat mass in Prdm13-knockdown mice were significantly higher than those in control mice (Fig.5F and G), whereas relative values of lean mass did not differ between control and Prdm13-knockdown mice (Fig.5H). We also observed that the amounts of daily food intake were indistinguishable between these two groups, suggesting that the increases in body weight and fat mass in Prdm13-knockdown mice are not due to increased daily food intake. These results indicate that DMH-specific Prdm13-knockdown mice exhibit progressive gain of adipose tissue when their sleep quality is affected.
Discussion
In our present study, we identified three DMH-enriched genes, Npvf,Prdm13, and Skor1, from microarray analysis using laser-microdissected hypothalamic nuclei. Intriguingly, Prdm13 is enriched in the DMC, where Sirt1 and Nkx2-1 are highly colocalized (Satoh et al., 2013). Indeed, the transcriptional activity of the Prdm13 promoter is upregulated by Nkx2-1, and the knockdown of Nkx2-1 decreases Prdm13 expression in primary hypothalamic neurons. Interestingly, the expression of Prdm13 increases under DR, whereas its expression declines with age. In addition, the expression level of Prdm13 in the DMH of aged BRASTO mice is significantly higher than the level in wild-type control mice. Furthermore, DMH-specific Prdm13-knockdown mice display reduced sleep quality and increased body weight, phenotypes very similar to old animals (Ancoli-Israel, 2009; Kuk et al., 2009). Given that Sirt1 and Nkx2-1 function together in the hypothalamus to counteract age-associated functional declines (Satoh et al., 2013), Prdm13 might also play an important role as one of their key target genes in the DMH to mediate age-associated pathophysiology, particularly sleep quality and body weight.
Different from other hypothalamic nuclei, it has been challenging to identify selective marker genes for the DMH. Lee et al. previously reported Gpr50,Pcsk5,Sulf1, Grp, and Rorβ as genes highly expressed in the DMV (Lee et al., 2012). Our microarray analysis did not identify those genes, except for Grp, which is highly expressed in other peripheral tissues. This is likely due to differences in sample collection between the two studies: DMV in their study versus DMH in our study. In fact, Prdm13 and Skor1 are highly expressed in the DMC, but not in the DMV. Npvf could have been detected in their study because Npvf is highly expressed in the DMV based on our results. Kasukawa et al. also reported Npvf as a potential candidate maker for the DMH, based on the BrainStars database that they created (http://brainstars.org/) (Kasukawa et al., 2011). Additionally, the localization of Npvf in the DMV can be confirmed in the Allen Brain Atlas (http://mouse.brain-map.org/). Taken together, Npvf and Prdm13 are specifically expressed in the DMV and DMC, respectively, potentially providing good selective markers for the DMH.
The physiological role of Prdm13 during adulthood is still poorly understood. Prdm13 belongs to the Prdm family of zinc finger transcriptional regulators (Fog et al., 2012; Hohenauer & Moore, 2012). Recently, it has been reported that Prdm13 has histone methyltransferase activity (Hanotel et al., 2014). During development, Prdm13 is directly downstream of Ptf1a, a basic helix–loop–helix transcription activator and is required for the specification of GABAergic and glutamatergic neurons (Chang et al., 2013). Remarkably, we demonstrated that Prdm13 in the DMH is important to maintain sleep quality and body weight associated with adiposity. DMH-specific Prdm13-knockdown mice displayed decreased wakefulness during the dark period and robust decline in NREM delta power through the entire diurnal cycle compared to those in control mice. They also displayed progressively increased body weight with adiposity. Because sleep quality and metabolism are highly associated (Morselli et al., 2012), poor sleep quality by knockdown of Prdm13 in the DMH might cause increased body weight secondarily. Alternatively, weight gain with adiposity might be regulated by the same set of DMH neurons but through an independent mechanism (see below). Sleep quality is regulated by circadian rhythmicity and sleep–wake homeostasis. Provided that NREM delta power is regulated independently of sleep duration (Davis et al., 2011), the observed abnormality of sleep in DMH-specific Prdm13-knockdown mice might be due to defects in sleep–wake homeostasis. The DMH has been suggested to be a relay point for signals going from the SCN to the locus coeruleus (Aston-Jones et al., 2001), the main site for synthesizing norepinephrine in the brain and facilitating arousal and sleep–wake homeostasis. Thus, it will be interesting to examine whether DMH-specific Prdm13-knockdown mice also display defects in sleep homeostasis.
It is likely that the Nkx2-1/Prdm13 pathway in the DMH has a significant contribution to the Sirt1-mediated control of aging and longevity (Fig.6). At the molecular level, Nkx2-1 is deacetylated by Sirt1 in the hypothalamus during the dark period when Sirt1 is activated (Satoh et al., 2013). In addition, Sirt1 cooperates with Nkx2-1 to upregulate Ox2r transcription (Satoh et al., 2013). Whether Sirt1 is also involved in the regulation of Prdm13 transcription and whether age-associated reduction of Prdm13 expression in the hypothalamus is due to a decline in Sirt1 activity still need to be investigated. It will also be important to examine potential roles of downstream targets of Prdm13, Nkx2-1, and Sirt1 in the DMH in the pathogenesis of age-associated sleep abnormality and metabolic complications. We demonstrated that BRASTO mice display delays in age-associated physiological declines and lifespan extension (Satoh et al., 2013). Maintaining quality of sleep is one of the striking phenotypes in aged BRASTO mice. In contrast, DMH/LH-specific Nkx2-1- or Sirt1-knockdown mice (Satoh et al., 2013) and DMH-specific Prdm13-knockdown mice display lower NREM delta power compared to controls. In addition, Sirt1 and Nkx2-1 are colocalized in the DMC, and Prdm13 is also localized in the same region. These results strongly imply that Prdm13, Nkx2-1, and Sirt1 together in the DMC play an important role in the regulation of sleep quality during the aging process. On the other hand, the role of DMH neurons expressing Nkx2-1 and/or Prdm13 in the regulation of body weight and adiposity is still unclear. Although DMH-specific Prdm13-knockdown mice display progressive weight gain, DMH/LH-specific Nkx2-1-knockdown mice do not display such phenotypes (unpublished finding). Much longer observation time or excessive energy intake might be necessary to produce definitive changes in body weight in DMH-specific Nkx2-1-knockdown mice. Alternatively, this phenotypic difference between Nkx2-1- and Prdm13-knockdown mice might be due to the heterogeneity of DMH neurons, particularly DMC neurons, expressing Nkx2-1,Prdm13, and/or Sirt1. In other words, Prdm13 in distinct subpopulations of DMC neurons, each of which are characterized by different neuropeptides or neurotransmitters, might differentially regulate sleep quality and body weight. These distinct neuronal subsets could mediate different functions through different projections but together contribute to the prevention against age-associated physiological decline and the promotion of longevity in mammals.
Figure 6.
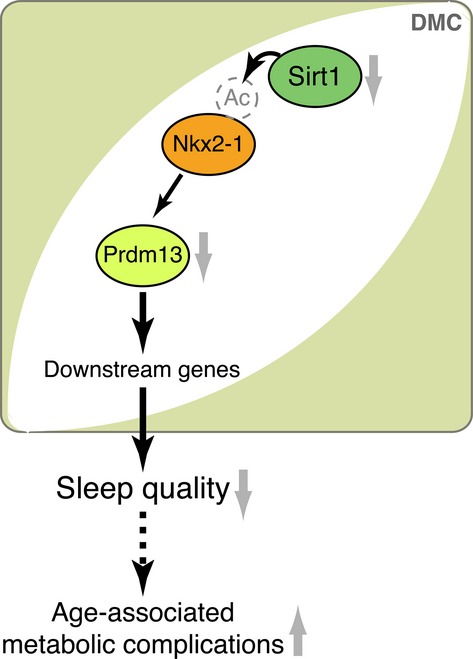
Model for the compact region of the DMH (DMC) in controlling sleep quality and metabolism during aging. The levels of Prdm13 decline with age in the DMC, causing altered expression of downstream target genes. This age-associated dysfunction of DMC neurons likely reduces sleep quality and causes metabolic complications such as increased adiposity during the aging process. Prdm13 transcription is upregulated by Nkx2-1, whose activity is regulated by Sirt1-mediated deacetylation. Therefore, the age-associated decline in Prdm13 expression might be due to the reduction of Sirt1 activity during the aging process.
In conclusion, our microarray-based gene expression profiling for hypothalamic nuclei provided valuable marker genes for the DMH. Investigation of the physiological roles of DMH-enriched genes in sleep homeostasis and metabolism will shed new light on the understanding of aging and longevity control in mammals.
Experimental procedures
Animal study
C57BL/6J female mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). All mice were maintained under 12-h light/12-h dark conditions and had free access to food and water. For DR, mice were fed 60% of control for 3 months as previously described (Satoh et al., 2010). The procedure for stereotactic delivery of lentiviruses was described previously (Satoh et al., 2013). Body composition was measured using EchoMRI™ 3-in-1 Body Composition Analyzer (EchoMRI LLC, Houston, TX, USA). BRASTO females in C57BL/6J background were described previously (Satoh et al., 2010). All animal procedures were approved by the Washington University Animal Studies Committee and were in accordance with NIH guidelines.
Sleep analysis
Briefly, isoflurane-anesthetized mice were surgically implanted with screw electrodes in the skull for electroencephalogram (EEG) and platinum wire electrodes in the nuchal muscle for electromyogram (EMG) recording. After a minimum of 2-week recovery, we conducted EEG/EMG recording under each physiological condition. The 10-s epochs of EEG/EMG signals were scored by visual examination as wake, lower amplitude delta (1–4 Hz) and theta (4–8 Hz) frequency with higher and variable amplitude EMG activity; NREM sleep, predominantly higher amplitude delta activity and low amplitude EMG activity; or REM sleep, predominantly low amplitude rhythmic theta activity and very low amplitude EMG activity. The 10-s EEG epochs then were processed by Fourier transformation to calculate the percentage of delta power relative to total power in the 0.1–25 Hz frequency range for each 10-s epoch. A Wilcoxon matched-pairs signed-rank test was used to compare differences of hourly data points through the light and dark periods between groups in sleep analysis.
Sample collection of hypothalamic nuclei
The Arc, VMH, DMH, LH, PVN, and SCN were dissected from two C57BL/6J females at 3 months of age by laser microdissection using the Leica LMD 6000 system (Leica Microsystems, Buffalo Grove, IL, USA). We confirmed that there was no contamination from surrounding hypothalamic nuclei by conducting quantitative real-time RT-PCR using probes Agrp,Nr5a1,hypocretin (Hcrt), and corticotropin-releasing hormone (Crh) (Fig. S1B). RNA was isolated from each hypothalamic nucleus using the Arcturus PicoPure RNA Isolation Kit (Life Technologies, Grand Island, NY, USA). The detailed procedure was described previously (Satoh et al., 2010). For microarray, the quality of total RNA was determined by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and two samples for each hypothalamic nucleus were used for microarray analysis. For quantitative real-time RT-PCR, RNA concentration was determined by NanoDrop, and cDNA was synthesized using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY, USA).
Agilent one color microarray and expression analysis
Total RNA was amplified by the WTA2 kit (Sigma-Aldrich, St. Louis, MO, USA), according to the manufacturer's protocol. cDNAs were chemically labeled with Kreatech ULS RNA Labeling Kit (Kreatech Diagnostics, Durham, NC, USA). Labeled cDNA was purified with Qiagen PCR purification columns, and cDNAs were quantitated on a NanoDrop spectrophotometer. cDNA samples were hybridized to Agilent Mouse_v1 8x60K microarrays (Design ID-028005, Agilent Technologies, Santa Clara, CA, USA). Slides were scanned on an Agilent C-class Microarray scanner to detect Cy5 fluorescence, according to the manufacturer's specifications. Gridding and analysis of images was performed using Feature Extraction software (v11.5.1.1, Agilent Technologies, Santa Clara, CA, USA). To identify differentially expressed genes in each hypothalamic nucleus, ratios of signal intensities to each hypothalamic nucleus were calculated, and genes whose expression levels were greater than or equal to 10-fold in each hypothalamic nucleus compared to the others were selected. The heat map for each log2-transformed gene expression level was generated using Partek Genomics Suite (Partek Inc., St. Louis, MO, USA). The microarray data were deposited into the NCBI GEO database (GEO Accession Number GSE58238).
Quantitative real-time RT-PCR
Total RNA was extracted from brain regions using the Ambion PureLink RNA Mini Kit (Life Technologies, Grand Island, NY, USA). Hypothalamic nuclei were collected by laser microdissection as described above. RNA was reverse-transcribed into cDNA as described above, and quantitative real-time RT-PCR was conducted as described in detail previously (Satoh et al., 2013). Relative expression levels of the genes of interest were normalized to Gapdh levels.
Luciferase assay
HEK293 cells were transfected with a firefly luciferase reporter driven by 360-bp Prdm13 promoter, pCMV-Nkx2-1 (a gift from Dr. Shioko Kimura, National Cancer Institute, Bethesda, MD, USA) or control vector, and a Renilla luciferase expression vector. Luciferase assays were conducted as described previously (Satoh et al., 2010). Firefly luciferase activity levels were normalized to Renilla luciferase activity of each sample.
Primary hypothalamic neuronal isolation and Nkx2-1 knockdown
Hypothalami were dissected from E15.5 C57BL/6J mice, and hypothalamic neurons were isolated as described previously (Satoh et al., 2013). Hypothalamic neurons were plated at 2.5 × 105 cells/well in 6-well plates coated with 100 μg ml-1 poly-L-lysine, and cells were maintained in Neurobasal media containing 10% FBS, 1 × B27, 2 mm L-glutamine, and penicillin/streptomycin. Cultures were treated with 10 μm Ara-C (Sigma-Aldrich, St. Louis, MO, USA) after 2 days in vitro (DIV). Media was replaced to Dulbecco's modified Eagle's media containing 50% F12, 5 mm HEPES, 1x B27, 1x N2, 0.5% L-glutamax, 0.5 mM sodium pyruvate, and penicillin/streptomycin after 4 DIV. On the next day, Nkx2-1 shRNA lentivirus was added, and cells were harvested after 7 DIV. RNA was isolated as described previously (Satoh et al., 2013).
Statistical analysis
Statistical analyses were carried out using unpaired or paired Student's t-tests for two groups and one-way ANOVA with the Tukey–Kramer post hoc test for more than two groups. The rhythmicity of gene expression was assessed by the JTK-Cycle (Hughes et al., 2010), which is available as a computationally efficient R script (http://openwetware.org/wiki/HughesLab:JTK_Cycle). In all analyses, we set the α level at 0.05. We used Microsoft Excel 2008 and SPSS 11.0 to conduct statistical analyses.
Acknowledgments
We thank Vicki Immethun, Mitsukuni Yoshida, Myeong Jin Yoon, E. J. Brace, and Michael Wong for their technical help and members of the Imai Laboratory for their help and discussions.
Funding
This work was supported by grants from the National Institute on Aging (AG02150, AG037457), the Ellison Medical Foundation, and the Washington University NORC grant NIH P30 DK56341 to S.I.
Conflict of interest
S.I. is a cofounder of Metro Midwest Biotech.
Supporting Information
Table S1 Genes selectively expressed in the Arc, VMH, DMH, and LH
Data S1 Supplementary Figure Legends and Experimental Procedures.
Fig. S1 Laser-microdissection of distinct hypothalamic nuclei.
Fig. S2 Nkx2-1 is colocalized with NeuN in the DMH.
References
- Acosta-Galvan G, Yi CX, van der Vliet J, Jhamandas JH, Panula P, Angeles-Castellanos M, Del Carmen Basualdo M, Escobar C, Buijs RM. Interaction between hypothalamic dorsomedial nucleus and the suprachiasmatic nucleus determines intensity of food anticipatory behavior. Proc. Natl Acad. Sci. USA. 2011;108:5813–5818. doi: 10.1073/pnas.1015551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto S, Miyasaka K. Age-associated changes of appetite-regulating peptides. Geriatr. Gerontol. Int. 2010;10(Suppl 1):S107–S119. doi: 10.1111/j.1447-0594.2010.00587.x. [DOI] [PubMed] [Google Scholar]
- Ancoli-Israel S. Sleep and its disorders in aging populations. Sleep Med. 2009;10(Suppl 1):S7–S11. doi: 10.1016/j.sleep.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Aston-Jones G, Chen S, Zhu Y, Oshinsky ML. A neural circuit for circadian regulation of arousal. Nat. Neurosci. 2001;4:732–738. doi: 10.1038/89522. [DOI] [PubMed] [Google Scholar]
- Bechtold DA, Sidibe A, Saer BR, Li J, Hand LE, Ivanova EA, Darras VM, Dam J, Jockers R, Luckman SM, Loudon AS. A role for the melatonin-related receptor GPR50 in leptin signaling, adaptive thermogenesis, and torpor. Curr. Biol. 2012;22:70–77. doi: 10.1016/j.cub.2011.11.043. [DOI] [PubMed] [Google Scholar]
- Chang JC, Meredith DM, Mayer PR, Borromeo MD, Lai HC, Ou YH, Johnson JE. Prdm13 mediates the balance of inhibitory and excitatory neurons in somatosensory circuits. Dev. Cell. 2013;25:182–195. doi: 10.1016/j.devcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CJ, Clinton JM, Jewett KA, Zielinski MR, Krueger JM. Delta wave power: an independent sleep phenotype or epiphenomenon? J. Clin. Sleep Med. 2011;7:S16–S18. doi: 10.5664/JCSM.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmquist JK, Elias CF, Saper CB. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron. 1999;22:221–232. doi: 10.1016/s0896-6273(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Enriori PJ, Sinnayah P, Simonds SE, Garcia Rudaz C, Cowley MA. Leptin action in the dorsomedial hypothalamus increases sympathetic tone to brown adipose tissue in spite of systemic leptin resistance. J. Neurosci. 2011;31:12189–12197. doi: 10.1523/JNEUROSCI.2336-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fog CK, Galli GG, Lund AH. PRDM proteins: important players in differentiation and disease. BioEssays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- Gooley JJ, Schomer A, Saper CB. The dorsomedial hypothalamic nucleus is critical for the expression of food-entrainable circadian rhythms. Nat. Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanotel J, Bessodes N, Thelie A, Hedderich M, Parain K, Van Driessche B, Brandao Kde O, Kricha S, Jorgensen MC, Grapin-Botton A, Serup P, Van Lint C, Perron M, Pieler T, Henningfeld KA, Bellefroid EJ. The Prdm13 histone methyltransferase encoding gene is a Ptf1a-Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev. Biol. 2014;386:340–357. doi: 10.1016/j.ydbio.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukawa T, Masumoto KH, Nikaido I, Nagano M, Uno KD, Tsujino K, Hanashima C, Shigeyoshi Y, Ueda HR. Quantitative expression profile of distinct functional regions in the adult mouse brain. PLoS ONE. 2011;6:e23228. doi: 10.1371/journal.pone.0023228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby ED, Geraghty AC, Ubuka T, Bentley GE, Kaufer D. Stress increases putative gonadotropin inhibitory hormone and decreases luteinizing hormone in male rats. Proc. Natl Acad. Sci. USA. 2009;106:11324–11329. doi: 10.1073/pnas.0901176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y, Ament SA, Eddy JA, Caballero J, Earls JC, Hood L, Price ND. Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc. Natl Acad. Sci. USA. 2013;110:3095–3100. doi: 10.1073/pnas.1222897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk JL, Saunders TJ, Davidson LE, Ross R. Age-related changes in total and regional fat distribution. Ageing Res. Rev. 2009;8:339–348. doi: 10.1016/j.arr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J. Comp. Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lee S, Bookout AL, Lee CE, Gautron L, Harper MJ, Elias CF, Lowell BB, Elmquist JK. Laser-capture microdissection and transcriptional profiling of the dorsomedial nucleus of the hypothalamus. J. Comp. Neurol. 2012;520:3617–3632. doi: 10.1002/cne.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavanji V, Teske JA, Billington CJ, Kotz CM. Elevated sleep quality and orexin receptor mRNA in obesity-resistant rats. Int. J. Obes. (Lond) 2010;34:1576–1588. doi: 10.1038/ijo.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SF, Nakamura K, Madden CJ. Central control of thermogenesis in mammals. Exp. Physiol. 2008;93:773–797. doi: 10.1113/expphysiol.2007.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli LL, Guyon A, Spiegel K. Sleep and metabolic function. Pflugers Arch. 2012;463:139–160. doi: 10.1007/s00424-011-1053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds MA, Ingram DK, Talan M. Relationship of body temperature stability to mortality in aging mice. Mech. Ageing Dev. 1985;30:143–152. doi: 10.1016/0047-6374(85)90003-x. [DOI] [PubMed] [Google Scholar]
- Satoh A, Imai S. Systemic regulation of mammalian ageing and longevity by brain sirtuins. Nat. Commun. 2014;5:4211. doi: 10.1038/ncomms5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal JP, Stallings NR, Lee CE, Zhao L, Socci N, Viale A, Harris TM, Soares MB, Childs G, Elmquist JK, Parker KL, Friedman JM. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J. Neurosci. 2005;25:4181–4188. doi: 10.1523/JNEUROSCI.0158-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Minamoto Y, Ono M, Watanabe S. Age-related impairment of food anticipatory locomotor activity in rats. Physiol. Behav. 1994;55:875–878. doi: 10.1016/0031-9384(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Soga T, Kitahashi T, Clarke IJ, Parhar IS. Gonadotropin-inhibitory hormone promoter-driven enhanced green fluorescent protein expression decreases during aging in female rats. Endocrinology. 2014;155:1944–1955. doi: 10.1210/en.2013-1786. [DOI] [PubMed] [Google Scholar]
- Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat. Rev. Neurosci. 2009;10:397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalen J, Buxbaum JN. Is older colder or colder older? The association of age with body temperature in 18,630 individuals. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:487–492. doi: 10.1093/gerona/glr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Lee I, Carlson G, Hood L, Galas D. Systems biology and the discovery of diagnostic biomarkers. Dis. Markers. 2010;28:199–207. doi: 10.3233/DMA-2010-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Scott KA, Hyun J, Tamashiro KL, Tray N, Moran TH, Bi S. Role of dorsomedial hypothalamic neuropeptide Y in modulating food intake and energy balance. J. Neurosci. 2009;29:179–190. doi: 10.1523/JNEUROSCI.4379-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretskaia MV, Zaretsky DV, DiMicco JA. Role of the dorsomedial hypothalamus in thermogenesis and tachycardia caused by microinjection of prostaglandin E2 into the preoptic area in anesthetized rats. Neurosci. Lett. 2003;340:1–4. doi: 10.1016/s0304-3940(03)00047-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kerman IA, Laque A, Nguyen P, Faouzi M, Louis GW, Jones JC, Rhodes C, Munzberg H. Leptin-receptor-expressing neurons in the dorsomedial hypothalamus and median preoptic area regulate sympathetic brown adipose tissue circuits. J. Neurosci. 2011;31:1873–1884. doi: 10.1523/JNEUROSCI.3223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li J, Purkayastha S, Tang Y, Zhang H, Yin Y, Li B, Liu G, Cai D. Hypothalamic programming of systemic ageing involving IKK-b, NF-kB and GnRH. Nature. 2013;497:211–216. doi: 10.1038/nature12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Genes selectively expressed in the Arc, VMH, DMH, and LH
Data S1 Supplementary Figure Legends and Experimental Procedures.
Fig. S1 Laser-microdissection of distinct hypothalamic nuclei.
Fig. S2 Nkx2-1 is colocalized with NeuN in the DMH.


