Abstract
Plasma membrane PI4P is an important direct regulator of many processes that occur at the plasma membrane and also a biosynthetic precursor of PI(4,5)P2 and its downstream metabolites. The majority of this PI4P pool is synthesized by an evolutionarily conserved complex, which has as its core the PI 4-kinase PI4KIIIα (Stt4 in yeast) and also comprises TTC7 (Ypp1 in yeast) and the peripheral plasma membrane protein EFR3. While EFR3 has been implicated in the recruitment of PI4KIIIα via TTC7, the plasma membrane protein Sfk1 was also shown to participate in this targeting and activity in yeast. Here, we identify a member of the TMEM150 family as a functional homologue of Sfk1 in mammalian cells and demonstrate a role for this protein in the homeostatic regulation of PI(4,5)P2 at the plasma membrane. We also show that the presence of TMEM150A strongly reduces the association of TTC7 with the EFR3-PI4KIIIα complex, without impairing the localization of PI4KIIIα at the plasma membrane. Collectively our results suggest a plasticity of the molecular interactions that control PI4KIIIα localization and function.
Keywords: phospholipase C, PI4KA, Rolling blackout, Ypp1
Introduction
Phosphoinositides are phospholipids derived from the phosphorylation of phosphatidylinositol at the 3-, 4- and 5-positions of the inositol ring 1, 2. The seven phosphoinositides are differentially localized on the cytosolic leaflets of cellular membranes, where they play a critical role in controlling interactions that occur at their surface. Via this mechanism, which helps generate a code of membrane identity, they regulate a multiplicity of cellular processes 1, 2. Among phosphoinositides, phosphatidylinositol 4-phosphate (PI4P) plays numerous fundamental roles in the Golgi complex, the endosomal system, and the plasma membrane 1, 2, 3.
PI4P is a biosynthetic precursor of PI(4,5)P2 and thus an upstream metabolite in metabolic pathways leading to diacylglycerol (DAG), IP3, and PI(3,4,5)P3 4. In addition, PI4P can contribute to the regulation of other membrane lipids, including sterols 5, 6, 7, 8, phosphatidylserine 9, and sphingolipids 10. Therefore, tight regulation of PI4P levels is important for a variety of cellular functions.
In mammals, four enzymes can phosphorylate phosphatidylinositol at the 4-position of the inositol ring to generate PI4P 2, 3. Of the four enzymes, PI 4-kinase type IIIα (PI4KIIIα; Stt4 in yeast) plays a major role in the generation of PI4P at the plasma membrane 11, 12, 13, 14. This enzyme is of fundamental physiological importance, as germline disruption of its gene in mice produces embryonic lethality, and its conditional disruption in mouse fibroblasts results in cell death 11, 15, 16. Interest in this enzyme has been recently enhanced by its identification as a critical host factor for the replication of the hepatitis C virus in humans 17, 18, 19.
PI4KIIIα/Stt4 localizes to the plasma membrane as part of an evolutionarily conserved complex comprising two adaptor proteins (each of them represented by two similar A and B isoforms), the peripheral membrane protein EFR3 and the scaffolding protein TTC7 (Ypp1 in yeast) 11, 12, 20. Studies in yeast have identified an additional regulator of Stt4, termed Sfk1 (suppressor of four-kinase 1), a multipass transmembrane protein localized at the plasma membrane, whose overexpression partially rescues defects in a temperature-sensitive Stt4 mutant strain 21. Sfk1 physically interacts with Stt4, and loss of Sfk1 compromises proper plasma membrane localization of Stt4 21. Despite the importance of Sfk1 in the regulation of Stt4 localization and function in yeast, no mammalian counterpart of Sfk1 has yet been reported, and major outstanding questions remain about how the PI4KIIIα complex is regulated at the plasma membrane. Here, we identify TMEM150A as a mammalian homologue of yeast Sfk1. TMEM150A forms a complex at the plasma membrane with PI4KIIIα and EFR3, but not TTC7, suggesting a plasticity of the protein interactions of PI4KIIIα. We also find that levels of TMEM150A impact the rate of PI(4,5)P2 re-synthesis following acute depletion of this lipid, possibly due to an impact of this protein on the pool of PI4P available for the synthesis of PI(4,5)P2.
Results and Discussion
Putative identification of TMEM150A as a mammalian homologue of yeast Sfk1
BLAST searches for amino acid sequences in higher eukaryotes related to regions of Sfk1 highly conserved among various species of yeast and fungi yielded members of the mammalian TMEM150/FRAG1/DRAM family, whose defining structural feature is the presence of six predicted transmembrane regions (Fig1A). DRAM (Damage-Regulated Autophagy Regulator) 1 and 2 were reported to be localized in lysosomal membranes and to function as p53-induced modulators of autophagy 22, 23. TMEM150A, TMEM150B, and TMEM150C remain uncharacterized. Thus, we explored whether TMEM150A proteins could be functional homologues of yeast Sfk1 in the control of the localization and function of PI4KIIIα, the mammalian homologue of yeast Stt4 21.
Figure 1.
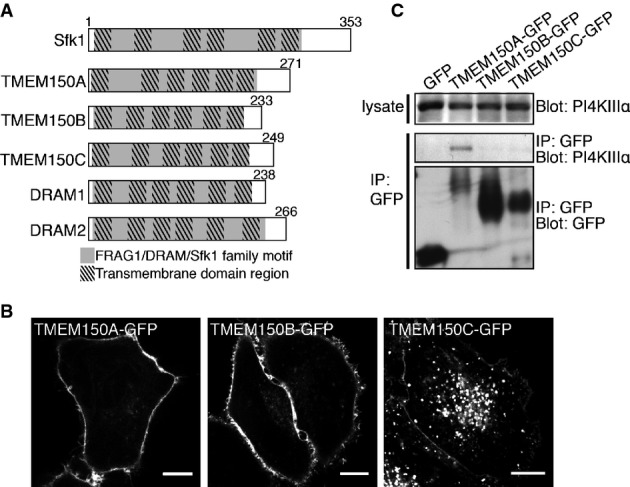
- A Domain cartoons of yeast Sfk1 and of its putative homologues in mammals. The light gray portions represent FRAG1/DRAM/Sfk1 family homology and the superimposed darker boxes represent transmembrane domain regions.
- B Confocal imaging of live HeLa cells transfected with TMEM150A-GFP, TMEM150B-GFP, and TMEM150C-GFP as indicated. Scale bars: 20 μm.
- C Western blot analysis for GFP and PI4KIIIα of anti-GFP immunoprecipitates from lysates of HeLa cells transfected with GFP or TMEM150A-GFP fusion proteins.
C-terminally GFP-tagged TMEM150A, TMEM150B, or TMEM150C were transiently expressed in HeLa cells. TMEM150A-GFP and TMEM150B-GFP were predominantly localized at the plasma membrane, and only a minor pool of these proteins was localized on intracellular structures. TMEM150C-GFP was localized at the plasma membrane, but a greater pool of this protein was localized on intracellular vesicular structures (Fig1B) that co-localized with RFP-tagged LAMP1 (a lysosomal marker) (Supplementary Fig S1). Western blot analysis of anti-GFP immunoprecipitates generated from these cells, and from cells expressing GFP alone as a control, revealed coprecipitation of PI4KIIIα with TMEM150A, although not B or C (Fig1C). Based on these results, we further explored the properties of TMEM150A and its potential function in PI4KIIIα regulation.
TMEM150A interacts with PI4KIIIα via its C-terminal cytoplasmic region
To determine the topology of TMEM150A within the plasma membrane, live HeLa cells expressing TMEM150A-GFP were exposed to an anti-GFP antibody at 4°C without permeabilization by detergent. While intrinsic GFP fluorescence was observed, anti-GFP immunostaining did not give any signal without prior fixation and permeabilization (Fig2A, upper panel), indicating that the C-terminal tail of TMEM150A is oriented toward the cytosol. By contrast, immunofluorescence analysis of a construct containing an HA tag in the middle of Loop 1 (termed HA-TMEM150A-GFP) produced fluorescence without any prior permeabilization, suggesting that Loop 1 is exposed to the extracellular medium (Fig2A, bottom panel). Based on these results, we hypothesized that TMEM150A has the topology illustrated in Fig2B, with a C-terminal cytosolic tail.
Figure 2.
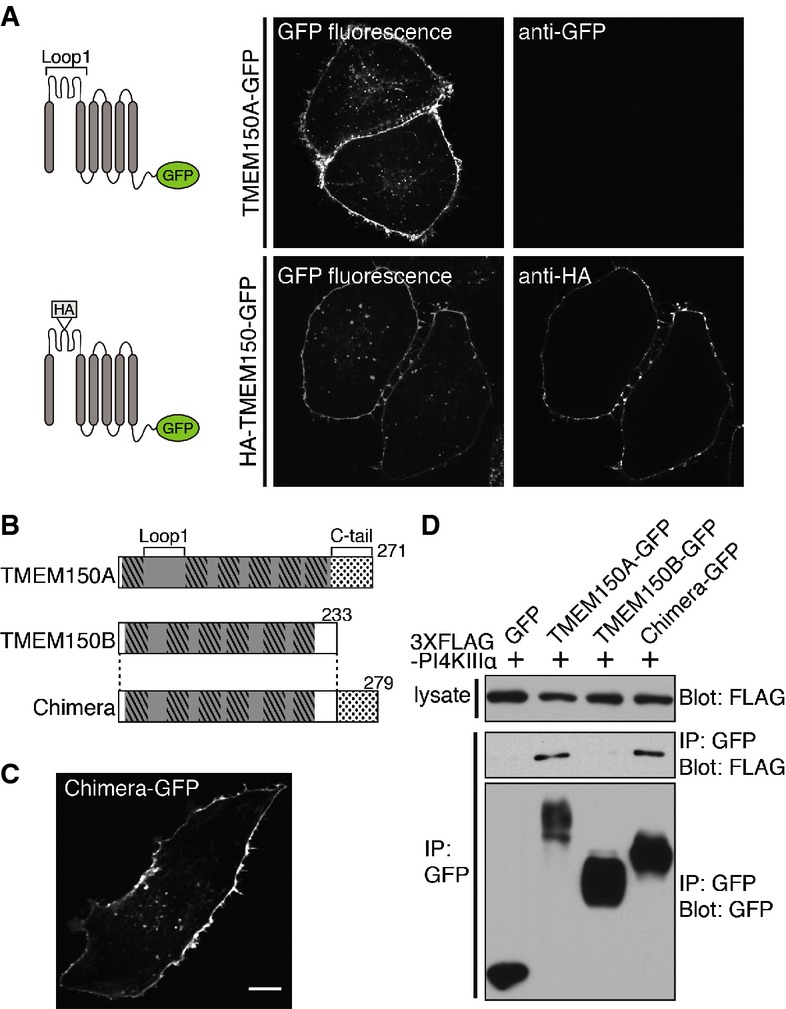
- A Fluorescence of HeLa cells expressing TMEM150A-GFP (top) or HA-TMEM150A-GFP (bottom) (see cartoons at left) and incubated with anti-GFP (top) or anti-HA (bottom) antibodies without permeabilization before fixation. Left panels represent intrinsic GFP fluorescence, and right panels represent anti-GFP or anti-HA immunofluorescence.
- B Schematic diagram of TMEM150A, TMEM150B, and Chimera. Chimera was generated by fusing the C-terminal 40 amino acids of TMEM150A (dotted box) to the C-terminus of full-length TMEM150B.
- C Snapshot of confocal live imaging of HeLa cell expressing Chimera-GFP. Scale bar: 20 μm.
- D Anti-FLAG and anti-GFP Western blots of immunoprecipitates generated from lysates of HeLa cells double-transfected with 3 × FLAG-PI4KIIIα and either TMEM150A-GFP, TMEM150B-GFP, or Chimera-GFP.
Next, we determined the region of TMEM150A responsible for the recovery of a pool of PI4KIIIα in anti-TMEM150A-GFP immunoprecipitates. The C-tail is the major portion of TMEM150A protruding toward the cytosol. Thus, we focused on this region. Because TMEM150B-GFP does not coprecipitate PI4KIIIα, we generated a chimeric protein (Chimera) in which the C-tail of TMEM150A was appended to the C-terminus of full-length TMEM150B (Fig2B). This Chimera was targeted to the plasma membrane, similar to both TMEM150A-GFP and TMEM150B-GFP, indicating that its longer C-tail did not affect its subcellular targeting (Fig2C). Anti-GFP immunoprecipitation experiments were then performed in cells coexpressing 3 × FLAG-PI4KIIIα and either TMEM150A-GFP, TMEM150B-GFP, or Chimera-GFP. As shown in Fig2D, anti-FLAG Western blotting revealed that PI4KIIIα coprecipitated to the same extent with either TMEM150A or with Chimera, while it did not coprecipitate with TMEM150B. We conclude that the portion of TMEM150A required for the formation of a complex with PI4KIIIα possibly indirectly is its cytosolic C-tail.
TMEM150A alone is not sufficient to recruit PI4KIIIα to the plasma membrane
We previously reported that PI4KIIIα forms a complex with EFR3B and TTC7B at the plasma membrane 11 and that both EFR3B and TTC7B are required for PI4KIIIα targeting to the plasma membrane (Supplementary Fig S2) 11, 12, 20: EFR3 is the plasma membrane-anchored component 11, 20, while PI4KIIIα is recruited to EFR3 indirectly by TTC7 (see 5Fig F) 11, 20. In view of the recovery of a pool endogenous PI4KIIIα in anti-TMEM150A-GFP immunoprecipitates, as described above, we explored whether overexpression of TMEM150A in HeLa cells could bypass the need for EFR3 and TTC7 in the plasma membrane recruitment of PI4KIIIα. As shown by Fig3A, this was not the case, as GFP-PI4KIIIα had a diffuse cytosolic distribution in cells expressing TMEM150A-mCherry, which was localized in the plasma membrane. This discrepancy between the two localizations is consistent with the low recovery of PI4KIIIα in anti-TMEM150A-GFP immunoprecipitates, in spite of the specificity of this recovery (Fig3B). Co-overexpression of tagged PI4KIIIα, EFR3B, TTC7B, and TMEM150A in different combinations revealed that plasma membrane targeting of PI4KIIIα occurred only when both TTC7 and EFR3 were also co-overexpressed (Fig3A). Thus, even in the presence of co-overexpressed TMEM150A, efficient plasma membrane targeting of GFP-PI4KIIIα requires co-overexpression of both EFR3 and TTC7. However, while overexpression of EFR3 increases the recovery of GFP-PI4KIIIα in anti-TMEM150A-GFP immunoprecipitates (see below in Fig3C), a small pool of GFP-PI4KIIIα is recovered in such immunoprecipitates also when only endogenous EFR3 is present or even after EFR3 (EFR3A and EFR3B) knockdown (Fig3B). Conversely, siRNA-mediated knockdown of TMEM150A did not change the subcellular localization pattern of PI4KIIIα, EFR3B, and TTC7B when each of these proteins was overexpressed alone or in various combinations (Supplementary Fig S3).
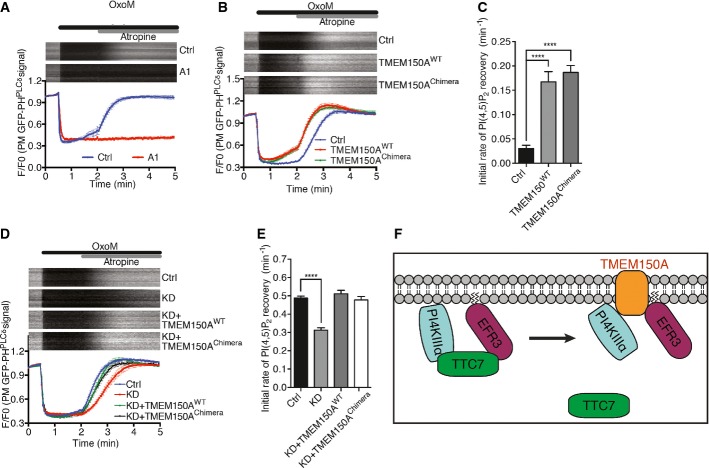
Figure 5.
- A Time course of GFP fluorescence, as assessed by TIRF microscopy, from HeLa cells transfected with GFP-PHPLCδ and muscarinic receptor (M1R) and pretreated with compound A1 (100 nM) for 10 min. Oxo-M (10 mM) and atropine (50 mM) were added at the indicated times. Kymographs of representative cells (top) and normalized average traces (bottom; n ≥ 10) are shown. Quantitative data are represented as mean ± SEM.
- B Time course of GFP fluorescence as in (A) from HeLa cells expressing the indicated proteins together with GFP-PHPLCδ and M1R.
- C Bar graph showing initial rate of PI(4,5)P2 recovery (before atropine) from (B), represented as mean ± SEM (n ≥ 10). Statistical significance was assessed by Student's t-test. ****P < 0.0001.
- D Time course of normalized GFP fluorescence, as in (A) from HeLa cells expressing the indicated proteins together with GFP-PHPLCδ and M1R, and in addition treated with control TMEM150A-specific siRNA 24 h prior to other transfection.
- E Bar graph showing initial rate of PI(4,5)P2 recovery (after atropine) from (D), represented as mean ± SEM (n ≥ 10). Statistical significance was assessed by Student's t-test. ****P < 0.0001.
- F Schematic representation of PI4KIIIα interactions at the plasma membrane. TTC7 interacts directly with both PI4KIIIα and EFR3. The presence of TMEM150A in a complex comprising PI4KIIIα is mutually exclusive to the presence in the complex of TTC7. The interactions of TMEM150A shown at the right were demonstrated biochemically and result in a positive regulation of PI4KIIIα but may be indirect.
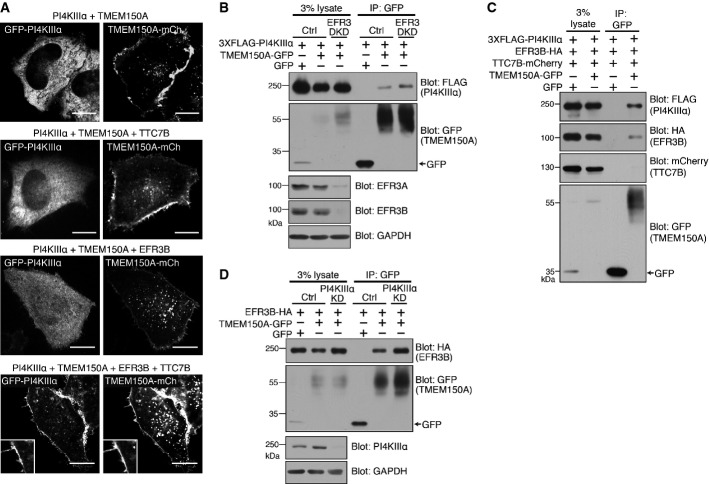
Figure 3.
- A Confocal imaging for GFP and mCherry fluorescence of live HeLa cells transfected with GFP-PI4KIIIα, TMEM150A-mCherry, EFR3B-HA, and TTC7B-FLAG in various combinations. Scale bars: 20 μm.
- B Western blot analysis for the epitopes and proteins indicated, of starting lysates and anti-GFP immunoprecipitates generated from HeLa cells treated with control or EFR3 (EFR3A and EFR3B)-specific siRNAs 24 h prior to transfection of 3 × FLAG-PI4KIIIα and TMEM150A-GFP as shown at the top.
- C Western blot analysis for the epitopes and proteins indicated, of starting lysates and anti-GFP immunoprecipitates generated from HeLa cells transfected with 3 × FLAG-PI4KIIIα, EFR3B-HA, TTC7B-mCherry, and TMEM150A-GFP as shown at the top.
- D Western blot analysis for the epitopes and protein indicated, of starting lysates and anti-GFP immunoprecipitates generated from HeLa cells treated with control and PI4KIIIα-specific siRNA 24 h prior to transfection of EFR3B-HA and TMEM150A-GFP as shown at the top.
Overexpression of TMEM150A affects the interactions of TTC7 with EFR3 and PI4KIIIα
As our data support a link of TMEM150A to PI4KIIIα, we explored whether TMEM150A interacts with EFR3 and TTC7 using biochemical experiments. HeLa cells were transiently transfected with 3 × FLAG-PI4KIIIα, EFR3B-HA, TTC7B-mCherry, and either TMEM150A-GFP or GFP. Extracts prepared from these cells were then immunoprecipitated with anti-GFP antibodies. Western blotting of immunoprecipitates from TMEM150A-GFP-expressing cells, but not of those from GFP-expressing cells, revealed robust presence of both PI4KIIIα and EFR3, but surprisingly, not of TTC7B (Fig3C), suggesting the occurence of a complex between TMEM150A, PI4KIIIα, and EFR3B that is distinct from the previously reported PI4KIIIα/EFR3B/TTC7B complex 11, 12, 20. Notably, when using cells expressing only TMEM150A-GFP and EFR3B-HA, a robust recovery of EFR3B-HA in TMEM150A-GFP immunoprecipitates, which was not affected by the siRNA-mediated knockdown of PI4KIIIα, was observed, revealing an interaction of these two plasma membrane-localized proteins (Fig3D). Lack of dependence of the TMEM150A-EFR3 interaction on the presence of PI4KIIIα was not unexpected because TTC7 is known to function as a bridge between PI4KIIIα and EFR3 11, 20, and the PI4KIIIα/EFR3B/TMEM150A-GFP complex does not include TTC7.
We further explored the surprising absence of TTC7 in PI4KIIIα-EFR3B-TMEM150A complexes, as TTC7 overexpression was needed to target efficiently PI4KIIIα to the plasma membrane under all conditions. As previously described, TTC7B-mCherry has a diffuse cytosolic localization when expressed alone 11, but is recruited to the plasma membrane when co-expressed with EFR3, its direct and plasma membrane-localized interactor 11 (Fig4A and B). However, the additional expression of TMEM150A-GFP in cells co-expressing TTC7B-mCherry and EFR3 abolished the plasma membrane localization of TTC7B (Fig4C). Thus, the interaction of EFR3 with either TTC7 or with TMEM150A appears to be mutually exclusive. We corroborated this microscopic finding with biochemical experiments. In cells transfected with TMEM150A-GFP, EFR3B-HA, and TTC7B-FLAG, analysis of anti-EFR3B (anti-HA) and anti-TTC7B (anti-FLAG) immunoprecipitates revealed that TTC7B did not co-immunoprecipitate with EFR3B (Fig4D, top panel, compare lanes 9 and 10). Vice versa, EFR3B did not coimmunoprecipitate with TTC7B (Fig4D, bottom panel, compare lanes 8 and 10), if TMEM150A-GFP was also present. The appearance of a TTC7 fragment in cells overexpressing TMEM150A (Fig4D, asterisk) may reflect an impact of TMEM150A on TTC7 stability.
Figure 4.
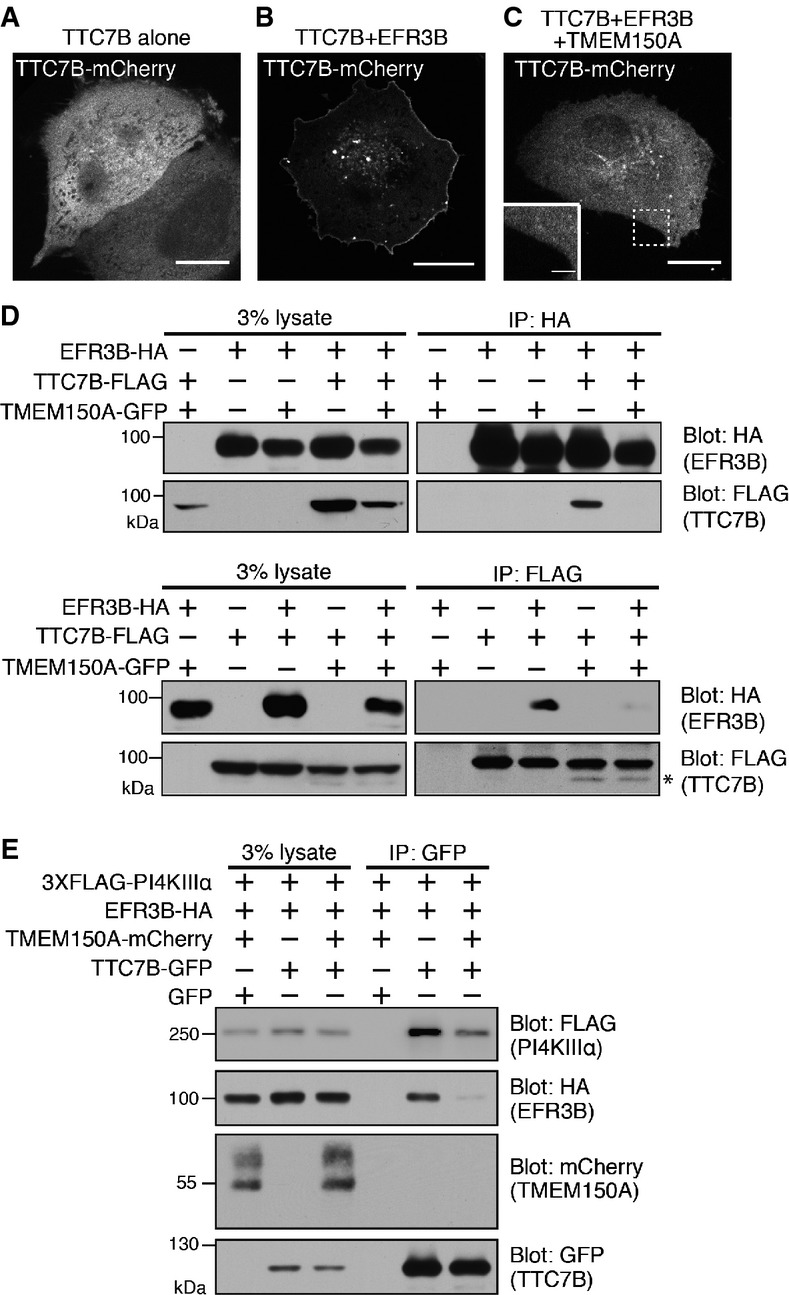
- A-C Confocal imaging of live HeLa cells transfected with TTC7B-mCherry alone or with EFR3B-HA and TMEM150A-GFP as indicated. Only the fluorescence of TTC7B-mCherry is shown. Scale bars: 20 μm.
- D Anti-HA and anti-FLAG Western blots of starting lysates and immunoprecipitates generated from lysates of HeLa cells triple-transfected with EFR3B-HA, TTC7B-FLAG, and TMEM150A-GFP. Protein complexes were immunoprecipitated by anti-HA (top panels) or anti-FLAG (bottom panels) antibodies. The asterisk indicates fragments of TTC7B just below the full-length TTC7B bands.
- E Western blot analysis for the epitopes and proteins indicated, of starting lysates and anti-GFP immunoprecipitates generated from HeLa cells transfected with 3 × FLAG-PI4KIIIα, EFR3B-HA, TMEM150A-mCherry, and TTC7B-GFP as shown at the top.
We also assessed more directly if TTC7 is only present in complexes that exclude TMEM150A in HeLa cells overexpression expressing various combinations of the four proteins (PI4KIIIα, EFR3, TMEM150A, and TTC7B), using TTC7-GFP as a bait in immunoprecipitation experiments. Both EFR3B and PI4KIIIα were recovered in the immunoprecipitates in the absence of TMEM150A overexpression (Fig4E, lane 5), but no EFR3B was recovered and the recovery of PI4KIIIα was strongly reduced when TMEM150A was overexpressed (Fig4E, lane 6).
Collectively, these results demonstrate that the interaction of EFR3 with TMEM150A is mutually exclusive with the interaction of TTC7 with EFR3 and that EFR3 and TMEM150A form a complex at the plasma membrane that does not include TTC7. Concerning PI4KIIIα, its recruitment and concentration in the plasma membrane is critically dependent on TTC7 expression even if the bulk of TTC7 is cytosolic in the presence of TMEM150A. We hypothesize that a complex of PI4KIIIα comprising TTC7 may represent an intermediate needed for the targeting/presentation of PI4KIIIα to plasma membrane. At this membrane, the EFR3-TTC7 interaction is competed by the EFR3-TMEM150A interaction, which in turn leads to a stabilization of the plasma membrane localization of PI4KIIIα even in the absence of TTC7, indicating that also the TTC7–PI4KIIIα is lost.
TMEM150A affects the resynthesis of PI(4,5)P2 at the plasma membrane following its acute depletion
If TMEM150A plays a role in anchoring PI4KIIIα at the plasma membrane, one may expect its presence to have an impact of PI4P production in this membrane. In yeast, deletion of Sfk1 increases the severity of PI4P and PI(4,5)P2 defects in a temperature-sensitive STT4 mutant background 21. To determine whether TMEM150A has a regulatory role in the metabolism of PI4P and its downstream metabolite PI(4,5)P2 in mammalian cells, we first examined whether modulation of TMEM150A levels, via either overexpression or down-regulation through RNA interference, affects total levels of these phosphoinositides. As quantified by HPLC, no significant differences in the total levels of PI4P and PI(4,5)P2 were observed in TMEM150A-overexpressing or knockdown cells (Supplementary Fig S4), although a modest decrease in the level of plasma membrane-associated PI4P, as detected by an immunofluorescence staining procedure 15, 24, 25, was observed in the TMEM150A knockdown cells (Supplementary Fig S5). This result prompted us to investigate whether an effect of TMEM150A overexpression or knockdown on phosphoinositide metabolism could be revealed by monitoring the response to an acute perturbation. To this aim, we examined the efficiency of PI(4,5)P2 resynthesis after triggered PLC-mediated hydrolysis.
In cells transiently transfected with the M1 muscarinic acetylcholine receptor (M1R), stimulation with the agonist oxotremorine-M (Oxo-M) results in phospholipase C (PLC) activation, leading to the rapid and massive cleavage of plasma membrane PI(4,5)P2 into IP3 and DAG 26. Subsequent application of the M1R antagonist atropine terminates PLC activation, allowing the cell to resynthesize PI(4,5)P2 from PI4P at the plasma membrane. Importantly, it has been well established that PI(4,5)P2 resynthesis under these conditions requires PI4P generation by PI4KIIIα 13, 15, 25, 27, 28, 29. We used this model and monitored PI(4,5)P2 changes in the plasma membrane by examining the fluorescence of GFP-PHPLCδ, a genetically encoded PI(4,5)P2 marker, using total internal reflection fluorescence (TIRF) microscopy 26, 30 (Fig5A).
In pilot experiments, we confirmed the requirement of PI4KIIIα for PI(4,5)P2 resynthesis, as this recovery was completely blocked by a very potent and highly specific PI4KIIIα inhibitor, the compound A1 15 (Fig5A). Transient expression of TMEM150A accelerated PI(4,5)P2 recovery (Fig5B), with an initial rate of PI(4,5)P2 recovery about 150% higher than that of control cells (Fig5C). A similar effect was observed with the transient expression of the TMEM150 Chimera (Fig5B), supporting the critical importance of the C-terminal region of TMEM150A, that is, its PI4KIIIα binding region, in this effect. Conversely, knockdown of TMEM150A resulted in a reduction of the initial recovery rate of PI(4,5)P2 levels, and the reduction was ‘rescued’ by co-expression of wild-type TMEM150A or the TMEM150 Chimera (Fig5D and E).
Concluding remarks
Collectively, our study corroborates evidence for an important role of an integral membrane protein conserved from yeast to humans, Sfk1/TMEM150A, in the function of PI4KIIIα to the plasma membrane. The occurence of a complex comprising PI4KIIIα and TTC7 that binds EFR3 is strongly supported by genetic, biochemical, and structural data 11, 12, 20. Our present findings support a critical role of TTC7 together with EFR3 in the targeting of PI4KIIIα to the plasma membrane. However, they also indicate that accumulation of TTC7 at the plasma membrane along with EFR3 and PI4KIIIα is under the control of TMEM150A, as TMEM150A overexpression prevents such accumulation. We suggest the occurrence of two separate PI4KIIIα-containing complexes: (1) a complex comprising PI4KIIIα, EFR3, and TTC7, which is required for initial targeting of PI4KIIIα to the plasma membrane and predominates in the absence of overexpressed TMEM150A; (2) another complex only in the plasma membrane that comprises PI4KIIIα, EFR3, and TMEM150A, but that excludes TTC7 and predominates when TMEM150A is overexpressed. Additional work is required to test this hypothesis. It is also possible that levels of TMEM150A may control interactions of TTC7 indirectly. Collectively, however, our data clearly reveal a plasticity of the PI4KIIIα complex. The mechanisms underlying this plasticity remain unknown.
Additionally, our data show that the presence of TMEM150A positively regulates PI4P production. As members of the Sfk1/TMEM150A comprise a highly conserved region containing six predicted transmembrane spans, they are unlikely to act simply as membrane-anchoring factors for Stt4/PI4KIIIα and may affect PI4KIIIα function both directly and indirectly. The expression of five TMEM150 family members with different localization and properties in mammalian cells further supports this possibility. Addressing these properties and their potential impact on the dynamics of PI4P is an important priority for future work in this field.
Materials and Methods
Cell extracts, immunoprecipitation, and immunoblotting
For immunoprecipitation, cell lysates were incubated with Chromotek GFP-trap agarose beads (Allele Biotech), anti-HA affinity matrix (Roche), or anti-FLAG M2 affinity gel (Sigma-Aldrich), and the solubilized bead-bound material was processed for SDS–PAGE and standard procedure of immunoblotting. Further details are described in the Supplementary Information.
Antibody accessibility assay
Briefly, to detect cell surface epitopes, HeLa cells transfected with proper constructs were cooled to 4°C and incubated with primary antibodies without fixation at 4°C (non-permeabilized conditions). Cells were then fixed with 4% paraformaldehyde/PBS. Further details are described in the Supplementary Information.
Immunostaining for the plasma membrane PI(4)P
Immunostaining of the plasma membrane PI(4)P was performed and quantified as described in the reference paper 16. Further details are described in the Supplementary Information.
Other methods are described in the Supplementary Information.
Acknowledgments
We would like to thank Michael Caplan and Christopher Burd for helpful discussions and Louise Lucast for help with lipid analysis. This work was supported in part by grants from the NIH (R37NS036251, DK082700, and DA018343 to PDC; T32GM007223 to JC; and K99GM110121 to JMB) and from the Simons Foundation to PDC and from a fellowship from the Jane Coffin Childs Fund to JMB.
Author contributions
JC, FN, JB, and PDC designed the experiments; JC performed the experiments; all authors contributed to writing the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Review Process File
References
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P - not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Irvine RF. Inositol phosphates and cell signalling. Nature. 1989;341:197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–1213. [PubMed] [Google Scholar]
- Raychaudhuri S, Prinz WA. The diverse functions of oxysterol-binding proteins. Annu Rev Cell Dev Biol. 2010;26:157–177. doi: 10.1146/annurev.cellbio.042308.113334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Wu WI, Pedretti J, Yates R, Voelker DR. A genetic screen for aminophospholipid transport mutants identifies the phosphatidylinositol 4-kinase, STT4p, as an essential component in phosphatidylserine metabolism. J Biol Chem. 1998;273:13189–13196. doi: 10.1074/jbc.273.21.13189. [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird D, Stefan C, Audhya A, Weys S, Emr SD. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J Cell Biol. 2008;183:1061–1074. doi: 10.1083/jcb.200804003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III alpha: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol Biol Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt FH, Brault M, Pilote L, Uyttersprot N, Gaillard ET, Stoltz JH, Knight BL, Pantages L, McFarland M, Breitfelder S, et al. Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target. J Virol. 2012;86:11595–11607. doi: 10.1128/JVI.01320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet M-S, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, CornellaTaracido I, et al. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chi RJ, Baskin JM, Lucast L, Burd CG, De Camilli P, Reinisch KM. Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex. Dev Cell. 2014;28:19–29. doi: 10.1016/j.devcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr S. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Park S-M, Kim K, Lee E-J, Kim B-K, Lee TJ, Seo T, Jang I-S, Lee S-H, Kim S, Lee J-H, et al. Reduced expression of DRAM2/TMEM77 in tumor cells interferes with cell death. Biochem Biophys Res Commun. 2009;390:1340–1344. doi: 10.1016/j.bbrc.2009.10.149. [DOI] [PubMed] [Google Scholar]
- Hammond GRV, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond GRV, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger BH, Jensen JB, Hille B. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J Gen Physiol. 2010;135:81–97. doi: 10.1085/jgp.200910344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz LF. Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. J Gen Physiol. 2005;126:243–262. doi: 10.1085/jgp.200509309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A, Kim YJ, Varnai P, Szentpetery Z, Knight Z, Shokat KM, Balla T. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIalpha. Mol Biol Cell. 2008;19:711–721. doi: 10.1091/mbc.E07-07-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson EJ, Jensen JB, Hille B. Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc Natl Acad Sci USA. 2014;111:E2281–E2290. doi: 10.1073/pnas.1407133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File