Abstract
BACKGROUND
Comprehensive molecular characterization of cancer that has metastasized to bone has proved challenging, which may limit the diagnostic and potential therapeutic opportunities for patients with bone-only metastatic disease.
METHODS
We describe successful tissue acquisition, DNA extraction, and whole-exome sequencing from a bone metastasis of a patient with metastatic, castration-resistant prostate cancer (PCa).
RESULTS
The resulting high-quality tumor sequencing identified plausibly actionable somatic genomic alterations that dysregulate the phosphoinostide 3-kinase pathway, as well as a theoretically actionable germline variant in the BRCA2 gene.
CONCLUSIONS
We demonstrate the feasibility of diagnostic bone metastases profiling and analysis that will be required for the widespread application of prospective ‘precision medicine’ to men with advanced PCa.
Keywords: bone biopsy, whole-exome sequencing, genomics, precision medicine
INTRODUCTION
Significance
The application of prospective massively parallel sequencing to tissue obtained from bone metastases is feasible in the clinical setting. The resulting data may be actionable in metastatic prostate cancer (PCa), and the approach may be applicable to many patients with metastatic cancer limited to the bone.
Background
The prospective use of massively parallel sequencing in oncology has started to impact medical decision making by promoting individualized therapeutic choices based on genomic information.1,2 In some instances, this may require fresh biopsies from patients with metastatic disease either before treatment, during treatment or upon relapse. However, obtaining sufficient high-quality tumor tissue for genomic studies can be technically and logistically challenging depending on the specific anatomic location of the metastases.
Metastatic PCa is emblematic of an epithelial malignancy that primarily metastasizes to the bone. For example, over 90% of patients enrolled in clinical trials of abiraterone acetate or enzalutamide (therapeutics that suppress androgen synthesis or androgen receptor (AR) signaling, respectively) had bone metastases at baseline, whereas only 50% had soft tissue metastases.3,4 In order to study resistance to castration-based therapies and/or guide clinical trial enrollment, obtaining tumor genetic material from bone metastases will be required for most patients.
Although castration-resistant PCas have undergone research-oriented genomics studies, most of these tumors were acquired from soft tissue metastases or autopsy specimens.5,6 Here, we describe successful bone biopsy tissue acquisition and whole-exome sequencing (WES) for a living patient with metastatic, castration-resistant PCa. Several plausibly actionable somatic and germline events were detected that may guide future clinical decision making. More generally, these results show the feasibility of applying clinical sequencing to cancer patients with extensive bony metastases.
MATERIALS AND METHODS
Ethics statement
Written informed consent was obtained from the patient to participate in this study. Patient tumor and matched normal samples were collected and sequenced under an Institution Review Board approved protocol (UCSF Committee on Human Research (CHR: http://www.research.ucsf.edu/chr/NewInv/chrNewInv.asp) protocol number 10-01418).
Radiographic biopsy methods
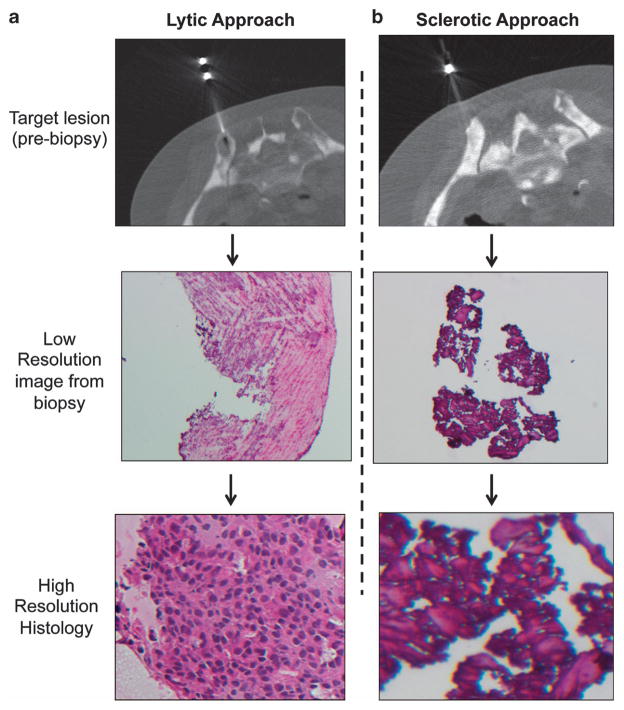
Written informed consent was obtained before the procedure from the patient. The risks and benefits were discussed, and the patient agreed to proceed. The patient was placed prone on the computed tomography gantry couch. Scout computed tomography images were obtained at a slice thickness of 5 mm. Two adjacent sites were marked overlying the right posterior superior ilium. The skin was prepped and draped in the usual sterile manner. In all, 1% lidocaine was used for local anesthesia. A small dermatotomy was made in the skin. A 16-gauge biopsy needle was used to obtain two core samples of a sclerotic lesion (Figure 1b). The biopsy needle was removed. Rapid hemostasis was achieved locally. Next, following local anesthesia with 1% lidocaine, a small dermotomy was made in the skin inferiorly. A 16-gauge Bonopty (AprioMed, Uppsala, Sweden) bone biopsy needle system was used to obtain two core samples of an adjacent lytic lesion (Figure 1a).
Figure 1.
Successful and unsuccessful approaches for the bone biopsy. (a, b) Radiographic images of successful and unsuccessful biopsies from the patient corresponding to lytic and sclerotic lesions (a, b), with subsequent biopsy and histology images obtained from frozen tissue.
Research biopsies were divided in two, surrounded in OCT (optimum cutting temperature frozen embedding medium) and frozen immediately after the biopsy. Tissue was subsequently stored in vapor phase liquid nitrogen before tissue collection. Samples were submitted to the research associate at the bedside. Samples for surgical pathology were submitted. Gauze and a Tegaderm were applied to the skin after it was cleaned. The patient tolerated the procedure well without observable complication. Total radiation dose as reported as a total dose-length product was 106.8 mGy-cm. The patient was discharged to the radiology nursing holding area in good condition for half an hour of observation.
Three serial sections were used on each slide to assess tissue variability through the core. Staining was done using a standard hematoxylin (Gill #2) and eosin protocol. Cryosectioning at −20 °C was performed on frozen biopsies without prior decalcification to obtain tissue sections of approximately 8–10 μm in thickness. Each tissue section was collected on charged glass slides and bone fragments present within the tissue sections were removed manually before laser capture microdissection (LCM). Multiple tissue sections were used to collect cells (Supplementary Table 2).
DNA extraction, library preparation, assembly and quality control
Each LCM tube was filled with 50 μl of lysing buffer from the Stratagene Absolutely RNA nanoprep isolation kit (Agilent Technologies, Santa Clara, CA, USA), containing β mercaptoethanol. This buffer removes cells from the LCM cap more rapidly than proteinase-K digestion alone. Following LCM, the sample caps were attached to tubes containing the Stratagene buffer, inverted, then incubated at room temperature for an hour. The tube was then spun to retain the buffer/sample and the cap was discarded. The same buffer was then used for a second sample LCM cap to maximize tissue/ buffer concentration. The contents of two buffer tubes were then combined (100 μl total) and added to 300 μl of digestion buffer (10 mM NaCl, 1.5 mM MgCl2, 50 mM KCl, 0.05% Tween-20) and 7 μl of proteinase-K (20 μg μl−1). The digestion and DNA isolation was carried out with the eight LCM slides distributed between two 400 μl digestion samples to maximize DNA retention in the columns. The digestion samples were placed in a 55 °C water bath and rocked overnight. In all, 7 μl of fresh proteinase-K was added to each tube following overnight incubation, and repeated for two more overnight incubations. After the third incubation, proteinase-K was deactivated at 95 °C for 10 min. Sample volumes were then loaded onto pre-wetted Amicon Ultra 30K filtration columns and spun for 8 min at 12 000 relative centrifugal force. Two 450 μl water washes were performed (8 min, 12 000 relative centrifugal force) before inverting the columns into a clean sample tube and extracting the concentrated DNA sample at 13 000 relative centrifugal force for 1 min. The two samples were combined and quantity was measured using quantitative PCR and the PicoGreen assay. DNA was isolated from blood using a standard Qiagen Blood Maxi kit (Qiagen, Hilden, Germany) according to protocol. Germline DNA was isolated from peripheral mononuclear blood cells to use for downstream analyses.
Exome capture and library construction were performed per standard Broad Sequencing Platform procedures,8 and libraries were sequenced on Illumina HiSeq 2000 machines (Illumina, San Diego, CA, USA). The resulting data obtained from the Illumina pipeline were assembled using the Picard pipeline (http://picard.sourceforge.net/) and aligned to the hg19 reference genome. Cross-contamination of samples was estimated using ContEst,24 to confirm that neither tumor nor germline sample had >1% contamination. Single-nucleotide polymorphism fingerprints from each lane of a tumor/ normal pair were cross-checked to confirm concordance, and non-matching lanes were removed from analysis. Sequencing metrics and coverage analyses were performed using the HS_Metrics tool from the Picard pipeline and DepthOfCoverage from the Genome Analysis Toolkit,25 respectively.
Identification of somatic alterations
Somatic single-nucleotide base-pair substitutions were identified using MuTect,26 and small insertions and deletions were identified with Indelocator (http://www.broadinstitute.org/cancer/cga/indelocator). Copy ratios were inferred from analysis of sequencing depth in tumor and normal samples. The resulting copy ratios were segmented using the circular binary segmentation algorithm. Genes in regions with segment means of >2 were evaluated for amplifications, and genes in regions with segment means of <−1 were evaluated for deletions. Rearrangements were determined using BreakPointer.9
Annotation of identified variants was done using Oncotator (http://www.broadinstitute.org/cancer/cga/oncotator). These algorithms were executing using the Broad Firehose Infrastructure (http://www.broadinstitute.org/cancer/cga/Firehose). Clinical interpretation of the collected somatic alterations was determined with a heuristic platform for the analysis and interpretation of cancer exome data.10
Identification of germline alterations
Single-nucleotide variants and insertion/deletions were identified on the hg19-aligned germline BAM (the compressed binary version of the Sequence Alignment/Map (SAM) format) file using Unified Genotyper25 with a pooled set of 50 additional publically available healthy adult germline BAMs. The variants were annotated using Oncotator, and those in the subset of genes identified as relevant to cancer risk2 were selected for further review. The estimated population frequencies of resulting variants were calculated using data from the Exome Variant Server project,14 and variants with <1% population frequency were selected for manual review.
RESULTS
Clinical background
The patient was a 57-year-old man who presented with an elevated PSA of 4.42 ng dl−1 during a screening evaluation. A prostate biopsy revealed localized prostate adenocarcinoma (Gleason 3 + 3 = 6) in 13/15 cores with perineural invasion (clinical staging was T3b, N0, M0). The patient received neoadjuvant androgen deprivation therapy (leuprolide and bicalutamide) followed by combined external beam radiation therapy and brachytherapy. His PSA became undetectable during treatment, but 4 years later the PSA began rising and his bone scan was suspicious for metastatic PCa. Androgen deprivation therapy was resumed, and his PSA fell from 7.59 to 0.5. After <12 months of therapy his PSA began to rise despite castrate levels of testosterone. He was treated with secondary hormonal therapy including ketoconazole with only modest and fleeting reductions in his PSA. When his PSA rapidly climbed to 91.6, he was treated with carboplatin and docetaxel on a clinical protocol and zoledronic acid to reduce skeletal-related events. After six cycles of chemotherapy, his PSA fell to 1.97 and continued to decline to a nadir of 1.22 approximately 2 months after stopping chemotherapy.
Eight months later, the patient’s PSA climbed to 28.2 and he started enzalutamide, experiencing a transient PSA response. He then developed signs of early spinal cord compression and received focal radiation therapy. Once stabilized, he enrolled in a clinical trial of cabozatinib and his PSA declined from 42.5 to 31.8 as his best response. Two months thereafter, his PSA increased to 80.4 and a chest computed tomography scan revealed multiple new pulmonary nodules. He re-started chemotherapy with carboplatin and docetaxel and again had a meaningful clinical response with a PSA decline to 55. At this time, because of limited remaining treatment options, the patient underwent a research computed tomography-guided bone biopsy and consented to additional tissue being obtained for research purposes (UCSF IRB 10-01418).
Acquisition of tumor tissue from bone metastases
Two distinct bone metastases in the right ilium were biopsied: a lytic lesion (Figure 1a) and a primarily sclerotic lesion (Figure 1b). Histological analysis of the biopsied tissue revealed clusters of malignant epithelial cells in frozen core 3 (lytic lesion) that appeared adequate for LCM. Immunohistochemical studies of prostatic acid phosphatase confirmed the prostatic origin of the epithelial cells (Supplementary Figure 1A). Using previously described methods,7 LCM was used to capture the malignant epithelial cells, and multiple slides of LCM-captured tissue were combined into a single nucleic acid isolation tube (Table 1). Two rounds of LCM produced >200 ng of genomic DNA (Supplementary Figure 1B).
Table 1.
Summary of sequencing results
| Somatic DNA | Germline DNA | |
|---|---|---|
| Concentration (ng μl−1) | 7 | 16 |
| Total volume (μl) | 46 | 700 |
| Total quantity (μg) | 0.322 | 11.2 |
| Mean target coverage (X) | 145 | 147 |
| Targets 30X (%) | 88 | 87 |
| Zero coverage targets (%) | 1 | 1 |
Adequate extraction of adequate genomic DNA from the tumor and germline DNA was subjected to exon capture and sequenced (see Materials and methods section). Standard sequencing metrics are shown, including mean target coverage, targets covered at a sequencing depth of ≥30-fold, and the percent of zero target coverage.
WES of bone metastasis-derived PCa cells
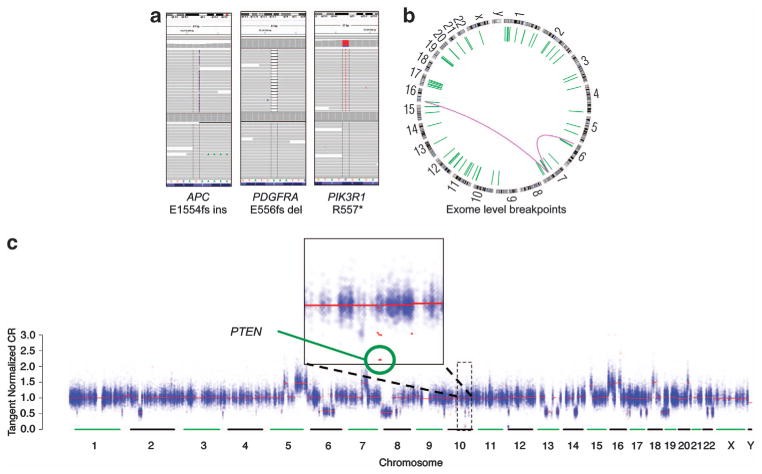
WES was performed on 50 ng of tumor and 85 ng of paired normal DNA (see Materials and methods section). In the tumor, the mean target sequencing coverage was 145-fold, with 88% of territory covered at ≥30-fold (Table 1); these parameters met or exceeded ‘conventional’ exome sequencing target goals.8 A total of 1955 unique somatic mutations, small insertions deletions and copy number alterations were observed. The overall mutation rate was 2.52 per megabase, comparable to advanced PCa5 (Figures 2a and c; Supplementary Table 2). Although many important DNA rearrangements occur outside of exon coding regions9 and are not robustly detected by WES, assessment of breakpoints within or near exons demonstrated multiple putative events (Figure 2b). Germline analysis revealed 23 856 single-nucleotide polymorphisms across the exome (Supplementary Table 3).
Figure 2.
Sequencing results yield multiple different categories of alterations. (a–c) Whole-exome sequencing from this patient’s bone biopsy using the defined protocol yields interpretable data. This includes the ability to identify non-synonymous mutations, short insertions and deletions (a), exome-level fusions and rearrangements (b) and chromosomal copy number alterations across the exome (c). For the copy number plot, the signal data corresponding to each exon target are colored blue, and the red lines denote the copy number segments defined by the circular binary segmentation algorithm. PTEN, phosphatase and tensin homolog deleted on chromosome 10.
Plausibly actionable somatic and germline alterations
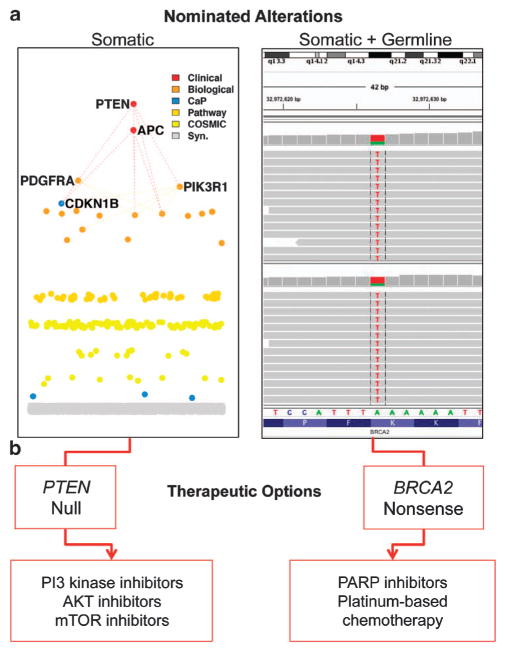
Analysis of variants from the tumor and normal exomes10 identified several genomic alterations of interest (Figures 3a and b). Several alterations were present in genes that encode members of the phosphoinostide 3-kinase (PI3K) pathway, including a homozygous phosphatase and tensin homolog deleted on chromosome 10 (PTEN) deletion and a PIK3R1 R557* nonsense mutation. The latter mutation was present at an allelic fraction of 0.82, suggesting biallelic inactivation in the setting of minimal stromal admixture. PIK3R1 encodes the P85α regulatory subunit of PI3K. In glioblastoma models, PIK3R1 mutations were shown to increase PI3K signaling and conferred increased sensitivity to AKT inhibitors.11
Figure 3.
Clinically actionable findings in the tumor. (a, b) Integrated clinical analysis and interpretation of the patient’s tumor and normal exome revealed multiple somatic and germline events warranting further review (a). These included a homozygous PTEN deletion and a PIK3R1 nonsense mutation. Remaining alterations were scored by rules as being biologically relevant, being relevant specifically to prostate cancer biology, being in the pathway of a cancer gene, or being present in COSMIC. Germline clinical analysis also revealed a BRCA2 variant of uncertain significance. These events may be linked to clinical actions, which are listed here (b). mTOR, mammalian target of rapamycin; PARP, poly(ADP-ribose) polymerase; PI3K, phosphoinostide 3-kinase; PTEN, phosphatase and tensin homolog deleted on chromosome 10.
Other salient somatic mutations included an APC E1554fs insertion. Loss-of-function APC mutations are common in gastrointestinal malignancies; however, APC mutations have also been observed in ~3.5% of PCas.12 Moreover, a PDGFRA E556fs deletion was observed that had not been reported previously. The biological significance of the APC and PDGFRA mutations is uncertain. Finally, the tumor harbored a deletion in CDKN1B, which was recently found to be significantly altered in primary PCa.13
We did not detect alterations in several genes known to govern AR function in PCa. Specifically, no somatic events were observed in AR, FOXA1, GATA2, NXK3-1, PXN, FASN or UGT2B17,5,13 despite there being adequate tumor and normal coverage to detect alterations in these genes (Supplementary Figure 2, Supplementary Table 4).
In the patient’s germline exome, we detected a BRCA2 K3226* nonsense variant. This variant occurs with an estimated population frequency of 0.6% per the Exome Variant Server.14 The significance of this event is uncertain, as it occurs near the carboxy-terminus of the protein and is not a known risk factor for breast or PCa susceptibility.15 However, inspection of this locus in the tumor WES data revealed loss of the normal allele and the suggestion of BRCA2 biallelic inactivation. Some BRCA2 mutations have been linked to increased platinum-based chemotherapy sensitivity in other disease contexts,16 and may predict response to poly(ADP-ribose) polymerase inhibitors.17
CONCLUSIONS
Multiple technical hurdles have limited detailed genomic studies of bone metastases from epithelial malignancies.6 We successfully achieved ‘clinical-grade’ WES of a bone metastasis biopsy specimen obtained from a living patient. Although these findings will have to be confirmed by a clinical laboratory before they can be used to guide therapy in this patient, we did identify actionable genomic alterations that may impact clinical decision making.
Although the mainstay of therapy for men with castration-resistant PCa is directed against the androgen signaling axis, this patient developed rapid resistance to primary hormonal therapy and had similarly short durations of response to newer AR targeting agents (enzalutamide). These poor clinical results are intriguing in light of the lack of genomic alterations involving either AR itself or other genes associated with AR activity.
The bone metastasis characterized here contained multiple genetic events predicted to activate the PI3K pathway in genes that were previously reported to be significantly altered in primary13 and metastatic5 PCa cohorts. PI3K dysregulation occurs in the majority of advanced PCas; however, the concomitant PTEN homozygous deletion and PIK3R1 mutation may conceivably confer enhanced dependency on this pathway. Although allosteric mammalian target of rapamycin inhibitors have met with limited success in the setting of metastatic PCa,18 these events may support enrollment in clinical trials of single-agent PI3K, AKT or mTOR kinase inhibitors given as single agents19 or together with AR-directed therapies.20 Moreover, should this patient (or patients like him) have an extraordinary response to one of these agents, these clinical observations may be linked to the observed genomic features to nominate new biomarkers for response or resistance to novel therapies that can be studied prospectively.
The germline sequencing data raise the possibility that BRCA2 activity may be reduced in this patient’s tumor. Notably, the patient’s clinical course bears important similarities to that seen in BRCA2 carriers who develop PCa. In particular, such men exhibit a relatively short duration of response to androgen deprivation and robust clinical responses to platinum-based chemotherapy regimens. Thus, both the somatic and germline WES data offered potentially actionable genomic information in this case.
Although our biopsy protocol was successful here and in other contexts,21 the patient characteristics that predict successful and unsuccessful bone biopsies remain unclear. Furthermore, this study was limited to an evaluation of the coding regions of the tumor and normal DNA. In PCa, this is pertinent because of the emergence of clinical trials testing distinct targeted therapies in E26 transformation-specific (ETS) fusion-positive22 or SPINK1 overexpressed23 PCas—neither of which could be observed with WES alone. In the future, additional sequencing modalities (for example, transcriptome sequencing or whole-genome sequencing) may be required to achieve a more complete view of clinically actionable PCa alterations.
In summary, we have successfully procured and sequenced tumor DNA from an epithelial bone metastasis. The approach described here may be applicable to other epithelial malignancies that metastasize to the bone, thus enabling expanded views into the biology and therapeutic vulnerabilities linked to this common and debilitating mode of tumor progression.
Supplementary Material
Acknowledgments
This work was supported by Stand Up To Cancer, Prostate Cancer Foundation, Prostate Dream Team Translational Cancer Research Grants, made possible by the generous support of the Movember Foundation (EMV, AF, LAG, PGF), the Prostate Cancer Foundation (EMV, PGF), the Starr Cancer Consortium (I4-A424; LAG), the NIH (5 U01 CA157703; PGF), and the Department of Defense (W81XWH-10-PCRP-SIDA; LAG). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. The authors acknowledge Deborah Farlow for her project management contribution to this effort.
Footnotes
CONFLICT OF INTEREST
Levi A Garraway is an equity holder and consultant in Foundation Medicine, a consultant to Novartis, Millenium/Takeda, and Boehringer Ingelheim, and a recipient of a grant from Novartis. Phillip G Febbo is the principal investigator on a Novartis supported trial and speaker and consultant for Janssen and Dendrion. The remaining authors declare no conflict of interest.
References
- 1.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Translational Med. 2011;3:111ra21. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehra R, Kumar-Sinha C, Shankar S, Lonigro RJ, Jing X, Philips NE, et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin Cancer Res. 2011;17:3924–3932. doi: 10.1158/1078-0432.CCR-10-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Febbo PG, Thorner A, Rubin MA, Loda M, Kantoff PW, Oh WK, et al. Application of oligonucleotide microarrays to assess the biological effects of neoadjuvant imatinib mesylate treatment for localized prostate cancer. Clin Cancer Res. 2006;12:152–158. doi: 10.1158/1078-0432.CCR-05-1652. [DOI] [PubMed] [Google Scholar]
- 8.Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Allen E, Wagle N, Kryukov G, Ramos A, Getz G, Garraway L. A heuristic platform for clinical interpretation of cancer genome sequencing data. J Clin Oncol. 2012;(suppl):abstr 10502. [Google Scholar]
- 11.Quayle SN, Lee JY, Cheung LW, Ding L, Wiedemeyer R, Dewan RW, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PloS One. 2012;7:e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA, USA: 2012. [Google Scholar]
- 15.Farrugia DJ, Agarwal MK, Pankratz VS, Deffenbaugh AM, Pruss D, Frye C, et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–3531. doi: 10.1158/0008-5472.CAN-07-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306(14):1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 18.Nakabayashi M, Werner L, Courtney KD, Buckle G, Oh WK, Bubley GJ, et al. Phase II trial of RAD001 and bicalutamide for castration-resistant prostate cancer. BJU Int. 2012;110:1729–1735. doi: 10.1111/j.1464-410X.2012.11456.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Allen EM, Pomerantz M. Moving toward personalized medicine in castration-resistant prostate cancer. Urol Clinics North Am. 2012;39:483–490. doi: 10.1016/j.ucl.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spritzer CE, Alfonso PD, Vinson EN, Turnbull JD, Morris KK, Foye A, et al. Bone marrow biopsy: RNA isolation with expression profiling in men with metastatic castration-resistant prostate cancer - factors affecting diagnostic success. Radiology. 2013;269:816–823. doi: 10.1148/radiol.13121782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ateeq B, Tomlins SA, Laxman B, Asangani IA, Cao Q, Cao X, et al. Therapeutic targeting of SPINK1-positive prostate cancer. Sci Translational Med. 2011;3:72ra17. doi: 10.1126/scitranslmed.3001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;2718:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.