Abstract
Genome-wide association studies (GWAS) have uncovered numerous genetic variants (SNPs) that are associated with blood pressure (BP). Genetic variants may lead to BP changes by acting on intermediate molecular phenotypes such as coded protein sequence or gene expression, which in turn affect BP variability. Therefore, characterizing genes whose expression is associated with BP may reveal cellular processes involved in BP regulation and uncover how transcripts mediate genetic and environmental effects on BP variability. A meta-analysis of results from six studies of global gene expression profiles of BP and hypertension in whole blood was performed in 7017 individuals who were not receiving antihypertensive drug treatment. We identified 34 genes that were differentially expressed in relation to BP (Bonferroni-corrected p<0.05). Among these genes, FOS and PTGS2 have been previously reported to be involved in BP-related processes; the others are novel. The top BP signature genes in aggregate explain 5%–9% of inter-individual variance in BP. Of note, rs3184504 in SH2B3, which was also reported in GWAS to be associated with BP, was found to be a trans regulator of the expression of 6 of the transcripts we found to be associated with BP (FOS, MYADM, PP1R15A, TAGAP, S100A10, and FGBP2). Gene set enrichment analysis suggested that the BP-related global gene expression changes include genes involved in inflammatory response and apoptosis pathways. Our study provides new insights into molecular mechanisms underlying BP regulation, and suggests novel transcriptomic markers for the treatment and prevention of hypertension.
Author Summary
The focus of blood pressure (BP) GWAS has been the identification of common DNA sequence variants associated with the phenotype; this approach provides only one dimension of molecular information about BP. While it is a critical dimension, analyzing DNA variation alone is not sufficient for achieving an understanding of the multidimensional complexity of BP physiology. The top loci identified by GWAS explain only about 1 percent of inter-individual BP variability. In this study, we performed a meta-analysis of gene expression profiles in relation to BP and hypertension in 7017 individuals from six studies. We identified 34 differentially expressed genes for BP, and discovered that the top BP signature genes explain 5%–9% of BP variability. We further linked BP gene expression signature genes with BP GWAS results by integrating expression associated SNPs (eSNPs) and discovered that one of the top BP loci from GWAS, rs3184504 in SH2B3, is a trans regulator of expression of 6 of the top 34 BP signature genes. Our study, in conjunction with prior GWAS, provides a deeper understanding of the molecular and genetic basis of BP regulation, and identifies several potential targets and pathways for the treatment and prevention of hypertension and its sequelae.
Introduction
Systolic and diastolic blood pressure (SBP and DBP) are complex physiological traits that are affected by the interplay of multiple genetic and environmental factors. Hypertension (HTN) is a critical risk factor for stroke, renal failure, heart failure, and coronary heart disease [1]. Genome-wide association studies (GWAS) have identified numerous loci associated with BP traits [2,3]. These loci, however, only explain a small proportion of inter-individual BP variability. In aggregate the 29 loci reported by the International Consortium of Blood Pressure (ICBP) consortium GWAS account for about one percent of BP variation in the general population [3]. Most genes near BP GWAS loci are not known to be mechanistically associated with BP regulation [3]. Therefore, further studies are needed to determine whether the genes implicated in GWAS demonstrate functional relations to BP physiology and to uncover the molecular actions and interactions of genetic and environmental factors involved in BP regulation.
Alterations in gene expression may mediate the effects of genetic variants on phenotype variability. We hypothesized that characterizing gene expression signatures of BP would reveal cellular processes involved in BP regulation and uncover how transcripts mediate genetic and environmental effects on BP variability. We additionally hypothesized that by integrating gene expression profiling with genetic variants associated with altered gene expression (eSNPs or eQTLs) and with BP GWAS results, we would be able to characterize the genetic architecture of gene expression effects on BP regulation.
Several previous studies have examined the association of global gene expression with BP [4,5] or HTN [6,7]. Most of these studies, however, were based on small sample sizes and lacked replication [4,5,6,7]. To address this challenge, we conducted an association study of global gene expression levels in whole blood with BP traits (SBP, DBP, and HTN) in six independent studies. In order to avoid the possibility that the differentially expressed genes we identified reflect drug treatment effects, we excluded individuals receiving anti-hypertensive treatment. The eligible study sample included 7017 individuals: 3679 from the Framingham Heart Study (FHS), 972 from the Estonian Biobank (EGCUT), 604 from the Rotterdam Study (RS) [8], 597 from the InCHIANTI Study, 565 from the Cooperative Health Research in the Region of Augsburg [KORA F4] Study [9], and 600 from the Study of Health in Pomerania [SHIP-TREND] [10]. We first identified differentially expressed BP genes in the FHS (n = 3679) followed by external replication in the other five studies (n = 3338). Subsequently, we performed a meta-analysis of all 7017 individuals from the six studies, and identified 34 differentially expressed genes associated with BP traits using a stringent statistical threshold based on Bonferroni correction for multiple testing of 7717 unique genes. The differentially expressed genes for BP (BP signature genes) were further integrated with eQTLs and with BP GWAS results in an effort to differentiate downstream transcriptomic changes due to BP from putatively causal pathways involved in BP regulation.
Results
Clinical characteristics
After excluding individuals receiving anti-hypertensive treatment, the eligible sample size was 7017 (FHS, n = 3679; EGCUT, n = 972; RS, n = 604; InCHIANTI, n = 597; KORA F4, n = 565 and SHIP-TREND, n = 600). Clinical characteristics of participants from the four studies are presented in Table 1. The mean age varied across the cohorts (FHS = 51, EGCUT = 36, RS = 58, InCHIANTI = 71, KORA F4 = 72 and SHIP-TREND = 46 years) as did the proportion of individuals with hypertension (11% in FHS, 19% in EGCUT, 35% in RS, 45% in InCHIANTI, 26% in KORA, and 12% in SHIP).
Table 1. Clinical characteristics of the study cohorts.
| FHS N = 3,679 | EGCUT N = 972 | RS N = 604 | InCHIANTI N = 597 | KORA F4 N = 565 | SHIP-TREND N = 600 | |
|---|---|---|---|---|---|---|
| Age (yr) | 51 ± 12 | 36 ± 14 | 58 ± 8 | 71 ± 16 | 72 ± 5 | 46 ± 13 |
| Sex, male (%) | 42 | 49 | 46 | 46 | 51 | 43 |
| Hypertension (%) | 11 | 19 | 35 | 45 | 26 | 12 |
| BMI (kg/m 2) | 27.2 ± 5.3 | 24.8 ± 4.4 | 26.8 ± 4.1 | 27.0 ± 4.2 | 29.8± 4.6 | 26 ± 4.2 |
| Systolic BP (mm Hg) | 118 ± 15 | 122 ± 16 | 132 ± 20 | 132 ± 20 | 129± 21 | 120 ± 15 |
| Diastolic BP (mm Hg) | 74 ±9 | 76 ± 10 | 82 ± 11 | 78 ±10 | 73±11 | 75 ± 9 |
Identification and replication of differentially expressed BP signature genes
At a Bonferroni corrected p<0.05, we identified 73, 31, and 8 genes that were differentially expressed in relation to SBP, DBP, and HTN, respectively in the FHS, which used an Affymetrix array for expression profiling, and 6, 1, and 1 genes in the meta-analysis of the 5 cohorts that used an Illumina array (Illumina cohorts): EGCUT, RS, InCHIANTI, KORA F4 and SHIP-TREND (S1 Table). For each differentially expressed BP gene in the FHS or in the Illumina cohorts, we attempted replication in the other group. At a replication p<0.05 (Bonferroni corrected), 13 unique genes that were identified in the FHS were replicated in the Illumina cohorts, including 10 for SBP (CD97, TAGAP, DUSP1, FOS, MCL1, MYADM, PPP1R15A, SLC31A2, TAGLN2, and TIPARP), 5 for DBP (CD97, BHLHE40, PRF1, CLC, and MYADM), and 2 for HTN (GZMB and MYADM) (Table 2). Each of the unique BP signature genes in the Illumina cohorts, 6 for SBP (TAGLN2, BHLHE40, MYADM, SLC31A2, DUSP1, and MCL1), 1 for DBP (BHLHE40) and 1 for HTN (SLC31A2), replicated in the FHS. All 6 Illumina cohorts BP signature genes that replicated in the FHS were among the 13 FHS BP signature genes that replicated in the Illumina cohorts. The BP signature genes identified in the FHS showed enrichment in the Illumina cohorts at pi1 = 0.88, 0.75, and 0.99 for SBP, DBP, and HTN respectively (pi1 value indicates the proportion of significant signals among the tested associations [11]; see details in the Methods section). Fig. 1 shows that the mean gene expression levels of the top BP signature genes were consistent with the BP phenotypic changes observed in the FHS and the Illumina cohorts.
Table 2. Differentially expressed genes associated with BP and hypertension at Bonferroni correction p<0.05 in meta-analysis of the six cohorts.
| Gene | Chr. | Gene Description | FHS Beta | FHS s.e. | FHS pvalue | Illumina Beta | Illumina s.e. | Illumina pvalue | Meta * | Meta s.e. | Meta pvalue |
|---|---|---|---|---|---|---|---|---|---|---|---|
| —SBP Signature genes | |||||||||||
| SLC31A2 | 9 | solute carrier family 31 (copper transporters), member 2 | 2.4E-03 | 3.3E-04 | 1.2E-13 | 2.1E-03 | 3.3E-04 | 9.9E-11 | 2.3E-03 | 2.3E-04 | <1E-16 |
| MYADM | 19 | myeloid-associated differentiation marker | 2.5E-03 | 3.2E-04 | 2.2E-14 | 2.7E-03 | 3.9E-04 | 2.2E-12 | 2.6E-03 | 2.5E-04 | <1E-16 |
| DUSP1 | 5 | dual specificity phosphatase 1 | 2.2E-03 | 3.9E-04 | 1.1E-08 | 2.1E-03 | 4.2E-04 | 3.7E-07 | 2.2E-03 | 2.9E-04 | 2.0E-14 |
| TAGLN2 | 1 | transgelin 2 | 2.0E-03 | 4.1E-04 | 1.0E-06 | 2.0E-03 | 4.0E-04 | 1.3E-06 | 2.0E-03 | 2.9E-04 | 5.8E-12 |
| CD97 | 19 | CD97 molecule | 1.7E-03 | 3.2E-04 | 1.4E-07 | 1.5E-03 | 3.5E-04 | 1.6E-05 | 1.6E-03 | 2.4E-04 | 1.0E-11 |
| BHLHE40 | 3 | basic helix-loop-helix family, member e40 | 1.5E-03 | 3.4E-04 | 4.3E-06 | 1.5E-03 | 3.0E-04 | 6.4E-07 | 1.5E-03 | 2.2E-04 | 1.2E-11 |
| MCL1 | 1 | myeloid cell leukemia sequence 1 (BCL2-related) | 1.0E-03 | 2.0E-04 | 7.5E-07 | 1.6E-03 | 3.2E-04 | 1.5E-06 | 1.2E-03 | 1.7E-04 | 1.4E-11 |
| PRF1 | 10 | perforin 1 (pore forming protein) | 2.5E-03 | 4.1E-04 | 2.5E-09 | 1.8E-03 | 5.3E-04 | 1.0E-03 | 2.2E-03 | 3.3E-04 | 1.6E-11 |
| GPR56 | 16 | G protein-coupled receptor 56 | 2.0E-03 | 3.4E-04 | 3.5E-09 | 1.7E-03 | 5.8E-04 | 3.0E-03 | 1.9E-03 | 2.9E-04 | 3.9E-11 |
| PPP1R15A | 19 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | 1.5E-03 | 2.6E-04 | 1.7E-09 | 1.3E-03 | 3.0E-04 | 2.8E-05 | 1.4E-03 | 2.4E-04 | 1.5E-08 |
| FGFBP2 | 4 | fibroblast growth factor binding protein 2 | 2.3E-03 | 5.0E-04 | 5.8E-06 | 2.0E-03 | 6.2E-04 | 1.5E-03 | 2.2E-03 | 3.9E-04 | 3.3E-08 |
| GNLY | 2 | granulysin | 2.6E-03 | 6.4E-04 | 3.6E-05 | 2.6E-03 | 7.2E-04 | 3.0E-04 | 2.6E-03 | 4.8E-04 | 4.0E-08 |
| FOS | 14 | FBJ murine osteosarcoma viral oncogene homolog | 1.7E-03 | 2.5E-04 | 1.6E-11 | 2.6E-03 | 6.3E-04 | 3.6E-05 | 2.3E-03 | 4.1E-04 | 4.8E-08 |
| NKG7 | 19 | natural killer cell group 7 sequence | 2.3E-03 | 5.3E-04 | 1.9E-05 | 1.4E-03 | 5.5E-04 | 8.8E-03 | 1.9E-03 | 3.8E-04 | 9.4E-07 |
| GRAMD1A | 19 | GRAM domain containing 1A | -6.0E-04 | 1.4E-04 | 2.1E-05 | -6.7E-04 | 2.8E-04 | 1.8E-02 | -6.2E-04 | 1.3E-04 | 1.1E-06 |
| GLRX5 | 14 | glutaredoxin 5 | 1.7E-03 | 3.9E-04 | 1.3E-05 | 1.3E-03 | 6.1E-04 | 3.5E-02 | 1.6E-03 | 3.3E-04 | 1.5E-06 |
| TMEM43 | 3 | transmembrane protein 43 | 7.5E-04 | 2.1E-04 | 3.0E-04 | 7.7E-04 | 2.5E-04 | 2.4E-03 | 7.6E-04 | 1.6E-04 | 2.3E-06 |
| TIPARP | 3 | TCDD-inducible poly(ADP-ribose) polymerase | 1.2E-03 | 2.3E-04 | 1.3E-07 | 8.6E-04 | 2.4E-04 | 3.3E-04 | 9.5E-04 | 2.0E-04 | 2.6E-06 |
| AHNAK | 11 | AHNAK Nucleoprotein | 9.1E-04 | 2.6E-04 | 4.1E-04 | 9.7E-04 | 3.4E-04 | 4.0E-03 | 9.3E-04 | 2.0E-04 | 5.2E-06 |
| PIGB | 15 | phosphatidylinositol glycan anchor biosynthesis, class B | 1.1E-03 | 3.1E-04 | 5.3E-04 | 6.7E-04 | 2.1E-04 | 1.9E-03 | 8.0E-04 | 1.8E-04 | 6.1E-06 |
| TAGAP | 6 | T-cell activation RhoGTPase activating protein | 1.7E-03 | 2.5E-04 | 5.7E-12 | 1.3E-03 | 3.7E-04 | 7.1E-04 | 1.4E-03 | 3.1E-04 | 6.4E-06 |
| —DBP Signature genes | |||||||||||
| BHLHE40 | 3 | basic helix-loop-helix family, member e40 | 2.4E-03 | 5.1E-04 | 2.3E-06 | 2.5E-03 | 5.2E-04 | 2.8E-06 | 2.4E-03 | 3.6E-04 | 2.7E-11 |
| ANXA1 | 9 | annexin A1 | 3.5E-03 | 5.7E-04 | 1.2E-09 | 2.1E-03 | 7.8E-04 | 6.3E-03 | 3.0E-03 | 4.6E-04 | 6.5E-11 |
| PRF1 | 10 | perforin 1 (pore forming protein) | 3.2E-03 | 6.2E-04 | 3.2E-07 | 3.2E-03 | 9.4E-04 | 5.7E-04 | 3.2E-03 | 5.2E-04 | 6.7E-10 |
| KCNJ2 | 17 | potassium inwardly-rectifying channel, subfamily J, member 2 | -2.6E-03 | 5.6E-04 | 3.9E-06 | -2.0E-03 | 5.5E-04 | 2.6E-04 | -2.3E-03 | 3.9E-04 | 4.9E-09 |
| CLC | 19 | Charcot-Leyden crystal protein | -4.1E-03 | 8.6E-04 | 2.6E-06 | -3.6E-03 | 1.0E-03 | 5.7E-04 | -3.9E-03 | 6.7E-04 | 5.8E-09 |
| CD97 | 19 | CD97 molecule | 2.3E-03 | 4.8E-04 | 1.6E-06 | 1.9E-03 | 5.8E-04 | 1.1E-03 | 2.1E-03 | 3.7E-04 | 7.4E-09 |
| IL2RB | 22 | interleukin 2 receptor, beta | 2.3E-03 | 4.9E-04 | 3.0E-06 | 2.2E-03 | 7.3E-04 | 2.4E-03 | 2.3E-03 | 4.1E-04 | 2.5E-08 |
| S100A10 | 1 | S100 calcium binding protein A10 | 3.2E-03 | 6.1E-04 | 2.4E-07 | 1.6E-03 | 6.2E-04 | 9.9E-03 | 2.4E-03 | 4.4E-04 | 4.0E-08 |
| GPR56 | 16 | G protein-coupled receptor 56 | 2.5E-03 | 5.2E-04 | 1.1E-06 | 2.4E-03 | 1.0E-03 | 1.7E-02 | 2.5E-03 | 4.6E-04 | 5.5E-08 |
| TIPARP | 3 | TCDD-inducible poly(ADP-ribose) polymerase | 1.3E-03 | 3.4E-04 | 1.3E-04 | 1.1E-03 | 3.1E-04 | 2.8E-04 | 1.2E-03 | 2.3E-04 | 1.4E-07 |
| HAVCR2 | 5 | Hepatitis A Virus Cellular Receptor 2 | 1.7E-03 | 4.6E-04 | 3.8E-04 | 1.8E-03 | 4.8E-04 | 1.8E-04 | 1.7E-03 | 3.3E-04 | 2.4E-07 |
| PTGS2 | 1 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | -2.1E-03 | 4.9E-04 | 2.2E-05 | -1.3E-03 | 5.1E-04 | 9.0E-03 | -1.7E-03 | 3.5E-04 | 1.0E-06 |
| MYADM | 19 | myeloid-associated differentiation marker | 2.8E-03 | 4.9E-04 | 1.7E-08 | 4.1E-03 | 1.0E-03 | 8.6E-05 | 3.6E-03 | 7.4E-04 | 1.1E-06 |
| ANTXR2 | 4 | anthrax toxin receptor 2 | 1.5E-03 | 3.3E-04 | 5.2E-06 | 8.3E-04 | 4.3E-04 | 5.5E-02 | 1.3E-03 | 2.6E-04 | 1.7E-06 |
| OBFC2A | 2 | nucleic acid binding protein 1 | -1.7E-03 | 3.9E-04 | 7.2E-06 | -9.6E-04 | 4.6E-04 | 3.8E-02 | -1.4E-03 | 3.0E-04 | 1.8E-06 |
| GRAMD1A | 19 | GRAM domain containing 1A | -9.3E-04 | 2.1E-04 | 1.4E-05 | -8.7E-04 | 5.0E-04 | 7.8E-02 | -9.2E-04 | 2.0E-04 | 2.8E-06 |
| ARHGAP15 | 2 | Rho GTPase activating protein 15 | -1.3E-03 | 4.1E-04 | 1.1E-03 | -1.4E-03 | 4.4E-04 | 1.5E-03 | -1.4E-03 | 3.0E-04 | 5.2E-06 |
| FBXL5 | 4 | F-box and leucine-rich repeat protein 5 | -1.6E-03 | 3.7E-04 | 2.1E-05 | -9.4E-04 | 4.9E-04 | 5.5E-02 | -1.3E-03 | 2.9E-04 | 5.3E-06 |
| SLC31A2 | 9 | solute carrier family 31 (copper transporters), member 2 | 2.8E-03 | 4.9E-04 | 1.0E-08 | 2.4E-03 | 8.1E-04 | 2.6E-03 | 2.6E-03 | 5.6E-04 | 5.4E-06 |
| VIM | 10 | vimentin | 1.7E-03 | 3.8E-04 | 5.5E-06 | 7.6E-04 | 5.9E-04 | 2.0E-01 | 1.4E-03 | 3.2E-04 | 6.2E-06 |
| —HTN Signature genes | |||||||||||
| SLC31A2 | 9 | solute carrier family 31 (copper transporters), member 2 | 5.9E-02 | 1.4E-02 | 1.9E-05 | 6.4E-02 | 1.4E-02 | 2.1E-06 | 6.1E-02 | 9.6E-03 | 1.8E-10 |
| MYADM | 19 | myeloid-associated differentiation marker | 7.8E-02 | 1.4E-02 | 1.2E-08 | 7.3E-02 | 2.1E-02 | 6.2E-04 | 7.4E-02 | 1.4E-02 | 3.0E-07 |
| TAGAP | 6 | T-cell activation RhoGTPase activating protein | 4.4E-02 | 1.1E-02 | 3.2E-05 | 3.2E-02 | 1.2E-02 | 5.3E-03 | 3.9E-02 | 7.8E-03 | 7.3E-07 |
| GZMB | 14 | granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 1.6E-01 | 2.3E-02 | 1.1E-11 | 1.1E-01 | 3.5E-02 | 9.6E-04 | 1.3E-01 | 2.6E-02 | 1.4E-06 |
| KCNJ2 | 17 | potassium inwardly-rectifying channel, subfamily J, member 2 | -5.2E-02 | 1.6E-02 | 8.4E-04 | -4.4E-02 | 1.3E-02 | 5.5E-04 | -4.7E-02 | 9.9E-03 | 1.7E-06 |
*Meta: meta-analysis of all six cohorts.
Fig 1. Effect size of differentially expressed BP genes in the Framingham Heart Study and the Illumina cohorts.
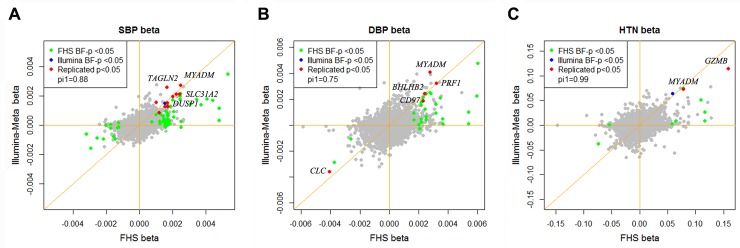
A) SBP; B) DBP; C) HTN. The x-axis is the effect size of the differentially expressed genes in the FHS cohort and the y-axis is the effect size in the Illumina cohorts. The BP signature genes identified both in the FHS and the Illumina cohorts at p<0.05 (Bonferroni corrected) are highlighted. pi1 values indicate the proportion of significant signals among the tested associations [11] (See details in the Methods section).
The 73 SBP signature genes in the FHS (55 of these 73 genes were measured in the Illumina cohorts) at a Bonferroni corrected p<0.05 in aggregate explained 9.4% of SBP phenotypic variance in the Illumina cohorts, and the 31 DBP signature genes from the FHS (22 of these 31 genes were measured in the Illumina cohorts) in aggregate explained 5.3% of DBP phenotypic variance in the Illumina cohorts. These results suggest that in contrast to common genetic variants identified by BP GWAS, which explain in aggregate only about 1% of inter-individual BP variation [3], changes in gene expression levels explains a considerably larger proportion of phenotypic variance in BP.
Meta-analysis of the six cohorts identifies differentially expressed BP signature genes
A meta-analysis of differential expression across all six cohorts revealed 34 differentially expressed BP genes at p<0.05 (Bonferroni corrected for 7717 genes that were measured and passed quality control in the FHS and Illumina cohorts), including 21 for SBP, 20 for DBP, and 5 for HTN (Table 2 and S2 Fig.). All of the 34 differentially expressed BP signature genes showed directional consistency in the FHS and the Illumina cohorts (Table 2). The 34 BP signature genes included all 13 genes that were cross-validated between the FHS and the Illumina cohorts. Of the 34 BP signature genes, 27 were positively correlated with BP and only 7 genes were negatively correlated. MYADM and SLC31A2 were top signature genes for SBP, DBP, and HTN. At FDR<0.2, 224 unique genes were differentially expressed in relation BP phenotypes including 142 genes for SBP, 137 for DBP, and 45 for HTN (details are reported in the S1–S2 Text, and S3–S5 Table).
Functional analysis of differentially expressed BP signature genes
We used gene set enrichment analysis (GSEA) to identify the biological process and pathways associated with gene expression changes in relation to SBP, DBP, and HTN in order to better understand the biological themes within the data. As shown in Table 3, the GSEA of genes whose expression was positively associated with BP showed enrichment for antigen processing and presentation (p<0.0001), apoptotic program (p<0.0001), inflammatory response (p<0.0001), and oxidative phosphorylation (p = 0.0018). The negatively associated genes showed enrichment for nucleotide metabolic process (p<0.0001), positive regulation of cellular metabolic process (p<0.0001), and positive regulation of DNA dependent transcription (p = 0.0021).
Table 3. Gene set enrichment analysis for BP associated gene expression changes.
| Name | Pos / Neg associated gene expression changes | Database | Number of genes in pathway | NES* | p value | FDR |
|---|---|---|---|---|---|---|
| - DBP signature | ||||||
| Antigen processing and presentation | Positive | KEGG | 37 | 2.0 | <1E-4 | 0.01 |
| Nature killer cell mediated cytotoxicity | Positive | KEGG | 71 | 1.8 | <1E-4 | 0.07 |
| Porphyrin and chlorophyll metabolism | Positive | KEGG | 15 | 1.7 | 0.01 | 0.13 |
| Rho protein signaling transduction | Negative | GO-BP | 18 | -1.8 | 3.9E-3 | 0.10 |
| Receptor mediated endocytosis | Negative | GO-BP | 16 | -1.8 | 3.9E-3 | 0.17 |
| Detection of stimulus | Negative | GO-BP | 18 | -1.9 | 9.8E-3 | 0.20 |
| - SBP signature | ||||||
| Natural killer cell mediated cytotoxicity | Positive | KEGG | 71 | 1.9 | 1.7E-3 | 0.05 |
| Apoptotic program | Positive | GO-BP | 37 | 1.9 | <1E-4 | 0.03 |
| Inflammatory response | Positive | GO-BP | 72 | 2.0 | <1E-4 | 0.05 |
| Nucleotide metabolic process | Negative | GO-BP | 32 | -1.9 | <1E-4 | 0.04 |
| Translation | Negative | GO-BP | 79 | -1.8 | <1E-4 | 0.05 |
| - HTN signature | ||||||
| Antigen processing and presentation | Positive | KEGG | 37 | 1.8 | <1E-4 | 0.04 |
| Oxidative phosphorylation | Positive | KEGG | 52 | 1.8 | 1.8E-3 | 0.05 |
| Apoptotic program | Positive | GO-BP | 37 | 1.9 | 1.8E-3 | 0.14 |
| Positive regulation of nucleic acid metabolic process | Negative | GO-BP | 71 | -1.9 | <1E-4 | 0.08 |
| Positive regulation of cellular metabolic process | Negative | GO-BP | 105 | -1.8 | <1E-4 | 0.08 |
| Positive regulation of transcription DNA dependent | Negative | GO-BP | 56 | -1.8 | 2.1E-3 | 0.09 |
*NES: normalized enrichment score;
GO-BP: Gene ontology- biological process;
KEGG: Kyoto encyclopedia of genes and genomes.
Genetic effects on expression of BP signature genes
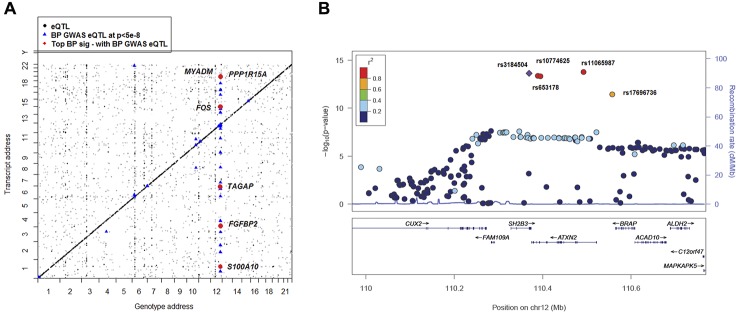
Among the 34 BP signatures genes from the meta-analysis of all 6 studies, 33 were found to have cis-eQTLs and 26 had trans-eQTLs (Fig. 2A and S2 Table) based on whole blood profiling [12,13]. Of these, six master trans-eQTLs mapped to either five or six BP signature genes (no master cis-eQTL was identified). Five master trans-eQTLs (rs653178, rs3184504, rs10774625, rs11065987, and rs17696736) were located on chromosome 12q24 within the same linkage disequilibrium (LD) block (r2 >0.8, Fig. 2B). We retrieved a peak cis- and trans-eQTL for each BP signature gene. The peak cis-eQTL explained 0.2–20% of the variance in the corresponding transcript levels, in contrast, the peak trans-eQTL accounted for very little (0.02–2%) of the corresponding transcript variance. Westra et al. also reported a similar small proportion of variance in transcript levels explained by trans-eQTLs [12].
Fig 2. Global view of BP eQTLs effects on differentially expressed BP signature genes.
A) 2-Dimensional plot of in whole blood eQTLs vs. transcript position genome wide. eQTL-transcript pairs at FDR<0.1 are shown in black dots; those that fall along the diagonal are cis eQTLs and all others are trans eQTLS. eQTL-transcript pair SNPs that are associated with BP in GWAS [3] are highlighted with blue triangles. eQTL-transcript pair genes that are BP signature genes from analysis of differential gene expression in relation to BP are depicted by red circles. B) Regional association plots for rs3184504 proxy QTLs that showing association with BP in ICBP GWAS [3]. −log10(p) indicated the −log10 transformed DBP association p values in ICBP GWAS [3]. Color coding indicates the strength (measured by r2) of LD of each SNP with the top SNP (rs3184504). Five master trans-eQTLs (also BP GWAS SNPs) for BP signature genes are labeled in the figure. This figure was drawn by LocusZoom [32].
We then linked the cis- and trans-eQTLs of the 34 BP signature genes with BP GWAS results from the ICBP Consortium [3] and the NHGRI GWAS Catalog [14] (Fig. 2 and S2 Table). We did not find any cis-eQTLs for the top BP signature genes that also were associated with BP in the ICBP GWAS [3]. However, the 6 master trans-eQTLs were all associated with BP at p<5e-8 in the ICBP GWAS [3] and were associated with multiple complex diseases or traits (Table 4). For example, rs3184504, a nonsynonymous SNP in SH2B3 that was associated in GWAS with BP, coronary heart disease, hypothyroidism, rheumatoid arthritis, and type 1 diabetes [12], is a trans-eQTL for 6 of our 34 BP signature genes from the meta-analysis (FOS, MYADM, PP1R15A, TAGAP, S100A10, and FGBP2; Fig. 2A-B and Table 4). These 6 genes are all highly expressed in neutrophils, and their expression levels are correlated significantly (average r2 = 0.04, p<1e-16). rs653178, intronic to ATXN2 and in perfect LD with rs3184504 (r2 = 1), also is associated with BP and multiple other diseases in the NHGRI GWAS Catalog [14]. It also is a trans-eQTL for the same 6 BP signature genes (Table 4). These two SNPs are cis-eQTLs for expression SH2B3 in whole blood (FDR<0.05), but not for ATXN2 (FDR = 0.4). We found that the expression of SH2B3 is associated with expression of MYADM, PP1R15A, and TAGAP (at Bonferroni corrected p<0.05), but not with FOS, S100A10, or FGBP2. The expression of ATXN2 was associated with expression of 5 of the 6 genes (PP1R15A was not associated). S3 Fig. shows the coexpression levels of the eight genes that were cis- or trans- associated with rs3184504 and rs653178 genotypes. These results suggest that there may be a pathway or gene co-regulatory mechanism underling BP regulation involving these genes that is driven by this common genetic variant (rs3184504; minor allele frequency 0.47) or its proxy SNPs.
Table 4. GWAS eQTLs for the top differentially expressed BP signature genes.
| SNP—Trait Association | SNP-Gene Association | Gene-Trait Association | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | SNP. Location | ICBP-SBP pval | ICBP-DBP pval | Other Traits in GWAS Catalog | Gene | Chr. Gene | Cis/Trans | SBP pval | DBP pval | HTN pval |
| rs3184504* | chr12 (missense, SH2B3) | 1.70E-09 | 2.30E-14 | Coronary heart disease; Rheumatoid arthritis; Type 1 diabetes | MYADM | chr19 | trans | <1e-16 & | 1.1e-6 | 3.0e-7 |
| FOS | chr14 | trans § | 4.9e-8 | 3.2e-4 | 7.9e-5 | |||||
| PPP1R15A | chr19 | trans § | 1.6e-8 | 1.2e-5 | 6.1e-4 | |||||
| TAGAP | chr6 | trans | 6.4e-6 | 1.3e-4 | 7.3e-7 | |||||
| S100A10 | chr1 | trans § | 2.6e-4 | 4.0e-8 | 7.0e-5 | |||||
| FGFBP2 | chr4 | trans § | 3.3e-8 | 1.8e-5 | 5.1e-3 | |||||
| rs10187424 | chr2 (intergenic) | - | - | Prostate cancer | GNLY | chr2 | cis § | 4.0e-8 | 2.8e-5 | 2.2e-4 |
| rs411174 | chr5 (intron, ITK) | - | - | Personality dimensions | HAVCR2 | chr5 | cis § | 1.6e-4 | 2.4e-7 | 1.5e-3 |
| rs3758354 | chr9 (intergenic) | - | - | Schizophrenia, bipolar disorder and depression | ANXA1 | chr9 | cis | 1.8e-3 | 6.5e-11 | 7.5e-3 |
| rs1950500 | chr14 (intergenic) | - | - | Height | GZMB | chr14 | cis | 7.8e-5 | 6.0e-5 | 1.4e-6 |
| rs8017377 | chr14 (missense, NYNRIN) | - | - | LDL cholesterol | GZMB | chr14 | cis | 7.8e-5 | 6.0e-5 | 1.4e-6 |
| rs8192917 | chr14 (missense, GZMB) | - | - | Vitiligo | GZMB | chr14 | cis | 7.8e-5 | 6.0e-5 | 1.4e-6 |
| rs2284033 | chr22 (intron, IL2RB) | - | - | Asthma | IL2RB | chr22 | cis § | 1.6e-4 | 2.5e-8 | 9.3e-3 |
| rs11724635 + | chr4 (intergenic) | - | - | Parkinsons disease | FBXL5 | chr4 | cis | 5.9e-5 | 5.3e-6 | 0.07 |
| rs4333130 $ | chr4 (intron, ANTXR2) | - | - | Ankylosing spondylitis | ANTXR2 | chr4 | cis | 2.8e-4 | 1.7e-6 | 0.04 |
| rs8005962 | chr14 (intergenic) | - | - | Tuberculosis | GLRX5 | chr14 | cis | 1.5e-6 | 0.13 | 0.09 |
| rs7995215 | chr13 (intron, GPC6) | - | - | Attention deficit hyperactivity disorder | TAGAP | chr6 | trans | 6.4e-6 | 1.3e-4 | 7.3e-7 |
| rs12047808 | chr1 (intron, C1orf125) | - | - | Multiple sclerosis (age of onset) | FOS | chr14 | trans § | 4.9e-8 | 3.2e-4 | 7.9e-5 |
| rs2894207 | chr6 (intergenic) | - | - | Nasopharyngeal carcinoma | AHNAK | chr11 | trans | 5.2e-6 | 6.8e-5 | 1.8e-3 |
| rs3763313 | chr6 (neargene 5, BTNL2) | - | - | HIV-1 control | PPP1R15A | chr19 | trans | 1.6e-8 | 1.2e-5 | 6.1e-4 |
| rs9376092 | chr6 (intergenic) | - | - | Beta thalassemia/hemoglobin E disease | GPR56 | Chr16 | trans | 3.9e-11 | 5.5e-8 | 4.9e-4 |
* rs653178, intronic to ATXN2 and in tight linkage disequilibrium with rs3184504 (r2 = 1), was also associated with BP in ICBP GWAS and all the 6 genes;
+ A proxy SNP rs4698412 at LD r2 = 1 associated with the same trait;
$ A proxy SNP rs4389526 at LD r2 = 1 associated with the same trait;
§ indicated eQTL were identified from[12].
& highlighted p values indicated passing transcriptome-wide significance at Bonferroni corrected p<0
We further checked whether the cis- or trans-eQTLs for the top 34 BP signature genes are associated with other diseases or traits in the NHGRI GWAS catalog [14]. We identified 12 cis-eQTLs (for 8 genes) and 6 trans-eQTLs (for 6 genes) that are associated with other diseases or traits in the NHGRI GWAS catalog [14] (Table 4).
Discussion
Our meta-analysis of gene expression data from 7017 individuals from six studies identified and characterized whole blood gene expression signatures associated with BP traits. Thirty-four BP signature genes were identified at Bonferroni corrected p<0.05 (224 genes were identified at FDR<0.2, reported in the S1 Text). Thirteen BP signature genes replicated between the FHS and Illumina cohorts. The top BP signature genes identified in the FHS (55 genes for SBP and 22 genes for DBP) explained 5–9% of interindividual variation in BP in the Illumina cohorts on average.
Among the 34 BP signature genes (at Bonferroni corrected p<0.05), only FOS [15] and PTGS2 [16] have been previously implicated in hypertension. We did not find literature support for a direct role of the remaining signature genes in BP regulation. However, we found several genes involved in biological functions or processes that are highly related to BP, such as cardiovascular disease (GZMB, ANXA1, TMEM43, FOS, KCNJ2, PTGS2, and MCL1), angiogenesis (VIM and TIPARP), and ion channels (CD97, ANXA1, S100A10, PRF1, ANTXR2, SLC31A2, TIPARP, and KCNJ2). We speculate that these genes may be important for BP regulation, but further experimental validation is needed.
Seven of the 34 signature genes, including KCNJ2, showed negative correlation of expression with BP. KCNJ2 is a member of the potassium inwardly-rectifying channel subfamily; it encodes the inward rectifier K+ channel Kir2.1, and is found in cardiac, skeletal muscle, and nervous tissue [17]. Most outward potassium channels are positively correlated with BP. Loss-of-function mutations in ROMK (KCNJ1, the outward potassium channel) are associated with Bartter's syndrome, and ROMK inhibitors are used in the treatment of hypertension [18,19]. Previous studies reported that greater potassium intake is associated with lower blood pressure [20,21,22,23]. These data suggest that KCNJ2 up-regulation may be a means of lowering BP.
By linking the BP signature genes with eQTLs and with BP GWAS results, we found several SNPs that are associated with BP in GWAS and that also are trans associated with several of our top BP signature genes. For example, rs3184504, a non-synonymous SNP located in exon 3 of SH2B3, is associated in GWAS with BP, coronary heart disease, hypothyroidism, rheumatoid arthritis, and type I diabetes [12]. rs3184504 is a common genetic variant with a minor allele frequency of approximately 0.47; the rs3184504-T allele is associated with an increment of 0.58 mm Hg in SBP and of 0.48 mm Hg in DBP [2]. rs3184504 is a cis-eQTL for SH2B3, expression of this gene was not associated with BP or hypertension in our data. However, rs3184504 also is a trans-eQTL for 6 of our 34 BP signature genes: FOS, MYADM, PP1R15A, TAGAP, S100A10, and FGBP2. These 6 genes are highly expressed in neutrophils [12], and are coexpressed. Prior studies have suggested an important role of neutrophils in BP regulation [24]. We speculate that these 6 BP signature genes, all driven by the same BP-associated eQTL, point to a critical and previously unrecognized mechanism involved in BP regulation. Further experimental validation is needed.
One limitation of our study is the use of whole blood derived RNA for transcriptomic profiling. GSEA showed that the top enriched biological processes for the differentially expressed BP genes include inflammatory response. Numerous studies have shown links between inflammation and hypertension [25,26,27]. The top ranked genes in inflammatory response categories provide a guide for further experimental work to recognize the contributions of inflammation to alterations in BP regulation. We speculate that using similar approaches in other tissues might identify additional differentially expressed BP signature genes.
In conclusion, we conducted a meta-analysis of global gene expression profiles in relation to BP and identified a number of credible gene signatures of BP and hypertension. Our integrative analysis of GWAS and gene expression in relation to BP can help to uncover the genetic and genomic architecture of BP regulation; the BP signature genes we identified may represent an early step toward improvements in the detection of susceptibility, and in the prevention and treatment of hypertension.
Materials and Methods
Study population and ethics statement
This investigation included six studies (the Framingham Heart Study (FHS), the Estonian Biobank (EGCUT), the Rotterdam Study (RS) [8], the InCHIANTI Study, the Cooperative Health Research in the Region of Augsburg (KORA F4) Study [9], and the Study of Health in Pomerania (SHIP-TREND) [10], each of which conducted genome-wide genotyping, mRNA expression profiling, and had extensive BP phenotype data. Each of the six studies followed the recommendations of the Declaration of Helsinki. The FHS: Systems Approach to Biomarker Research (SABRe) in cardiovascular disease is approved under the Boston University Medical Center’s protocol H-27984. Ethical approval of EGCUT was granted by the Research Ethics Committee of the University of Tartu (UT REC). Ethical approval of the InCHIANTI study was granted by the Instituto Nazionale Riposo e Cura Anziani institutional review board in Italy. Ethical approval of RS was granted by the medical ethics committee of the Erasmus Medical Center. The study protocol of SHIP-TREND was approved by the medical ethics committee of the University of Greifswald. KORA F4 is a population-based survey in the region of Augsburg in Southern Germany which was performed between 2006 and 2008. KORA F4 was approved by the local ethical committees. Informed consent was obtained from each study participant.
Hypertension (HTN) was defined as SBP ≥140 mm Hg or DBP ≥90 mm Hg. We excluded individuals receiving anti-hypertensive treatment because of the possibility that some of the differentially expressed genes we identified would reflect treatment effects. The eligible study sample included 7017 individuals: 3679 from FHS, 972 from EGCUT, 604 from RS, 597 from InCHIANTI, 565 from KORA F4, and 600 from SHIP-TREND.
Gene expression profiling
RNA was isolated from whole blood samples that were collected in PaxGene tubes (PreAnalytiX, Hombrechtikon, Switzerland) in FHS, RS, InCHIANTI, KORA F4 and SHIP-TREND, and in Blood RNA Tubes (Life Technologies, NY, USA) in EGCUT. Gene expression in the FHS samples used the Affymetrix Exon Array ST 1.0. EGCUT, RS, InCHANTI, KORA F4, and SHIP-TREND used the Illumina HT12v3 (EGCUT, InCHANTI, KORA F4, and SHIP-TREND) or HT12v4 (RS) array. Raw data from gene expression profiling are available online (FHS [http://www.ncbi.nlm.nih.gov/gap; accession number phs000007], EGCUT [GSE48348], RS [GSE33828], InCHIANTI [GSE48152], KORA F4 [E-MTAB-1708] and SHIP-TREND [GSE36382]). The details of sample collection, microarrays, and data processing and normalization in each cohort are provided in the S2 Text.
Identification and replication of differentially expressed genes associated with BP
The association of gene expression with BP was analyzed separately in each of the six studies (Equation 1). A linear mixed model was used in the FHS in order to account for family structure. Linear regression models were used in the other five studies. In each study, gene expression level, denoted by geneExp, was included as the dependent variable, and explanatory variables included blood pressure phenotypes (SBP, DBP, and HTN), and covariates included age, sex, body mass index (BMI), cell counts, and technical covariates. A separate regression model was fitted for each gene. The general formula is shown below, and the details of analyses for each study are provided in the S2 Text and S6 Table.
The overall analysis framework is provided in S1 Fig.. We first identified differentially expressed genes associated with BP (BP signature genes) in the FHS samples (Set 1) and attempted replication in the meta-analysis results from the Illumina cohorts (Set 2, see Methods, Meta-analysis). We next identified BP signature genes in the Illumina cohorts (Set 2), and then attempted replication in the FHS samples (Set 1). The significance threshold for pre-selecting BP signature genes in discovery was at Bonferroni corrected p = 0.05 (in FHS, corrected for 17,318 measured genes [17,873 transcripts], and in illumina cohorts, corrected for 12,010 measured genes [14,222 transcripts] that passed quality control). Replication was established at Bonferroni corrected p = 0.05, correcting for the number of pre-selected BP signatures genes in the discovery set. We computed the pi1 value to estimate the enrichment of significant p values in the replication set (the Illumina cohorts) for BP signatures identified in the discovery set (the FHS) by utilizing the R package Qvalue [11]. Pi1 is defined as 1-pi0. Pi0 value provided by the Qvalue package, represents overall probability that the null hypothesis is true. Therefore, pi1 value represents the proportion of significant results. For genes passing Bonferroni corrected p<0.05 in the discovery set for SBP, DBP and HTN, we calculated pi1 values for each gene set in the replication set.
Meta-analysis
We performed meta-analysis of the five Illumina cohorts (for discovery and replication purposes), and then performed meta-analysis of all six cohorts. An inverse variance weighted meta-analysis was conducted using fixed-effects or random-effects models by the metagen() function in the R package Meta (http://cran.r-project.org/web/packages/meta/index.html). At first, we tested heterogeneity for each gene using Cochran’s Q statistic. If the heterogeneity p value is significant (p<0.05), we will use a random-effects model for the meta-analysis, otherwise use a fixed-effects model. The Benjamini-Hochberg (BH) method [28] was used to calculate FDR for differentially expressed genes in relation to BP following the meta-analysis of all six cohorts. We also used a more stringent threshold to define BP signature genes by utilizing p<6.5e-6 (Bonferroni correction for 7717 unique genes [7810 transcript] based on the overlap of FHS and illumina cohort interrogated gene sets).
Estimating the proportion of variance in BP attributable to BP signature genes
To estimate the proportion of variances in SBP or DBP explained by a group of differentially expressed BP signature genes (gene 1, gene 2, …, gene n), we used the following two models:
Full model:
Null model:
The proportion of variance in BP attributable to the group of differentially expressed BP signature genes () was calculated as:
where is the total phenotypic variance of SBP or DBP, and are the variance and error variance when modeling with the tested group of gene expression traits (gene 1, gene 2, …, gene n), and and are the variance and error variance when modeling without the tested group of gene expression traits.
The proportion of the variance in BP phenotypes attributable to the FHS BP signature genes was estimated in the five Illumina cohorts, respectively, and then the average proportion values were reported. In turn, the proportion of the variance in BP phenotypes attributable to the Illumina BP signature genes was estimated in the FHS.
Identifying eQTLs and estimating the proportion of variance in gene expression attributable to single cis- or trans-eQTLs
SNPs associated with altered gene expression (i.e. eQTLs) were identified using genome-wide genotype and gene expression data in all available FHS samples (n = 5257) at FDR<0.1 (Joehanes R, submitted, 2014, and a brief summary of methods and results are provided in the S2 Text). A cis-eQTL was defined as an eQTL within 1 megabase (MB) flanking the gene. Other eQTLs were defined as trans-eQTLs. We combined the eQTL list generated in the FHS with the eQTLs generated by meta-analysis of seven other studies (n = 5300) that were also based on whole blood expression[12].
For every BP signature gene, we estimated the proportion of variance in the transcript attributable to the corresponding cis- or trans-eQTLs () using the formula:
where was the total phenotypic variance of a gene expression trait; and were the variance and the residual error, respectively, when modeling with the tested eQTL; and were the variance and the residual error when modeling without the tested eQTL.
Functional category enrichment analysis
In order to understand the biological themes within the global gene expression changes in relation to BP, we performed gene set enrichment analysis[29] to test for enrichment of any gene ontology (GO) biology process[30] or KEGG pathways[31]. “Metric for ranking gene” parameters were configured to the beta value of the meta-analysis, in order to look at the top enriched functions for BP associated up-regulated and down-regulated gene expression changes respectively. One thousand random permutations were conducted and the significance level was set at FDR≤ 0.25 to allow for exploratory discovery [29].
Members of International Consortium for Blood Pressure GWAS (ICBP)
Steering Committee (alphabetical)
Gonçalo Abecasis, Murielle Bochud, Mark Caulfield (co-chair), Aravinda Chakravarti, Dan Chasman, Georg Ehret (co-chair), Paul Elliott, Andrew Johnson, Louise Wain, Martin Larson, Daniel Levy (co-chair), Patricia Munroe (co-chair), Christopher Newton-Cheh (co-chair), Paul O'Reilly, Walter Palmas, Bruce Psaty, Kenneth Rice, Albert Smith, Harold Snider, Martin Tobin, Cornelia Van Duijn, Germaine Verwoert.
Members
Georg B. Ehret1,2,3, Patricia B. Munroe4, Kenneth M. Rice5, Murielle Bochud2, Andrew D. Johnson6,7, Daniel I. Chasman8,9, Albert V. Smith10,11, Martin D. Tobin12, Germaine C. Verwoert13,14,15, Shih-Jen Hwang6,16,7, Vasyl Pihur1, Peter Vollenweider17, Paul F. O'Reilly18, Najaf Amin13, Jennifer L Bragg-Gresham19, Alexander Teumer20, Nicole L. Glazer21, Lenore Launer22, Jing Hua Zhao23, Yurii Aulchenko13, Simon Heath24, Siim Sõber25, Afshin Parsa26, Jian'an Luan23, Pankaj Arora27, Abbas Dehghan13,14,15, Feng Zhang28, Gavin Lucas29, Andrew A. Hicks30, Anne U. Jackson31, John F Peden32, Toshiko Tanaka33, Sarah H. Wild34, Igor Rudan35,36, Wilmar Igl37, Yuri Milaneschi33, Alex N. Parker38, Cristiano Fava39,40, John C. Chambers18,41, Ervin R. Fox42, Meena Kumari43, Min Jin Go44, Pim van der Harst45, Wen Hong Linda Kao46, Marketa Sjögren39, D. G. Vinay47, Myriam Alexander48, Yasuharu Tabara49, Sue Shaw-Hawkins4, Peter H. Whincup50, Yongmei Liu51, Gang Shi52, Johanna Kuusisto53, Bamidele Tayo54, Mark Seielstad55,56, Xueling Sim57, Khanh-Dung Hoang Nguyen1, Terho Lehtimäki58, Giuseppe Matullo59,60, Ying Wu61, Tom R. Gaunt62, N. Charlotte Onland-Moret63,64, Matthew N. Cooper65, Carl G.P. Platou66, Elin Org25, Rebecca Hardy67, Santosh Dahgam68, Jutta Palmen69, Veronique Vitart70, Peter S. Braund71,72, Tatiana Kuznetsova73, Cuno S.P.M. Uiterwaal63, Adebowale Adeyemo74, Walter Palmas75, Harry Campbell35, Barbara Ludwig76, Maciej Tomaszewski71,72, Ioanna Tzoulaki77,78, Nicholette D. Palmer79, CARDIoGRAM consortium80, CKDGen Consortium80, KidneyGen Consortium80, EchoGen consortium80, CHARGE-HF consortium80, Thor Aspelund10,11, Melissa Garcia22, Yen-Pei C. Chang26, Jeffrey R. O'Connell26, Nanette I. Steinle26, Diederick E. Grobbee63, Dan E. Arking1, Sharon L. Kardia81, Alanna C. Morrison82, Dena Hernandez83, Samer Najjar84,85, Wendy L. McArdle86, David Hadley50,87, Morris J. Brown88, John M. Connell89, Aroon D. Hingorani90, Ian N.M. Day62, Debbie A. Lawlor62, John P. Beilby91,92, Robert W. Lawrence65, Robert Clarke93, Rory Collins93, Jemma C Hopewell93, Halit Ongen32, Albert W. Dreisbach42, Yali Li94, J H. Young95, Joshua C. Bis21, Mika Kähönen96, Jorma Viikari97, Linda S. Adair98, Nanette R. Lee99, Ming-Huei Chen100, Matthias Olden101,102, Cristian Pattaro30, Judith A. Hoffman Bolton103, Anna Köttgen104,103, Sven Bergmann105,106, Vincent Mooser107, Nish Chaturvedi108, Timothy M. Frayling109, Muhammad Islam110, Tazeen H. Jafar110, Jeanette Erdmann111, Smita R. Kulkarni112, Stefan R. Bornstein76, Jürgen Grässler76, Leif Groop113,114, Benjamin F. Voight115, Johannes Kettunen116,126, Philip Howard117, Andrew Taylor43, Simonetta Guarrera60, Fulvio Ricceri59,60, Valur Emilsson118, Andrew Plump118, Inês Barroso119,120, Kay-Tee Khaw48, Alan B. Weder121, Steven C. Hunt122, Yan V. Sun81, Richard N. Bergman123, Francis S. Collins124, Lori L. Bonnycastle124, Laura J. Scott31, Heather M. Stringham31, Leena Peltonen119,125,126,127, Markus Perola125, Erkki Vartiainen125, Stefan-Martin Brand128,129, Jan A. Staessen73, Thomas J. Wang6,130, Paul R. Burton12,72, Maria Soler Artigas12, Yanbin Dong131, Harold Snieder132,131, Xiaoling Wang131, Haidong Zhu131, Kurt K. Lohman133, Megan E. Rudock51, Susan R Heckbert134,135, Nicholas L Smith134,136,135, Kerri L Wiggins137, Ayo Doumatey74, Daniel Shriner74, Gudrun Veldre25,138, Margus Viigimaa139,140, Sanjay Kinra141, Dorairajan Prabhakaran142, Vikal Tripathy142, Carl D. Langefeld79, Annika Rosengren143, Dag S. Thelle144, Anna Maria Corsi145, Andrew Singleton83, Terrence Forrester146, Gina Hilton1, Colin A. McKenzie146, Tunde Salako147, Naoharu Iwai148, Yoshikuni Kita149, Toshio Ogihara150, Takayoshi Ohkubo149,151, Tomonori Okamura148, Hirotsugu Ueshima152, Satoshi Umemura153, Susana Eyheramendy154, Thomas Meitinger155,156, H.-Erich Wichmann157,158,159, Yoon Shin Cho44, Hyung-Lae Kim44, Jong-Young Lee44, James Scott160, Joban S. Sehmi160,41, Weihua Zhang18, Bo Hedblad39, Peter Nilsson39, George Davey Smith62, Andrew Wong67, Narisu Narisu124, Alena Stančáková53, Leslie J. Raffel161, Jie Yao161, Sekar Kathiresan162,27, Chris O'Donnell163,27,9, Stephen M. Schwartz134, M. Arfan Ikram13,15, W. T. Longstreth Jr.164, Thomas H. Mosley165, Sudha Seshadri166, Nick R.G. Shrine12, Louise V. Wain12, Mario A. Morken124, Amy J. Swift124, Jaana Laitinen167, Inga Prokopenko51,168, Paavo Zitting169, Jackie A. Cooper69, Steve E. Humphries69, John Danesh48, Asif Rasheed170, Anuj Goel32, Anders Hamsten171, Hugh Watkins32, Stephan J.L. Bakker172, Wiek H. van Gilst45, Charles S. Janipalli47, K. Radha Mani47, Chittaranjan S. Yajnik112, Albert Hofman13, Francesco U.S. Mattace-Raso13,14, Ben A. Oostra173, Ayse Demirkan13, Aaron Isaacs13, Fernando Rivadeneira13,14, Edward G Lakatta174, Marco Orru175,176, Angelo Scuteri174, Mika Ala-Korpela177,178,179, Antti J Kangas177, Leo-Pekka Lyytikäinen58, Pasi Soininen177,178, Taru Tukiainen180,181,177, Peter Würtz177,18,180, Rick Twee-Hee Ong56,57,182, Marcus Dörr183, Heyo K. Kroemer184, Uwe Völker20, Henry Völzke185, Pilar Galan186, Serge Hercberg186, Mark Lathrop24, Diana Zelenika24, Panos Deloukas119, Massimo Mangino28, Tim D. Spector28, Guangju Zhai28, James F. Meschia187, Michael A. Nalls83, Pankaj Sharma188, Janos Terzic189, M. J. Kranthi Kumar47, Matthew Denniff71, Ewa Zukowska-Szczechowska190, Lynne E. Wagenknecht79, F. Gerald R. Fowkes191, Fadi J. Charchar192, Peter E.H. Schwarz193, Caroline Hayward70, Xiuqing Guo161, Charles Rotimi74, Michiel L. Bots63, Eva Brand194, Nilesh J. Samani71,72, Ozren Polasek195, Philippa J. Talmud69, Fredrik Nyberg68,196, Diana Kuh67, Maris Laan25, Kristian Hveem66, Lyle J. Palmer197,198, Yvonne T. van der Schouw63, Juan P. Casas199, Karen L. Mohlke61, Paolo Vineis200,60, Olli Raitakari201, Santhi K. Ganesh202, Tien Y. Wong203,204, E Shyong Tai205,57,206, Richard S. Cooper54, Markku Laakso53, Dabeeru C. Rao207, Tamara B. Harris22, Richard W. Morris208, Anna F. Dominiczak209, Mika Kivimaki210, Michael G. Marmot210, Tetsuro Miki49, Danish Saleheen170,48, Giriraj R. Chandak47, Josef Coresh211, Gerjan Navis212, Veikko Salomaa125, Bok-Ghee Han44, Xiaofeng Zhu94, Jaspal S. Kooner160,41, Olle Melander39, Paul M Ridker8,213,9, Stefania Bandinelli214, Ulf B. Gyllensten37, Alan F. Wright70, James F. Wilson34, Luigi Ferrucci33, Martin Farrall32, Jaakko Tuomilehto215,216,217,218, Peter P. Pramstaller30,219, Roberto Elosua29,220, Nicole Soranzo119,28, Eric J.G. Sijbrands13,14, David Altshuler221,115, Ruth J.F. Loos23, Alan R. Shuldiner26,222, Christian Gieger157, Pierre Meneton223, Andre G. Uitterlinden13,14,15, Nicholas J. Wareham23, Vilmundur Gudnason10,11, Jerome I. Rotter161, Rainer Rettig224, Manuela Uda175, David P. Strachan50, Jacqueline C.M. Witteman13,15, Anna-Liisa Hartikainen225, Jacques S. Beckmann105,226, Eric Boerwinkle227, Ramachandran S. Vasan6,228, Michael Boehnke31, Martin G. Larson6,229, Marjo-Riitta Järvelin18,230,231,232,233, Bruce M. Psaty21,135*, Gonçalo R Abecasis19*, Aravinda Chakravarti1, Paul Elliott18,233*, Cornelia M. van Duijn13,234*, Christopher Newton-Cheh27,115, Daniel Levy6,16,7, Mark J. Caulfield4, Toby Johnson4
Affiliations
Center for Complex Disease Genomics, McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Institute of Social and Preventive Medicine (IUMSP), Centre Hospitalier Universitaire Vaudois and University of Lausanne, Bugnon 17, 1005 Lausanne, Switzerland
Cardiology, Department of Specialties of Internal Medicine, Geneva University Hospital, Rue Gabrielle-Perret-Gentil 4, 1211 Geneva 14, Switzerland
Clinical Pharmacology and The Genome Centre, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK
Department of Biostatistics, University of Washington, Seattle, WA, USA
Framingham Heart Study, Framingham, MA, USA
National Heart Lung, and Blood Institute, Bethesda, MD, USA
Division of Preventive Medicine, Brigham and Women's Hospital, 900 Commonwealth Avenue East, Boston MA 02215, USA
Harvard Medical School, Boston, MA, USA
Icelandic Heart Association, Kopavogur, Iceland
University of Iceland, Reykajvik, Iceland
Department of Health Sciences, University of Leicester, University Rd, Leicester LE1 7RH, UK
Department of Epidemiology, Erasmus Medical Center, PO Box 2040, 3000 CA, Rotterdam, The Netherlands
Department of Internal Medicine, Erasmus Medical Center, Rotterdam, The Netherlands
Netherlands Consortium for Healthy Aging (NCHA), Netherland Genome Initiative (NGI), The Netherlands
Center for Population Studies, National Heart Lung, and Blood Institute, Bethesda, MD, USA
Department of Internal Medicine, Centre Hospitalier Universitaire Vaudois, 1011 Lausanne, Switzerland
Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK
Center for Statistical Genetics, Department of Biostatistics, University of Michigan School of Public Health, Ann Arbor, MI 48103, USA
Interfaculty Institute for Genetics and Functional Genomics, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Cardiovascular Health Research Unit, Departments of Medicine, Epidemiology and Health Services, University of Washington, Seattle, WA, USA
Laboratory of Epidemiology, Demography, Biometry, National Institute on Aging, National Institutes of Health, Bethesda, Maryland 20892, USA
MRC Epidemiology Unit, Institute of Metabolic Science, Cambridge CB2 0QQ, UK
Centre National de Génotypage, Commissariat à L'Energie Atomique, Institut de Génomique, Evry, France
Institute of Molecular and Cell Biology, University of Tartu, Riia 23, Tartu 51010, Estonia
University of Maryland School of Medicine, Baltimore, MD, USA, 21201, USA
Center for Human Genetic Research, Cardiovascular Research Center, Massachusetts General Hospital, Boston, Massachusetts, 02114, USA
Department of Twin Research & Genetic Epidemiology, King's College London, UK
Cardiovascular Epidemiology and Genetics, Institut Municipal d'Investigacio Medica, Barcelona Biomedical Research Park, 88 Doctor Aiguader, 08003 Barcelona, Spain
Institute of Genetic Medicine, European Academy Bozen/Bolzano (EURAC), Viale Druso 1, 39100 Bolzano, Italy—Affiliated Institute of the University of Lübeck, Germany
Department of Biostatistics, Center for Statistical Genetics, University of Michigan, Ann Arbor, Michigan, 48109, USA
Department of Cardiovascular Medicine, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK
Clinical Research Branch, National Institute on Aging, Baltimore MD 21250, USA
Centre for Population Health Sciences, University of Edinburgh, EH89AG, UK
Centre for Population Health Sciences and Institute of Genetics and Molecular Medicine, College of Medicine and Vet Medicine, University of Edinburgh, EH8 9AG, UK
Croatian Centre for Global Health, University of Split, Croatia
Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, SE-751 85 Uppsala, Sweden
Amgen, 1 Kendall Square, Building 100, Cambridge, MA 02139, USA
Department of Clinical Sciences, Lund University, Malmö, Sweden
Department of Medicine, University of Verona, Italy
Ealing Hospital, London, UB1 3HJ, UK
Department of Medicine, University of Mississippi Medical Center, USA
Genetic Epidemiology Group, Epidemiology and Public Health, UCL, London, WC1E 6BT, UK
Center for Genome Science, National Institute of Health, Seoul, Korea
Department of Cardiology, University Medical Center Groningen, University of Groningen, The Netherlands
Departments of Epidemiology and Medicine, Johns Hopkins University, Baltimore MD, USA
Centre for Cellular and Molecular Biology (CCMB), Council of Scientific and Industrial Research (CSIR), Uppal Road, Hyderabad 500 007, India
Department of Public Health and Primary Care, University of Cambridge, CB1 8RN, UK
Department of Basic Medical Research and Education, and Department of Geriatric Medicine, Ehime University Graduate School of Medicine, Toon, 791-0295, Japan
Division of Community Health Sciences, St George's University of London, London, SW17 0RE, UK
Epidemiology & Prevention, Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC 27157, USA
Division of Biostatistics and Department of Genetics, School of Medicine, Washington University in St. Louis, Saint Louis, Missouri 63110, USA
Department of Medicine, University of Eastern Finland and Kuopio University Hospital, 70210 Kuopio, Finland
Department of Preventive Medicine and Epidemiology, Loyola University Medical School, Maywood, IL, USA
Department of Laboratory Medicine & Institute of Human Genetics, University of California San Francisco, 513 Parnassus Ave. San Francisco CA 94143, USA
Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, 138672, Singapore
Centre for Molecular Epidemiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 117597, Singapore
Department of Clinical Chemistry, University of Tampere and Tampere University Hospital, Tampere, 33521, Finland
Department of Genetics, Biology and Biochemistry, University of Torino, Via Santena 19, 10126, Torino, Italy
Human Genetics Foundation (HUGEF), Via Nizza 52, 10126, Torino, Italy
Department of Genetics, University of North Carolina, Chapel Hill, NC, 27599, USA
MRC Centre for Causal Analyses in Translational Epidemiology, School of Social & Community Medicine, University of Bristol, Bristol BS8 2BN, UK
Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Heidelberglaan 100, 3508 GA Utrecht, The Netherlands
Complex Genetics Section, Department of Medical Genetics—DBG, University Medical Center Utrecht, 3508 GA Utrecht, The Netherlands
Centre for Genetic Epidemiology and Biostatistics, University of Western Australia, Crawley, WA, Australia
HUNT Research Centre, Department of Public Health and General Practice, Norwegian University of Science and Technology, 7600 Levanger, Norway
MRC Unit for Lifelong Health & Ageing, London, WC1B 5JU, UK
Occupational and Environmental Medicine, Department of Public Health and Community Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, 40530 Gothenburg, Sweden
Centre for Cardiovascular Genetics, University College London, London WC1E 6JF, UK
MRC Human Genetics Unit and Institute of Genetics and Molecular Medicine, Edinburgh, EH2, UK
Department of Cardiovascular Sciences, University of Leicester, Glenfield Hospital, Leicester, LE3 9QP, UK
Leicester NIHR Biomedical Research Unit in Cardiovascular Disease, Glenfield Hospital, Leicester, LE3 9QP, UK
Studies Coordinating Centre, Division of Hypertension and Cardiac Rehabilitation, Department of Cardiovascular Diseases, University of Leuven, Campus Sint Rafaël, Kapucijnenvoer 35, Block D, Box 7001, 3000 Leuven, Belgium
Center for Research on Genomics and Global Health, National Human Genome Research Institute, Bethesda, MD 20892, USA
Columbia University, NY, USA
Department of Medicine III, Medical Faculty Carl Gustav Carus at the Technical University of Dresden, 01307 Dresden, Germany
Epidemiology and Biostatistics, School of Public Health, Imperial College, London, W2 1PG, UK
Clinical and Molecular Epidemiology Unit, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece
Wake Forest University Health Sciences, Winston-Salem, NC 27157, USA
A list of consortium members is supplied in the Supplementary Materials
Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 48109, USA
Division of Epidemiology, Human Genetics and Environmental Sciences, School of Public Health, University of Texas at Houston Health Science Center, 12 Herman Pressler, Suite 453E, Houston, TX 77030, USA
Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20892, USA
Laboratory of Cardiovascular Science, Intramural Research Program, National Institute on Aging, NIH, Baltimore, Maryland, USA
Washington Hospital Center, Division of Cardiology, Washington DC, USA
ALSPAC Laboratory, University of Bristol, Bristol, BS8 2BN, UK
Pediatric Epidemiology Center, University of South Florida, Tampa, FL, USA
Clinical Pharmacology Unit, University of Cambridge, Addenbrookes Hospital, Hills Road, Cambridge CB2 2QQ, UK
University of Dundee, Ninewells Hospital &Medical School, Dundee, DD1 9SY, UK
Genetic Epidemiology Group, Department of Epidemiology and Public Health, UCL, London WC1E 6BT, UK
Pathology and Laboratory Medicine, University of Western Australia, Crawley, WA, Australia
Molecular Genetics, PathWest Laboratory Medicine, Nedlands, WA, Australia
Clinical Trial Service Unit and Epidemiological Studies Unit, University of Oxford, Oxford, OX3 7LF, UK
Department of Epidemiology and Biostatistics, Case Western Reserve University, 2103 Cornell Road, Cleveland, OH 44106, USA
Department of Medicine, Johns Hopkins University, Baltimore, USA
Department of Clinical Physiology, University of Tampere and Tampere University Hospital, Tampere, 33521, Finland
Department of Medicine, University of Turku and Turku University Hospital, Turku, 20521, Finland
Department of Nutrition, University of North Carolina, Chapel Hill, NC, 27599, USA
Office of Population Studies Foundation, University of San Carlos, Talamban, Cebu City 6000, Philippines
Department of Neurology and Framingham Heart Study, Boston University School of Medicine, Boston, MA, 02118, USA
Department of Internal Medicine II, University Medical Center Regensburg, 93053 Regensburg, Germany
Department of Epidemiology and Preventive Medicine, University Medical Center Regensburg, 93053 Regensburg, Germany
Department of Epidemiology, Johns Hopkins University, Baltimore MD, USA
Renal Division, University Hospital Freiburg, Germany
Département de Génétique Médicale, Université de Lausanne, 1015 Lausanne, Switzerland
Swiss Institute of Bioinformatics, 1015 Lausanne, Switzerland
Division of Genetics, GlaxoSmithKline, Philadelphia, Pennsylvania 19101, USA
International Centre for Circulatory Health, National Heart & Lung Institute, Imperial College, London, UK
Genetics of Complex Traits, Peninsula Medical School, University of Exeter, UK
Department of Community Health Sciences & Department of Medicine, Aga Khan University, Karachi, Pakistan
Medizinische Klinik II, Universität zu Lübeck, Lübeck, Germany
Diabetes Unit, KEM Hospital and Research Centre, Rasta Peth, Pune-411011, Maharashtra, India
Department of Clinical Sciences, Diabetes and Endocrinology Research Unit, University Hospital, Malmö, Sweden
Lund University, Malmö 20502, Sweden
Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, Massachusetts, 02139, USA
Department of Chronic Disease Prevention, National Institute for Health and Welfare, FIN-00251 Helsinki, Finland
William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK
Merck Research Laboratory, 126 East Lincoln Avenue, Rahway, NJ 07065, USA
Wellcome Trust Sanger Institute, Hinxton, CB10 1SA, UK
University of Cambridge Metabolic Research Labs, Institute of Metabolic Science Addenbrooke's Hospital, CB2 OQQ, Cambridge, UK
Division of Cardiovascular Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI, USA
Cardiovascular Genetics, University of Utah School of Medicine, Salt Lake City, UT, USA
Department of Physiology and Biophysics, Keck School of Medicine, University of Southern California, Los Angeles, California 90033, USA
National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland 20892,USA
National Institute for Health and Welfare, 00271 Helsinki, Finland
FIMM, Institute for Molecular Medicine, Finland, Biomedicum, P.O. Box 104, 00251 Helsinki, Finland
Broad Institute, Cambridge, Massachusetts 02142, USA
Leibniz-Institute for Arteriosclerosis Research, Department of Molecular Genetics of Cardiovascular Disease, University of Münster, Münster, Germany
Medical Faculty of the Westfalian Wilhelms University Muenster, Department of Molecular Genetics of Cardiovascular Disease, University of Muenster, Muenster, Germany
Division of Cardiology, Massachusetts General Hospital, Boston, MA, USA
Georgia Prevention Institute, Department of Pediatrics, Medical College of Georgia, Augusta, GA, USA
Unit of Genetic Epidemiology and Bioinformatics, Department of Epidemiology, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands
Department of Biostatical Sciences, Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC 27157, USA
Department of Epidemiology, University of Washington, Seattle, WA, 98195, USA
Group Health Research Institute, Group Health Cooperative, Seattle, WA, USA
Seattle Epidemiologic Research and Information Center, Veterans Health Administration Office of Research & Development, Seattle, WA 98108, USA
Department of Medicine, University of Washington, 98195, USA
Department of Cardiology, University of Tartu, L. Puusepa 8, 51014 Tartu, Estonia
Tallinn University of Technology, Institute of Biomedical Engineering, Ehitajate tee 5, 19086 Tallinn, Estonia
Centre of Cardiology, North Estonia Medical Centre, Sütiste tee 19, 13419 Tallinn, Estonia
Division of Non-communicable disease Epidemiology, The London School of Hygiene and Tropical Medicine London, Keppel Street, London WC1E 7HT, UK
South Asia Network for Chronic Disease, Public Health Foundation of India, C-1/52, SDA, New Delhi 100016, India
Department of Emergency and Cardiovascular Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, 41685 Gothenburg, Sweden
Department of Biostatistics, Institute of Basic Medical Sciences, University of Oslo, 0317 Oslo, Norway
Tuscany Regional Health Agency, Florence, Italy
Tropical Medicine Research Institute, University of the West Indies, Mona, Kingston, Jamaica
University of Ibadan, Ibadan, Nigeria
Department of Genomic Medicine, and Department of Preventive Cardiology, National Cerebral and Cardiovascular Research Center, Suita, 565-8565, Japan
Department of Health Science, Shiga University of Medical Science, Otsu, 520-2192, Japan
Department of Geriatric Medicine, Osaka University Graduate School of Medicine, Suita, 565-0871, Japan
Tohoku University Graduate School of Pharmaceutical Sciences and Medicine, Sendai, 980-8578, Japan
Lifestyle-related Disease Prevention Center, Shiga University of Medical Science, Otsu, 520-2192, Japan
Department of Medical Science and Cardiorenal Medicine, Yokohama City University School of Medicine, Yokohama, 236-0004, Japan
Department of Statistics, Pontificia Universidad Catolica de Chile, Vicuña Mackena 4860, Santiago, Chile
Institute of Human Genetics, Helmholtz Zentrum Munich, German Research Centre for Environmental Health, 85764 Neuherberg, Germany
Institute of Human Genetics, Klinikum rechts der Isar, Technical University of Munich, 81675 Munich, Germany
Institute of Epidemiology, Helmholtz Zentrum Munich, German Research Centre for Environmental Health, 85764 Neuherberg, Germany
Chair of Epidemiology, Institute of Medical Informatics, Biometry and Epidemiology, Ludwig-Maximilians-Universität, 81377 Munich, Germany
Klinikum Grosshadern, 81377 Munich, Germany
National Heart and Lung Institute, Imperial College London, London, UK, W12 0HS, UK
Medical Genetics Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA
Medical Population Genetics, Broad Institute of Harvard and MIT, 5 Cambridge Center, Cambridge MA 02142, USA
National Heart, Lung and Blood Institute and its Framingham Heart Study, 73 Mount Wayte Ave., Suite #2, Framingham, MA 01702, USA
Department of Neurology and Medicine, University of Washington, Seattle, USA
Department of Medicine (Geriatrics), University of Mississippi Medical Center, Jackson, MS, USA
Department of Neurology, Boston University School of Medicine, USA
Finnish Institute of Occupational Health, Finnish Institute of Occupational Health, Aapistie 1, 90220 Oulu, Finland
Wellcome Trust Centre for Human Genetics, University of Oxford, UK
Lapland Central Hospital, Department of Physiatrics, Box 8041, 96101 Rovaniemi, Finland
Center for Non-Communicable Diseases Karachi, Pakistan
Atherosclerosis Research Unit, Department of Medicine, Karolinska Institute, Stockholm, Sweden
Department of Internal Medicine, University Medical Center Groningen, University of Groningen, The Netherlands
Department of Medical Genetics, Erasmus Medical Center, Rotterdam, The Netherlands
Gerontology Research Center, National Institute on Aging, Baltimore, MD 21224, USA
Istituto di Neurogenetica e Neurofarmacologia, Consiglio Nazionale delle Ricerche, Cittadella Universitaria di Monserrato, Monserrato, Cagliari, Italy
Unita`Operativa Semplice Cardiologia, Divisione di Medicina, Presidio Ospedaliero Santa Barbara, Iglesias, Italy
Computational Medicine Research Group, Institute of Clinical Medicine, University of Oulu and Biocenter Oulu, 90014 University of Oulu, Oulu, Finland
NMR Metabonomics Laboratory, Department of Biosciences, University of Eastern Finland, 70211 Kuopio, Finland
Department of Internal Medicine and Biocenter Oulu, Clinical Research Center, 90014 University of Oulu, Oulu, Finland
Institute for Molecular Medicine Finland FIMM, 00014 University of Helsinki, Helsinki, Finland
Department of Biomedical Engineering and Computational Science, School of Science and Technology, Aalto University, 00076 Aalto, Espoo, Finland
NUS Graduate School for Integrative Sciences & Engineering (NGS) Centre for Life Sciences (CeLS), Singapore, 117456, Singapore
Department of Internal Medicine B, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute of Pharmacology, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute for Community Medicine, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
U557 Institut National de la Santé et de la Recherche Médicale, U1125 Institut National de la Recherche Agronomique, Université Paris 13, Bobigny, France
Department of Neurology, Mayo Clinic, Jacksonville, FL, USA
Imperial College Cerebrovascular Unit (ICCRU), Imperial College, London, W6 8RF, UK
Faculty of Medicine, University of Split, Croatia
Department of Internal Medicine, Diabetology, and Nephrology, Medical University of Silesia, 41-800, Zabrze, Poland
Public Health Sciences section, Division of Community Health Sciences, University of Edinburgh, Medical School, Teviot Place, Edinburgh, EH8 9AG, UK
School of Science and Engineering, University of Ballarat, 3353 Ballarat, Australia
Prevention and Care of Diabetes, Department of Medicine III, Medical Faculty Carl Gustav Carus at the Technical University of Dresden, 01307 Dresden, Germany
University Hospital Münster, Internal Medicine D, Münster, Germany
Department of Medical Statistics, Epidemiology and Medical Informatics, Andrija Stampar School of Public Health, University of Zagreb, Croatia
AstraZeneca R&D, 431 83 Mölndal, Sweden
Genetic Epidemiology & Biostatistics Platform, Ontario Institute for Cancer Research, Toronto
Samuel Lunenfeld Institute for Medical Research, University of Toronto, Canada
Faculty of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, UK
Department of Epidemiology and Public Health, Imperial College, Norfolk Place London W2 1PG, UK
Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku and the Department of Clinical Physiology, Turku University Hospital, Turku, 20521, Finland
Department of Internal Medicine, Division of Cardiovascular Medicine, University of Michigan Medical Center, Ann Arbor, Michigan, USA
Singapore Eye Research Institute, Singapore, 168751, Singapore
Department of Ophthalmology, National University of Singapore, Singapore, 119074, Singapore
Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 119074, Singapore
Duke-National University of Singapore Graduate Medical School, Singapore, 169857, Singapore
Division of Biostatistics, Washington University School of Medicine, Saint Louis, MO, 63110, USA
Department of Primary Care & Population Health, UCL, London, UK, NW3 2PF, UK
BHF Glasgow Cardiovascular Research Centre, University of Glasgow, 126 University Place, Glasgow, G12 8TA, UK
Epidemiology Public Health, UCL, London, UK, WC1E 6BT, UK
Departments of Epidemiology, Biostatistics, and Medicine, Johns Hopkins University, Baltimore MD, USA
Division of Nephrology, Department of Internal Medicine, University Medical Center Groningen, University of Groningen, The Netherlands
Division of Cardiology, Brigham and Women's Hospital, 900 Commonwealth Avenue East, Boston MA 02215, USA
Geriatric Rehabilitation Unit, Azienda Sanitaria Firenze (ASF), Florence, Italy
National Institute for Health and Welfare, Diabetes Prevention Unit, 00271 Helsinki, Finland
Hjelt Institute, Department of Public Health, University of Helsinki, 00014 Helsinki, Finland
South Ostrobothnia Central Hospital, 60220 Seinäjoki, Finland
Red RECAVA Grupo RD06/0014/0015, Hospital Universitario La Paz, 28046 Madrid, Spain
Department of Neurology, General Central Hospital, 39100 Bolzano, Italy
CIBER Epidemiología y Salud Pública, 08003 Barcelona
Department of Medicine and Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA
Geriatric Research and Education Clinical Center, Veterans Administration Medical Center, Baltimore, MD, USA
U872 Institut National de la Santé et de la Recherche Médicale, Centre de Recherche des Cordeliers, Paris, France
Institute of Physiology, Ernst-Moritz-Arndt-University Greifswald, 17487 Greifswald, Germany
Institute of Clinical Medicine/Obstetrics and Gynecology, University of Oulu, Finland
Service of Medical Genetics, Centre Hospitalier Universitaire Vaudois, 1011 Lausanne, Switzerland
Human Genetics Center, 1200 Hermann Pressler, Suite E447 Houston, TX 77030, USA
Division of Epidemiology and Prevention, Boston University School of Medicine, Boston, MA, USA
Department of Mathematics, Boston University, Boston, MA, USA
Institute of Health Sciences, University of Oulu, BOX 5000, 90014 University of Oulu, Finland
Biocenter Oulu, University of Oulu, BOX 5000, 90014 University of Oulu, Finland
National Institute for Health and Welfare, Box 310, 90101 Oulu, Finland
MRC-HPA Centre for Environment and Health, School of Public Health, Imperial College London, Norfolk Place, London W2 1PG, UK
Centre of Medical Systems Biology (CMSB 1–2), NGI Erasmus Medical Center, Rotterdam, The Netherlands
Supporting Information
At first, we identified BP differentially expressed genes in six cohorts (FHS, EGCUT, RS, InCHIANT, KORA F4 and SHIP-TREND) respectively. Second, we conducted a meta-analysis of the Illumina cohorts (EGCUT, RS, InCHIANT, KORA F4 and SHIP-TREND). Third, for discovery and replication purpose, we replicated the BP signature genes identified in the FHS cohort in the Illumina cohorts. And in turn, we replicated the BP signature genes identified in Illumina cohorts in FHS cohort. Fourth, we conducted a meta-analysis in the six cohorts and reported the BP signature genes passing Bonferroni corrected p<0.05 (corrected for 7717 genes). And finally, we cross-analyzed the BP signature genes with blood eQTLs as well as with BP GWAS results to identify the BP signature genes having BP GWAS eQTLs.
(TIF)
A) SBP; B) DBP; C) HTN. The x-axis is the effect size (beta values) of meta-analysis and the y-axis is the −log10 transformed p values.
(TIF)
The numbers in the Heatmap indicate Pearson correlations between pairs of genes.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
We thank the field staff in Augsburg who was involved in the conduct of the studies. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. We thank Marjolein Peters, MSc, Ms. Mila Jhamai, Ms. Jeannette M. Vergeer-Drop, Ms. Bernadette van Ast-Copier, Mr. Marijn Verkerk and Jeroen van Rooij, BSc for their help in creating the RNA array expression database.
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD. (http://biowulf.nih.gov).
Data Availability
Raw data from gene expression profiling are available online (FHS [http://www.ncbi.nlm.nih.gov/gap; accession number phs000007], EGCUT [GSE48348], RS [GSE33828], InCHIANTI [GSE48152], KORA F4 [E-MTAB-1708] and SHIP-TREND [GSE36382]).
Funding Statement
The Framingham Heart Study is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and the Center for Information Technology, National Institutes of Health, Bethesda, MD. EGCUT is supported by targeted financing from the Estonian Ministry of Science and Education [SF0180142s08]; the Development Fund of the University of Tartu (grant SP1GVARENG); the European Regional Development Fund to the Centre of Excellence in Genomics (EXCEGEN; grant 3.2.0304.11-0312); and through FP7 grant 313010. AD is supported by NWO grant (veni, 916.12.154) and the EUR Fellowship. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The generation and management of RNA-expression array data for the Rotterdam Study was executed and funded by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The InCHIANTI study was supported in part by the Intramural Research Program, National Institute on Aging. DM was generously supported by a Wellcome Trust Institutional Strategic Support Award (WT097835MF). The KORA research platform and the KORA Augsburg studies are financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, which is funded by the BMBF and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig Maximilians-Universität, as part of the LMU innovative and in part by a grant from the BMBF to the German Center for Diabetes Research (DZD). The German Diabetes Center is funded by the German Federal Ministry of Health and the Ministry of School, Science and Research of the State of North-Rhine-Westphalia. Additional support was obtained from the BMBF (National Genome Research Network NGFN plus Atherogenomics, 01GS0834), by the DZHK (Deutsches Zentrum für Herz-Kreislauf-Forschung – German Centre for Cardiovascular Research) and from the European Commission’s Seventh Framework Programme (FP7/2007-2013, HEALTH-F2-2011, grant agreement No. 277984, TIRCON and HEALTH-F2-2013, grant agreement No. 603288, SysVasc). SHIP is supported by the BMBF (German Ministry of Education and Research) and by the DZHK (German Centre for Cardiovascular Research) within the framework of the MetaXpress consortium. SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by BMBF (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the network ‘Greifswald Approach to Individualized Medicine (GANI_MED)’ funded by the BMBF (grant 03IS2061A). Generation of genome-wide data has been supported by the BMBF (grant no. 03ZIK012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, et al. (2003) Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 42: 1206–1252. [DOI] [PubMed] [Google Scholar]
- 2. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, et al. (2009) Genome-wide association study of blood pressure and hypertension. Nat Genet 41: 677–687. 10.1038/ng.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leonardson AS, Zhu J, Chen Y, Wang K, Lamb JR, et al. (2010) The effect of food intake on gene expression in human peripheral blood. Hum Mol Genet 19: 159–169. 10.1093/hmg/ddp476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, et al. (2010) Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5: e10693 10.1371/journal.pone.0010693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bull TM, Coldren CD, Moore M, Sotto-Santiago SM, Pham DV, et al. (2004) Gene microarray analysis of peripheral blood cells in pulmonary arterial hypertension. Am J Respir Crit Care Med 170: 911–919. [DOI] [PubMed] [Google Scholar]
- 7. Korkor MT, Meng FB, Xing SY, Zhang MC, Guo JR, et al. (2011) Microarray analysis of differential gene expression profile in peripheral blood cells of patients with human essential hypertension. Int J Med Sci 8: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, et al. (2011) The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol 26: 657–686. 10.1007/s10654-011-9610-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schurmann C, Heim K, Schillert A, Blankenberg S, Carstensen M, et al. (2012) Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PloS one 7: e50938 10.1371/journal.pone.0050938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Volzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, et al. (2011) Cohort profile: the study of health in Pomerania. Int J Epidemiol 40: 294–307. 10.1093/ije/dyp394 [DOI] [PubMed] [Google Scholar]
- 11. Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Westra H-J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics 45: 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joehanes R., Huan T., C Yao, X Zhang, S Ying, et al. (2013) Genome-wide Expression Quantitative Trait Loci: Results from the NHLBI’s SABRe CVD Initiative. the American Society of Human Genetics (ASHG) conference. Boston Convention Ctr. Boston, MA.
- 14. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106: 9362–9367. 10.1073/pnas.0903103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rowland NE, Li BH, Fregly MJ, Smith GC (1995) Fos induced in brain of spontaneously hypertensive rats by angiotensin II and co-localization with AT-1 receptors. Brain Res 675: 127–134. [DOI] [PubMed] [Google Scholar]
- 16. Beetz N, Harrison MD, Brede M, Zong X, Urbanski MJ, et al. (2009) Phosducin influences sympathetic activity and prevents stress-induced hypertension in humans and mice. J Clin Invest 119: 3597–3612. 10.1172/JCI38433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, et al. (2010) Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev 90: 291–366. 10.1152/physrev.00021.2009 [DOI] [PubMed] [Google Scholar]
- 18. Felix JP, Priest BT, Solly K, Bailey T, Brochu RM, et al. (2012) The inwardly rectifying potassium channel Kir1.1: development of functional assays to identify and characterize channel inhibitors. Assay Drug Dev Technol 10: 417–431. 10.1089/adt.2012.462 [DOI] [PubMed] [Google Scholar]
- 19. Fang L, Li D, Welling PA (2010) Hypertension resistance polymorphisms in ROMK (Kir1.1) alter channel function by different mechanisms. Am J Physiol Renal Physiol 299: F1359–1364. 10.1152/ajprenal.00257.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cappuccio FP, MacGregor GA (1991) Does potassium supplementation lower blood pressure? A meta-analysis of published trials. J Hypertens 9: 465–473. [DOI] [PubMed] [Google Scholar]
- 21. Geleijnse JM, Kok FJ, Grobbee DE (2003) Blood pressure response to changes in sodium and potassium intake: a metaregression analysis of randomised trials. J Hum Hypertens 17: 471–480. [DOI] [PubMed] [Google Scholar]
- 22. Fulgoni VL 3rd (2007) Limitations of data on fluid intake. J Am Coll Nutr 26: 588S−591S. [DOI] [PubMed] [Google Scholar]
- 23. Koliaki C, Katsilambros N (2013) Dietary sodium, potassium, and alcohol: key players in the pathophysiology, prevention, and treatment of human hypertension. Nutr Rev 71: 402–411. 10.1111/nure.12036 [DOI] [PubMed] [Google Scholar]
- 24. Morton J, Coles B, Wright K, Gallimore A, Morrow JD, et al. (2008) Circulating neutrophils maintain physiological blood pressure by suppressing bacteria and IFNgamma-dependent iNOS expression in the vasculature of healthy mice. Blood 111: 5187–5194. 10.1182/blood-2007-10-117283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrison DG, Guzik TJ, Lob HE, Madhur MS, Marvar PJ, et al. (2011) Inflammation, immunity, and hypertension. Hypertension 57: 132–140. 10.1161/HYPERTENSIONAHA.110.163576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrison DG, Marvar PJ, Titze JM (2012) Vascular inflammatory cells in hypertension. Front Physiol 3: 128 10.3389/fphys.2012.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harrison DG, Vinh A, Lob H, Madhur MS (2010) Role of the adaptive immune system in hypertension. Curr Opin Pharmacol 10: 203–207. 10.1016/j.coph.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological): 289–300.
- 29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene Ontology: tool for the unification of biology. Nature genetics 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
At first, we identified BP differentially expressed genes in six cohorts (FHS, EGCUT, RS, InCHIANT, KORA F4 and SHIP-TREND) respectively. Second, we conducted a meta-analysis of the Illumina cohorts (EGCUT, RS, InCHIANT, KORA F4 and SHIP-TREND). Third, for discovery and replication purpose, we replicated the BP signature genes identified in the FHS cohort in the Illumina cohorts. And in turn, we replicated the BP signature genes identified in Illumina cohorts in FHS cohort. Fourth, we conducted a meta-analysis in the six cohorts and reported the BP signature genes passing Bonferroni corrected p<0.05 (corrected for 7717 genes). And finally, we cross-analyzed the BP signature genes with blood eQTLs as well as with BP GWAS results to identify the BP signature genes having BP GWAS eQTLs.
(TIF)
A) SBP; B) DBP; C) HTN. The x-axis is the effect size (beta values) of meta-analysis and the y-axis is the −log10 transformed p values.
(TIF)
The numbers in the Heatmap indicate Pearson correlations between pairs of genes.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
Raw data from gene expression profiling are available online (FHS [http://www.ncbi.nlm.nih.gov/gap; accession number phs000007], EGCUT [GSE48348], RS [GSE33828], InCHIANTI [GSE48152], KORA F4 [E-MTAB-1708] and SHIP-TREND [GSE36382]).