Summary
An increasingly powerful approach for studying brain circuits relies on targeting genetically encoded sensors and effectors to specific cell types. However, current approaches for this are still limited in functionality and specificity. Here we utilize several intersectional strategies to generate multiple transgenic mouse lines expressing high levels of novel genetic tools with high specificity. We developed driver and double reporter mouse lines and viral vectors using the Cre/Flp and Cre/Dre double recombinase systems, and established a new, retargetable genomic locus, TIGRE, which allowed the generation of a large set of Cre/tTA dependent reporter lines expressing fluorescent proteins, genetically encoded calcium, voltage, or glutamate indicators, and optogenetic effectors, all at substantially higher levels than before. High functionality was shown in example mouse lines for GCaMP6, YCX2.60, VSFP Butterfly 1.2, and Jaws. These novel transgenic lines greatly expand the ability to monitor and manipulate neuronal activities with increased specificity.
Introduction
The brain comprises a large number of neuronal and non-neuronal cell types, whose connections and interactions are fundamental to its function. To observe and manipulate their activities selectively, the best available approach is genetic targeting of protein-based sensors and effectors to specific cell types (Huang and Zeng, 2013). In mice, the Cre/lox recombination system is the most widely used approach to access specific cell types, utilizing gene promoters or loci with specific expression patterns (Gerfen et al., 2013; Gong et al., 2007; Madisen et al., 2010; Taniguchi et al., 2011). However, cell populations defined by Cre driver lines are often heterogeneous, encompassing multiple brain regions and/or multiple cell types (Harris et al., 2014). Fundamentally, cell types are rarely defined by single genes, but rather by intersectional expression of multiple genes. Thus, it is imperative to develop intersectional genetic targeting approaches, combining regulatory elements from two or more genes to increase specificity of transgene expression. Important efforts have been made to develop transgenic intersectional approaches, most successfully with the combination of Cre and Flp site specific recombinases (SSRs) (Dymecki and Kim, 2007; Dymecki et al., 2010; Kranz et al., 2010; Ray et al., 2011; Robertson et al., 2013). However, thus far, intersectional approaches have not been widely used in functional studies, due to the limited number of validated transgenic tools available.
The ongoing development of increasingly effective sensors and effectors offers extraordinary opportunities for studies of neuronal interactions and functions (Fenno et al., 2011; Huang and Zeng, 2013; Knopfel, 2012). One issue with the practical utility of these tools is that they require high-level expression in cell populations of interest. Such high levels of expression can be obtained with techniques that result in high transgene copy numbers in individual cells, such as in utero electroporation and adeno-associated virus (AAV) infection. However, these approaches have limitations, including invasive surgical delivery, incomplete coverage of the desired cell population, variable levels of expression in different cells, and, in the case of viruses, potential cytotoxicity associated with long-term viral infection and/or uncontrolled gene expression.
Transgenic mouse lines that express high and heritable patterns of genetic tools in specific cell populations provide an alternative that can overcome at least some of these limitations (Zeng and Madisen, 2012; Zhao et al., 2011). We previously establish a standardized Cre-reporter system in which transgene expression was driven by a strong, ubiquitous CAG promoter targeted to the Rosa26 locus (Madisen et al., 2010; Muzumdar et al., 2007), expressing fluorescent proteins, calcium sensor GCaMP3, and optogenetic activator ChR2(H134R) and silencers Arch and eNpHR3.0 (Madisen et al., 2012; Madisen et al., 2010; Zariwala et al., 2012). Although proved useful in many applications (Ackman et al., 2012; Haddad et al., 2013; Issa et al., 2014; Jackman et al., 2014; Kheirbek et al., 2013; Lee et al., 2014; Nguyen-Vu et al., 2013; Pi et al., 2013), we and others have also identified limitations in the sensitivity or functionality of these reporters in other situations.
Currently the only transgenic mouse approach demonstrated to reliably achieve AAV-like high-level expression is the use of the Thy1.2 promoter in randomly integrated transgenes (Arenkiel et al., 2007; Dana et al., 2014; Feng et al., 2000; Zhao et al., 2008), presumably with multiple copies at the insertion site. Although powerful in driving tool gene expression, this approach also has drawbacks. Expression of transgenes driven by the Thy1.2 promoter is strongly position dependent, necessitating a screen of multiple founder lines to find potentially useful ones. Furthermore, adding Cre-dependent control, e.g. a floxed-stop cassette, to a multi-copy transgene is problematic because Cre could then induce recombination both within and between the different copies, resulting in reduced transgene copy numbers and variability of transgene expression among different cells.
We therefore undertook a systematic evaluation of multiple approaches aiming at more specific and more robust transgene expression. To complement existing Cre driver lines, we focused on the intersection of Cre with another recombinase or with a transcriptional activator. In addition, we built and validated a new docking site in a permissive genomic locus, the TIGRE locus (Zeng et al., 2008), which supports repeated targeting. By introducing a tTA-based transcriptional amplification approach to the TIGRE locus, all reporter lines doubly regulated by Cre and tTA drove robust expression of sensors and effectors at levels substantially higher than those in comparable Rosa-CAG based reporters. Functional characterization of lines carrying representative optical tools under Cre and tTA control demonstrates their enhanced efficiency for studies of neuronal activity, both in vitro and in vivo.
Results
In our effort to improve upon current strategies for cell type-specific transgene expression, we explored three strategies for intersectional control: (1) reporter expression that depends on two independent SSRs from the ubiquitous Rosa26 locus, (2) Cre-dependent reporter expression from an endogenous locus targeted because of its cell type-specific expression pattern, (3) reporter expression dependent on Cre and the transcriptional transactivator tTA from another ubiquitous genomic locus, TIGRE. Since the third strategy resulted in the most strongly enhanced transgene expression, we created a series of TIGRE reporter lines that show high-level expression of novel calcium, voltage and glutamate sensors and optogenetic effectors. The complete list of new intersectional transgenic mouse lines and AAVs introduced in this paper (17 reporter lines, 4 driver lines and 10 AAVs) is shown in Table 1. All other previously published mouse lines used in this study are listed in Table S1.
Table 1. Newly generated transgenic mouse lines and AAVs.
| Namea | Knock-in locus | Promoter used | Expression control | Gene expressed | Function | JAX Stock # |
|---|---|---|---|---|---|---|
| Ai65(RCFL-tdT) | Rosa26 | CAG | Cre and Flp dependent | tdTomato | Fluorescent labeling | 021875 |
| Ai66(RCRL-tdT) | Rosa26 | CAG | Cre and Dre dependent | tdTomato | Fluorescent labeling | 021876 |
| Ai57(RCFL-Jaws) | Rosa26 | CAG | Cre and Flp dependent | Jaws-GFP-ER2 | Optogenetic silencer | (not deposited) |
| Ai72(RCL-VSFPB) | Rosa26 | CAG | Cre dependent | VSFP-Butterfly 1.2 | Voltage indicator | (not deposited) |
| Ai87(RCL-iGluSnFR) | Rosa26 | CAG | Cre dependent | iGluSnFR | Glutamate indicator | (not deposited) |
| Ai95(RCL-GCaMP6f) | Rosa26 | CAG | Cre dependent | GCaMP6f | Calcium indicator | 024105 |
| Ai96(RCL-GCaMP6s) | Rosa26 | CAG | Cre dependent | GCaMP6s | Calcium indicator | 024106 |
| Ai62(TITL-tdT) | TIGRE | TRE | Cre and tTA dependent | tdTomato | Fluorescent labeling | 022731 |
| Ai82(TITL-GFP) | TIGRE | TRE | Cre and tTA dependent | EGFP | Fluorescent labeling | 023532 |
| Ai79(TITL-Jaws) | TIGRE | TRE | Cre and tTA dependent | Jaws-GFP-ER2 | Optogenetic silencer | 023529 |
| Ai93(TITL-GCaMP6f) | TIGRE | TRE | Cre and tTA dependent | GCaMP6f | Calcium indicator | 024103 |
| Ai94(TITL-GCaMP6s) | TIGRE | TRE | Cre and tTA dependent | GCaMP6s | Calcium indicator | 024104 |
| Ai92(TITL-YCX2.60) | TIGRE | TRE | Cre and tTA dependent | YCX2.60 | Calcium indicator | In process |
| Ai78(TITL-VSFPB) | TIGRE | TRE | Cre and tTA dependent | VSFP-Butterfly 1.2 | Voltage indicator | 023528 |
| Ai85(TITL-iGluSnFR) | TIGRE | TRE | Cre and tTA dependent | iGluSnFR | Glutamate indicator | In process |
| Snap25-LSL-2A-GFP | Snap25 | Snap25 | Pan-neuronal promoter, Cre dependent | EGFP | Fluorescent labeling | 021879 |
| Snap25-2A-GCaMP6s | Snap25 | Snap25 | Pan-neuronal promoter | GCaMP6s | Calcium indicator | 025111 |
| Pvalb-2A-Flpe | Pvalb | Pvalb | Driver line | Enhanced Flp | Recombination | 021191 |
| Pvalb-2A-Flpo | Pvalb | Pvalb | Driver line | Mammalianized Flp | Recombination | 022730 |
| Pvalb-2A-Dre | Pvalb | Pvalb | Driver line | Mammalianized Dre | Recombination | 021190 |
| Nr4a2-SA-IRES-Dre | Nr4a2 | Nr4a2 | Driver line | Mammalianized Dre | Recombination | (not deposited) |
| AAV pmSyn1-EBFP-Cre-bGHpA | Mouse Syn1 | Driver | EBFP-Cre | Recombination | ||
| AAV phSyn1-Dre-bGHpA | Human Syn1 | Driver | Mammalianized Dre | Recombination | ||
| AAV phSyn1-Flpo-bGHpA | Human Syn1 | Driver | Mammalianized Flp | Recombination | ||
| AAV phSyn1-Flpe-bGHpA | Human Syn1 | Driver | Enhanced Flp | Recombination | ||
| AAV phSyn1-FSF-FLEX-ChR2(H134R)-EYFP-WPRE-bGHpA | Human Syn1 | Cre and Flp dependent | ChR2(H134R)-EYFP | Optogenetic activator | ||
| AAV phSyn1-RSR-FLEX-ChR2(H134R)-EYFP-WPRE-bGHpA | Human Syn1 | Cre and Dre dependent | ChR2(H134R)-EYFP | Optogenetic activator | ||
| AAV pCAG-FSF-FLEX-EGFP-WPRE-bGHpA | CAG | Cre and Flp dependent | EGFP | Fluorescent labeling | ||
| AAV pCAG-FLEX2-tTA2-WPRE-bGHpA | CAG | Cre dependent driver | tTA2 | Transcriptional activation | ||
| AAV pTRE-FLEX-EGFP-WPRE-bGHpA | TRE | Cre and tTA dependent | EGFP | Fluorescent labeling | ||
| AAV pTRE-FSF-FLEX-EGFP-WPRE-bGHpA | TRE | Cre, Flp and tTA dependent | EGFP | Fluorescent labeling |
Acronyms: RCFL is Rosa26 – CAG promoter – FRT-STOP-FRT – LoxP-STOP-LoxP. RCRL is Rosa26 – CAG promoter – Rox-STOP-Rox – LoxP-STOP-LoxP. RCL is Rosa26 – CAG promoter – LoxP-STOP-LoxP. TITL is TIGRE – Insulators – TRE promoter – LoxP-STOP-LoxP. LSL is LoxP-STOP-LoxP. SA is Splice Acceptor. tdT is tdTomato. The 2A sequences used were F2A for Snap25-LSL-2A-GFP, and T2A for Snap25-2A-GCaMP6s, Pvalb-2A-Flpe, Pvalb-2A-Flpo and Pvalb-2A-Dre.
Dual-recombinase intersectional strategies for transgene regulation increase specificity
An intersectional approach that has proven useful in transgenic studies is to use both Cre and Flp, expressed from separate driver lines using distinct regulatory sequences, to turn on a doubly controlled reporter allele (Dymecki et al., 2010). To incorporate this into our Rosa-CAG reporter system (Madisen et al., 2010) and to test additional recombinases such as Dre (Anastassiadis et al., 2009; Sauer and McDermott, 2004), we generated Cre/Flp or Cre/Dre double-dependent reporters, the Ai65(RCFL-tdT) line which contains FRT-stop-FRT (FRT: Flp recognition site) and LoxP-stop-LoxP double cassettes in front of tdTomato, and the Ai66(RCRL-tdT) line containing Rox-stop-Rox (Rox: Dre recognition site) and LoxP-stop-LoxP double cassettes (Fig. 1A). We also generated driver lines expressing these recombinases in the Pvalb gene locus, Pvalb-2A-Flpe (Buchholz et al., 1998), Pvalb-2A-Flpo (Raymond and Soriano, 2007) and Pvalb-2A-Dre, through recombinase-mediated cassette exchange (RMCE) into our originally targeted Pvalb-2A-Cre line (Fig. S1A). To evaluate the recombination efficiency and specificity, we generated triple transgenic (Tg) mice whose genealogy included a pan-GABAergic Cre line Slc32a1-IRES-Cre (Slc32a1 is also known as VGAT, a GABA vesicular transporter), one of the Pvalb-SSR lines, and Ai65 or Ai66 reporter.
Figure 1.
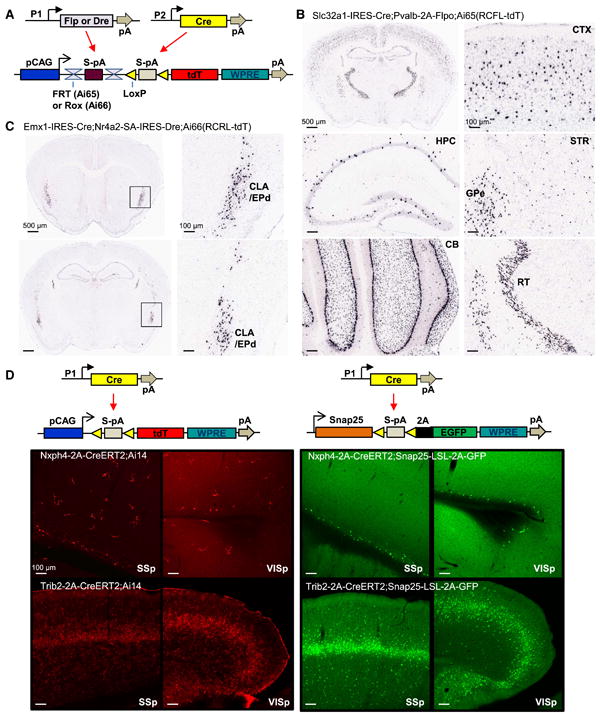
Intersectional strategies using dual recombinases or Cre regulation of a direct neuronally-restricted reporter. (A) Schematic diagram of intersectional control by either Cre/Flp or Cre/Dre recombinases, driven by different promoters (P1 or P2), on a doubly regulated reporter line: the Cre and Flp-dependent Ai65(RCFL-tdT), or the Cre and Dre-dependent Ai66(RCRL-tdT). (B) ISH images of restricted tdTomato expression in Slc32a1+/Pvalb+ GABAergic neurons in the Slc32a1-IRES-Cre;Pvalb-2A-Flpo;Ai65(RCFL-tdT) mouse. CTX, cortex. HPC, hippocampus. STR, striatum. GPe, globus pallidus, external segment. CB, cerebellum. RT, reticular nucleus of the thalamus. (C) ISH images of restricted tdTomato expression in Emx1+/Nr4a2+ neurons in the claustrum (CLA) and endopiriform nucleus dorsal part (EPd) in the Emx1-IRES-Cre;Nr4a2-SA-IRES-Dre;Ai66(RCRL-tdT) mouse. (D) Direct neuronally-restricted reporter gene expression by targeting Cre-dependent reporter gene to the pan-neuronal Snap25 gene locus. TdTomato expression in cortex in both neuronal and non-neuronal cells of Nxph4-2A-CreERT2;Ai14 and Trib2-2A-CreERT2;Ai14 mice (left panels) compared with neuronally-specific EGFP expression in Nxph4-2A-CreERT2;Snap25-LSL-2A-GFP and Trib2-2A-CreERT2;Snap25-LSL-2A-GFP mice (right panels). SSp, primary somatosensory cortex. VISp, primary visual cortex. (See also Figures S1-S4.)
Cre and Flp (both Pvalb-2A-Flpe and Pvalb-2A-Flpo) efficiently restricted transgene expression to cell populations with overlapping patterns of driver expression. In the Slc32a1-IRES-Cre;Pvalb-2A-Flpo;Ai65(RCFL-tdT) triple Tg mouse, tdTomato expression was observed in a subset of cells in multiple areas, consistent with Slc32a1+/Pvalb+ expression pattern (Fig. 1B). Reporter expression depended strictly on the presence of both Cre and Flp drivers (Fig. S2A). Importantly, these triple Tg mice showed higher selectivity in expression than double Tg, Pvalb-2A-Cre;Ai14 mice, where Ai14 is a Rosa-CAG based tdTomato reporter line (Madisen et al., 2010). In those mice, but not in the triple Tg mice, we found expression in cortical layer 5 pyramidal neurons and other non-interneuron cells, presumably arising from low-level or transient expression of the Pvalb gene (Fig. S2B, C). Thus, the intersectional approach depending on two drivers provides increased specificity for targeting Pvalb+ GABAergic neurons.
We also combined Cre and Dre to produce an effective intersectional approach in Slc32a1-IRES-Cre;Pvalb-2A-Dre;Ai66(RCRL-tdT) triple Tg mice (Fig. S3A). Control mice lacking Cre showed no reporter expression, but occasional tdTomato positive cells in Cre+/Dre-/Ai66+ mice were observed. To further evaluate the Cre/Dre intersection, we generated another Dre-driver line, Nr4a2-SA-IRES-Dre (Nr4a2 is also known as Nurr1, a nuclear receptor with expression in dopamine neurons and in claustrum), and crossed it with Emx1-IRES-Cre and Ai66. The resulting triple Tg mice showed specific, dense expression in claustrum and endopiriform nucleus (Fig. 1C), consistent with adult brain areas known to express both Emx1 and Nr4a2. Again, we observed no reporter expression in the absence of Cre but did detect sparse tdTomato positive cells in Cre+/Dre-/Ai66+ mice (Fig. S3B). This result suggests that while Dre specifically recombines Rox sites over Lox sites, Cre is slightly promiscuous towards Rox sites.
To complement our transgenic lines that incorporate dual recombinase control, we also generated a set of recombinant AAVs expressing Cre, Flpe, Flpo and Dre drivers, as well as Cre/Flp and Cre/Dre doubly-regulated reporters (Fig. S4A-D). Consistent with previous reports (Fenno et al., 2014; Kranz et al., 2010), AAV-Flpo and AAV-Dre induced recombination comparable to AAV-Cre, whereas the recombination efficiency of AAV-Flpe was significantly lower. Flp and Dre viruses exhibited high specificity for FRT and Rox sites, respectively, whereas AAV-Cre recombined Rox sites at low frequency, with recombination most prevalent at the injection site, indicating increased promiscuity with high levels of Cre.
Direct neuronally restricted reporter gene expression
To determine whether a simple intersectional strategy could restrict Cre-induced reporter expression to a specific cell type, we took advantage of the Snap25 gene. Snap25 encodes a synaptosome associated protein. It is pan-neuronal and is among the most highly transcribed genes in the brain based on microarray data analysis (data not shown). We generated a Snap25-LSL-2A-GFP knock-in line, where a floxed-stop cassette and T2A-linked EGFP reporter gene is targeted to the stop codon of Snap25 (Fig. 1D). We then assessed fluorescent reporter expression in double Tg mice containing Cre drivers, Nxph4-2A-CreERT2 or Trib2-2A-CreERT2, crossed to either Ai14 or Snap25-LSL-2A-GFP reporters.
In both sets of animals crossed to Ai14, we found both neuronal and non-neuronal expression (Fig. 1D, left). In Nxph4-2A-CreERT2;Ai14 (Nxph4 is neurexophilin 4), we observed sparse cortical layer 6b expression, but also significant expression in small, possibly glial cells. Likewise, in Trib2-2A-CreERT2;Ai14 (Trib2 is tribbles homolog 2) we saw enriched expression in layer 5a cortical neurons, but also strong and widespread fluorescence in vasculature. When crossed to Snap25-LSL-2A-GFP, however, EGFP expression in both lines was restricted to neurons, thereby increasing uniformity in the labeled cell populations (Fig. 1D, right). Snap25-LSL-2A-GFP also showed stronger GFP fluorescence than the Rosa-CAG based Ai3-EYFP reporter (Madisen et al., 2010) (data not shown).
The pan-neuronal expression strategy can also be employed to restrict the functionality of other genetic tools to facilitate more precise observation or manipulation of cell populations. To this end, we generated a Snap25-2A-GCaMP6s knock-in line, which expresses a calcium indicator GCaMP6s (Chen et al., 2013) pan-neuronally and independently of any driver line (Table 1). This line can be used to monitor neuronal calcium activity throughout the brain (Michael Crair and H.Z., unpublished results) while being readily combined with Cre or another driver dependent, cell-type-specific genetic manipulation.
Cre/tTA dependent reporters targeted to the TIGRE locus are more highly expressed than Rosa-CAG reporters
The tetracycline (Tet)-regulated expression system has been used in mice for inducible expression of genes from the TRE (tetracycline response element, also called tetO) promoter in response to tTA (Tet-Off) or rtTA (Tet-On) activation (Garner et al., 2012; Gossen and Bujard, 1992; Mayford et al., 1996; Reijmers et al., 2007; Urlinger et al., 2000). Previous work showed that TRE-driven genes targeted (via a retrovirus) to the mouse TIGRE locus, situated on Chromosome 9 between the AB124611 (HIDE1) and Carml loci, could be expressed in most tissues in the presence of tTA (Zeng et al., 2008). However, targeting of TRE driven transgenic cassette to the Rosa26 locus resulted in silencing or mosaicism of the transgene expression (Tasic et al., 2012). To determine if tTA/TRE can drive high-level expression in a defined locus, we created an expression platform in the TIGRE locus by homologous recombination. To establish feasibility, we first generated a tdTomato reporter allele, whose expression depends on both activation of the TRE promoter and Cre recombinase activity (Fig. 2A). To prevent undesirable interactions with nearby chromatin, we flanked the reporter allele with two copies of the chicken β-globin HS4 insulator element (Chung et al., 1997; Gaszner and Felsenfeld, 2006) on each side. We named this new reporter line Ai62(TITL-tdT), to represent the TIGRE-Insulators-TRE promoter-LSL-tdTomato components.
Figure 2.
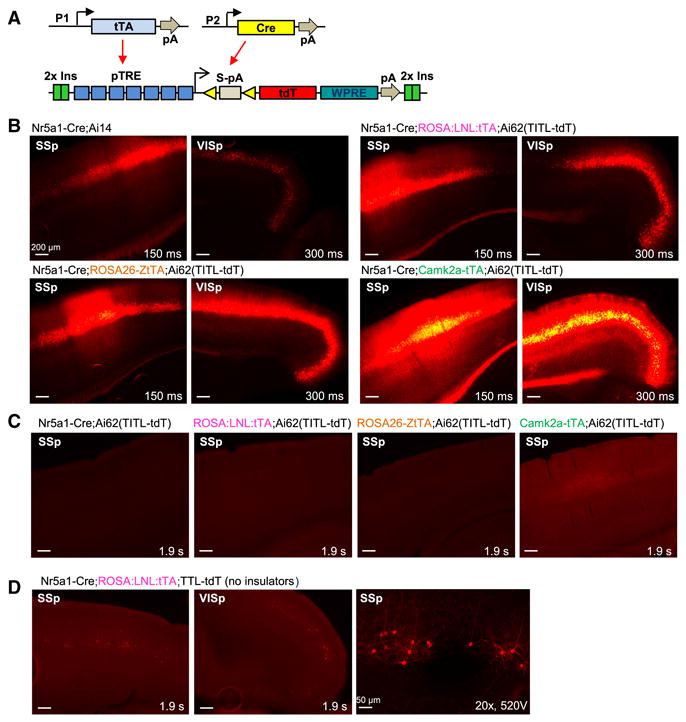
Cre and tTA dependent intersectional strategy at the TIGRE locus produces tightly regulated and high-level expression. (A) Schematic diagram of intersectional control by Cre and tTA, driven by different promoters (P1 or P2), on a double reporter line based in the TIGRE locus, Ai62(TITL-tdT). (B) Comparison of tdTomato fluorescence in 4 transgenic mouse lines carrying either Ai14 or Ai62 reporter alleles. (C) No detectable tdTomato expression in the Cre+/tTA- control Nr5a1-Cre;Ai62(TITL-tdT), or in Cre-/tTA+ controls ROSA:LNL:tTA;Ai62(TITL-tdT) and ROSA26-ZtTA;Ai62(TITL-tdT). Very weak tdTomato fluorescence was seen in the barrel cortex and hippocampus of Camk2a-tTA;Ai62(TITL-tdT) control mice (Cre-/tTA+). (D) Poor tdTomato expression in similar triple Tg mice with a TIGRE reporter (TTL-tdT) that lacks chromatin insulators (compare to B). Right, a higher-magnification confocal image of somatosensory cortex shows that the sparsely labeled cells are layer 4 neurons. For B-D, exposure time for each epifluorescence image is shown for comparison. (See also Figures S4-S6.)
To compare Ai62 with Ai14, we generated a series of double and triple Tg mice containing tdTomato, Cre and/or tTA expressing alleles and evaluated native fluorescence in brain sections (Fig. 2B). We used three different tTA lines with our TIGRE mice: Camk2a-tTA (Mayford et al., 1996), ROSA:LNL:tTA (Wang et al., 2008), and ROSA26-ZtTA (Li et al., 2010) (Table S1), to test if their varying tTA expression levels would influence the amount of reporter produced. Camk2a-tTA is restricted to forebrain excitatory neurons and striatal medium spiny neurons. The other two tTA lines are both targeted to the Rosa26 locus and contain floxed-stop cassettes, and are thus presumed to have Cre-dependent ubiquitous expression. For a common Cre line we used Nr5a1-Cre, which drives specific expression in cortical layer 4 neurons.
We observed robust tdTomato fluorescence in cortical layer 4 cells in all types of mice (Fig. 2B). However the fluorescence levels are different: lowest in Nr5a1-Cre;Ai14, followed by Nr5a1-Cre;ROSA:LNL:tTA;Ai62(TITL-tdT), then by Nr5a1-Cre;ROSA26-ZtTA;Ai62(TITL-tdT), and finally by Nr5a1-Cre;Camk2a-tTA;Ai62(TITL-tdT). Fluorescence-activated cell sorting (FACS) analysis of individual neurons isolated from cortical tissue of age-matched mice confirmed this result and revealed that tdTomato fluorescence per cell in each of the Ai62 triple Tg mice was 2-4 fold higher than that in Ai14 reporter cells (Fig. S5). We tested specificity of expression of the TIGRE reporter Ai62 by generating mice that lacked either Cre or tTA. In these mice we did not detect any fluorescence except for very weak fluorescence detected with Camk2a-tTA only. These results indicate that the Ai62 TIGRE reporter is tightly regulated (Fig. 2C). Interestingly, we found that inclusion of the chromatin insulators is essential for high-level reporter expression, as a TIGRE reporter allele that lacks these insulators is poorly expressed in similar triple Tg animals (Fig. 2D). The above comparison indicates that the TIGRE-targeted reporter is advantageous over Rosa26-CAG reporter in more robust transgene expression and the potential for additional specificity mediated through restricted tTA expression.
The new TIGRE allele differs from the Rosa26 allele in three key aspects: genomic location, use of the TRE promoter instead of the CAG promoter, and the flanking insulators. To begin to define which of these features contribute to the higher level of transgene expression apparent in Ai62 mice, we compared the expression of a pCAG-LSL-EGFP cassette that was targeted either to the TIGRE locus (with insulators) or to the Rosa26 locus (without insulators) in mouse ES cells (Fig. S6). Following transient transfection of Cre into targeted ES clones to delete the stop cassettes, FACS analysis of two independent clones with each targeted locus showed comparable levels of EGFP fluorescence in all. Thus, we conclude that the difference in expression levels between Rosa26 and TIGRE mice is not simply due to the genomic location and the insulators, rather it is more likely the result of the tTA/TRE-mediated transcriptional amplification (Iyer et al., 2001).
In addition to the transgenic reporter, we also generated AAV vectors to test the feasibility of Cre/tTA-and Cre/Flp/tTA-dependent regulation of virally-encoded intersectional drivers and reporters. All reporters demonstrated the expected specificity of expression when paired with double or triple drivers, thereby further expanding the repertoire of viral tools that can be used for highly specific and versatile genetic control (Fig. S4E-G).
New TIGRE reporter lines with strong expression of sensors and effectors
Based on the enhanced expression in Ai62, we proceeded to generate a series of TIGRE reporter lines carrying a variety of fluorescent sensors and optogenetic effectors (Table 1). This process was facilitated by our initial targeting strategy, which allows for subsequent rapid modification of the locus by RMCE (Fig. S1B). In parallel, we generated Rosa26-based reporter lines for some of the same genes, including calcium indicators GCaMP6f and GCaMP6s (Chen et al., 2013), voltage indicator VSFP-Butterfly 1.2 (VSFPB) (Akemann et al., 2012), glutamate sensor iGluSnFR (Marvin et al., 2013), and red-light optogenetic inhibitor Jaws (Chuong et al., 2014), for further direct comparison between the Rosa-CAG and TIGRE-TRE expression systems.
We evaluated native fluorescence expression by confocal microscopy using identical imaging parameters for pairs of age-matched reporter mice crossed to the same Cre lines (Fig. 3). Although the fluorescence level varies among different proteins, in all cases the same protein was more highly expressed from the TIGRE allele than from the Rosa26 allele. In addition, GCaMP6s expression in the TIGRE allele Ai94 was not only stronger than the Rosa-CAG allele Ai96 (Fig. 3B), but also stronger than the Snap25-2A-GCaMP6s line which, like Ai96, showed no basal level fluorescence (data not shown).
Figure 3.
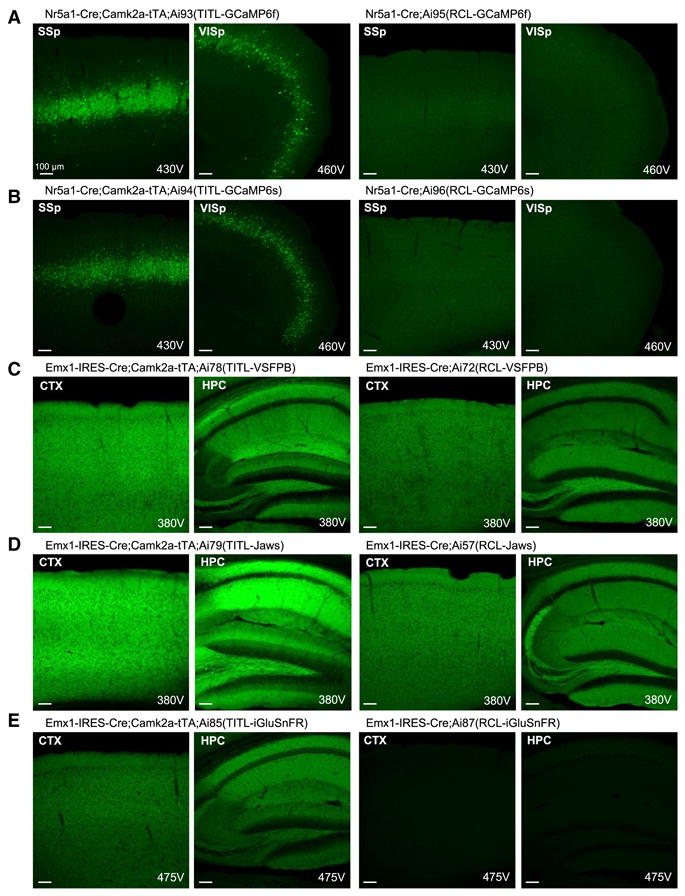
TIGRE reporter lines have higher-level transgene expression than Rosa reporter lines in multiple direct comparisons. Native fluorescence in each pair of lines was compared by confocal microscopy using identical imaging parameters. All images were taken under a 10× objective using 10% laser power, and the PMT gain (in voltage) is indicated on each image. Mouse names are shown above each set of images taken from the same mouse. Comparisons are shown for (A) GCaMP6f, (B) GCaMP6s, (C) VSFP-Butterfly 1.2, (D) Jaws-GFP-ER2, and (E) iGluSnFR. Ai57(RCL-Jaws) was created by crossing Ai57(RCFL-Jaws) with a Flp-deleter mouse, FLPeR, to delete the FSF cassette. (See also Figure S1.)
We observed strong expression of reporter transgenes when combining TIGRE alleles with a variety of Cre drivers and either Camk2a-tTA or ROSA26-ZtTA (Fig. 4). The EGFP reporter line (Ai82) exhibited superior cytoplasmic labeling (Fig. 4A) over previous Rosa-CAG reporter Ai3 (Madisen et al., 2010) and Snap25-LSL-2A-GFP (Fig. 1D). We saw strong expression and proper localization to the plasma membrane in lines expressing membrane proteins VSFPB (Ai78, Fig. 4B) and Jaws (Ai79, Fig. 4F). The calcium indicator YCX2.60 (Ai92, Fig. 4C) also displayed strong expression in both excitatory neurons and inhibitory interneurons. Likewise, cytoplasmic GCaMP6f (Ai93, Fig. 4D) and GCaMP6s (Ai94, Fig. 4E) were readily seen in cortical layer 4 and layer 2/3 excitatory neurons. In particular, long-term, sustained expression of GCaMP6f in the Ai93 brain did not result in nuclear invasion (Fig. 4G). This result contrasts the common observation associated with viral infection of GCaMP where transgene expression continues to rise and leads to unhealthy cells whose nuclei become filled with fluorescence within weeks. Thus, these stable transgenic lines may allow longer-term repeated experimentation within the same animal. Nonetheless, animals expressing these engineering proteins should still be used cautiously and monitored for possible unexpected adverse effects.
Figure 4.
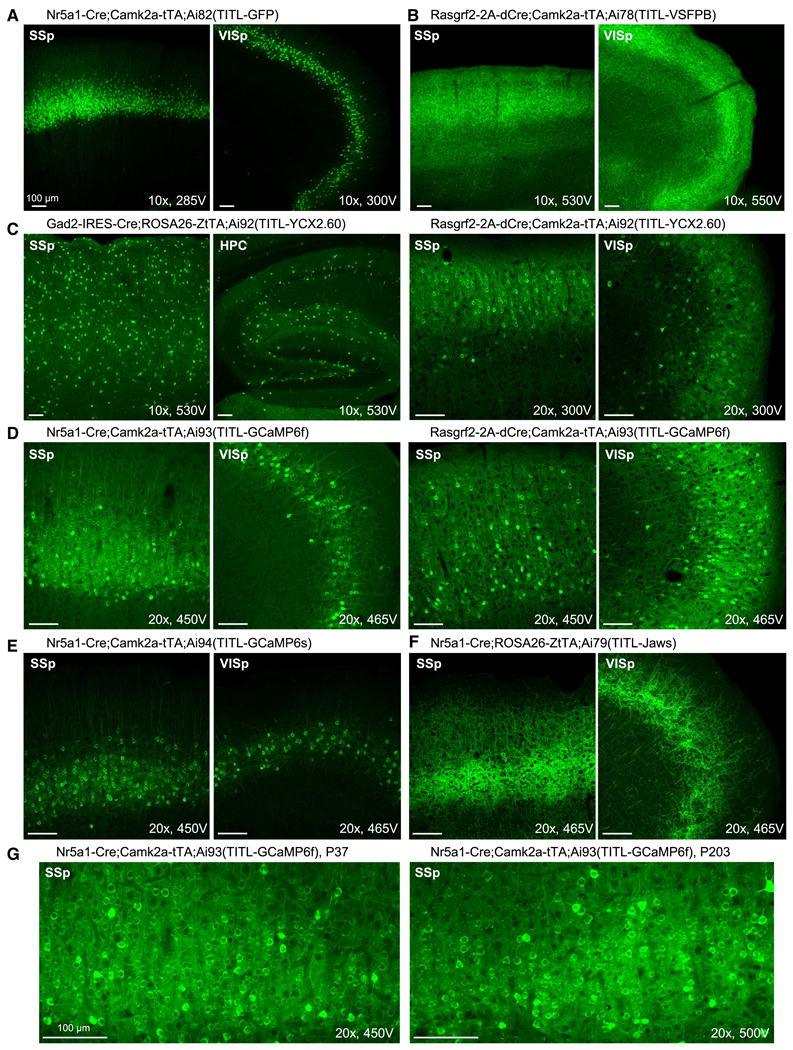
Strong transgene expression in all TIGRE-based reporter lines, driven by a variety of Cre lines combined with Camk2a-tTA or ROSA26-ZtTA. Confocal images of native fluorescence are shown. All images were taken under a 10× or 20× objective using 10% laser power, and the PMT gain (in voltage) is indicated on each image for comparison. (A) Very bright cytoplasmic GFP labeling of cortical layer 4 neurons in Nr5a1-Cre;Camk2a-tTA;Ai82(TITL-GFP) mouse. (B) Bright membrane VSFP-Butterfly 1.2 labeling of cortical layer 2/3 neurons in Rasgrf2-2A-dCre;Camk2a-tTA;Ai78(TITL-VSFPB) mouse. (C) Very bright cytoplasmic YCX2.60 labeling of interneurons in Gad2-IRES-Cre;ROSA26-ZtTA;Ai92(TITL-YCX2.60) mouse, and of cortical layer 2/3 neurons in Rasgrf2-2A-dCre;Camk2a-tTA;Ai92(TITL-YCX2.60) mouse. (D) Bright cytoplasmic GCaMP6f labeling of cortical layer 4 neurons in Nr5a1-Cre;Camk2a-tTA;Ai93(TITL-GCaMP6f) mouse, and of cortical layer 2/3 neurons in Rasgrf2-2A-dCre;Camk2a-tTA;Ai93(TITL-GCaMP6f) mouse. (E) Bright cytoplasmic GCaMP6s labeling of cortical layer 4 neurons in Nr5a1-Cre;Camk2a-tTA;Ai94(TITL-GCaMP6s) mouse. (F) Bright membrane Jaws-GFP-ER2 labeling of cortical layer 4 neurons in Nr5a1-Cre;Rosa26-ZtTA;Ai79(TITL-Jaws) mouse. (G) Comparison of young and old Ai93 mice shows no or little nuclear invasion of transgene proteins with time. The ages (postnatal days) at which the mice were sacrificed are shown. (See also Figure S7.)
To further compare gene expression patterns in Rosa26 and TIGRE reporter lines, we evaluated mRNA expression in multiple lines by in situ hybridization (ISH) (Fig. S7). All lines examined exhibited widespread and high-density expression in cortex when under the control of Emx1-IRES-Cre alone (for Rosa26 lines), or in combination with Camk2a-tTA or ROSA26-ZtTA (for TIGRE lines) (Fig. S7A). There was no significant difference in the numbers of expressing cells either between different lines or across ages. When regulated by some more specific Cre drivers, however, we found that although general expression patterns are similar between Rosa26 and TIGRE reporters, variations can occur depending on which tTA line is used for activation, with ROSA26-ZtTA tending to drive expression in fewer cells compared to Ai14, especially in subcortical regions (Fig. S7B-D). These findings indicate that the interplay between Cre, tTA and reporter alleles among different cells in different mouse lines may not always be predictable, thus reporter expression needs to be carefully assessed in new lines to ensure their applicability.
Imaging of membrane voltage in TIGRE-VSFPB mice
To evaluate the TIGRE lines in functional measurements of neural activity, we tested transgenic line Ai78, which delivers a highly sensitive, genetically encoded voltage indicator (GEVI), VSFP-Butterfly 1.2 (Akemann et al., 2012). VSFP-Butterfly 1.2 yields strong voltage signals in mouse sensory cortex, both under anesthesia and in wakefulness (Akemann et al., 2012; Carandini et al., 2015; Scott et al., 2014). It exhibits voltage-dependent Förster resonance energy transfer (FRET) between a pair of green and red fluorophores: mCitrine (donor) and mKate2 (acceptor). By imaging both fluorophores and taking the ratio of their fluorescence, one can obtain a good estimate of the underlying membrane voltage.
To study the functionality of the indicator in vitro, we imaged hippocampal slices from triple Tg mice Rasgrf2-2A-dCre;Camk2a-tTA;Ai78(TITL-VSFPB), which express the VSFP in the dentate granule cells of the hippocampus (Fig. S8A). In these slices, a single shock to the perforant path evoked robust VSFP responses imaged over the dendritic field of dentate granule cells (Fig. S8B). These responses could be observed both as increased acceptor signals and as decreased donor signals (Akemann et al., 2012). We readily resolved VSFP transients above baseline noise in single sweeps (amplitude ∼3-4× baseline noise, not shown). Multiple stimulations (5×) at 100 Hz increased the VSFP response amplitude. The increase was sub-linear, consistent with the temporal summation and well-known frequency dependent depression of the synapses from medial perforant path to dentate granule cells (Petersen et al., 2013).
To address how well the indicator functions in vivo, we imaged a large portion of cerebral cortex on the left hemisphere (Fig. 5A) of triple Tg mice Rasgrf2-2A-dCre;Camk2a-tTA;Ai78(TITL-VSFPB), where layer 2/3 pyramidal cells express the VSFP (Fig. 4B). The cortex exhibited approximately uniform expression (Fig. 5B, apparent variations in brightness are due largely to inhomogeneous illumination; see also Fig. S8C). This wide pattern of expression allowed us to image distinct sensory regions responsive to somatosensory, visual, or auditory stimuli in head-fixed, awake mice (Fig. 5C). Showing visual stimuli in different horizontal and vertical positions (Fig. 5D, F) allowed us to obtain maps of retinotopy covering multiple visual areas (Fig. 5E, G), including V1, LM, and AL (Wang and Burkhalter, 2007). Presenting tones of different frequencies (Fig. 5H) yielded maps of tonotopy in at least two auditory areas (A1 and AAF) (Hackett et al., 2011) (Fig. 5I).
Figure 5.
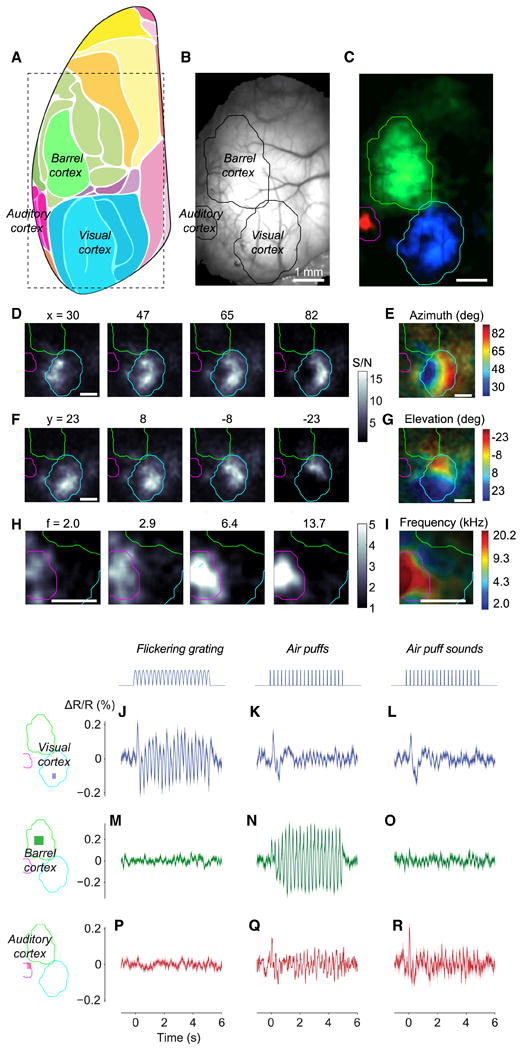
Wide-field imaging of sensory cortices of Ai78 mice expressing VSFP-Butterfly 1.2 in cortical layer 2/3 excitatory neurons. (A) Diagram showing mouse cortical regions observed at an angle of 30° laterally with a vertical optical axis. Adopted and modified from (Kirkcaldie, 2012). The rectangle shows the approximate extent of our imaging area. (B) Fluorescence of mCitrine imaged through the thinned skull. (C) Maps of VSFP signals (acceptor-donor ratio) to auditory (red), somatosensory (green), and visual (blue) stimuli. Sensory regions are mapped as 4 Hz amplitude in response to 4 Hz train of tones, 4 Hz train of air puffs directed to whole whisker field, and 2 Hz flickering visual stimulus. Response amplitude was divided for each modality by amplitude measured in the absence of stimulation. The three maps came from experiments performed on different days, and the resulting maps were aligned based on the blood vessel pattern. Overlaid contour lines show the outlines of visual cortex, barrel cortex, and auditory cortex. (D) Amplitude maps for 4 Hz responses to bars reversing in contrast at 2 Hz, presented at different horizontal positions (azimuths). (E) The resulting maps of azimuth preference (retinotopy). (F-G) Same as D and E for stimulus elevation (vertical position). (H) Amplitude maps for 6 Hz responses to tones in 6 Hz trains, for different tone frequencies. (I) The resulting maps of tone frequency preference (tonotopy). (J-R) Unisensory and multisensory signals in visual cortex (J-L), barrel cortex (M-O) and auditory cortex (P-R). Stimuli were contrast-reversing visual gratings (J, M, P), air puffs delivered to the whiskers (K, N, Q), and (sham) air puffs delivered away from the whiskers to replicate the sound but not the somatosensory stimulation (L, O, R). ΔR/R is calculated after normalization using data during the prestimulus period, and high-pass filtering above 0.5 Hz. (See also Figure S8.)
To measure signal/noise (S/N) ratios of voltage signals, we drove specific regions of sensory cortex with a periodic stimulus, making their activity oscillate. We then imaged this oscillating activity, and divided the amplitude of the oscillation observed during stimulation with that observed in the absence of stimuli (Benucci et al., 2007). In response to optimally placed visual stimuli, visual cortex gave S/N ratios of 11.8 in the example mouse (Fig. 5J), and similar values in five other mice (S/N = 12.1 ± 2.7 SEM). S/N ratios in these triple Tg mice were not statistically different (p = 0.05, Mann-Whitney test) from those seen in mice that expressed VSFP-Butterfly 1.2 following successful in utero electroporation (S/N = 5.54 ± 0.44, n = 3 mice) (Carandini et al., 2015).
Visual cortex responded faithfully to the visual stimuli, following the 4 Hz neural oscillations caused by reversals in visual contrast (Fig. 5J). Visual cortex, however, also responded to the onset of air puffs, whether they were delivered towards the whiskers (Fig. 5K) or away from them (Fig. 5L), indicating that these responses were due to the sound of the air puffs, and specifically by the first puff in the train. Such cross-modal interactions have been described previously (Iurilli et al., 2012), and in our data may be compounded by occasional blinks. In contrast, somatosensory cortex responded only to the air puffs delivered to the whiskers (Fig. 5N), and not to visual stimuli or to the sounds of the air puffs (Fig. 5M, O). Finally, auditory cortex responded approximately equally to air puffs, whether they were delivered towards or away from the whiskers (Fig. 5Q, R), and did not respond to visual stimuli (Fig. 5P). These data indicate that VSFPB expressed by the Ai78 reporter is sufficiently sensitive to reliably and specifically record the membrane potential of neuronal populations, with fine temporal resolution.
In vivo recording of calcium signals in GCaMP6f and YCX2.60 mice
GCaMP6 variants constitute the latest generation of genetically encoded calcium indicators (GECIs) (Chen et al., 2013) with fundamentally improved sensitivity and performance over previous generations, e.g. GCaMP3 and GCaMP5. We conducted in vivo two-photon imaging (Andermann et al., 2011; Bonin et al., 2011) through a cranial window over visual cortex in head-fixed, running GCaMP6 reporter mice, and were able to simultaneously image hundreds of neurons relatively deep in the cortex (∼300 μm below the pia surface, corresponding to visual cortical layer 4). Stimuli consisted of drifting sinusoidal gratings of 5 spatial frequencies (SF), 5 temporal frequencies (TF), and 8 orientations (0° to 315° in 45° steps). In Scnn1a-Tg3-Cre;Camk2a-tTA;Ai93(TITL-GCaMP6f) triple Tg mice (Fig. 6A-C), GCaMP6f-labeled layer 4 neurons exhibited low baseline fluorescence in vivo (Fig. 6A), while visual stimuli evoked responses with ΔF/F reaching 500% or higher (Fig. 6A-C). Visual responses selective for various orientations, SFs and TFs can be seen in different neurons (an example shown in Fig. 6C).
Figure 6.
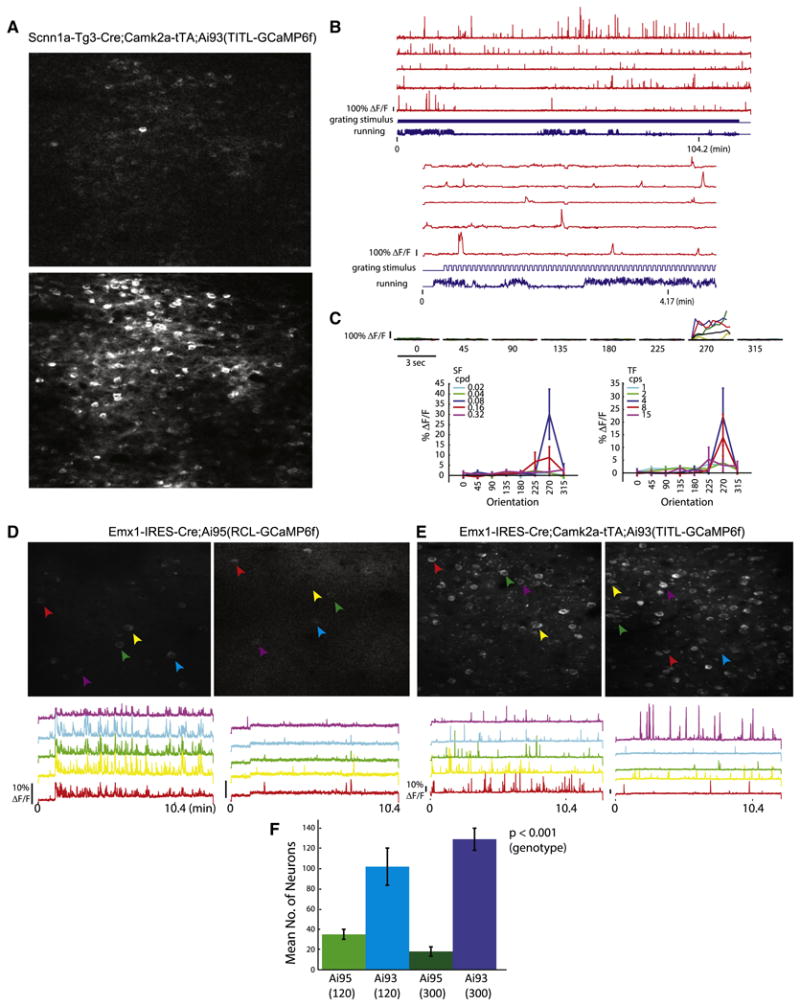
In vivo two-photon imaging of calcium signals in GCaMP6f reporter mice. (A-C) Calcium imaging in cortical layer 4 of Scnn1a-Tg3-Cre;Camk2a-tTA;Ai93(TITL-GCaMP6f) mice. (A) Images of baseline fluorescence with a single active cell (upper panel) and Z projection (time series) of the same field of view showing all active cells (lower panel). (B) Raw traces of 5 example neurons imaged during stimulus presentation. Upper panel shows the duration of the entire experiment. Lower panel shows the same 5 cells over a shorter time scale. The bottom cell trace is the cell analyzed for tuning properties shown in C. (C) Visually evoked responses of an example cell. Top, Peri-Stimulus-Time-Histogram (PSTH) of the cell's response at each stimulus orientation at optimal SF and TF. Colored lines represent individual trials and the black line represents the mean. Bottom left, mean response at each SF (averaged over all TFs) as a function of orientation. Bottom right, mean response at each TF (averaged over all SFs) as a function of orientation. (D-F) Comparison of Emx1-IRES-Cre;Ai95(RCL-GCaMP6f) and Emx1-IRES-Cre;Camk2a-tTA;Ai93(TITL-GCaMP6f) mice at two imaging depths, 120 μm and 300 μm below the pia, corresponding to cortical layers 2/3 and 4 respectively. (D) Z projection of 2-photon acquisition frames 120 μm (left) and 300 μm (right) below the pia of Ai95 mice. Raw traces of neural activity during ∼10 minutes of visual stimulus presentation for 5 representative cells are shown below each image panel. (E) Same as D for Ai93 mice. (F) Mean number of cells from which activity could be observed during a 10-min imaging period at 120 μm and 300 μm depths within a 250 × 150 μm imaging area. All values represent mean ± SEM. (See also Movies S1-S4).
Because of the low GCaMP6f fluorescence from the Rosa-CAG based Ai95 line (Fig. 3A), we investigated whether Ai95 would still allow effective calcium imaging in comparison with Ai93. We measured neural activity from Ai95 and Ai93 under control of the pan-cortical driver Emx1-IRES-Cre, i.e. in Emx1-IRES-Cre;Ai95(RCL-GCaMP6f) (Fig. 6D) and Emx1-IRES-Cre;Camk2a-tTA;Ai93(TITL-GCaMP6f) (Fig. 6E) mice. In both types of mice, robust fluorescent signal changes indicating neuronal activities were observed, however, signals were stronger in Ai93 than in Ai95 mice (compare bottom panels in Fig. 6D and 6E). Additionally, the number of active neurons observable was significantly lower in Ai95 mice compared to Ai93 mice, especially deeper into the tissue (Fig. 6F and Movies S1-S4). Two-way ANOVA revealed a significant difference between genotypes, but not by depths and with no interaction (cells at 120-μm depth: Ai95 35 ± 5, Ai93 102 ± 18; cells at 300-μm depth: Ai95 18 ± 5, Ai93 129 ± 11, n = 3 mice each, p = 4.6 × 10-5 between genotypes). In addition, activity-related signal flashes can also be easily seen in processes corresponding to dendrites and axons in Ai93 mice (Movies S3-S4). These results indicate that GCaMP6f expressed from both Ai95 and Ai93 mice yields signals responding to neuronal activities, but that due to its higher expression level Ai93 may be more capable of detecting weaker neuronal activities.
Ratiometric imaging using FRET-based calcium indicators, such as yellow cameleon (YC) indicators (Horikawa et al., 2010; Nagai et al., 2004; Yamada et al., 2011), is another approach that has the advantage of reduced sensitivity to motion artifacts and essentially permits quantification of calcium concentration levels. YCX2.60, a new version of YC indicator with an expanded dynamic range (Fig. 7A), also displayed strong expression in both excitatory neurons and inhibitory interneurons (Ai92, Fig. 4C). We performed two-photon calcium imaging in layer 2/3 of somatosensory cortex in Rasgrf2-2A-dCre;Camk2a-tTA;Ai92(TITL-YCX2.60) mice, and observed spontaneous and sensory-evoked neuronal activity with high sensitivity (Fig. 7B-D), confirming the functionality of the Ai92 mice.
Figure 7.
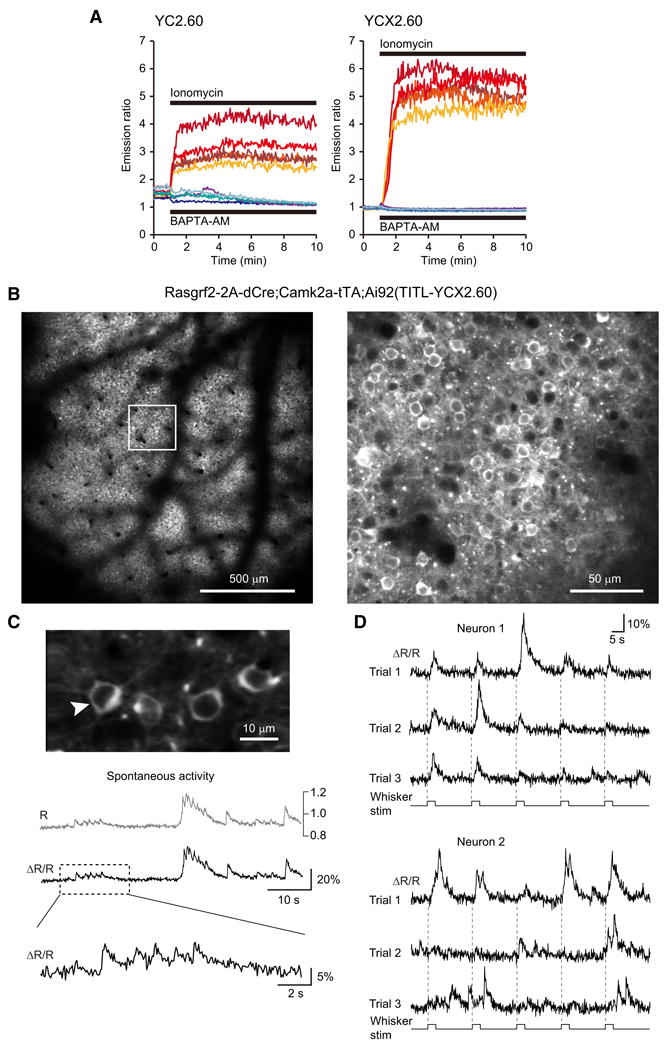
Calcium measurements with yellow cameleon YCX2.60. (A) Responses in individual HeLa cells to ionomycin applied to saturate yellow cameleons (YCs) with calcium (n = 5), and to BAPTA-AM applied to deplete YCs of calcium (n = 4). The dynamic range of YCX2.60 is twice as large as that of YC2.60 (Rmax/Rmin = 6.20 vs 3.17). The apparent dissociation constants of YC2.60 and YCX2.60 for calcium were calculated as 80 nM and 220 nM, respectively (Y.N. and A.M., unpublished results). (B-D) In vivo two-photon imaging of calcium signals in Rasgrf2-2A-dCre;Camk2a-tTA;Ai92(TITL-YCX2.60) mice. (B) Visualization of the uniform expression in layer 2/3 at 190-μm depth within the cranial window one week after TMP induction (field of view size 1.7 × 1.7 mm). Note the shadows from surface blood vessels. The image on the right is a magnified view of the boxed area in the left image, showing neuronal somata labeled with YCX2.60. (C) Representative 60-s example of spontaneous activity in the neuron marked with an arrow. Raw, unfiltered calcium transients are expressed once as YFP:CFP ratio R, which in principle can be calibrated in terms of absolute calcium concentration, or as relative percentage change of the ratio R (ΔR/R). The expanded view of the trace segment in the box highlights fast calcium transients presumably evoked by few or single action potentials. (D) Evoked activity in two example neurons following whisker stimulation. The principal whisker was repeatedly stimulated at 10 Hz for 2 seconds (onsets indicated with dashed lines). YCX2.60 AR/R traces are shown for two example layer 2/3 neuron for three trials, each comprising five stimulation periods. Note responses at stimulus onset, during stimulation, and at stimulus offset as well as spontaneous activity in between. Large calcium transients likely correspond to bursts of action potentials whereas small-amplitude transients may reflect occurrence of only few or single action potentials.
Red light-mediated optogenetic inhibition in Jaws mice
Jaws is the first red light drivable optogenetic inhibitor, engineered from H. salinarum (strain Shark) cruxhalorhodopin (Chuong et al., 2014). Compared with other photo-activated inhibitors such as eNpHR3.0, Arch or ArchT, Jaws responds robustly to red light with high sensitivity, large photocurrent and fast kinetics, and thus enables better tissue penetrance and more effective optogenetic silencing of large volumes (Chuong et al., 2014). We compared the performance of a modified trafficking variant of Jaws, Jaws-GFP-ER2, expressed from the Rosa-CAG allele, i.e. Emx1-IRES-Cre;Ai57(RCL-Jaws) mice (simplified as Ai57), and the TIGRE allele, i.e. Emx1-IRES-Cre;Camk2a-tTA;Ai79(TITL-Jaws) mice (simplified as Ai79), via slice and in vivo electrophysiology, using virally expressed Jaws and Arch-expressing mice Ai35 as references (Chuong et al., 2014; Madisen et al., 2012) (Fig. 8).
Figure 8.
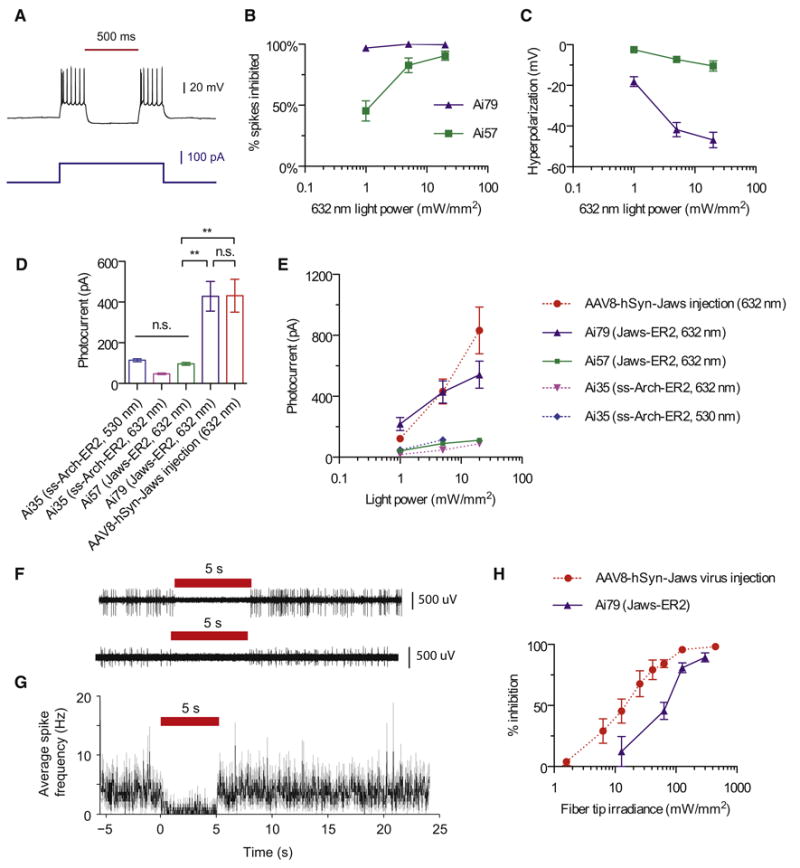
Optogenetic inhibition of neural activity in reporter lines expressing Jaws. (A) Representative current-clamp recording of a Jaws-expressing neuron from Ai79 undergoing optically evoked (632 nm, 5 mW/mm2) hyperpolarization in an acute cortical slice. (B) Comparison of red light induced inhibition of electrically evoked spiking in slices from Ai79 and Ai57 mice. Spiking was induced by a current injection of 1.5× the rheobase. (C) Comparison of red light induced hyperpolarization in slices from Ai79 and Ai57 mice. (D) Comparison of red or green light (632 or 530 nm, 5 mW/mm2) induced photocurrents in slices from Ai79, Ai57, Ai35 and AAV-Jaws virus injected mice. (E) Comparison of light induced photocurrents in slices from Ai79, Ai57, Ai35 and AAV-Jaws virus injected mice across different light intensities for red or green light. (F-G) Representative extracellular recordings (F) in awake Ai79 mice demonstrate the in vivo inhibition of spontaneous firing activities (G) of Jaws-expressing neurons. (H) Comparison of in vivo inhibition of spontaneous firing activities over a range of red light intensities between Ai79 and AAV-Jaws injected mice. All values represent mean ± SEM. ** p < 0.01, n.s. not significant. (See also Figure S9.)
In brain slices, Jaws-expressing cells showed normal membrane properties (n = 13 cells in Ai57 mice, n = 17 in Ai79, n = 12 in Ai35, Fig. S9A). Red light illumination (632 nm) induced strong hyperpolarizing currents in Jaws-expressing neurons in Ai79 mice and effectively blocked depolarization-induced spiking, whereas the light effect was small to moderate in Ai57 mice (Fig. 8A-C). Photocurrents were comparable between Ai79 and virally expressed Jaws at all irradiance levels tested, but were significantly smaller in Ai57 cells (Fig. 8D, E), consistent with the observation of stronger native fluorescence in Ai79 (Fig. 3D). We also noticed that the Jaws-GFP-ER2 in Ai57 displayed lower protein fluorescence than the Arch-GFP-ER2 in the similar Rosa-CAG allele Ai35 (Fig. S9C), possibly due to different protein properties (see Discussion below), but the low photocurrents generated were similar between the two (Fig. 8D, E and S9B).
Next we assessed Jaws performance in vivo. We conducted extracellular recordings in the primary visual cortex of awake, head-fixed mice using glass pipettes and with a 200-μm optical fiber in place (Chuong et al., 2014). Due to the low photocurrents of Ai35 and Ai57 in slices, only Ai79 was tested in vivo. Ai79 showed strong red light (637 nm) induced suppression of spontaneous neuronal firing (Fig. 8F, G; n = 23 units from 2 mice, n = 3 units showed no change). Light delivery through the fiber tip inhibited neuronal firing in a light intensity-dependent manner in these mice, comparable to the performance of virally-expressed Jaws (Fig. 8H).
Discussion
We sought to develop transgenic strategies for intersectional control and to compare performance of these strategies under native conditions in the mouse brain. In addition to striving for high-level and highly specific expression of genetic tools, we aimed to generate standardized expression platforms, in a few defined genomic loci, that would allow rapid modification for future improvements and applications. Such efforts are more laborious and time-consuming than other in vivo approaches, such as random transgenesis or viral infection, but they can provide more definitive evaluations and more consistent platforms for incorporating additional genetic tools and expansion into other cell types.
For example, we observed significant, heritable variation in protein expression among the various sensors and effectors (Figs. 3, 4), despite their having the same configuration within the TIGRE or Rosa26 alleles. We think this variation is most likely due to intrinsic protein properties, as they originate from diverse non-mammalian species. Protein engineering, such as codon optimization and addition of membrane-targeting tags (Gradinaru et al., 2010), is essential for optimizing expression in the mammalian brain. However, since many of the proteins we worked with had already undergone such engineering, the observed variation suggests that intrinsic differences still exist. Therefore expression in mice as a stably-integrated transgene is unpredictable and always needs to be examined experimentally in vivo. Our data indicate that while the Rosa-CAG approach could work well for some proteins with exceptional expressibility and/or functionality, e.g. in Ai14-tdTomato, Ai32-ChR2(H134R) (Madisen et al., 2012) as well as Ai95-GCaMP6f mice, our new TIGRE approach, with its much enhanced gene expression capability via tTA-assisted transcriptional amplification, will allow a wider spectrum of genetic tools to achieve functional levels and/or to perform better in stable transgenic settings. This amplification proves to be significant even in comparison with expression from transgenes targeting one of the brain's most highly expressed endogenous genes, Snap25.
On the other hand, in our effort to create more cell type-specific tTA driver lines, we found that several knock-in tTA driver lines we made exhibit no or low-level tTA-induced reporter expression (Table S2). This may be because the Tet-regulatory system is prone to epigenetic silencing (Tasic et al., 2012; Zhu et al., 2007). Further investigation and optimization will be needed to expand the repertoire of tTA drivers to take full advantage of the potential of TIGRE-based reporters. Nevertheless, our work validates the currently available Camk2a-tTA and ROSA26-ZtTA lines as effective components of a TIGRE-based approach in driving robust transgene expression.
We have established the TIGRE locus as a new permissive docking site for insertion of exogenous promoters and transgenes. This is significant since TRE-driven transgene cassettes are often silenced when inserted into the mouse genome randomly or even targeted to specific loci such as Rosa26 (B.T., L.M. and H.Z., unpublished results; see also Li et al., 2010; Tasic et al., 2012; Zhu et al., 2007). Our expression studies also showed that both TRE and CAG promoters worked well in TIGRE, suggesting that this locus can also serve as a docking site for insertion of other promoters and regulatory sequences. This could be particularly useful, for example, for the screening of enhancer elements or promoter variants. To facilitate repeated use of these expression platforms, we have built a cassette exchange mechanism into each, based on reconstitution of a split-hygromycin selectable marker (Fig. S1). By doing so, the swapping of either alternative drivers in targeted lines or new genetic tools in the TIGRE allele becomes a rapid and straightforward process, thereby extending the utility of these systems.
Besides the intersectional strategies already shown here, we also tested additional recombinases B3 and KD and the Gal4/UAS transcriptional activation system, but obtained undesirable outcomes (see Table S2). However, there are still other approaches with unique merits that we have not tested, such as split-Cre (Casanova et al., 2003; Hirrlinger et al., 2009; Wang et al., 2012) or the creation of Flp, Dre or tTA-dependent Cre driver lines, which could be applied to the many existing floxed alleles. Developing these alternative intersectional approaches will further expand our capability in cell type specific control.
The validated transgenic strategies and new transgenic lines reported here will likely benefit a variety of applications. More Flpo, Dre and tTA driver lines can be generated that, in conjunction with Cre driver lines, enable the targeting to highly specific cell types and populations. The dual-driver approaches allow for not only an “A and B” type of intersection, but also for other types of simultaneous differential control, such as “A or B” or “A not B” (Fenno et al., 2014; Huang and Zeng, 2013), when appropriate double reporter mice or viruses are developed. Even the low-level promiscuous recombination of Cre on Rox sites can be used to create extremely sparse labeling of individual cells by combining Rox-stop-Rox reporters with Cre drivers. An issue with using intersectional transgenic strategies is the laborious breeding involved in generating triple Tg mice. We found that both Rosa26 and TIGRE homozygous Tg mice are fertile, and because all these are knock-in alleles, PCR-genotyping methods can be designed to distinguish wild-type, heterozygous and homozygous alleles. So using homozygous alleles and/or double Tg × double Tg breeding schemes will increase breeding efficiency.
The collection of reporter lines should facilitate various ways of observing and manipulating cell type functions, having advantage over viral expressions in large-area, uniform, and long-term monitoring and manipulation of neuronal activities. In particular, stable transgenic expression of GCaMP6f and GCaMP6s (Ai93-96 and Snap25-2A-GCaMP6s), YCX2.60 (Ai92), VSFPB (Ai78), and iGluSnFR (Ai85, Tim Murphy and H.Z., unpublished results) provides various options of imaging neuronal activity with increased sensitivity, complementing other recently reported transgenic lines PC::G5-tdT (Gee et al., 2014) and Thy1-GCaMP6 (Dana et al., 2014). These examples indicate that the TIGRE platform is suitable for expressing existing or new genetic tools, and thus expanding the arsenal of genetic approaches to further our understanding of diverse biological systems.
Experimental Procedures
All experimental procedures related to the use of mice were conducted according to NIH guidelines, the UK Animals Scientific Procedures Act (1986), the guidelines of the Veterinary Office of Switzerland, and the New Zealand Animal Welfare Act (1999). Experiments performed at the Allen Institute were approved by the Institutional Animal Care and Use Committee (IACUC) of the Allen Institute for Brain Science. Experiments performed at University College London were under personal and project licenses released by the Home Office following appropriate ethics review. Experiments performed at the University of Zurich were approved by the Cantonal Veterinary Office in Zurich. Additional details of experimental procedures are available in Supplemental Experimental Procedures online.
Transgenic mice generation and expression characterization
Transgenic mice were generated by inserting transgene cassettes into endogenous genomic loci via homologous recombination as previously described (Madisen et al., 2010) and, in many cases, subsequent Flp-mediated RMCE. Expression of the reporter genes was assessed by epifluorescence or laser-scanning confocal imaging of native fluorescence (without antibody staining), by FACS of single cell suspensions, or by ISH. All ISH data can be found at the Transgenic Characterization database (http://connectivity.brain-map.org/transgenic/search/basic).
In vitro and in vivo wide-field voltage imaging
Hippocampal slice recording was done at room temperature. The dentate gyrus was imaged with a cooled CCD camera. In vivo imaging was done through thinned skull of head fixed animal allowed to move freely on a spherical treadmill. Stimuli were trains of visual, somatosensory, or auditory stimuli, delivered to elicit periodic responses in cortex.
In vivo two-photon calcium imaging
In visual cortex imaging experiments, the animal was allowed to move freely on a rotatable disc while head fixed. Visual stimuli consisted of drifting sinusoidal gratings with varying spatial frequencies, temporal frequencies, and orientations. In somatosensory cortex imaging experiments, mice were anesthetized and neuronal responses were measured in layer 2/3 upon mechanical stimulation of the identified principal whisker. Image data were acquired using custom-built two-photon microscopes with resonant and galvo scanners.
In vitro and in vivo electrophysiology of optogenetic silencing
In vivo extracellular recordings were conducted in the visual cortex of awake, head-fixed mice using a glass microelectrode attached with a 200-μm diameter optical fiber that is coupled to a 637-nm laser. Whole-cell patch-clamp recording in slice was carried out at 32°C with added Picrotoxin and kynurenic acid in ACSF to block GABAergic and glutamatergic synaptic transmission. Light pulses were delivered through the objective lens using a 530-nm LED or a 625-nm LED plus a 632±11 nm filter.
Supplementary Material
Acknowledgments
We are grateful to the Research and Development, Structured Science and Technology teams at the Allen Institute for their technical support in stereotaxic injections, mouse colony management, and ISH gene expression characterization. We thank Robert Hunter for coordinating transgenic mice production and Charu Reddy and Wolfgang Omlor for technical support. We thank Douglas Kim for providing the GCaMP6f and GCaMP6s constructs, Loren Looger for providing the iGluSnFR construct, Gary Felsenfeld for providing the chicken β-globin HS4 insulator element construct, Philippe Soriano for providing the FLPo construct via Addgene, and Anton Maximov for providing the DHFR-Cre construct. This work was funded by the Allen Institute for Brain Science, NIH grant MH085500 to H.Z. and A.N., NIH grant DA028298 to H.Z., and Wellcome Trust grant 095669 to M.C. M.C. holds the GlaxoSmithKline / Fight for Sight Chair in Visual Neuroscience. The authors wish to thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support.
Footnotes
Author Contributions: L.M. and H.Z. designed the transgenic strategies. L.M., H.G. and M.M. generated all new transgenic mouse lines except Nr4a2-SA-IRES-Dre, and some of the AAVs. C.M. and A.N. generated the Nr4a2-SA-IRES-Dre mouse line. D.S., A.B., T.K. and M.C. conducted in vivo wide-field voltage imaging on Ai78 mice. R.M.E. conducted slice physiology on Ai78 mice. A.R.G., A.C. and R.C.R. conducted in vivo 2-photon calcium imaging on Ai93 and Ai95 mice. A.v.B., L.E. and F.H. conducted in vivo 2-photon calcium imaging on Ai92 mice. N.C.K., A.S.C., L.L. and E.S.B. conducted optogenetic silencing studies on Ai35, Ai57 and Ai79 mice. Y.N. and A.M. developed YCX2.60. B.T. and T.N.N. conducted FACS analysis and made some of the AAVs. S.M.S. provided project management for this work. L.M., A.R.G., D.S., A.S.C., L.L., R.M.E., T.K., M.C. and H.Z. were main contributors to data analysis and manuscript writing, with input from other co-authors.
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackman JB, Burbridge TJ, Crair MC. Retinal waves coordinate patterned activity throughout the developing visual system. Nature. 2012;490:219–225. doi: 10.1038/nature11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akemann W, Mutoh H, Perron A, Park YK, Iwamoto Y, Knopfel T. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Models Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- Andermann ML, Kerlin AM, Roumis DK, Glickfeld LL, Reid RC. Functional specialization of mouse higher visual cortical areas. Neuron. 2011;72:1025–1039. doi: 10.1016/j.neuron.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benucci A, Frazor RA, Carandini M. Standing waves and traveling waves distinguish two circuits in visual cortex. Neuron. 2007;55:103–117. doi: 10.1016/j.neuron.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin V, Histed MH, Yurgenson S, Reid RC. Local diversity and fine-scale organization of receptive fields in mouse visual cortex. J Neurosci. 2011;31:18506–18521. doi: 10.1523/JNEUROSCI.2974-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Carandini M, Shimaoka D, Rossi LF, Sato TK, Benucci A, Knopfel T. Imaging the awake visual cortex with a genetically encoded voltage indicator. J Neurosci. 2015;35:53–63. doi: 10.1523/JNEUROSCI.0594-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova E, Lemberger T, Fehsenfeld S, Mantamadiotis T, Schutz G. Alpha complementation in the Cre recombinase enzyme. Genesis. 2003;37:25–29. doi: 10.1002/gene.10227. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc Natl Acad Sci USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong AS, Miri ML, Busskamp V, Matthews GA, Acker LC, Sorensen AT, Young A, Klapoetke NC, Henninger MA, Kodandaramaiah SB, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–1129. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Chen TW, Hu A, Shields BC, Guo C, Looger LL, Kim DS, Svoboda K. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS ONE. 2014;9:e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Kim JC. Molecular neuroanatomy's “Three Gs”: a primer. Neuron. 2007;54:17–34. doi: 10.1016/j.neuron.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gee JM, Smith NA, Fernandez FR, Economo MN, Brunert D, Rothermel M, Morris SC, Talbot A, Palumbos S, Ichida JM, et al. Imaging activity in neurons and glia with a Polr2a-based and cre-dependent GCaMP5G-IRES-tdTomato reporter mouse. Neuron. 2014;83:1058–1072. doi: 10.1016/j.neuron.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett TA, Barkat TR, O'Brien BM, Hensch TK, Polley DB. Linking topography to tonotopy in the mouse auditory thalamocortical circuit. J Neurosci. 2011;31:2983–2995. doi: 10.1523/JNEUROSCI.5333-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R, Lanjuin A, Madisen L, Zeng H, Murthy VN, Uchida N. Olfactory cortical neurons read out a relative time code in the olfactory bulb. Nat Neurosci. 2013;16:949–957. doi: 10.1038/nn.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS ONE. 2009;4:e4286. doi: 10.1371/journal.pone.0004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K, Yamada Y, Matsuda T, Kobayashi K, Hashimoto M, Matsu-ura T, Miyawaki A, Michikawa T, Mikoshiba K, Nagai T. Spontaneous network activity visualized by ultrasensitive Ca(2+) indicators, yellow Cameleon-Nano. Nat Methods. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- Issa JB, Haeffele BD, Agarwal A, Bergles DE, Young ED, Yue DT. Multiscale optical ca(2+) imaging of tonal organization in mouse auditory cortex. Neuron. 2014;83:944–959. doi: 10.1016/j.neuron.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurilli G, Ghezzi D, Olcese U, Lassi G, Nazzaro C, Tonini R, Tucci V, Benfenati F, Medini P. Sound-driven synaptic inhibition in primary visual cortex. Neuron. 2012;73:814–828. doi: 10.1016/j.neuron.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir SS. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci USA. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman SL, Beneduce BM, Drew IR, Regehr WG. Achieving high-frequency optical control of synaptic transmission. J Neurosci. 2014;34:7704–7714. doi: 10.1523/JNEUROSCI.4694-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, Zeng H, Fenton AA, Hen R. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron. 2013;77:955–968. doi: 10.1016/j.neuron.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkcaldie MTK. Neocortex. In: Watson C, Paxinos G, Puelles L, editors. The Mouse Nervous System. San Diego, CA: Academic Press; 2012. [Google Scholar]
- Knopfel T. Genetically encoded optical indicators for the analysis of neuronal circuits. Nat Rev Neurosci. 2012;13:687–700. doi: 10.1038/nrn3293. [DOI] [PubMed] [Google Scholar]
- Kranz A, Fu J, Duerschke K, Weidlich S, Naumann R, Stewart AF, Anastassiadis K. An improved Flp deleter mouse in C57B1/6 based on Flpo recombinase. Genesis. 2010;48:512–520. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- Lee SH, Marchionni I, Bezaire M, Varga C, Danielson N, Lovett-Barron M, Losonczy A, Soltesz I. Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron. 2014;82:1129–1144. doi: 10.1016/j.neuron.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tasic B, Micheva KD, Ivanov VM, Spletter ML, Smith SJ, Luo L. Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS ONE. 2010;5:e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ, 3rd, Gu X, Zanella S, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods. 2013;10:162–170. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Vu TD, Kimpo RR, Rinaldi JM, Kohli A, Zeng H, Deisseroth K, Raymond JL. Cerebellar Purkinje cell activity drives motor learning. Nat Neurosci. 2013;16:1734–1736. doi: 10.1038/nn.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RP, Moradpour F, Eadie BD, Shin JD, Kannangara TS, Delaney KR, Christie BR. Electrophysiological identification of medial and lateral perforant path inputs to the dentate gyrus. Neuroscience. 2013;252:154–168. doi: 10.1016/j.neuroscience.2013.07.063. [DOI] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RS, Corcoran AE, Brust RD, Kim JC, Richerson GB, Nattie E, Dymecki SM. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333:637–642. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- Robertson SD, Plummer NW, de Marchena J, Jensen P. Developmental origins of central norepinephrine neuron diversity. Nat Neurosci. 2013;16:1016–1023. doi: 10.1038/nn.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G, Fagerholm ED, Mutoh H, Leech R, Sharp DJ, Shew WL, Knopfel T. Voltage imaging of waking mouse cortex reveals emergence of critical neuronal dynamics. J Neurosci. 2014;34:16611–16620. doi: 10.1523/JNEUROSCI.3474-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Miyamichi K, Hippenmeyer S, Dani VS, Zeng H, Joo W, Zong H, Chen-Tsai Y, Luo L. Extensions of MADM (mosaic analysis with double markers) in mice. PLoS ONE. 2012;7:e33332. doi: 10.1371/journal.pone.0033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, Roos RP. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis. 2008;29:400–408. doi: 10.1016/j.nbd.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen T, Sakurai K, Han BX, He Z, Feng G, Wang F. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci Rep. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Burkhalter A. Area map of mouse visual cortex. J Comp Neurol. 2007;502:339–357. doi: 10.1002/cne.21286. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Michikawa T, Hashimoto M, Horikawa K, Nagai T, Miyawaki A, Hausser M, Mikoshiba K. Quantitative comparison of genetically encoded Ca indicators in cortical pyramidal cells and cerebellar Purkinje cells. Front Cell Neurosci. 2011;5:18. doi: 10.3389/fncel.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala HA, Borghuis BG, Hoogland TM, Madisen L, Tian L, De Zeeuw CI, Zeng H, Looger LL, Svoboda K, Chen TW. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci. 2012;32:3131–3141. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Horie K, Madisen L, Pavlova MN, Gragerova G, Rohde AD, Schimpf BA, Liang Y, Ojala E, Kramer F, et al. An inducible and reversible mouse genetic rescue system. PLoS Genet. 2008;4:e1000069. doi: 10.1371/journal.pgen.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Madisen L. Mouse transgenic approaches in optogenetics. Prog Brain Res. 2012;196:193–213. doi: 10.1016/B978-0-444-59426-6.00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, Deisseroth K, Augustine GJ, Feng G. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36:141–154. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Ting JT, Atallah HE, Qiu L, Tan J, Gloss B, Augustine GJ, Deisseroth K, Luo M, Graybiel AM, et al. Cell type-specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nat Methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Aller MI, Baron U, Cambridge S, Bausen M, Herb J, Sawinski J, Cetin A, Osten P, Nelson ML, et al. Silencing and un-silencing of tetracycline-controlled genes in neurons. PLoS ONE. 2007;2:e533. doi: 10.1371/journal.pone.0000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


