Abstract
Prediction of human pharmacokinetics (PK) can be challenging for monoclonal antibodies (mAbs) exhibiting target-mediated drug disposition (TMDD). In this study, we performed a quantitative analysis of a diverse set of six mAbs exhibiting TMDD to explore translational rules that can be utilized to predict human PK. A TMDD model with rapid-binding approximation was utilized to fit PK and PD (i.e., free and/or total target levels) data, and average absolute fold error (AAFE) was calculated for each model parameter. Based on the comparative analysis, translational rules were developed and applied to a test antibody not included in the original analysis. AAFE of less than two-fold was observed between monkey and human for baseline target levels (R0), body-weight (BW) normalized central elimination rate (Kel/BW−0.25) and central volume (Vc/BW1.0). AAFE of less than three-fold was estimated for the binding affinity constant (KD). The other four parameters, i.e., complex turnover rate (Kint), target turnover rate (Kdeg), central to peripheral distribution rate constant (Kpt) and peripheral to central rate constant (Ktp) were poorly correlated between monkey and human. The projected human PK of test antibody based on the translation rules was in good agreement with the observed nonlinear PK. In conclusion, we recommend a TMDD model-based prediction approach that integrates in vitro human biomeasures and in vivo preclinical data using translation rules developed in this study.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-014-9690-8) contains supplementary material, which is available to authorized users.
KEY WORDS: ADME of biologics, human translation, monoclonal antibodies, PK/PD modeling, TMDD
INTRODUCTION
Biotech revolution over the last three decades has created substantial interest and opportunities in the development of proteins, peptides and antibody therapeutics. More than 30 monoclonal antibodies (mAbs) have already been approved by the US FDA with hundreds more in queue (1). However, clinical development of mAbs can be challenging due to safety risks arising from exaggerated pharmacology and exposure nonlinearities caused by target-mediated drug disposition (2–5). Several recent cases highlight this challenge such as progressive multifocal leukoencephalopathy (PML) observed with natalizumab (6), reactivation of Epstein-Barr virus (EBV) with anti-CD3 mAb (7), and the occurrence of cytokine storm with an anti-CD28 super agonist (8). Thus, successful testing of novel biotherapeutic in clinic requires the selection of an optimum dose range that maximizes the probability of observing pharmacological effects while minimizing the unintended safety consequences. Design of such optimum dose range requires accurate prediction of human PK parameters, e.g., maximum drug concentration (Cmax) and area under the curve (AUC). These PK parameters enable the determination of safety margins and the selection of a starting dose that is low enough to avoid exaggerated pharmacology but high enough to allow efficient escalation of clinical doses to the top dose, resulting in faster regulatory approvals with lower development costs.
For antibodies and protein with linear PK characteristics, simple allometric power models have extensively been used to predict human PK (2). Use of power models was first applied by Mordenti et al. (9) to scale up human clearance (CL) and volume of distribution at steady state (Vss) for five therapeutic proteins. Subsequently, Mahmood et al. (10) expanded the allometric scaling to 15 protein drugs. Wang et al. (11) performed a meta-analysis on 34 therapeutic proteins and scaled the clearance values using three different approaches; i.e., simple allometry, allometry with brain weight correction, and fixed exponent method. Similarly, Ling et al. performed an analysis of 14 mAbs where clearance was scaled from nonhuman primates as preclinical species using fixed exponent method (12). Deng et al. have also showed recently that simple allometry of monkey CL alone with an exponent of 0.85 provided a preferable prediction of human clearance (13). More recent work by Oitate et al. (14,15) using data from 24 mAbs showed that the human CL and Vss were predicted reasonably well only from the monkey data alone with estimated exponents of Vss and CL.
Although these reports suggest that simple allometry is a promising tool for antibodies with linear PK profile, principles of allometric power models alone are inadequate for biologics exhibiting nonlinear PK. Many of the mAbs currently on the market or in the development pipeline exhibit TMDD wherein a significant fraction of drug amount is cleared through high affinity interaction with the intended target. Such mAbs exhibit nonlinear PK as a function of dose that becomes linear at a dose high enough to saturate the TMDD process. For such antibodies, concentrations in the plasma are not in rapid equilibrium with the concentrations in the tissue and hence one cannot calculate and scale up Vss for these compounds (2,3). In a recent modeling study with six mAbs exhibiting nonlinear PK, Dong and coworkers showed that predictions of human PK parameters such as Cmax and AUC is challenging within the dose range where significant nonlinearities are seen (16). Thus, a successful prediction of human PK for mAbs with TMDD requires a mechanistic model-based approach that explicitly incorporates target-specific parameters such as baseline target levels (Ro), free target turnover rate (Kdeg), complex turnover rate (Kint), and equilibrium binding constant (KD). One example of this approach is the study by Kagan et al. (17) that involved interspecies scaling of IFN-β1a, IFN-β1b, and IFN-α2a based on the quasi-equilibrium approximation of TMDD model. Authors reported that the human PK/PD profiles of IFN-β1a can be predicted using the same target-specific rate constants derived for monkeys. Luu et al. (18) also demonstrated an application of TMDD modeling for an IgG2 antibody directed against human ALK1 (activin receptor-like kinase 1).
Despite the publication of few TMDD model-based studies as mentioned above, no systematic evaluation has been done to explore the monkey-to-human translation of target-specific parameters incorporated in a TMDD model. There is a significant lack of scholarship around general translational rules for target-specific parameters that are valid across a diverse range of targets. The objective of current analysis was to systematically apply TMDD modeling and explore the translatability of estimated parameters from monkey to human for mAbs exhibiting TMDD. Our analysis involved a diverse set of six mAbs with preclinical (nonhuman primates (NHPs)) and clinical (phase 1 and/or phase 2a) PK and/or PD data. Monkey and human specific parameters were estimated using the rapid-binding approximation of TMDD models, and a comparative analysis was performed to establish translational rules. In a prospective fashion, these rules were applied to predict nonlinear human PK of a new antibody not included in original analysis.
MATERIALS AND METHODS
Datasets
PK/PD data of the following six mAbs was included in the modeling and correlation analysis: mAb-1 (Efalizumab, anti-CD11a), mAb-2 (TRX-1, anti-CD4), mAb-3 (MTRX-1011A, anti-CD4), mAb-4, mAb-5, and mAb-6. One additional mAb, mAb-7, was used for the validation. Table I presents a brief description of the different mAbs and study designs included in this meta-analysis. In the case of Efalizumab, TRX-1, MTRX-1011A and mAb-6, both the PK and target expression data were available, and utilized to estimate the model parameters. PK data sets for mAb-4, mAb-5, and mAb-6 came from Pfizer in-house programs. Data for remaining mAbs were extracted from the literature and digitized using Engauge Digitizer (http://digitizer.sourceforge.net/). The data was transformed into molar units across different case-studies.
Table I.
Datasets Included in the Translational Analysis
| Drug | Study | Study description | Subjects | Dosing regimen |
|---|---|---|---|---|
| mAb-1 (Efalizumab) |
Preclinical | DES; PK and %CD11a expression levels |
Healthy chimpanzees | Single dose of 0.5–10 mg/kg (only 8 mg/kg available) administered intravenously |
| Phase 1 | DES, MC, DB; PK and %CD11a expression levels |
31 patients | Single dose of 0.03, 0.1, 0.3, 0.6, 1, 2, 3 and 10 mg/kg administered intravenously as a short-term infusion (1–2 h) | |
| mAb-2 (TRX-1) |
Preclinical | SC, PC, toxicokinetics study; PK, free and total %CD4 expression levels |
16 antibody-naïve olive baboons | 40 mg/kg administered twice a week for two weeks as a short-term infusion (1–2 h) |
| Phase 1 | DES, SC, DB, PC; PK, free and total %CD4 expression levels |
12 healthy volunteers | Single doses of TRX-1 administered intravenously at dose-levels of 1, 5 and 10 mg/kg as short-term infusion (1–2 h) | |
| mAb-3 (MTRX-1011A) |
Preclinical | SC, PC, toxicokinetics study; PK, free and total %CD4 expression levels |
16 Antibody-naïve olive baboons | 40 mg/kg administered twice a week for two weeks as a short-term infusion (1–2 h) |
| Phase 1 | SC, DB, PC, DES; PK, free and total %CD4 expression levels |
20 patients | Single doses of 0.3, 1, 3.5 and 7.0 mg/kg administered intravenously | |
| mAb-4 | Preclinical | MC, PC, single and multiple DES; PK |
30 healthy cynomolgus monkeys | Single doses of 1, 5 mg/kg and multiple doses of 1, 5, and 10 mg/kg/week administered intravenously |
| Phase 1 | SC, DES; PK | 40 healthy volunteers | Single doses of 3, 10, 30, 100 and 300 mg administered intravenously | |
| mAb-5 | Preclinical | MC, DES, multiple-dose toxicokinetics study; PK |
30 healthy cynomolgus monkey | Single doses of 0.01, 0.05 and 0.1 mg/kg and multiple doses of 1, 10 and 100 mg/kg administered intravenously |
| Phase 1/2a | SC, DB, PC, single and multiple DES; PK |
80 patients | Single doses of 0.03, 0.1, 0.3, 1 and 10 mg/kg and multiple-doses of 0.1, 0.3 and 3.0 mg/kg administered intravenously | |
| mAb-6 | Preclinical | SC, PC, DES; PK and total % receptor expression levels. |
16 healthy cynomolgus monkeys | Single doses of 1, 10 and 100 mg/kg administered intravenously |
| Phase 1 | SC, DB, PC, DES; PK and total % receptor expression levels |
81 healthy volunteers | Single doses of 0.3, 1, 3, 10, 30, and 100 mg administered intravenously | |
| mAb-7 (Test drug) |
Preclinical | DES; PK | 17 healthy cynomolgus monkeys | Single doses of 0.3 mg/kg, 1 mg/kg, 5 mg/kg administered intravenously and 5 mg/kg administered subcutaneously |
| Phase 1 | SC, DB; PK | 30 healthy subjects | Single doses of 3 mg, 10 mg, 30 mg, 60 mg and 120 mg administered subcutaneously |
mAbs 1–6 were used for translation rule building, and mAb-7 dataset was used for validation
DES dose escalation, SC single center, MC multiple center, PC placebo controlled, DB double blinded, PK pharmacokinetics, PD pharmacodynamics
TMDD Model
Data fitting and parameter estimation was performed using rapid-binding (RB) approximation of the TMDD model (Fig. 1) (19,20). Analysis was done on naïve-pooled and naïve-averaged data rather than population approach to estimate TMDD parameters. The model was implemented in Monolix 4.12s using the SAEM algorithm (21). The M3 method described by Beal et al. was employed to handle the data below the limit of quantification (BLQ) (22). This method is based on simultaneous modeling of continuous and categorical data where the BLQ observations are treated as categorical data. The likelihood for BLQ observations is maximized with respect to the model parameters.
Fig. 1.
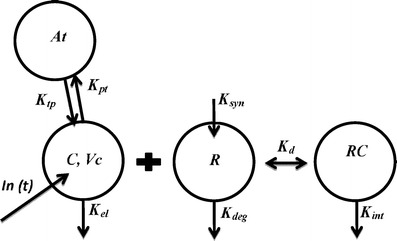
Schematic of a target-mediated disposition (TMDD) model with rapid-binding (RB) approximation. Antibody levels in central and peripheral compartment is denoted by C and At respectively. V c represents central volume of distribution. The K el, K deg and K int are first-order rate constants representing antibody elimination, target turnover, and complex turnover rates, respectively. K syn denotes endogenous synthesis rate of target. The drug distribution from central-to-peripheral and peripheral-to-central compartment is represented by K pt and K tp respectively. K D is the equilibrium binding constant
The equations for TMDD model with rapid-binding approximation are written below. It is assumed that the association (Kon) and dissociation (Koff) rate constants are significantly faster than other kinetic rates such that the free drug, free target, and complex are in rapid equilibrium determined by the equilibrium constant (KD).
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
Ctot represents the total mAb concentration (unbound and bound) in the central compartment and Rtot represents the total receptor expression levels. Kel represents the first-order nonspecific elimination rate constant of mAb from the central compartment “C”. Kpt represents the first-order distributional rate constant of mAb from plasma to tissue compartment “At” whereas Ktp represents the first-order distributional rate constant of mAb from tissue to plasma compartment. Vc represents the volume of distribution of mAb in the central compartment. Ksyn and Kdeg are the zero- and first-order rate constants associated with the turnover of the target whereas Kint represents the first-order degradation rate constant of mAb-target complex. Initial conditions for Eqs. (1), (2), and (3) are DoseIV/Vc, 0 and R0 (or Ksyn/Kdeg).
Monkey-to-Human Translation Analysis
Monkey-to-human translation and correlation were explored for all 8 TMDD parameters. The point estimates and the associated confidence intervals of Kel and the Vc were body-weight-normalized with the commonly used allometric exponents of −0.25 and 1, respectively prior to the correlation analysis (16,18). Average absolute fold error (AAFE) between monkey and human for each parameter across all antibodies was calculated using following equation:
| 6 |
Pearson correlation coefficients (R values) were calculated using Minitab 16 software package and associated p values were used as a metric in assessing the statistical significance of correlation among different parameters. Each data point represented mean parameter estimate from one of the six mAbs.
Allometric Scaling
Where applicable, the first-order rate parameter Ki and volume parameter Vi were allometrically scaled with exponents of −0.25 and 1.0 as described below:
| 7 |
| 8 |
Prediction of Test Antibody PK
Based on the obtained relationship of TMDD parameters between the two species, translation rules were generated and applied to predict the human PK of the test drug mAb-7 in nonlinear exposure regime. Cynomolgus PK of mAb-7 at single IV dose of 0.3, 1, 5 mg/kg IV, and single SC dose of 5 mg/kg was modeled using the RB approximation of TMDD model (described above) and model parameters were estimated. Availability of 5 mg/kg SC dose group allowed the estimation of SC bioavailability and first-order absorption rate along with the eight TMDD model parameters. To generate 90% confidence intervals related to human PK predictions for the test drug, Monte-Carlo simulations were conducted for both TMDD model-based approach and empirical method using a sample size of 5000. All the pharmacokinetic parameters were assumed to be log-normally distributed except for bioavailability (F), which was assumed to be logit-normally distributed. The PK simulations for 5000 monkeys were conducted with means equal to parameter estimates and standard deviation equal to two-fold of standard error on that parameter estimate. Model predictions from 5000 monkeys were then translated to 5000 human predictions using the two methods and 5th, median, and 95th percentiles were obtained.
RESULTS
Monkey and Human Parameter Estimates
PK and PD (i.e., free and/or total target) data in monkeys and humans for six mAbs were fitted using a TMDD model with rapid-binding approximation (Fig. 1), followed by a correlation analysis of estimated parameters to assess monkey-to-human translation. Table I presents the study details and dosing regimen information for each antibody. For mAb-1 (anti-CD11a), mAb-2(anti-CD4), mAb-3 (anti-CD4), and mAb-6, both PK and target expression data were available from literature (23–26), whereas only PK was available for mAb-4 and mAb-5. The mAb-7 antibody was chosen as the “test antibody” for the validation, and not used at the stage of translation rule building.
The rapid-binding approximation of TMDD model well characterized the PK/PD data across different datasets as indicated by the goodness of fit plots (Fig. 2). The data fell around the line of unity for most of studies except for slight deviations seen in the case of mAb-1 (human fit) and mAb-3 (monkey fit). Based on the model fits of individual antibody datasets (Supplementary information), the TMDD structural model was able to capture the trend of the data very well. Table II presents the estimated values of all eight model parameters for both monkeys and humans. In the case of baboon data of mAb-3, R0 and KD were already available from the published reports and used as such (25,26). Majority of the parameter estimates were obtained with reasonable precision (%RSE <50%) across different studies but there were few parameter estimates with a %RSE of >50%. This is explained by insufficient data available within the dataset to inform about the parameter estimate in question.
Fig. 2.
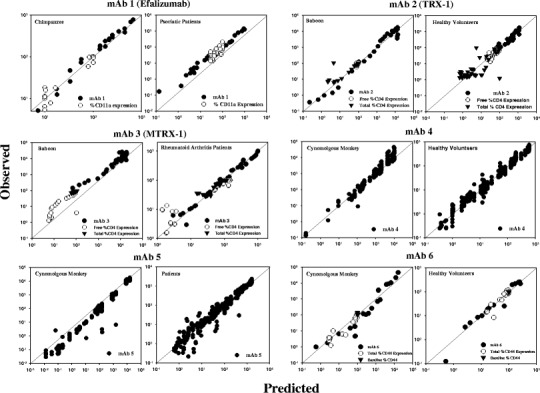
Goodness-of-fit plots for mAbs 1–6 with PK/PD data from NHPs and humans. Individual panels show observed vs. predicted for each antibody. Solid diagonal line represents perfect agreement between observed data and model fits. Individual fits of PK/PD time courses at each dose for every antibody are included in supplementary information
Table II.
TMDD Model Estimated Parameter for the mAbs 1–6
| Drug | Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | R 0 (nM) | K D (nM) | K el (day−1) | V c (L) | K pt (day−1) | K tp (day−1) | K int (day−1) | K deg (day−1) | |
| mAb-1 (Efalizumab) | Chimpanzees | 6.11 (25) | 0.427 (12) | 0.123 (11) | 2.92 (11) | 0.395 (24) | 0.383 (8) | 3.17 (9) | 0.303 (8) |
| Patients | 5.41 (13) | 0.516 (21) | 0.112 (11) | 4.47 (9) | 0.286 (18) | 0.506 (11) | 2.06 (9) | 0.597 (6) | |
| mAb-2 (TRX-1) |
Baboons | 32.4 (27) | 398 (75) | 2.04 (69) | 0.931 (17) | 0.84 (106) | 4.57 (143) | 2.04 (69) | 1.46 (74) |
| Healthy volunteers | 28.4 (24) | 30.1 (15) | 4.25 (12) | 3.79 (11) | 0.14 (100) | 0.413 (85) | 4.25 (12) | 0.867 (14) | |
| mAb-3 (MTRX-1011A) | Baboons | 34.3 (fixed) | 398 (fixed) | 0.116 (19) | 0.61 (20) | 2.62 (26) | 9.4 (68) | 0.103 (31) | 0.069 (32) |
| Patients | 18.7 (10) | 42.4 (10) | 0.109 (7) | 2.85 (16) | 0.549 (13) | 1.35 (21) | 5.17 (4) | 1.35 (2) | |
| mAb-4 | Cynomolgus monkeys | 5.49 (52) | 0.707 (1) | 0.115 (17) | 0.089 (5) | 0.159 (26) | 0.155 (32) | 2.23 (51) | 2.5 (56) |
| Healthy volunteers | 1.62 (7) | 0.381 (20) | 0.0693 (12) | 3.39 (2) | 0.031 (19) | 0.046 (41) | 0.441 (10) | 4.42 (4) | |
| mAb-5 | Cynomolgus monkeys | 2.72 (16) | 0.294 (18) | 0.08 (30) | 0.184 (4) | 2.56 (8) | 9.14 (18) | 0.579 (36) | 2.47 (27) |
| Patients | 1.57 (11) | 0.153 (36) | 0.06 (7) | 3.53 (4) | 0.115 (30) | 0.159 (25) | 0.014 (49) | 0.351 (30) | |
| mAb-6 | Cynomolgus monkeys | 145 (48) | 59 (62) | 0.565 (31) | 0.119 (44) | 1.47 (46) | 1.32 (30) | 8.83 (22) | 0.251 (11) |
| Healthy volunteers | 130 (31) | 5.61 (39) | 0.221 (135) | 1.6 (22) | 1.32 (32) | 0.341 (49) | 1.83 (15) | 0.239 (23) | |
Mean parameters for each species are provided along with %relative standard error (%RSE) in parenthesis
R 0 baseline target levels, K D equilibrium binding constant, K el central elimination rate constant, V c central volume of distribution, K pt central to peripheral distribution rate constant, K tp peripheral to central rate constant, K deg target turnover rate constant, K int complex turnover rate constant
Average Absolute Fold Error (AAFE) Between Monkey and Human
AAFE between monkey and human for each parameter across all antibodies was calculated to explore the translation relationships between monkeys and human. Fitting of monkey and human PK/PD data for each antibody resulted in the estimation of 4 drug-specific model parameters (e.g., Vc, Kel, Kpt, Ktp) as well as four target-specific model parameters (e.g., KD, Ro, Kdeg, Kint). The parameter estimates with %RSE > 50% were not included in the AAFE calculations due to lack of precision. Figure 3 shows the correlation plots of all eight model parameters with respective AAFE values. AAFE values for R0, KD, Kel/BW−0.25 and Vc/BW1.0 were estimated to be 1.4, 2.5, 1.3, and 1.6, respectively. Note that Kel and Vc parameters were body weight (BW)-normalized (Kel/BW−0.25 and Vc/BW1.0) as these parameters represent linear component of the drug PK and have been shown to scale allometrically with coefficients of −0.25 and 1.0, respectively (10,16). The associated p values indicate a statistically significant correlation between monkey and human except for Vc where p value (0.194) was higher than 0.05. The second row in Fig. 3 shows the results for the remaining four parameters, i.e., Kint, Kdeg, Kpt, and Ktp with AAFE values within 3.9–10.1-fold. The parameters were not highly correlated between monkey to human with the respective R values of −0.21, −0.45, 0.19, and −0.30. The associated p values (>0.05) indicate that the obtained R values are not statistically different from zero.
Fig. 3.

Monkey to human correlation plots for the eight TMDD parameters estimated for mAbs 1–6. Solid diagonal line is the line of unity. The dotted lines represent two-fold range above and below the line of unity. Average absolute fold error (AAFE), correlation coefficient (R), and associated p values are presented for each parameter. Mean estimated values are shown along with 95% confidence interval for both monkey and human. Data with open circles were not included in AAFE or R value calculation as these referred to cases where the corresponding parameter was not estimated with precision (%RSE > 50%), with confidence intervals containing zero
In vitro to In vivo Correlation (IVIVC) for Human KD
To explore the utility of in vitro measurements to predict in vivo estimates, a correlation analysis was conducted between human in vivo KD (TMDD model estimated) vs. human in vitro KD values available for five mAbs. In vitro data were available for only KD and similar evaluation could not be performed for other parameters. In the case of mAb-1 (Efalizumab), in vitro KD value was obtained from purified PMBCs (23) whereas BIAcore assays using recombinant proteins were used to measure KD values for mAb-2, 4, 5 and 6. Figure 4 presents the comparison between the in vitro human to in vivo human KD. An AAFE value of 12.4 indicated significant disconnect between in vitro vs. in vivo. Especially, for antibodies with KD values obtained from BIAcore experiments, in vitro values significantly underpredicted in vivo KD. In contrast, in the case of mAb-1 (Efalizumab), in vitro KD of 0.76 nM was estimated in physiological relevant conditions (i.e., PBMCs derived from human whole blood) and correlated well with the in vivo estimate of 0.52 nM.
Fig. 4.

In vitro to in vivo correlation plot for human K D. The human in vitro K D was not available for mAb-4. Solid diagonal line represents line of unity. The gray lines represent two-fold range above and below the line of unity
Development of Translation Rules
Based on the observed AAFE and correlation for eight TMDD parameters between monkeys and human, translation rules for each parameter were developed to guide human prediction based on the monkey data. Table III summarizes the translational rules for all eight TMDD parameters. Detailed rationale specific to each parameter is provided within the discussion section.
Table III.
Translation Rules Recommended for TMDD Model Parameters Towards Predicting Human PK of Antibodies Exhibiting TMDD
| Parameters | Definition | Translation rules to obtain human parameter |
|---|---|---|
| Drug-specific | ||
|
V
c
K el |
Central volume of distribution Antibody elimination rate constant |
Allometric scaling of monkey estimates Allometric scaling of monkey estimates |
|
K
pt
K tp |
Plasma to tissue distribution rate constant Tissue to plasma distribution rate constant |
Sensitivity analysis within a range (same as monkeys or allometric scaling of monkey estimates) |
| Target-specific | ||
| R o | Baseline target expression level | Use monkey estimate; Adjust for relative expression differences between healthy vs. disease state |
| K D | Equilibrium binding constant | Use monkey estimate; Adjust if in vitro assays indicate significant potency differences (e.g., BIAcore assays) |
|
K
deg
K int |
Target turnover rate constant Complex degradation rate constant |
Sensitivity analysis within a range (same as monkeys or allometric scaling of monkey estimates); If feasible, use experimentally derived values (e.g., human in vitro assays using primary cells) |
Application of Translation Rules to mAb-7
As a validation step, the translation rules in Table III were applied to predict human PK of a new “test” antibody (mAb-7) that exhibited TMDD in monkeys. The TMDD model-based predictions were compared with observed human data from few healthy volunteer cohorts (3–120 mg SC) to assess the model performance. For the validation process, we first modeled the monkey IV PK data using TMDD model to estimate monkey parameters. The monkey data also included a SC arm at 5 mg/kg doses. This additional piece of data allowed estimation of SC bioavailability (F) and first-order absorption rate constant (Ka). As shown in Fig. 5, the model fitted the data well. All monkey parameters including distributional rate constants (i.e., Kpt and Ktp) were estimated with high precision (Table IV).
Fig. 5.

TMDD model-based fitting of monkey PK data for test-antibody (mAb-7). Individual panels show model fits corresponding to 0.3 mg/kg IV, 1 mg/kg IV, 5 mg/kg IV, and 5 mg/kg SC. Observed data is depicted by solid circles whereas model fits are shown by solid lines
Table IV.
Estimated Monkey Parameter for Test Antibody (mAb-7) and Corresponding Human Parameters Derived Using Translation Rules Presented in Table III
| Parameters | Units | Cynomolgus monkeys, mean (%RSE) | Translated human parameter (mean) |
|---|---|---|---|
| TMDD model | |||
| R 0 | nM | 1.17 (1) | 1.17 |
| K deg | day−1 | 15.5 (8) | 15.5 |
| K D | nM | 9.37 (4) | 9.37 |
| K int | day−1 | 5.07 (3) | 5.07 |
| K el | day−1 | 0.14 (11) | 0.066 |
| K pt | day−1 | 1.25 (10) | 0.625 |
| K tp | day−1 | 0.941 (1) | 0.471 |
| V c | L | 0.132 (4) | 2.64 |
| K a | day−1 | 0.657 (7) | 0.311 |
| F | – | 0.802 (5) | 0.802 |
R 0 baseline target levels, K D equilibrium binding constant, K el central elimination rate constant, V c central volume of distribution, K pt central to peripheral distribution rate constant, K tp peripheral to central rate constant, K deg target turnover rate constant, K int complex turnover rate constant, K a absorption rate constant for SC route, F bioavailability for SC route, CL central clearance rate
For TMDD model-based prediction of mAb-7 human PK, the translation rules presented in Table III were used as a guide to scale monkey parameters to humans. The drug-specific model parameters (e.g., Vc, Kel, Kpt, Ktp) were allometrically scaled to humans. The target-specific model parameters (e.g., KD, Ro, Kdeg, Kint) were scaled 1:1 between monkeys and healthy human volunteers due to high sequence homology, similar target expression, and absence of any experimental data which indicates substantial difference between monkeys and human. Given the SC route of administration, two additional PK parameters, SC bioavailability (F) and absorption rate constant (Ka), needed to be scaled to humans. The systemic bioavailability with SC route was estimated to be 80.2% in cynomolgus monkeys and was assumed to be the same for humans. The Ka was allometrically scaled with an exponent of −0.25.
Most of the data was well predicted by TMDD model within both the linear and the nonlinear zone (Supplementary Figure S11). Figure 6 presents the performance of TMDD model in predicting human AUC/Cmax values observed in phase 1 trials with healthy volunteers. The predicted AUC and Cmax using the TMDD model correlate well with the observed mean AUC and Cmax at different dose levels.
Fig. 6.
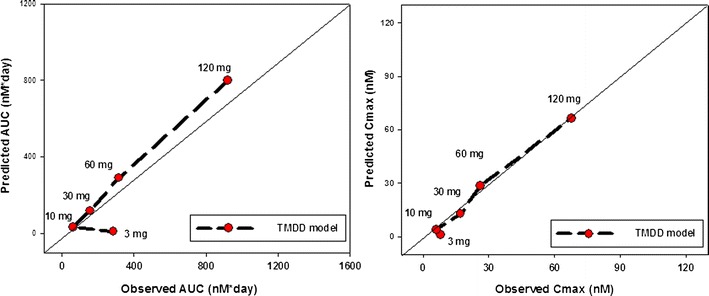
Application of translation rules to predict human PK of test antibody (mAb-7). Predicted AUC (a) and C max (b) at each dose (3–120 mg SC) are shown and compared against the observed data. Solid diagonal line represents perfect agreement
DISCUSSION
Several mAbs currently in the market or in development phase exhibit nonlinear behavior in their pharmacokinetic profile due to TMDD. Our ability to successfully translate this behavior from the preclinical to clinical space is critical for the efficient design and conduct of FIH trials and speedy approval. Dong et al. (16) were the first group to attempt the scaling of mAbs exhibiting nonlinear PK from NHP’s to humans using an empirical Michaelis-Menten approach utilizing Vmax/Km model. In their study, Vmax and CL parameters were scaled with an exponent of 0.75, and rate constants with an exponent of −0.25 while assuming similar Km between human and monkeys. While this approach performed well in the linear regime, human exposures were predicted poorly in the nonlinear regime (e.g., Cmax was estimated up to 5.3-fold higher than observed).
In light of shortcomings with the empirical approaches, we performed a comprehensive analysis of a dataset containing six mAbs and a test antibody exhibiting nonlinear PK. Monkey and human PK/PD data were fit separately to estimate TMDD model parameters, and translation from monkey-to-human was explored systematically for every model parameter. Results from the AAFE calculations and correlation analysis of each of these parameters led to the development of translation rules (Table III) that can be used to enable human PK predictions of a new antibody or biologic with a potential to exhibit TMDD profile in clinic. The following sections provide detailed rationale behind the translation rules specific to each model parameter.
Baseline Target Expression Levels (R0)
Model estimated receptor expression parameter (R0) was found to be similar between the two species (AAFE =1.4). Thus monkey estimate of target expression can be used for predictions in healthy volunteers. For cases where biological evidence suggests significant differences in target expression between monkey and human (disease vs. healthy), human specific values should be utilized for model prediction. For some targets expressed in soluble form or solely on circulating blood cells, it may be possible to measure baseline levels or turnover rates in human whole blood or purified PBMCs using techniques such as ELISA or flow cytometry (27–30). On the other hand, for targets expressed mainly in tissues, there are experimental limitations to accurately determine the total target abundance in tissues.
Our recommendations are consistent with the approach taken by two recent studies, first by Luu et al. towards human PK prediction of mAb targeting ALK1 (18), and second by Betts et al. towards predicting human PK of anti-DKK1 antibody (31). In the first study, the target expression (R0) of ALK1 was assumed to be the same between the two species. In the second study, mice data indicated five-fold higher DKK-1 baseline levels in disease state compared to healthy animals. Thus, soluble DKK-1 target levels were obtained in human serum of postmenopausal women and osteoporosis patients, and directly utilized for model predictions.
Equilibrium Binding Constant (KD)
AAFE = 2.5 was observed between human and monkey estimated KD values (Fig. 3). A less than three-fold difference in potencies between the two species is reasonable as our dataset included fully humanized antibodies designed to be cross-reactive between NHPs and humans. Thus, we recommend the use of monkey in vivo KD towards human predictions for antibodies with good cross-reactivity. Generally, many targets show high sequence homology between monkeys and humans, resulting in similar binding epitopes and high cross-reactivity. However, monkey and human binding epitopes can be very different for some targets. In such cases, monkey KD should be adjusted prior to use in human, based on the relative potency differences based on in vitro assays such as BIAcore. However, caveats apply when using in vitro KD values since BIAcore experiments or similar techniques use recombinant receptor targets in soluble forms unlike their physiologically relevant cell-surface forms. Our own results suggest lack of correlation between human in vitro KD to human in vivo KD (Fig. 4). As a best practice approach, KD should be measured under physiological conditions such as purified PBMCs or primary tissue cells derived from healthy volunteers or patients.
Target (Kdeg) and Complex (Kint) Turnover Rates
AAFE values of 4.1 and 10.1 were estimated for Kdeg and Kint, respectively. Additionally, there was no correlation observed for Kdeg and Kint between the two species (Fig. 3). Large AAFE values and a lack of unifying correlation between six mAbs do not allow us to make firm recommendations on scaling monkey Kdeg and Kint to human. Literature precedence suggests using same as monkeys (18) or their allometrically scaled values (31). Taking a cautious approach, our recommendation is to perform human predictions using a sensitivity analysis for Kdeg and Kint within a range (same as monkeys vs. allometrically scaled). If feasible, experiments should be performed to generate human-specific values using assays under physiological conditions such as purified PBMCs or primary tissue cells derived from healthy volunteers or patients as appropriate.
Drug-Specific Parameters (Kel, Vc, Kpt, Ktp)
A reasonably good agreement between monkey and human was observed for the body-weight-normalized Kel and Vc parameters, based on the AAFE values of 1.3 and 1.6, respectively. Thus, monkey-derived values of Kel and Vc (body-weight-normalized) are recommended for human projection. IgGs share general elimination processes such as proteolysis and protein catabolism following endocytosis. Furthermore, for IgG-based drugs such as mAbs/Fc fusion proteins, it is known that human Fc binds with similar affinity to human and monkey neonatal Fc receptor (FcRn) that salvages IgG from catabolism by lysosomal enzymes and hence contributes to the nonspecific clearance mechanism (32).
In contrast, high AAFE values and poor correlation were observed for the two distributional rate constants (i.e., Kpt and Ktp). It should be noted that estimation of these parameters is highly sensitive to the accurate sampling of the fast distribution phase during early time points, and digital extraction of the data for mAb-1 (Efalizumab), mAb-2 (TRX-1), and mAb-3 (MTRX-1011A) in this phase from the published reports may not be accurate. Thus, a cautious approach is required for interpreting current results for Kpt and Ktp. Based on the learnings from previous studies (18,31), it may be reasonable to use same monkey values or allometric scaling of monkey values if precise estimates are available.
An alternative approach for Kpt and Ktp parameters is also suggested based on the comprehensive study by Dirks and Meibohm (5). This report presented a population analysis of a wide range of therapeutic mAbs currently in market, including fully humanized and chimeric mouse antibodies. To derive fully human IgG-relevant PK parameters, we further refined the population estimates by excluding the data from chimeric mouse antibodies (manuscript under preparation). This refinement led to a population estimate of 0.26 L/day for the central clearance (CL), 0.56 L/day for the distributive clearance (Q), 3.1 L for the central volume of distribution (Vc), and 3.06 L for the peripheral volume of distribution (Vp). Using these values derived for a typical fully human IgG antibody, the mean value of Kpt and Ktp can be calculated to be 0.181 and 0.183 day−1 respectively.
In conclusion, the analysis presented here is the first comprehensive study exploring translation rules for TMDD model parameters using a range of mAbs. Based on our analysis and results from application of the translation rules (Table III) to a test antibody, we recommend a TMDD model-based approach that integrates available experimental data on human parameters in conjunction with the learnings from preclinical species such as NHPs. Future analysis should include a population modeling-based approach on larger datasets to estimate interindividual variability for model parameters. Additionally, translation of SC absorption-specific parameters such as Ka and F should also be explored.
Electronic supplementary material
mAb-1 (Efalizumab) pharmacokinetics and %CD11a receptor modulation in chimpanzees fitted with a rapid-binding approximation of TMDD model. (DOCX 35 kb)
mAb-1 (Efalizumab) pharmacokinetics and %CD11a receptor modulation in Psoriatic patients fitted with a rapid-binding approximation of a TMDD model. (DOCX 101 kb)
mAb-2 (TRX-1) pharmacokinetics, free and total % CD4+ cells modulation in healthy baboons fitted with a rapid-binding approximation of TMDD model. (DOCX 108 kb)
mAb-2 (TRX-1) pharmacokinetics, free and total % CD4+ cells modulation in healthy volunteers in phase 1 clinical trial fitted with a rapid-binding approximation of TMDD model. (DOCX 104 kb)
mAb-3 (MTRX-1) pharmacokinetics, free and total %CD4+ cells modulation in baboons fitted with a rapid-binding approximation of TMDD model. (DOCX 60 kb)
mAb-3 (MTRX-1) pharmacokinetics, free and total %CD4+ cells modulation fitted with a rapid-binding approximation of TMDD model. (DOCX 67 kb)
mAb-4 pharmacokinetics in cynomolgus monkeys and healthy volunteers in phase 1 trial fitted with a rapid-binding approximation of a TMDD model. (DOCX 353 kb)
mAb-5 pharmacokinetics in cynomolgus monkeys and in humans fitted with a rapid-binding approximation of a TMDD model. (DOCX 163 kb)
mAb-6 pharmacokinetics and total %target modulation in cynomolgus monkeys fitted with a rapid-binding approximation of TMDD model. (DOCX 50 kb)
mAb-6 pharmacokinetics and total %target modulation in healthy volunteers in a phase-1 trial fitted with a rapid-binding approximation of TMDD model. (DOCX 52 kb)
TMDD model based predictions for the test drug (mAb-7) PK in a phase-1 clinical trial in healthy volunteers. Note that most of the PK data (except for one subject) for 3 mg SC dose was below the limit of quantification of the assay and could not be presented in Figure 6. Consistent with these findings, model predictions for 3mg SC group were below LLOQ. (DOCX 103 kb)
Vmax/Km model based predictions for the test drug (mAb-7) PK in a phase-1 clinical trial in healthy volunteers. (DOCX 105 kb)
Comparative performance of translation rule based predictions vs. empirical (V max/K m) approach. Predicted AUC (A) and Cmax (B) at each dose (3-120mg SC) are shown for the two approaches and compared against the observed data. Solid diagonal line represents perfect agreement. (DOCX 217 kb)
ACKNOWLEDGMENTS
We thank Dr. Donald E. Mager, University at Buffalo, for helping in the development and review of this manuscript. This work was partially supported by the stipend Aman P. Singh received as a Pharmacokinetic/Pharmacodynamic Summer Intern at PDM Department in Pfizer and partially supported by the NIH grant GM 57980
REFERENCES
- 1.Rodrigues ME, Costa AR, Henriques M, Azeredo J, Oliveira R. Technological progresses in monoclonal antibody production systems. Biotechnol Prog. 2010;26(2):332–351. doi: 10.1002/btpr.348. [DOI] [PubMed] [Google Scholar]
- 2.Lobo ED, Hansen RJ, Balthasar JP. Antibody pharmacokinetics and pharmacodynamics. J Pharm Sci. 2004;93(11):2645–2668. doi: 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Wang EQ, Balthasar JP. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2008;84(5):548–558. doi: 10.1038/clpt.2008.170. [DOI] [PubMed] [Google Scholar]
- 4.Mould DR, Green B. Pharmacokinetics and pharmacodynamics of monoclonal antibodies: concepts and lessons for drug development. BioDrugs. 2010;24(1):23–39. doi: 10.2165/11530560-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Dirks NL, Meibohm B. Population pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;49(10):633–659. doi: 10.2165/11535960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 6.Yousry TA, Major EO, Ryschkewitsch C, Fahle G, Fischer S, Hou J, et al. Evaluation of patients treated with natalizumab for progressive multifocal leukoencephalopathy. N Engl J Med. 2006;354(9):924–933. doi: 10.1056/NEJMoa054693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keymeulen B, Vandemeulebroucke E, Ziegler AG, Mathieu C, Kaufman L, Hale G, et al. Insulin needs after CD3-antibody therapy in new-onset type 1 diabetes. N Engl J Med. 2005;352(25):2598–2608. doi: 10.1056/NEJMoa043980. [DOI] [PubMed] [Google Scholar]
- 8.Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355(10):1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- 9.Mordenti J, Chen SA, Moore JA, Ferraiolo BL, Green JD. Interspecies scaling of clearance and volume of distribution data for five therapeutic proteins. Pharm Res. 1991;8(11):1351–1359. doi: 10.1023/A:1015836720294. [DOI] [PubMed] [Google Scholar]
- 10.Mahmood I. Interspecies scaling of protein drugs: prediction of clearance from animals to humans. J Pharm Sci. 2004;93(1):177–185. doi: 10.1002/jps.10531. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Prueksaritanont T. Prediction of human clearance of therapeutic proteins: simple allometric scaling method revisited. Biopharm Drug Dispos. 2010;31(4):253–263. doi: 10.1002/bdd.708. [DOI] [PubMed] [Google Scholar]
- 12.Ling J, Zhou H, Jiao Q, Davis HM. Interspecies scaling of therapeutic monoclonal antibodies: initial look. J Clin Pharmacol. 2009;49(12):1382–1402. doi: 10.1177/0091270009337134. [DOI] [PubMed] [Google Scholar]
- 13.Deng R, Iyer S, Theil FP, Mortensen DL, Fielder PJ, Prabhu S. Projecting human pharmacokinetics of therapeutic antibodies from nonclinical data: what have we learned? MAbs. 2011;3(1):61–66. doi: 10.4161/mabs.3.1.13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oitate M, Masubuchi N, Ito T, Yabe Y, Karibe T, Aoki T, et al. Prediction of human pharmacokinetics of therapeutic monoclonal antibodies from simple allometry of monkey data. Drug Metab Pharmacokinet. 2011;26(4):423–430. doi: 10.2133/dmpk.DMPK-11-RG-011. [DOI] [PubMed] [Google Scholar]
- 15.Oitate M, Nakayama S, Ito T, Kurihara A, Okudaira N, Izumi T. Prediction of human plasma concentration-time profiles of monoclonal antibodies from monkey data by a species-invariant time method. Drug Metab Pharmacokinet. 2012;27(3):354–359. doi: 10.2133/dmpk.dmpk-11-sh-059. [DOI] [PubMed] [Google Scholar]
- 16.Dong JQ, Salinger DH, Endres CJ, Gibbs JP, Hsu CP, Stouch BJ, et al. Quantitative prediction of human pharmacokinetics for monoclonal antibodies: retrospective analysis of monkey as a single species for first-in-human prediction. Clin Pharmacokinet. 2011;50(2):131–142. doi: 10.2165/11537430-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Kagan L, Abraham AK, Harrold JM, Mager DE. Interspecies scaling of receptor-mediated pharmacokinetics and pharmacodynamics of type I interferons. Pharm Res. 2010;27(5):920–932. doi: 10.1007/s11095-010-0098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luu KT, Bergqvist S, Chen E, Hu-Lowe D, Kraynov E. A model-based approach to predicting the human pharmacokinetics of a monoclonal antibody exhibiting target-mediated drug disposition. J Pharmacol Exp Ther. 2012;341(3):702–708. doi: 10.1124/jpet.112.191999. [DOI] [PubMed] [Google Scholar]
- 19.Mager DE, Jusko WJ. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokinet Pharmacodyn. 2001;28(6):507–532. doi: 10.1023/A:1014414520282. [DOI] [PubMed] [Google Scholar]
- 20.Mager DE, Krzyzanski W. Quasi-equilibrium pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Pharm Res. 2005;22(10):1589–1596. doi: 10.1007/s11095-005-6650-0. [DOI] [PubMed] [Google Scholar]
- 21.Lavielle M, Mentre F. Estimation of population pharmacokinetic parameters of saquinavir in HIV patients with the MONOLIX software. J Pharmacokinet Pharmacodyn. 2007;34(2):229–249. doi: 10.1007/s10928-006-9043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beal SL. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn. 2001;28(5):481–504. doi: 10.1023/A:1012299115260. [DOI] [PubMed] [Google Scholar]
- 23.Bauer RJ, Dedrick RL, White ML, Murray MJ, Garovoy MR. Population pharmacokinetics and pharmacodynamics of the anti-CD11a antibody hu1124 in human subjects with psoriasis. J Pharmacokinet Biopharm. 1999;27(4):397–420. doi: 10.1023/A:1020917122093. [DOI] [PubMed] [Google Scholar]
- 24.Ng CM, Stefanich E, Anand BS, Fielder PJ, Vaickus L. Pharmacokinetics/pharmacodynamics of nondepleting anti-CD4 monoclonal antibody (TRX1) in healthy human volunteers. Pharm Res. 2006;23(1):95–103. doi: 10.1007/s11095-005-8814-3. [DOI] [PubMed] [Google Scholar]
- 25.Scheerens H, Su Z, Irving B, Townsend MJ, Zheng Y, Stefanich E, et al. MTRX1011A, a humanized anti-CD4 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: a phase I randomized, double-blind, placebo-controlled study incorporating pharmacodynamic biomarker assessments. Arthritis Res Ther. 2011;13(5):R177. doi: 10.1186/ar3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Y, Scheerens H, Davis JC, Jr, Deng R, Fischer SK, Woods C, et al. Translational pharmacokinetics and pharmacodynamics of an FcRn-variant anti-CD4 monoclonal antibody from preclinical model to phase I study. Clin Pharmacol Ther. 2011;89(2):283–290. doi: 10.1038/clpt.2010.311. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa M, Fujimoto M, Kikuchi K, Takehara K. Elevated serum levels of interleukin 4 (IL-4), IL-10, and IL-13 in patients with systemic sclerosis. J Rheumatol. 1997;24(2):328–332. [PubMed] [Google Scholar]
- 28.Hasegawa M, Sato S, Fujimoto M, Ihn H, Kikuchi K, Takehara K. Serum levels of interleukin 6 (IL-6), oncostatin M, soluble IL-6 receptor, and soluble gp130 in patients with systemic sclerosis. J Rheumatol. 1998;25(2):308–313. [PubMed] [Google Scholar]
- 29.Machy P, Truneh A. Differential half-life of major histocompatibility complex encoded class I molecules in T and B lymphoblasts. Mol Immunol. 1989;26(8):687–696. doi: 10.1016/0161-5890(89)90027-8. [DOI] [PubMed] [Google Scholar]
- 30.Truneh A, Machy P. Detection of very low receptor numbers on cells by flow cytometry using a sensitive staining method. Cytometry. 1987;8(6):562–567. doi: 10.1002/cyto.990080605. [DOI] [PubMed] [Google Scholar]
- 31.Betts AM, Clark TH, Yang J, Treadway JL, Li M, Giovanelli MA, et al. The application of target information and preclinical pharmacokinetic/pharmacodynamic modeling in predicting clinical doses of a Dickkopf-1 antibody for osteoporosis. J Pharmacol Exp Ther. 2010;333(1):2–13. doi: 10.1124/jpet.109.164129. [DOI] [PubMed] [Google Scholar]
- 32.Tabrizi MA, Tseng CM, Roskos LK. Elimination mechanisms of therapeutic monoclonal antibodies. Drug Discov Today. 2006;11(1–2):81–88. doi: 10.1016/S1359-6446(05)03638-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mAb-1 (Efalizumab) pharmacokinetics and %CD11a receptor modulation in chimpanzees fitted with a rapid-binding approximation of TMDD model. (DOCX 35 kb)
mAb-1 (Efalizumab) pharmacokinetics and %CD11a receptor modulation in Psoriatic patients fitted with a rapid-binding approximation of a TMDD model. (DOCX 101 kb)
mAb-2 (TRX-1) pharmacokinetics, free and total % CD4+ cells modulation in healthy baboons fitted with a rapid-binding approximation of TMDD model. (DOCX 108 kb)
mAb-2 (TRX-1) pharmacokinetics, free and total % CD4+ cells modulation in healthy volunteers in phase 1 clinical trial fitted with a rapid-binding approximation of TMDD model. (DOCX 104 kb)
mAb-3 (MTRX-1) pharmacokinetics, free and total %CD4+ cells modulation in baboons fitted with a rapid-binding approximation of TMDD model. (DOCX 60 kb)
mAb-3 (MTRX-1) pharmacokinetics, free and total %CD4+ cells modulation fitted with a rapid-binding approximation of TMDD model. (DOCX 67 kb)
mAb-4 pharmacokinetics in cynomolgus monkeys and healthy volunteers in phase 1 trial fitted with a rapid-binding approximation of a TMDD model. (DOCX 353 kb)
mAb-5 pharmacokinetics in cynomolgus monkeys and in humans fitted with a rapid-binding approximation of a TMDD model. (DOCX 163 kb)
mAb-6 pharmacokinetics and total %target modulation in cynomolgus monkeys fitted with a rapid-binding approximation of TMDD model. (DOCX 50 kb)
mAb-6 pharmacokinetics and total %target modulation in healthy volunteers in a phase-1 trial fitted with a rapid-binding approximation of TMDD model. (DOCX 52 kb)
TMDD model based predictions for the test drug (mAb-7) PK in a phase-1 clinical trial in healthy volunteers. Note that most of the PK data (except for one subject) for 3 mg SC dose was below the limit of quantification of the assay and could not be presented in Figure 6. Consistent with these findings, model predictions for 3mg SC group were below LLOQ. (DOCX 103 kb)
Vmax/Km model based predictions for the test drug (mAb-7) PK in a phase-1 clinical trial in healthy volunteers. (DOCX 105 kb)
Comparative performance of translation rule based predictions vs. empirical (V max/K m) approach. Predicted AUC (A) and Cmax (B) at each dose (3-120mg SC) are shown for the two approaches and compared against the observed data. Solid diagonal line represents perfect agreement. (DOCX 217 kb)


