Abstract
Successful performance and execution of rapid diagnostics in a clinical laboratory hinges heavily on careful validation, accurate and timely communication of results, and real-time quality monitoring. Laboratories must develop strategies to integrate diagnostics with stewardship and evidence-based clinical practice guidelines. We present a collaborative SUCCESS model for execution and monitoring of rapid sepsis diagnostics to facilitate timely treatment. Six months after execution of the Verigene Gram-Positive Blood Culture (BC-GP) and the AdvanDx PNA-FISH assays, data were collected on 579 and 28 episodes of bacteremia and fungemia, respectively. Clinical testing was executed using a SUCCESS model comprising the following components: stewardship, utilization of resources, core strategies, concierge services, education, support, and surveillance. Stewardship needs were identified by evaluating the specialty services benefiting from new testing. Utilization of resources was optimized by reviewing current treatment strategies and antibiogram and formulary options. Core strategies consisted of input from infectious disease leadership, pharmacy, and laboratory staff. Concierge services included automated Micro-eUpdate and physician-friendly actionable reports. Education modules were user-specific, and support was provided through a dedicated 24/7 microbiology hotline. Surveillance was performed by daily audit by the director. Using the SUCCESS model, the turnaround time for the detailed report with actionable guidelines to the physician was ∼3 hours from the time of culture positivity. The overall correlation between rapid methods and culture was 94% (546/579). Discrepant results were predominantly contaminants such as a coagulase-negative staphylococci or viridans streptococci in mixed cultures. SUCCESS is a cost-effective and easily adaptable model for clinical laboratories with limited stewardship resources.
Bacteremia is a major cause of severe sepsis and septic shock, accounting for 30% to 40% of cases, with an estimate of about 250,000 cases occurring annually in the United States (1). A significant proportion of causative organisms are gram-positive bacteria, most commonly Staphylococcus species (2). Multiple studies have established that timely administration of appropriate antibiotics significantly reduces the mortality of severe sepsis and septic shock. Use of inappropriate empiric antibiotics is a common factor associated with mortality rates as high as 75% (3, 4). Delays in initiating antimicrobial treatment are correlated with a progressive increase in mortality (5). The choice of initial antibiotics for treatment of bacteremia must currently be determined empirically. A reduction in time to an accurate identification and susceptibility results may lead to improved patient outcomes, although literature on the magnitude of such an effect is mixed (6, 7).
Current standard blood culture procedures consist of inoculating a blood culture bottle and placing it on an automated continuous monitoring and alert platform (8). Upon positivity, the contents are Gram stained, plated on appropriate media, and allowed to grow for 18 to 42 hours or longer, with subsequent subcultures and susceptibility testing as appropriate. The temporal delay between collection of a blood sample from a patient and availability of traditional identification and susceptibility results has obvious implications regarding patient care.
Newer technologies such as nucleic acid amplification tests, fluorescence in situ hybridization (FISH), and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) provide rapid identification of pathogens and codetection of key resistance markers directly from positive blood cultures. The Verigene Gram-Positive (BC-GP) and Gram-Negative (BC-GN) blood culture assays are approved by the Food and Drug Administration (FDA) to detect common gram-positive and gram-negative organisms, respectively, and associated resistance markers within 3 hours from positive blood cultures (9). The Verigene assays are nonamplified tests that rely on nucleic acid extraction from positive blood cultures followed by microarray-based detection using capture and detection probes. BC-GP is specific for 12 gram-positive bacterial identification targets and 3 associated resistance markers (mecA, vanA, and vanB), while BC-GN is specific for 8 gram-negative bacterial identification targets and 6 resistance markers (blaCTX-M, blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA). The turnaround time from positive blood culture to results can be markedly reduced compared with traditional methods, potentially providing clinically useful data hours or days before traditional methods. Peptide nucleic acid (PNA) FISH is also an FDA-approved technology that uses a PNA probe that hybridizes to the target rRNA when present in the sample and allows visualization of bacteria (such as Staphylococcus aureus/coagulase-negative staphylococci, Enterococcus faecalis/other enterococci, Escherichia coli, Klebsiella and Pseudomonas spp.) and yeasts (Candida spp.) in positive blood cultures within 1.5 hours of positivity (10). Rapid identification and resistance reporting may allow de-escalation of empiric coverage to appropriate targeted therapy and reduction in length of hospital stay.
In this prospective study, we evaluated the laboratory performance of two rapid molecular tests, Verigene BC-GP and Yeast Traffic Light PNA FISH, on a cohort of inpatients from Baylor University Medical Center at Dallas and regional hospitals in Dallas, Texas. In addition, we developed a logical execution protocol to ensure clinical “SUCCESS” of the laboratory testing.
MATERIALS AND METHODS
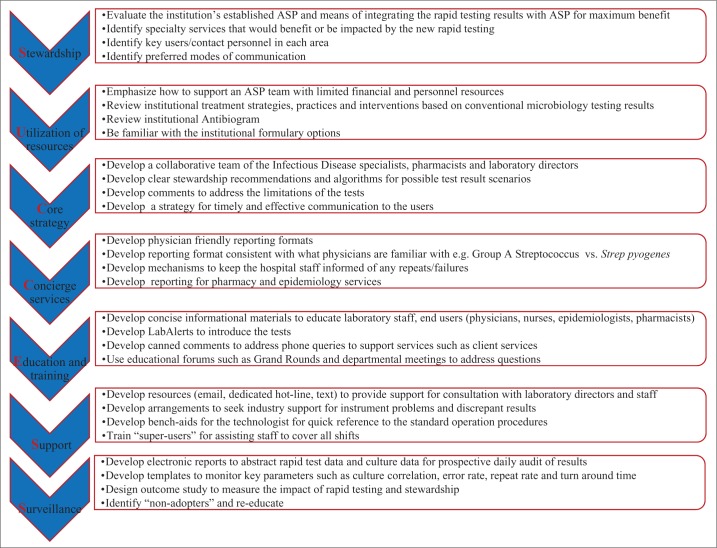
Laboratory performance and execution of rapid diagnostics was based on seven key components of the SUCCESS model (Figure 1). Standardized treatment guidelines were developed by a collaborative team of infectious disease specialists, pharmacists, and laboratory directors. To support an antimicrobial stewardship program team with limited financial and personnel resources, a strategy to bypass pharmacy intervention and include treatment recommendations on the report was chosen for timely and effective communication of the results to the users. Stewardship recommendations and algorithms for possible test result scenarios were formulated after the review of institutional treatment strategies, practices and interventions based on conventional microbiology testing results, and antibiogram and formulary options. In addition, comments were developed to address possible limitations of the assay to avoid adverse patient outcomes. Extensive materials were developed for training and education. Electronic tools were developed for daily updates, surveillance, and audit of results.
Figure 1.
The seven key components of the SUCCESS model.
For monitoring the performance of the executed tests in routine three-shift microbiology laboratories, results were audited for a period of 6 months after the go-live date. A total of 15,793 blood cultures were performed during this period. Blood cultures with positive alerts from the BacT/ALERT system (bioMérieux, Inc., Durham, NC) containing gram-positive cocci (n = 579) and/or yeast (n = 28) were tested according to manufacturer procedures using the FDA-approved BC-GP (Nanosphere, Northbrook, IL) and Yeast PNA FISH assay (AdvanDX, Woburn, MA), respectively. Any results indicating no organisms detected or an internal control failure were reflexively repeated. Concurrent with the BC-GP and PNA-FISH testing, traditional laboratory procedures were also used to identify causative organisms, including plating on appropriate media and the routine biochemical and antibiotic susceptibility tests.
Results from rapid testing were compared with biochemical testing for concordance in identification and antibiotic susceptibility. Turnaround time analysis was measured from the time of Gram stain following culture bottle positivity to the availability of rapid assay results.
RESULTS
The execution model was direct communication of a detailed report with actionable guidelines to the physicians within 3 hours from the time of culture positivity using the laboratory and hospital information systems in addition to critical calls. Tables 1 and 2 outline the comments that were incorporated in the Verigene and PNA-FISH reports, respectively. Briefly, the initial Gram stain was reported with a critical call as routine. The report was updated with the Verigene or PNA-FISH results and the applicable stewardship comment within 1.5 hours for yeasts and 3 hours for bacteria. A second critical call was initiated at this time to communicate the result update to the physicians. Subsequently, the identification by conventional methods from culture and susceptibility results was communicated when available. Careful consideration was given to communicate the limitations of the assay with appropriate canned comments. For example, in an event of codetection of S. aureus, S. epidermidis, and mecA targets, a comment was added to specify that methicillin resistance was detected but could not be assigned to either S. aureus or S. epidermidis, and the recommendation was made to refer to the final pathogen identification and sensitivities to prevent premature change in management. Similarly, a comment was added to address the cross-reactivity of S. pneumoniae with other members of the S. mitis group. All not-detect calls also went out with a recommendation to refer to final identification and sensitivities to prevent misinterpreting a not-detect call as negative for targets that were not present on the BC-GP panel (Table 1).
Table 1.
Stewardship guidance for Verigene reportinga
| Verigene result | Reported organism | Comment |
|---|---|---|
| S. aureus | S. aureus | Methicillin susceptible. Consider de-escalating to oxacillin or cefazolin if no severe allergy. |
| S. epidermidis ± mecA | S. epidermidis (1 of 2 sets) | |
| Staphylococcus spp. | Coagulase-negative staphylococcus, NOT S. epidermidis or S. lugdunensis (1 of 2 sets) | Common contaminant, often does not require treatment.b |
| S. aureus + S. epidermidis + mecA |
S. aureus S. epidermidis |
Methicillin resistance detected but cannot be assigned to either S. aureus or S. epidermidis. Refer to final ID and sensitivities.c |
| E. faecalis + vanA and/or vanB | E. faecalis, vancomycin resistant; initiate VRE contact precaution | Consider de-escalating to ampicillin, ampicillin/sulbactam or piperacillin/tazobactam; synergistic gentamicin may be required with ampicillin in some circumstances. |
| E. faecalis | E. faecalis | |
| E. faecium + vanA and/or vanB | E. faecium, vancomycin resistant | Initiate VRE contact precaution. Consider daptomycin or linezolid. |
|
S. pyogenes S. agalactiae |
β-hemolytic strep group A β-hemolytic strep group B |
Consider de-escalating to oxacillin, cefazolin, or other penicillin if no severe allergy. |
| S. anginosis | S. anginosis | |
| S. pneumoniae | S. pneumoniae | Other members of Streptococcus mitis group may also give a positive result.c |
| Listeria spp. | Listeria spp. | Ampicillin is the preferred drug therapy. Consider switching to ampicillin if no severe allergy. For severe beta-lactam allergy, consider trimethoprim-sulfamethoxazole. |
| Not-detect | The Gram-positive organism(s) seen on Gram stain will be identified by routine culture and susceptibility methods. The organism(s) could not be identified by the Verigene Molecular Assay.c | |
| Verigene culture discordant result | Corrected report | This isolate was originally Not Detected by Verigene Molecular Assay. |
The stewardship guidance is based on institutional practice and guidelines and may not be generalizable.
Remove comment if second bottle turns positive for same morphology.
Remove comment when culture results are updated.
VRE indicates vancomycin-resistant enterococci.
Table 2.
Stewardship guidance for PNA-FISH reportinga
| PNA-FISH result | Reported organism | Comment |
|---|---|---|
| C. albicans/parapsilosis | C. albicans/parapsilosis | Unable to differentiate between C. albicans and C. parapsilosis. Both organisms are typically susceptible to fluconazole. Culture identification to follow.b |
| C. glabrata/krusei | C. glabrata/ krusei | Unable to differentiate between C. glabrata and C. krusei. Culture identification to follow. Use Micafungin until final susceptibilities are available.c |
| C. tropicalis | C. tropicalis | Typically susceptible to fluconazole. |
| Not-detect | The Yeast seen on Gram stain will be identified by routine culture. The yeast could not be identified by the PNA FISH.b | |
| PNA-FISH culture discordant result | Corrected report | This isolate was originally Not Detected by the PNA FISH. |
The stewardship guidance is based on institutional practice and guidelines and may not be generalizable.
Remove comment when culture results are updated.
Remove comment when sensitivity results are updated. C. krusei is intrinsically resistant to fluconazole.
PNA indicates peptide nucleic acid; FISH, fluorescence in situ hybridization.
For periodic cumulative updates to the physicians, an automated personalized Micro eUpdate service was provided. This service sent a summary of updated microbiology results by physician/physician group every 6 hours via a secure email. This provided easy access to results when the electronic medical record might not be easily accessible. A cumulative electronic report on Verigene and PNA-FISH results from the prior 24 hours was also sent to the pharmacist every morning. This allowed the pharmacist to monitor compliance with treatment recommendations and identify and target “nonadopters” for additional education. Using this approach, we were able to target 64% uptake at execution and ∼80% uptake after 3 months of execution after additional education (data not shown).
Performance of the tests was evaluated by daily audit and correlation with the conventional results. During the 6-month period, 579 blood cultures were assayed, of which 525 were monomicrobial and 54 were polymicrobial in culture. The correlation between the results by conventional methods and the Verigene BC-GP assay were 97% (508/525) for monomicrobial (Table 3) and 70% (38/54) for polymicrobial cultures (Table 4), with an overall correlation rate of 94% (546/579). The average turnaround time from Gram stain to Verigene reporting was 3.1 ± 1.1 hours.
Table 3.
Concordance between monomicrobial cultures and the isolates detected by BC-GP assay
| No. (%) of isolates |
Discrepant results | |||
|---|---|---|---|---|
| Monomicrobial culture results | Total | Correct calls | Comments | |
| Enterococcus faecalis | 14 (2.7%) | 14 (100%) | ||
| Enterococcus faecalis, vancomycin resistant | 1 (0.2%) | 1 (100%) | ||
| Enterococcus faecium | 2 (0.4%) | 2 (100%) | ||
| Enterococcus faecium, vancomycin resistant | 15 (2.9%) | 15 (100%) | ||
| Staphylococcus aureus | 51 (9.7%) | 48 (94.1%) | 3 | 2 S. epidermidis, 1 Staphylococcus spp. |
| Staphylococcus aureus, methicillin resistant | 51 (9.7%) | 50 (98.0%) | 1 | Missed mecA |
| Staphylococcus epidermidis | 193 (36.8%) | 192 (98.9%) | 1 | 1 Staphylococcus spp. |
| Staphylococcus lugdunensis | 1 (0.2%) | 1 (100%) | ||
| Staphylococcus spp. NOT S. epidermidis or S. lugdunensis | 105 (20.0%) | 102 (97.1%) | 3 | 3 S. epidermidis |
| Streptococcus agalactiae | 15 (2.9%) | 15 (100%) | ||
| Streptococcus anginosus group | 7 (1.3%) | 7 (100%) | ||
| Streptococcus spp. NOT S. pyogenes, S. agalactiae, S. pneumoniae, or S. anginosus group | 38 (7.2%) | 32 (84.2%) | 6 | 5 not detected, 1 S. pneumoniae |
| Streptococcus pneumoniae | 1 (0.2%) | 1 (100%) | ||
| Streptococcus pyogenes | 6 (1.1%) | 3 (50%) | 3 | 3 IC failure |
| Non–BC-GP target | 25 (4.8%) | 25 (100%) | ||
| Total | 525 | 508 | 17 | |
Sensitivity: 96.8%. BC-GP indicates gram-positive blood culture; IC, internal control.
Table 4.
Concordance between polymicrobial cultures and BC-GP assay
| No. of isolates |
|||||
|---|---|---|---|---|---|
| Verigene BC-GP call | Total | Correct reads | Discrepant results | Culture results | |
| Not detected | 16 | ||||
| 11 | 1 or 2 non–BC-GP targets | ||||
| 3 | CoNS + 1 or 2 non–BC-GP targets | ||||
| 1 | Viridans streptococci + CoNS | ||||
| 1 | Viridans streptococci + 1 or 2 non–BC-GP targets | ||||
| Staphylococcus spp. NOT S. epidermidis or S. lugdunensis | 11 | ||||
| 5 | CoNS + 1 or 2 non–BC-GP targets | ||||
| 5 | CoNS multiple morphotypes | ||||
| 1 | CoNS + viridans streptococci + 1 or 2 non–BC-GP targets | ||||
| Staphylococcus epidermidis | 15 | ||||
| 11 | S. epidermidis + 1 or 2 non–BC-GP targets | ||||
| 4 | S. epidermidis + CoNS | ||||
| Streptococcus spp. NOT S. pyogenes, S. agalactiae, S. pneumoniae, or S. anginosus group | 5 | ||||
| 2 | Streptococcus spp. + 1 or 2 non–BC-GP targets | ||||
| 2 | Streptococcus spp. + CoNS | ||||
| 1 | Streptococcus spp. + MRSA | ||||
| Staphylococcus aureus | 1 | ||||
| 1 | MSSA + 1 or 2 non–BC-GP targets | ||||
| Staphylococcus aureus + mecA | 4 | ||||
| 2 | MRSA + MSSA | ||||
| 2 | MSSA + CoNS | ||||
| Streptococcus pneumoniae | 1 | ||||
| 1 | S. pneumoniae + MSSA + 1 or 2 non–BC-GP targets | ||||
| Enterococcus faecalis + vanA | 1 | ||||
| 1 | E. faecalis (not VRE) + 1 or 2 non–BC-GP targets | ||||
| Total | 54 | 38 | 16 | ||
Sensitivity: 70.4%. BC-GP indicates gram-positive blood culture; CoNS, coagulase-negative Staphylococcus; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
There were seven errors with major clinical impact in the monomicrobial cultures, which included miscalling S. aureus as coagulase-negative staphylococci (n = 3), failure to identify the mecA marker in an S. aureus isolate (n = 1), and miscalling S. agalactiae as coagulase-negative staphylococci (n = 3). The highest proportion (47.06%) of errors was due to the inability to detect viridans streptococci (n = 5) or S. pyogenes (n = 3) (Table 3).
For polymicrobial cultures, the majority of the misses were coagulase-negative staphylococci and/or viridans streptococci with or without a non–BC-GP target. There was one mixed culture with Streptococcus spp. and methicillin-resistant Staphylococcus aureus, where Verigene missed S. aureus and the associated mecA target. Another mixed culture miscalled the vanA marker in a mix of E. faecalis and a non–BC-GP target, Enterobacter cloacae (Table 4).
In 41 instances the BC-GP molecular assay failed to identify any organisms, including the non–BC-GP targets such as Abiotrophia spp., Bacillus spp., Aerococcus spp., Actinomyces spp., Acinetobacter spp., anaerobic gram-positive cocci, Corynebacterium spp., Clostridium perfringens, Escherichia coli, Enterococcus casseliflavus, E. gallinarum, and Micrococcus spp. The most common organism associated with the not-detected call was Micrococcus spp. (n = 25).
PNA FISH analysis of 28 blood cultures containing yeast on Gram stain yielded identification of C. albicans/parapsilosis (n = 21), C. glabrata/krusei (n = 4), and C. tropicalis (n = 1). Two specimens were not-detect call by PNA-FISH and were identified as Cryptococcus neoformans after routine laboratory culture and identification methods. PNA-FISH gave accurate results for the two specimens mixed with bacterial targets. The average turnaround time from Gram stain to PNA-FISH reporting was ∼1.5 hours.
DISCUSSION
The validation of newer techniques is a vital component in the endless process of laboratory improvement. There is substantial data in the literature to support the superior laboratory performance and better turnaround time of new diagnostics such as Verigene and PNA-FISH compared to conventional methods for sepsis (11–15). However, limited guidance is available for integration of such techniques into the laboratory workflow, and the subsequent introduction to the clinical setting reveals a separate set of challenges. In this study, we sought to evaluate both the validity of the laboratory portion of the molecular assay in a true clinical setting along with the execution and uptake of the results by the end users.
The laboratory performance for BC-GP in our study was comparable to that of published reports. The overall concordance between the Verigene BC-GP assay and the expected results (i.e., correct identification of targeted organisms and susceptibility) using conventional testing was 94%. Many of the discrepancies were related to organisms of little relative clinical significance, such as S. epidermidis or another coagulase-negative staphylococcus in a single blood culture set, or where there was another accurately reported pathogenically dominant organism in a mixed culture (such as pneumococcus alongside coagulase-negative staphylococcus). Other discrepancies included clinically relevant gram-positive rods, which were not the targets of the molecular assay. One notable discrepancy in our study was a 50% (3 of 6 cultures) failure rate of the BC-GP assay to detect Group A streptococcus. This was realized as a limitation of the assay. Group A streptococcus harbors cell-wall–located DNase, which serves as an important virulence factor in pathogenesis (16). The DNases are also known to degrade the internal processing control that comprises a nontarget organism, Bacillus subtilis, which invalidated the result.
Despite significant literature on analytical and laboratory validation of rapid diagnostics, there are a handful of reports in the literature that have looked at the clinical and economic outcomes for patients after successful execution. Sango et al (17) evaluated the impact of Enterococcus identification and resistance detection using Verigene BC-GP. The intervention by an infectious disease and/or critical care pharmacist on 74 patients with enterococcal bacteremia led to a significant decrease in the mean time to appropriate antimicrobial therapy in the postintervention group (23.4 h; P = 0.005) compared with the preintervention group. Alby et al (18) developed a treatment algorithm for streptococci and enterococci identified with the Verigene BC-GP assay in collaboration with their institutional antimicrobial stewardship program. However, the execution plan and algorithm utilization still relied on effective manual communication of the BC-GP results directly to an on-call pharmacist, who in turn used the treatment algorithm as a guide when recommending therapy at the bedside. Bauer et al (19) also used direct phone contact with the infectious diseases pharmacist as an effective mode of communication with results of the rapid PCR for methicillin-resistant S. aureus/S. aureus bacteremia. Clinical and economic outcome evaluation on 156 patients showed that the mean time to switch from empiric vancomycin to cefazolin or nafcillin in patients with methicillin-susceptible S. aureus bacteremia was 1.7 days shorter (P = 0.002), the mean length of stay was 6.2 days shorter (P = 0.07), and the mean hospital costs were $21,387 less (P = 0.02) after PCR. However, none of these studies demonstrated an effective execution plan for an in-hospital or reference laboratory serving a hospital with limited staff available for routine stewardship. In addition, the additional cost incurred due to extended stewardship resources is not accounted for in the cost evaluation.
To our knowledge, this is a first communication that describes the effective execution of rapid molecular diagnostics for sepsis without direct involvement of an infectious disease pharmacist or physician for routine stewardship recommendations. Using our SUCCESS approach, we have eliminated the additional cost and time related to a pharmacist's intervention. Using our model, the execution of the molecular testing procedures into the laboratory and clinical reporting workflow went smoothly, with technologist training, bench-aids, daily audits, and easy access to technical and scientific guidance all contributing to smooth integration of the new methods. In general, the ordering clinicians viewed the newly available type of data generated by the molecular assay as positive and useful in the patient management workflow, based on feedback provided to the microbiology hotline, as well as email and verbal communications. The high rate of correlation between rapid methods and culture validates the usefulness of the BC-GP molecular assay in a clinical laboratory workflow for providing proper and rapid patient management. The impact on clinical outcomes from this study is addressed in a separate communication (20). Briefly, we demonstrated a significant decrease (P < 0.05) in time from collection to the first dose of appropriate antibiotics for patients with methicillin-sensitive Staphylococcus aureus and vancomycin-resistant enterococci. Additionally, the percent of patients on empiric therapy who were placed on appropriate antibiotics after the Gram stain result was available increased from 64% pre-BC-GP to 80% post-BC-GP (20).
As with any other technology, there are several limitations of the BC-GP assay. First, it can only report from a small set of organisms, and some of those are called only to genus level. While the majority of bacteremia cases are due to specifically targeted organisms, the clinical reality is that there may be a nontrivial amount of bacteremia due to other nonpredominating organisms. Second, the resistance markers cannot be assigned to a particular marker in a mixed culture. This is particularly an issue when S. aureus, S. epidermidis, and mecA are concurrently detected. A resolution by conventional testing is required for such cases. Lastly, the assay cannot differentiate between S. pneumoniae and the S. mitis/oralis group. False-positive S. pneumoniae does not limit the utility of the assay, as treatment algorithms can be developed around this limitation by effective communication of the results and assay limitations (18).
Acknowledgments
We would like to thank all the lab personnel that helped to collect this data.
References
- 1.Bearman GM, Wenzel RP. Bacteremias: a leading cause of death. Arch Med Res. 2005;36(6):646–659. doi: 10.1016/j.arcmed.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Laupland KB, Zygun DA, Davies HD, Church DL, Louie TJ, Doig CJ. Population-based assessment of intensive care unit-acquired bloodstream infections in adults: incidence, risk factors, and associated mortality rate. Crit Care Med. 2002;30(11):2462–2467. doi: 10.1097/00003246-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Gaieski DF, Mikkelsen ME, Band RA, Pines JM, Massone R, Furia FF, Shofer FS, Goyal M. Impact of time to antibiotics on survival in patients with severe sepsis or septic shock in whom early goal-directed therapy was initiated in the emergency department. Crit Care Med. 2010;38(4):1045–1053. doi: 10.1097/CCM.0b013e3181cc4824. [DOI] [PubMed] [Google Scholar]
- 4.Lueangarun S, Leelarasamee A. Impact of inappropriate empiric antimicrobial therapy on mortality of septic patients with bacteremia: a retrospective study. Interdiscip Perspect Infect Dis. 2012;2012:765205. doi: 10.1155/2012/765205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34(6):1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 6.Khatib R, Saeed S, Sharma M, Riederer K, Fakih MG, Johnson LB. Impact of initial antibiotic choice and delayed appropriate treatment on the outcome of Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis. 2006;25(3):181–185. doi: 10.1007/s10096-006-0096-0. [DOI] [PubMed] [Google Scholar]
- 7.Lodise TP, McKinnon PS, Swiderski L, Rybak MJ. Outcomes analysis of delayed antibiotic treatment for hospital-acquired Staphylococcus aureus bacteremia. Clin Infect Dis. 2003;36(11):1418–1423. doi: 10.1086/375057. [DOI] [PubMed] [Google Scholar]
- 8.Lee DH, Kim SC, Bae IG, Koh EH, Kim S. Clinical evaluation of BacT/Alert FA plus and FN plus bottles compared with standard bottles. J Clin Microbiol. 2013;51(12):4150–4155. doi: 10.1128/JCM.01935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott LJ. VerigeneR gram-positive blood culture nucleic acid test. Mol Diagn Ther. 2013;17(2):117–122. doi: 10.1007/s40291-013-0021-z. [DOI] [PubMed] [Google Scholar]
- 10.Stender H, Williams B, Coull J. PNA fluorescent in situ hybridization (FISH) for rapid microbiology and cytogenetic analysis. Methods Mol Biol. 2014;1050:167–178. doi: 10.1007/978-1-62703-553-8_14. [DOI] [PubMed] [Google Scholar]
- 11.Beal SG, Ciurca J, Smith G, John J, Lee F, Doern CD, Gander RM. Evaluation of the Nanosphere Verigene gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J Clin Microbiol. 2013;51(12):3988–3992. doi: 10.1128/JCM.01889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchan BW, Ginocchio CC, Manii R, Cavagnolo R, Pancholi P, Swyers L, Thomson RB, Jr, Anderson C, Kaul K, Ledeboer NA. Multiplex identification of gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med. 2013;10(7):e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan KV, Turner NN, Roundtree SS, Young S, Brock-Haag CA, Lacey D, Abuzaid S, Blecker-Shelly DL, Doern CD. Rapid detection of gram-positive organisms by use of the Verigene gram-positive blood culture nucleic acid test and the BacT/Alert Pediatric FAN system in a multicenter pediatric evaluation. J Clin Microbiol. 2013;51(11):3579–3584. doi: 10.1128/JCM.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, Procop GW, Richter SS. Evaluation of the Verigene gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2013;51(7):2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mestas J, Polanco CM, Felsenstein S, Dien Bard J. Performance of the Verigene gram-positive blood culture assay for direct detection of gram-positive organisms and resistance markers in a pediatric hospital. J Clin Microbiol. 2014;52(1):283–287. doi: 10.1128/JCM.02322-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa T, Minami M, Okamoto A, Tatsuno I, Isaka M, Ohta M. Characterization of a virulence-associated and cell-wall-located DNase of Streptococcus pyogenes. Microbiology. 2010;156(Pt 1):184–190. doi: 10.1099/mic.0.031955-0. [DOI] [PubMed] [Google Scholar]
- 17.Sango A, McCarter YS, Johnson D, Ferreira J, Guzman N, Jankowski CA. Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol. 2013;51(12):4008–4011. doi: 10.1128/JCM.01951-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alby K, Daniels LM, Weber DJ, Miller MB. Development of a treatment algorithm for streptococci and enterococci from positive blood cultures identified with the Verigene gram-positive blood culture assay. J Clin Microbiol. 2013;51(11):3869–3871. doi: 10.1128/JCM.01587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer KA, West JE, Balada-Llasat JM, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program's impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis. 2010;51(9):1074–1080. doi: 10.1086/656623. [DOI] [PubMed] [Google Scholar]
- 20.Beal SG, Thomas C, Dhiman N, Nguyen D, Qin H, Hawkins JM, Dekmezian M, Benavides R. Antibiotic utilization improvement with the Nanosphere Verigene Gram-Positive Blood Culture assay. Proc (Bayl Univ Med Cent) 2015;28(2):139–143. doi: 10.1080/08998280.2015.11929214. [DOI] [PMC free article] [PubMed] [Google Scholar]