Abstract
We evaluated four dengue diagnostic devices from Alere, including the SD Bioline Dengue Duo (nonstructural [NS] 1 Ag and IgG/IgM), the Panbio Dengue Duo Cassette (IgM/IgG) rapid diagnostic tests (RDTs), and the Panbio dengue IgM and IgG capture enzyme-linked immunosorbent assays (ELISAs) in a prospective, controlled, multicenter study in Peru, Venezuela, Cambodia, and the United States, using samples from 1,021 febrile individuals. Archived, well-characterized samples from an additional 135 febrile individuals from Thailand were also used. Reference testing was performed on all samples using an algorithm involving virus isolation, in-house IgM and IgG capture ELISAs, and plaque reduction neutralization tests (PRNT) to determine the infection status of the individual. The primary endpoints were the clinical sensitivities and specificities of these devices. The SD Bioline Dengue Duo had an overall sensitivity of 87.3% (95% confidence interval [CI], 84.1 to 90.2%) and specificity of 86.8% (95% CI, 83.9 to 89.3%) during the first 14 days post-symptom onset (p.s.o.). The Panbio Dengue Duo Cassette demonstrated a sensitivity of 92.1% (87.8 to 95.2%) and specificity of 62.2% (54.5 to 69.5%) during days 4 to 14 p.s.o. The Panbio IgM capture ELISA had a sensitivity of 87.6% (82.7 to 91.4%) and specificity of 88.1% (82.2 to 92.6%) during days 4 to 14 p.s.o. Finally, the Panbio IgG capture ELISA had a sensitivity of 69.6% (62.1 to 76.4%) and a specificity of 88.4% (82.6 to 92.8%) during days 4 to 14 p.s.o. for identification of secondary dengue infections. This multicountry prospective study resulted in reliable real-world performance data that will facilitate data-driven laboratory test choices for managing patient care during dengue outbreaks.
INTRODUCTION
Dengue fever is the most important arthropod-borne viral disease in terms of human morbidity, mortality, and economic impact (1). Dengue fever is caused by the dengue virus (DENV), a flavivirus that can be classified into four predominant serotypes (DENV-1, -2, -3, and -4) (2). DENV comprises three structural proteins (capsid, membrane, and envelope) and seven nonstructural (NS) proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5). DENV is transmitted by mosquitoes, principally Aedes aegypti and Aedes albopictus. Clinically, dengue fever is characterized by fever, headache, myalgia, arthralgia, rash, leukopenia, and sometimes thrombocytopenia (3, 4). The severity of the disease can range from asymptomatic or mild to severe with high fever, hemorrhage, and shock (2, 5). Severe dengue can sometimes lead to shock and even death, especially in the absence of fluid replacement and modern supportive care (2). There is no vaccine or antiviral drug to prevent or cure dengue fever (6). The only available treatment options are supportive therapies, including bed rest, fluids, and symptomatic relief using analgesics (7, 8). The accurate diagnosis of dengue followed by attentive supportive care to manage severe dengue can demonstrably improve outcomes (5). Managing the consequences of hemorrhage can save lives and decrease hospitalization costs (9). A timely diagnosis of the disease also enables health care professionals to exclude other causes of febrile illness which presents with similar clinical symptoms in the acute phase of disease in areas where dengue is endemic (9–12). Early and rapid dengue identification is equally important for epidemiologists and public health officers, providing a means of monitoring dengue transmission dynamics in real time and allowing for a more rapid response to dengue outbreaks. Rapid case confirmation can also inform timely and focused vector control measures, the most effective responses to outbreaks (13, 14).
One traditional method of diagnosing dengue infections involves the incubation of acute-phase patient serum samples with a permissive cell line in tissue culture, followed by an immunofluorescence assay (IFA) using serotype-specific monoclonal antibodies to identify any growing virus (15). Although this method requires several days, it is still regarded as a gold standard. More frequently, an enzyme-linked immunosorbent assay (ELISA) is used to detect an immune response to DENV infection in the form of anti-dengue IgM or IgG antibodies (16). Serological responses are detectable following onset of symptoms, and a rise in the titer is determined with greater accuracy when paired samples from both acute- and convalescent-phase time points are used (17). Newer generations of dengue diagnostic devices also detect the NS1 protein, which is released into the serum early on in a DENV infection and may facilitate acute-phase diagnosis (18–21). A traditional ELISA can take several hours to complete and often involves an overnight incubation step for special reagent preparation (10, 22). Thus, these techniques are not only time-consuming but also labor-intensive, requiring trained personnel and specialized equipment. A delay in obtaining laboratory results can hinder evidence-based patient care decisions, leading physicians to rely on symptomatic diagnosis, the accuracy of which varies with physician experience (23–27).
The purpose of the current study was to evaluate the diagnostic accuracy of four devices fulfilling this critical need by conducting a comprehensive prospective multicountry evaluation of two dengue rapid diagnostic tests (RDTs) and two ELISAs: the SD Bioline Dengue Duo, the Panbio Dengue Duo Cassette, the Panbio dengue IgM capture ELISA, and the Panbio dengue IgG capture ELISA (Alere Inc., Waltham, MA). The primary endpoints of our trial were clinical sensitivity and specificity. The study was undertaken using quality systems approaching good clinical laboratory practices (GCLP) suitable for data submission to the U.S. Food and Drug Administration (FDA) for clearance, generating the most reliable and rigorous performance data on these dengue diagnostic devices to date.
(Some of these data were presented in a poster at the 63rd Annual Meeting of the American Society of Tropical Medicine and Hygiene, November 2014.)
MATERIALS AND METHODS
Human use statement.
The procedures undertaken in this study were done in accordance with the ethical standards of the Naval Medical Research Unit No. 6 (NAMRU-6) institutional review board (IRB) in compliance with all applicable federal regulations governing the protection of human subjects. The following study protocols were approved for this study: NMRC.2010.0021 for the Naval Medical Research Center (NMRC) (Silver Spring, MD, USA), NMRCD.2010.0005 for the Naval Medical Research Unit No. 6 (Lima, Peru), NAMRU2.2010.0003 for the Naval Medical Research Unit-2 (NAMRU-2 PP) (Phnom Penh, Cambodia), Walter Reed Army Institute of Research (WRAIR) protocol 1770 for the Armed Forces Research Institute of Medical Sciences (AFRIMS) (Bangkok, Thailand), CBIIB(UC)-015 for Laboratorio Regional de Diagnostico e Investigación del Dengue y otras Enfermedades Virales (LARDIDEV), Instituto de Investigaciones Biomédicas de la Universidad de Carabobo (BIOMED-UC) (Maracay, Venezuela), and IRB 09-240 for the University of Texas Medical Branch (UTMB) (Galveston, TX, USA). Study protocols were also reviewed by public health authorities in Peru, Thailand, Cambodia, and Venezuela, and permission was obtained in writing prior to study commencement. Written informed consent was obtained from subjects 18 years of age and older. In Texas, verbal assent was obtained from all participants; the requirement for written consent was waived due to the minimal risk associated with this study. For younger participants (<18 years of age), written consent was obtained from a parent or legal guardian, and written assent was obtained from the participant when appropriate.
Study sites.
Prospective subject recruitment took place at participating clinics and hospitals in Peru, Cambodia, Venezuela, and the United States. In Peru, this included 12 hospitals and clinics around Iquitos in collaboration with the NAMRU-6 laboratories in Lima and Iquitos, in Venezuela, this included two major general hospitals (Hospital Central de Maracay and Hospital Instituto Venezolano de los Seguros Sociales [IVSS] Jose Maria Carabaño Tosta) and two outpatient clinics (Ambulatorio 23 de Enero and Ambulatorio Hospital Civil), in Cambodia, this included one health clinic from a province outside Phnom Penh and testing at the field laboratory at NAMRU-2, and in Texas, this included the UTMB Emergency Department and an outpatient general medicine clinic.
Study design. (i) Quality systems.
The entire study was performed under quality systems described in title 21 of the Code of Federal Regulations, part 58 describing GLP, and title 42 of the Code of Federal Regulations, part 493 describing Clinical Laboratory Improvement Amendments (CLIA) laboratory requirements. Manufacturer-provided positive and negative controls were run every day to ensure experimental device viability in accordance with a quality control plan. A failure of the internal and/or external controls was reported to the study monitor as invalid results, and the specimen was retested if volume permitted. An external regulatory consultant (MDC Associates LLC, Beverly, MA) in consultation with the site principal investigators monitored the study quality systems.
(ii) Subject recruitment.
All subjects presenting to local clinics and hospitals with high fever and suspicion of dengue were invited to participate in the study and presented with informed consent forms. The inclusion criteria for participation included fever symptoms consistent with possible dengue (≥38°C oral, tympanic, or rectal; ≥37.5°C axillary) accompanied with headache, muscle and ocular and/or joint pain, and the availability of paired samples. The acute-phase sample had to be collected within the first 6 days post-symptom onset (p.s.o.) and the convalescent-phase sample between 2 and 30 days after the acute-phase sample. The majority of convalescent-phase samples were collected day 15 p.s.o. or later (90.1%). Exclusion criteria included the following: any person not meeting the inclusion criteria, persons with severe or acute mental or physical disabilities, persons from whom insufficient (<750 μl) sera and/or plasma volume was obtained, samples with any visible or documented problems (hemolysis, lipemia, microbial growth, failure to maintain a sample storage temperature <10°C, or >5 freeze-thaws), or any person for whom a convalescent-phase sample was not available (with the exception of persons in Texas). All withdrawals were reported to the study monitor and documented. Symptoms and demographic information were collected. The acceptable age of enrollment was determined by the individual study site in collaboration with local ministries of health (where applicable).
(iii) Test performance.
Four dengue diagnostic devices manufactured by Alere Inc. were evaluated in this clinical study: the SD Bioline Dengue Duo (catalog no. 11FK45), the Panbio Dengue Duo Cassette (catalog no. R-DEN03D), the Panbio dengue IgM capture ELISA (catalog no. E-DEN01M), and the Panbio dengue IgG capture ELISA (catalog no. E-DEN02G). All devices were provided by Alere and were run with strict adherence to the manufacturer's instructions. For all Panbio products, 10 μl of whole blood, serum, or plasma was used, while the SD Bioline Dengue Duo required 3 drops (approximately 100 μl) and 10 μl of serum for the NS1 Ag and IgM/IgG portions of the test, respectively. Positive controls (pooled human IgM and IgG positive samples and contrived pooled NS1) were run every day. Device failure, as indicated by a failure of the positive controls to be reactive on the RDTs, did not occur in this study. Device failure would have invalidated any data collected on that day and required corrective action. ELISAs came with manufacturer-provided positive and negative controls and a calibrator sample. Tests with equivocal (ELISAs only) or invalid results (failure of internal and/or external controls) were repeated when specimen was available.
Whole blood samples via fingersticks were used to test the Panbio Dengue Duo Cassette only. This testing was performed by a health care provider at the enrollment site. Venous blood specimens, which were then taken to the individual central laboratories and processed into serum daily, were collected at all prospective site. Venous blood was also processed into plasma in Peru and Texas to evaluate the effect of different matrices on diagnostic performance. Serum (and plasma where applicable) samples were used to interrogate the Panbio Dengue Duo Cassette at the site laboratory. All other devices were tested using only patient sera. The matrices evaluated on each device were chosen based on the data required to support the individual product claims. Matrix inclusion was also influenced by logistical limitations at the clinical trial sites. Enrollees were asked to return to the clinic 2 to 30 days after the first blood draw only at the study sites where dengue is endemic (Peru, Cambodia, and Venezuela), and the specimen from this visit was also used to interrogate the dengue diagnostic devices using the same workflow as that described for the acute-phase specimens. Samples from an additional 135 individuals from Thailand were selected for evaluation of the devices. These retrospective specimens were selected from a larger archive of previously characterized dengue-positive and -negative samples, with a special emphasis on capturing late time points (days 4 to 14 p.s.o.) and primary infections, as well as all four DENV serotypes.
Reference testing.
Blind coded aliquots of all specimens meeting the inclusion criteria were shipped on dry ice to a central reference laboratory in Thailand (AFRIMS). A comprehensive gold standard testing algorithm utilizing both acute- and convalescent-phase specimens was used to determine the overall dengue infection status for each individual enrollee (Fig. 1). Virus isolation was attempted for all acute-phase samples in C6/36 cells and visualized using a dengue ELISA as previously described (17, 28, 29); any individual with a confirmed DENV virus culture was considered to be dengue positive. In-house dengue IgM and IgG capture ELISAs were also performed on all paired samples using the methods previously described (30). These titers were interpreted by subject matter experts, using an established algorithm. Briefly, at least 40 U of anti-dengue IgM (wherein anti-dengue IgM must also be greater than anti-Japanese encephalitis virus [JEV] IgM) in an acute-phase sample was considered evidence of recent dengue infection. When an acute-phase sample was determined to be IgM positive, a dengue IgM-to-IgG ratio of ≥1.8 was defined as a primary dengue virus infection, while a ratio of <1.8 was defined as a secondary dengue virus infection. In the absence of dengue IgM (<40 U), subjects could still be classified as having a secondary infection if their IgG titers exceeded 100 U and demonstrably rose (>2-fold) between the acute- and convalescent-phase blood draws. Recent dengue exposure and primary and secondary infection status were determined as previously described (30, 31). Any individuals whose IgM and IgG ELISA profiles were suggestive of recent dengue infection yet who did not have isolatable DENV were further characterized using a plaque reduction neutralization test (PRNT50) (32, 33). A 4-fold rise in the PRNT titers against DENV between acute- and convalescent-phase samples was interpreted as confirmation of recent DENV infection, a rise between 2- and 4-fold was considered to be equivocal, and any rise of <2-fold was considered to be dengue negative (34). In summary, individuals determined to be dengue positive via our reference standard had either isolatable DENV in their acute-phase samples or were confirmed to be serology positive for dengue exposure based on an in-house capture ELISA and PRNT. Individuals who tested negative for isolatable DENV as well as ELISA serology were classified as dengue negative. A small number of individuals (1%) could not be conclusively identified as dengue positive or negative by this reference algorithm and were classified as equivocal and excluded from the calculations of device sensitivity. Retrospective specimens utilized in this evaluation were previously characterized using the same techniques described above, with the exception of PCR being used to detect acute viremia in lieu of virus culture (35).
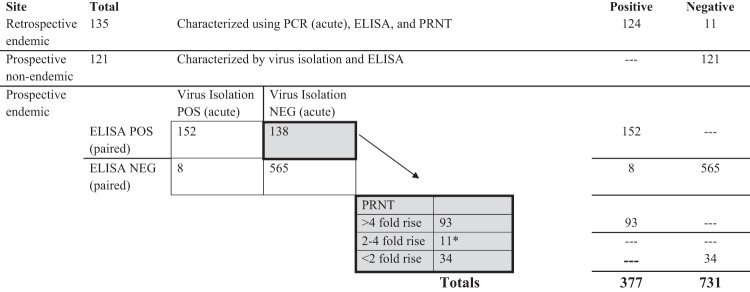
FIG 1.
Reference testing results. *, number of samples showing a 2- to 4-fold rise in PRNT titers between acute- and convalescent-phase samples were classified as equivocal and not used for calculating device sensitivity or specificity.
Statistical methods.
Sensitivity, specificity, and agreement were calculated with reference to the gold standard reference methodology using widely accepted definitions (36). Confidence intervals (CIs) for sensitivity and specificity were calculated using the exact binomial method (37). The Z-ratio for the significance of the difference between two independent ratios was calculated to determine whether a given sensitivity (or specificity) was statistically different from another, and a P value of <0.05 (two-tailed) was considered to be statistically significant (38). Cohen's kappa (κ) was used to describe the degree of agreement between populations or tests (39).
RESULTS
Patient demographics and dengue prevalence by site.
Prospective subject recruitment for this study occurred between March 2010 and April 2012 (Table 1). A total of 1,247 subjects were prospectively recruited; of these specimens from 1,156 subjects met all inclusion and exclusion criteria. The primary reason for withdrawal from the analysis was a failure to appear for a convalescent-phase blood draw. Other reasons for withdrawal included the following: samples collected too late for acute- or convalescent-phase classification (>6 days for acute-phase or >30 days for convalescent-phase), subjects withdrawing consent, insufficient devices to run quality control testing on the day of subject enrollment, insufficient sample volume collected from the subject, and/or significant hemolysis in serum samples. No adverse events due to the study participation were reported. A majority of the subjects with symptoms suggestive of dengue fever in Peru, Cambodia, Venezuela, and Texas presented to the clinic for care within days 1 to 4 p.s.o. (88.3%), with the highest number (29%) self-reporting that they had experienced high fever symptoms for 2 days (Fig. 2). Very few individuals reported to the clinic for an acute-phase blood draw on days 5 to 7 p.s.o. (5%). Symptoms at presentation included fever and headache and occasionally included pain, rash, chills, nausea, diarrhea, or vomiting. Approximately equal numbers of male and female subjects were recruited at all sites (Table 1). A majority of subjects reporting to the clinic in Cambodia with suspicions of dengue were children <18 years of age (ages ranged from 2 to 80, with a median of 8 years), while Peru and Venezuela had a more uniform distribution of both children and adults (5 to 85 and 1 to 61 years of age with a median of 26 and 17 years of age, respectively). Enrollees in Texas were all adults (≥18 years of age), as specified in their study protocol, with an age range from 18 to 97 years and median age of 45 years. This study captured naturally circulating variants of all 4 DENV serotypes from the sites where dengue is endemic (Table 2). Peru observed only DENV-2 and DENV-4 circulating during their study enrollment period (71% and 29%, respectively), while Cambodia predominated with DENV-1 and DENV-2 serotypes (69% and 30%, respectively), underscoring the importance of multicountry participation in order to capture serotype diversity. Venezuela had a balance of all 4 circulating serotypes, with DENV-3 being the predominant serotype (DENV-1, 22%; DENV-2, 14%; DENV-3, 50%; and DENV-4, 14%). Retrospective samples from AFRIMS were selected to represent all DENV serotypes (DENV-1, 35%; DENV-2, 24%; DENV-3, 31%; and DENV-4, 10%).
TABLE 1.
Subject recruitment informationa
| Study site | No. of subjects | Sample dates | No. of adults ≥18 yr | No. of children <18 yr | % male |
|---|---|---|---|---|---|
| Thailand | 135 | 3/2009−7/2011 | 2 | 133 | |
| Cambodia | 383 | 7/2011−9/2011 | 100 | 283 | 48 |
| Peru | 357 | 9/2010−4/2012 | 277 | 80 | 46 |
| Texas | 137 | 3/2010−12/2011 | 137 | 0 | 37 |
| Venezuela | 144 | 1/2011−2/2012 | 66 | 78 | 54 |
| Totals | 1,156 | 582 | 574 |
Numbers of subjects at each site are listed. Archived sample collection dates for Thailand and prospective enrollment dates for Cambodia, Texas, Peru, and Venezuela are shown. At the Texas site, individuals provided only one sample during acute febrile infection. At all other sites, paired samples were obtained and used in evaluating all four diagnostic devices and reference testing. Numbers of adults and children and the percentage of the population that was male are presented.
FIG 2.
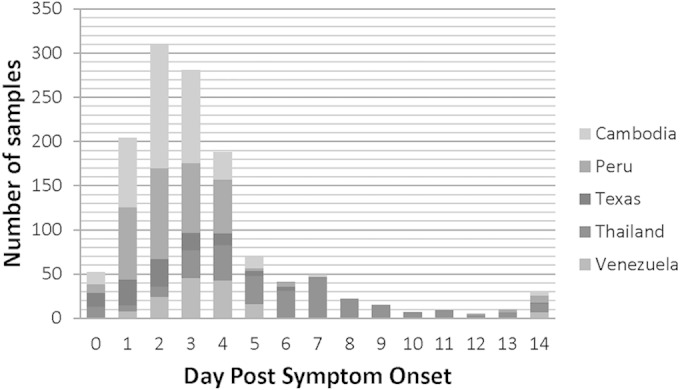
Numbers of samples collected (y axis) on each day p.s.o. (x axis) at each site from days 0 to 14 p.s.o. are shown. At prospective sites, this represents the times when patients with fever symptoms reported to the participating medical centers for diagnosis and treatment.
TABLE 2.
Characterization of study participants based on reference testinga
| Site | DENV positive | Infection status |
Serotype |
DENV negative | Equivocal | Prevalence (%) | Primary infection (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary | Secondary | DENV1 | DENV2 | DENV3 | DENV4 | ||||||
| Venezuela | 30 | 3 | 26 | 14 | 3 | 2 | 7 | 104 | 4 | 22 | 10 |
| Texas | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 121 | 0 | 0 | - |
| Peru | 149 | 0 | 145 | 0 | 58 | 0 | 24 | 188 | 5 | 44 | 0 |
| Cambodia | 74 | 14 | 57 | 44 | 19 | 0 | 1 | 307 | 2 | 19 | 19 |
| Thailand | 124 | 44 | 80 | 41 | 24 | 17 | 5 | 11 | 0 | ||
| Total | 377 | 61 | 308 | 88 | 103 | 24 | 32 | 731 | 11 | ||
The numbers of individuals considered DENV positive, DENV negative, or equivocal based on reference testing are shown. These numbers were used to calculate the prevalence of dengue among febrile patients meeting the inclusion criteria and the rate of primary infections among DENV-positive individuals. When the virus is isolatable from the acute-phase sample, the serotype of DENV isolated is shown.
Among the paired, retrospective samples obtained in Thailand from 135 individuals, 124 had been previously characterized as DENV positive, while 11 had been characterized as DENV negative (Fig. 1). All 121 study subjects in Texas were determined to be dengue negative by virus isolation and capture ELISA. Among all specimens prospectively collected from sites where dengue is endemic (acute and convalescent phases), 1,108 were characterized with complete reference data. Among these, 565 individuals were negative for both virus isolation and capture ELISA and were therefore classified as dengue negative (Fig. 1). The etiology of fever in dengue-negative patients was not characterized further. Another 152 individuals were determined to be dengue positive by both virus isolation and anti-DENV IgM and IgG ELISA results, consistent with recent dengue exposure. Interestingly, 8 individuals had isolatable DENV but no serological response in a comparison of IgM and IgG titers in both acute- and convalescent-phase specimens; these samples were classified as DENV positive, but their primary or secondary infection status remained uncharacterized. Another 138 subjects were determined to be positive by capture ELISA, but no virus could be isolated from the acute-phase specimen. These samples were further characterized using PRNT; among these, 93 individuals demonstrated a rise in neutralizing titers of >4-fold and were called dengue positive. Thirty-four individuals demonstrated a rise in neutralizing antibody titers of <2-fold between acute- and convalescent-phase specimens and were classified as dengue negative. Those individuals with rises between 2- and 4-fold were considered to be equivocal (11 individuals), and these subjects could not be conclusively characterized as dengue positive or negative based on ELISA or PRNT reference results (Fig. 1).
According to the reference standard classification, the site-specific breakdown of dengue-positive and -negative individuals is summarized in Table 2. The prevalence of laboratory-confirmed dengue among febrile participants from sites where dengue is endemic ranged from 19.4% (Cambodia) and 22.4% (Venezuela) to 44.2% (Peru). Interestingly, none of the individuals enrolled in Peru during the study were characterized as having a primary infection, while in Venezuela and Cambodia, 10% and 19%, respectively, of their subjects were characterized as such.
Rapid devices.
We determined the overall diagnostic accuracy of the SD Bioline Dengue Duo by comparing the dengue classification of an individual, based on reference testing, to the device result. According to the product insert, the presence of any line (NS1, IgM, or IgG) was interpreted as positive for dengue. The device had a sensitivity of 80.2% for days 0 to 3 p.s.o., after which the sensitivity rose to 89.5% on days 4 to 7 and to 98.5% for days 8 to 14 p.s.o. (Fig. 3A). Over days 0 to 14 p.s.o., the SD Bioline Dengue Duo demonstrated a clinical sensitivity of 87.3% (95% CI, 84.4 to 90.2%) and specificity of 86.8% (95% CI, 84.1 to 89.4%) when serum samples were used. The Panbio IgM/IgG Dengue Duo Cassette, on the other hand, had a lower sensitivity of 48% for days 0 to 3 p.s.o., but the sensitivity at later time points rose to 89.5% for days 4 to 7 p.s.o. and to 98.5% for days 8 to 14 p.s.o. (Fig. 3A). We selected days 4 to 14 p.s.o. as a clinically useful and relevant window for diagnosis using IgM and IgG responses. During this period, we observed a clinical sensitivity of 92.1% (95% CI, 87.8 to 95.2%) when serum samples were used. The test demonstrated comparable sensitivity using fingerstick whole-blood (87.7%; 95% CI, 76.3 to 94.9%) or plasma (97.0%; 95% CI, 84.2 to 99.9%) samples for the same time period, and any differences in sensitivities were not statistically significant (see Table S1 in the supplemental material). The clinical specificity of the Panbio Dengue Duo Cassette was 62.2% (95% CI, 55.0 to 69.5%) when serum samples were used and remained comparable for other matrices: 63.5% (55.9 to 71.0%) for whole blood and 74.6% (64.2 to 85.0%) for plasma samples. Agreement (Cohen's κ) between the matrices was uniformly high: 74.3% (κ = 0.484) between serum and whole blood; 92.8% (κ = 0.856) between serum and plasma; and 80.6% (κ = 0.612) between plasma and whole blood.
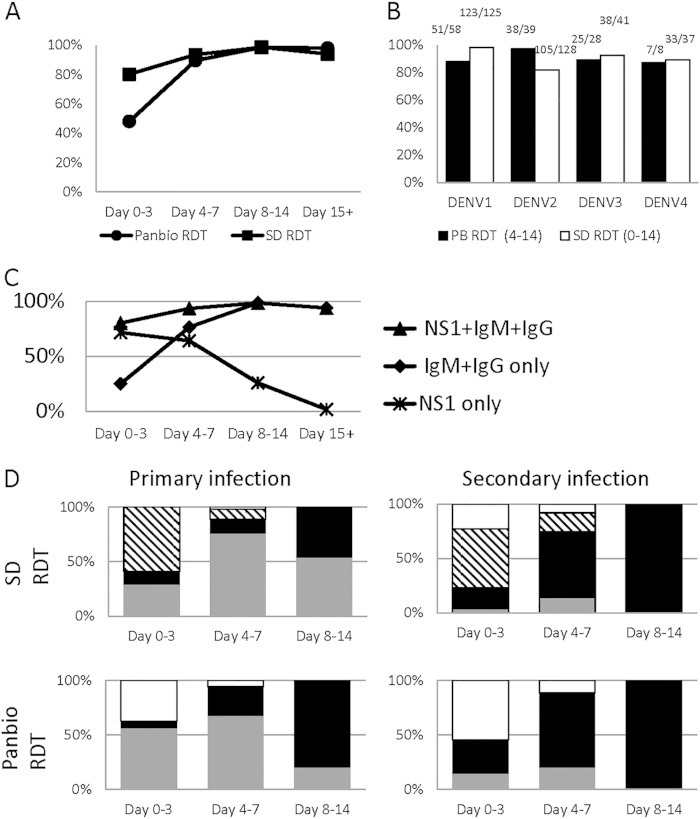
FIG 3.
Rapid diagnostic test performance. (A) Sensitivities of the Panbio and SD RDTs over time are shown. (B) When virus characterization was possible, the sensitivities of the Panbio RDT for days 4 to 14 p.s.o. and SD RDT for days 0 to 14 p.s.o. for each serotype are shown. The number of positive test results over the total number of samples of the given serotype that were tested using the device is provided. (C) The SD Bioline Dengue Duo is reactive to DENV NS-1, IgM, and IgG; the sensitivity of the device if the NS-1 line only, the IgM and IgG lines only, or the presence of any line (NS1/IgM/IgG) is used to interpret DENV positivity is shown to demonstrate the temporal effectiveness of the individual DENV biomarkers alone and in conjunction. (D) The RDT articles can be used to interpret whether an infection is primary (the presence of the IgM line without IgG) or secondary (the presence of IgG). Samples that were characterized as primary (first column) or secondary (second column) by the reference method were further stratified based on day p.s.o. (x axis). The RDT results are denoted using the following colors: white, the absence of any line on the RDT suggesting a negative readout; black and white stripes, the presence of the NS-1 line only, suggesting dengue infection; gray, the appearance of the IgM line on the RDT without an IgG line, suggesting a primary infection readout; black, the presence of the IgG line on the RDT, suggesting a secondary infection readout. The RDT results are shown for these samples to demonstrate the effectiveness of these RDTs in correctly characterizing primary or secondary infections.
Minor variations in serotype sensitivities were observed for the SD Bioline Dengue Duo, with clinical sensitivities of 98.4% (95% CI, 94.3 to 99.8%) for DENV-1, 82% (74.3 to 88.3%) for DENV-2, 92.7% (80.1 to 98.5%) for DENV-3, and 89.2% (74.6 to 97.0%) for DENV-4 (Fig. 3B). The Panbio Dengue Duo Cassette also performed equivalently on all 4 serotypes, with sensitivities of 87.9% (76.7 to 95.0%), 97.4% (86.5 to 99.9%), 89.3% (71.8 to 97.7%), and 87.5% (47.4 to 99.7%) for DENV-1, -2, -3, and -4, respectively, on days 4 to 14 p.s.o.
For all dengue-positive specimens collected on days 0 to 3 p.s.o., the NS1 line was reactive in approximately 72% of the samples (Fig. 3C). Consistent with previous literature reports, the inclusion of the IgM line in the interpretation of positivity increased the overall sensitivity to >80% during this time period (40, 41). Over time, the sensitivity of the NS1 line alone fell to 64% on days 4 to 7 p.s.o. and to 25% on days 8 to 14, reaching <2% for convalescent-phase samples collected past day 15 p.s.o. Inclusion of IgM and IgG complemented NS1 detection to produce higher overall sensitivities (93.5% for days 4 to 7, 98.6% for days 8 to 14, and 93.9% for day 15 onward). NS1 levels are known to correlate with and follow DENV viremia (42), and agreement was observed at 83.2% (κ = 0.507) between the appearance of the NS1 line and the ability to isolate DENV from the sample.
Some studies have demonstrated improved sensitivity of dengue diagnostic devices during primary versus secondary dengue infections (43). We saw a similar trend, with the NS1 line failing to appear in early samples (days 0 to 3 p.s.o.) more often in secondary infections than in primary infections. Overall, for the SD Bioline Dengue Duo, the clinical sensitivities of primary and secondary infections were 99.0% (94.61 to 100.0%) and 84.5% (80.6 to 88.0%), respectively (Fig. 3D). The Panbio Dengue Duo Cassette was also slightly better for detection of primary versus secondary infections (95.2% versus 90.9% sensitivity for days 4 to 14 p.s.o.). The Panbio Dengue Duo Cassette “intended use” claims to correctly identify primary and secondary infections: the appearance of the IgM line in the absence of IgG signifies a primary dengue infection, while the appearance of an IgG line is indicative of a secondary dengue infection. For samples where reference testing determined the primary/secondary status, we found that the Panbio Dengue Duo Cassette correctly identified primary infections 67% of the time when read on days 4 to 8 p.s.o. (Fig. 3D). However, if the device was used after the first 8 days p.s.o., the eventual appearance of the IgG line on the test article misclassified the subject as having a secondary DENV infection, leading to only 20% correct classification at these later time points. Conversely, secondary infections were correctly classified 91% of the time from days 4 to 14 and 100% of the time past day 8. Although the SD Bioline Dengue Duo does not claim to distinguish between primary and secondary infections, we found that using the exclusive appearance of the IgM line led to correct identification of primary infections 71% of the time (days 4 to 14 p.s.o.) (Fig. 3D). As with the Panbio Dengue Duo Cassette, primary infections were more likely to be misclassified as secondary infections if the test was performed after day 8 p.s.o. The secondary sensitivity for the SD Bioline Dengue Duo was 68% for days 4 to 14 p.s.o., generally improving with time. With both RDTs, the highest accuracy in determining primary or secondary infection appears to be between days 4 to 8 p.s.o. Overall, the SD Bioline Dengue Duo demonstrated significantly higher specificity than the Panbio Dengue Duo Cassette (two-tailed Z-ratio, P < 0.0002), in addition to significantly higher sensitivity on the first 3 days of illness (two-tailed Z-ratio, P < 0.0002) (Table 3).
TABLE 3.
Device performance using serum for all sites, stratified by days p.s.o.a
| Day p.s.o. | Test article | No. of specimens that were: |
Sensitivity (95% CI) (%) | Specificity (95% CI) (%) | |||
|---|---|---|---|---|---|---|---|
| TP | FN | FP | TN | ||||
| 0–3 | Panbio RDT | 120 | 130 | 213 | 371 | 48.0 (41.7–54.4) | 63.5 (59.5–67.4) |
| 4–8 | 145 | 17 | 56 | 90 | 89.5 (83.7–93.8) | 61.6 (53.2–69.6) | |
| 9–14 | 64 | 1 | 9 | 17 | 98.5 (91.7–100.0) | 65.4 (44.3–82.8) | |
| 15+ | 248 | 5 | 242 | 339 | 98.0 (95.5–99.4) | 58.3 (54.2–62.4) | |
| 4–14 | 209 | 18 | 65 | 107 | 92.1 (87.8–95.2) | 62.2 (54.5–69.5) | |
| 0–3 | SD RDT | 206 | 51 | 57 | 429 | 80.2 (74.8–84.9) | 88.3 (85.1–91.0) |
| 4–8 | 159 | 11 | 24 | 93 | 93.5 (88.7–96.7) | 79.5 (71.0–86.4) | |
| 9–14 | 69 | 1 | 2 | 23 | 98.6 (92.3–100.0) | 92.0 (74.0–99.0) | |
| 15+ | 232 | 15 | 73 | 495 | 93.9 (90.2–96.6) | 87.1 (84.1–89.8) | |
| 0–14 | 434 | 63 | 83 | 545 | 87.3 (84.1–90.1) | 86.8 (83.9–89.3) | |
| 0–3 | IgM ELISA | 83 | 166 | 41 | 540 | 33.3 (27.5–39.6) | 92.9 (90.6–94.9) |
| 4–8 | 143 | 28 | 19 | 124 | 83.6 (77.2–88.8) | 86.7 (80.0–91.8) | |
| 9–14 | 68 | 2 | 1 | 24 | 97.1 (90.1–99.7) | 96.0 (79.7–99.9) | |
| 15+ | 199 | 48 | 41 | 530 | 80.6 (75.1–85.3) | 92.8 (90.4–94.8) | |
| 4–14 | 211 | 30 | 20 | 148 | 87.6 (82.7–91.4) | 88.1 (82.2–92.6) | |
| 0–3 | IgG ELISA | 40 | 216 | 45 | 530 | 15.6 (11.4–20.7) | 92.2 (89.7–94.2) |
| 4–8 | 79 | 90 | 18 | 127 | 46.7 (39.0–54.6) | 87.6 (81.1–92.5) | |
| 9–14 | 56 | 14 | 2 | 25 | 80.0 (68.7–88.6) | 92.6 (75.7–99.1) | |
| 15+ | 226 | 24 | 56 | 508 | 90.4 (86.1–93.8) | 90.1 (87.3–92.4) | |
| 4–14 | 135 | 104 | 20 | 152 | 56.5 (49.9–62.9) | 88.4 (82.6–92.8) | |
The numbers of true positives (TP), false negatives (FN), false positives (FP), and true negatives (TN) of the test article when compared with the reference method are provided, and sensitivity [TP/(TP+FN)] and specificity [TN/(TN+FP)] are calculated. The upper and lower bounds for confidence intervals (95%) are also shown.
IgM and IgG ELISAs.
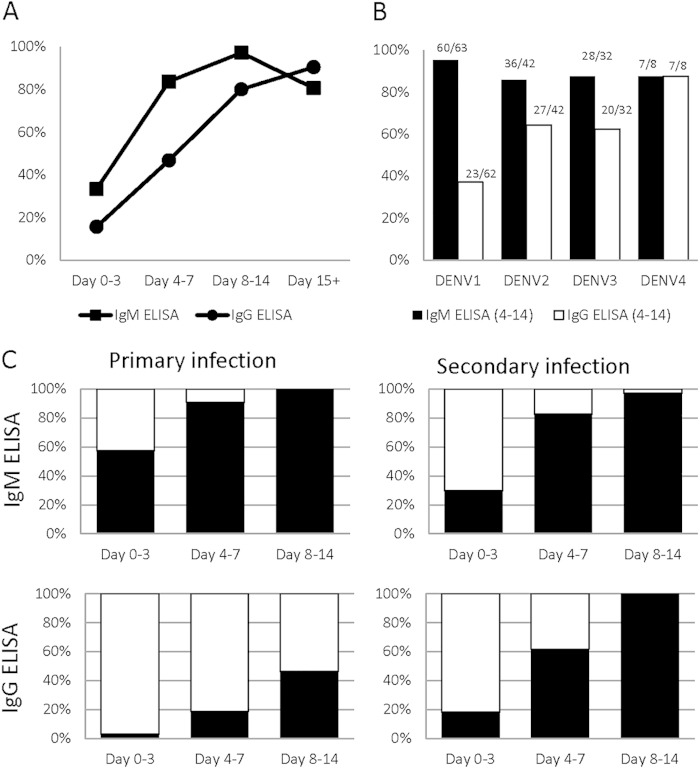
Our evaluation of the Panbio IgM ELISA demonstrated improved sensitivity over time until it reached nearly 100% for samples collected on days 8 to 14 (Fig. 4A and Table 3). For samples collected between days 4 and 14 p.s.o., the Panbio IgM ELISA had a clinical sensitivity of 87.6% (95% CI, 83.4 to 91.7%) (Table 1). The clinical specificity of the IgM ELISA from days 4 to 14 p.s.o. was 88.1% (82.2 to 92.6%). The Panbio IgG ELISA demonstrated a clinical sensitivity of 56.5% (95% CI, 50.2 to 62.8%) from days 4 to 14 p.s.o. The clinical specificity for the Panbio IgG ELISA from days 4 to 14 p.s.o. was 88.4% (82.6 to 92.8%). The Panbio IgG ELISA had low sensitivities to nearly all DENV serotypes (Fig. 4B), particularly DENV1, coincident with an overall lower sensitivity than the IgM ELISA. The Panbio IgM ELISA demonstrated similar sensitivities during both primary and secondary DENV infections (92.5% and 85.5%, respectively) (Fig. 4C). For secondary dengue infections, the clinical sensitivity of the IgG ELISA was 69.6% (62.7% to 76.5%), according to the intended use of the product. After day 8 p.s.o., the Panbio IgG ELISA was reactive to all secondary infections.
FIG 4.
ELISA performance. (A) Sensitivities of the IgM and IgG ELISAs over time are shown. (B) When virus characterization was possible, sensitivities of the IgM and IgG ELISAs for days 4 to 14 p.s.o. for each serotype are shown. The number of positive test results over the total number of samples of the given serotype that were tested using the device is provided. (C) Samples that were characterized as primary (first column) or secondary (second column) by the reference method were further stratified based on day p.s.o. (x axis). The ELISA results are denoted using the following colors: white, negative ELISA result; black, positive ELISA result. The ELISA results are shown for these samples to demonstrate the effectiveness in identifying primary and secondary infections.
Since IgM and IgG rise following dengue exposure, another method of assessing the effectiveness of the Panbio ELISAs was to compare their agreement with a reference ELISA. For this analysis, the reference ELISA was interpreted using fixed cutoffs for positivity for a given sample and not using a reference algorithm for paired sera: an ELISA titer >10 was considered positive for the IgM ELISA and >21 was considered positive for the IgG ELISA. Positive and negative percent agreements of 64.9% and 61.2%, respectively, were observed between the Panbio IgM and the reference IgM ELISAs (κ =0.22). The Panbio IgG ELISA had a positive percent agreement of 63.0% compared to in-house IgG ELISA positive specimens classified as secondary and a negative percent agreement of 68.7% (κ = 0.27).
DISCUSSION
The evaluation of infectious disease diagnostic tests using retrospective samples often leads to an inaccurate assessment of their utility at the point of need. Other groups that have evaluated the SD Bioline Dengue Duo RDT have found sensitivities ranging from 83.7% to 93.9% and specificities ranging from 83.9% to 98.8% (18, 43–48). The Panbio Dengue Duo Cassette RDT has shown a reported sensitivity between 83.9% and 100% and specificity between 75% and 100% (47, 49–51). The Panbio IgM ELISA has a reported sensitivity between 45% and 96.8% and specificity between 87.8% and 100% (46, 52–54). The Panbio IgG ELISA has a sensitivity of around 39.8% and a specificity of around 56.4% (46). While most of these reported ranges are in broad agreement with our findings, there is considerable heterogeneity in the way the studies were designed, the inclusion criteria used for the samples, and the reference testing algorithms used. The objective of this study was to obtain reliable performance data under field use conditions by using GCLP quality systems in locations where dengue is endemic and nonendemic. A global distribution of field sites also ensured the capture of a variety of circulating DENV strains and serotypes and the heterogeneity of the patient population with regard to genetic backgrounds and previous flavivirus exposures. Importantly, our study included point-of-need testing by health care providers and testing by laboratory workers without product-specific training, a factor which has been previously shown to affect device accuracy (45). Special emphasis was placed on having a comprehensive reference methodology involving isolation and confirmation of DENV from an acute-phase sample in addition to a serological response indicative of recent dengue exposure. Some samples testing negative by virus culture may have tested positive using a more sensitive reverse transcriptase quantitative PCR (RT-qPCR) protocol. However, regulatory agencies for which these data were intended regarded virus culture to be the definitive method of identifying acute dengue infection. As a result, we did not use RT-qPCR to identify any prospectively collected acute-phase specimens. Despite the limitations inherent to virus culture, by adding serological diagnosis in the reference algorithm (Fig. 1), we feel confident that all recent DENV exposures have been identified accurately. When needed, confirmation of serology was performed using PRNT, the most specific serological test currently available for identifying dengue infections. This inclusive approach allows for an accurate determination of the dengue infection status of the individual and constituted a true clinical gold standard against which the diagnostic tests under evaluation were compared.
Overall device performance was generally consistent between sites and operators. For example, the sensitivity and specificity of the Panbio Dengue Duo Cassette using serum samples for each site varied no more than 10%. With whole-blood samples, the Panbio Dengue Duo Cassette also demonstrated consistent performance at nearly all sites with one noticeable outlier: the test demonstrated a lower sensitivity (54.5%) and a higher specificity (74.3%) in Cambodia for day 4 to 14 p.s.o. (see Table S1 in the supplemental material); this difference was not statistically significant. The SD Bioline Dengue Duo, however, demonstrated significantly higher sensitivity in Cambodia than in Peru using serum samples on days 0 to 14 p.s.o., (83.7% versus 71.7%, P = 0.0471). Such differences may be related to site-specific interpretation of the manufacturer's instructions. A loss in sensitivity at an individual field site could also be reflective of serotype- or strain-specific differences at that particular site, as not all serotypes were seen at all sites, and serotype-specific variations have been previously reported (46–48, 55). This variability demonstrates the importance of prospective real-world trials of diagnostic devices, particularly RDTs which are interpreted using the naked eye, as different operators and/or clinic conditions can affect the perceived product performance.
Despite occasional variability, all test articles demonstrated improved sensitivity over time. Use of the RDTs and ELISAs may therefore be implemented with sequential sampling strategies to enhance diagnostic accuracy. The effectiveness of certain markers (e.g., NS1 or viral nucleic acid quantified by qPCR) wanes even as that of others (e.g., IgM followed by IgG) rise (Fig. 3C and 4A). These temporal patterns have been reported before, and in agreement with previous literature reports, we found that the most sensitive and useful tests are those that use a combination of NS1, IgM, and IgG (56). We found that regardless of country, most patients seek medical attention for dengue-like symptoms on days 0 to 3 p.s.o. (92.3%), and during this period, the SD Bioline Dengue Duo performed significantly (P < 0.0002, two-tailed Z-ratio) better than all other devices. This was true for both primary and secondary infections and can be attributed to the inclusion of NS1 detection in this device. The SD Bioline Dengue Duo was also more specific than the Panbio Dengue Duo Cassette (P < 0.0002), making it the more accurate RDT. Although this study is the most comprehensive evaluation of these products to date, previous groups have assessed the sensitivities and specificities of these devices. The results for the SD Bioline Dengue Duo in this study are in agreement with the results from these studies (previously observed sensitivities and specificities of 80.7 to 93.9% and 83.9 to 97.9%, respectively), despite slight differences in sample timing and reference testing methodologies (18, 43, 45, 47, 48). This study is also the first to demonstrate the SD Bioline Dengue Duo's effectiveness in distinguishing between a primary and secondary infection when read on days 4 to 8 p.s.o. Secondary infection has been correlated with a higher risk of proceeding to severe dengue, and correct identification may improve triage decisions in limited-resource settings, especially when a secondary case presents with other CDC warning signs (such as stomach ache, bloody stool, lethargy, etc.) (57).
The Panbio IgM ELISA also demonstrated high accuracy in identifying acute DENV infection past day 4 p.s.o. Its specificity was better than that of the Panbio Dengue Duo Cassette RDT, but comparable to that of the SD Bioline Dengue Duo rapid test. The Panbio IgM ELISA may serve as an effective tool for laboratory-based diagnosis of hospitalized subjects. Sequential samples collected through the progression of illness may be tested, with enhanced sensitivity for later time points. The Panbio IgG ELISA, on the other hand, demonstrated poor sensitivity (56.5%) and did not appear to be effective for identifying acute dengue infections. It was more reactive to secondary dengue infections (sensitivity of 69.6%) and therefore may demonstrate better clinical sensitivity when used between days 4 to 14 p.s.o. in settings where dengue is endemic. This format may have utility in epidemiological studies for identifying exposure-related seroconversion, but evaluation of that application is beyond the scope of this study. The ELISA format enables testing of multiple samples on a single plate and can provide economies of scale when used for hospital-based testing during larger outbreaks. However, compared to rapid tests, ELISAs do pose a higher logistical burden on the laboratory in terms of time and training requirements. In most low-incidence outbreaks, the RDTs demonstrated high negative predictive values (NPV): the SD Bioline Dengue Duo demonstrated NPV of 95.9% in Cambodia, 79.6% in Peru, and 97.5% in Venezuela (see Table S1 in the supplemental material), making RDTs an effective test to screen out nondengue infections. In this scenario, the positive test results may benefit from laboratory confirmation. During large outbreaks and especially when the testing is performed after day 3 p.s.o., the IgM ELISA is likely to provide a good balance between positive predictive values (PPV) and negative predictive values (PPV of 81.4% for days 4 to 8 p.s.o. and 96.6% for days 8 to 14 p.s.o.). The decision to adopt a specific diagnostic solution will depend on various factors, including device characteristics (see Table S2 in the supplemental material), cost, availability of cold chain transportation, days p.s.o. when patient samples are being tested, insurance reimbursement, and/or physicians' preferences.
One notable limitation of representing stratified data is that any stratification may also result in biased sampling based on a different variable. For example, the majority (88 out of 122 specimens) of the primary dengue specimens were retrospectively collected. Some devices demonstrated improved performance when archived specimens were utilized. For example, the SD Bioline Dengue Duo had a sensitivity of 97.5% when evaluated using retrospective specimens (AFRIMS) selected from days 0 to 14 p.s.o. compared to sensitivities of 83.8%, 71.7%, or 93.3% for prospectively collected specimens from Cambodia, Peru, or Venezuela, respectively (see Table S1 in the supplemental material). This apparent increase could be due to the overrepresentation of primary infections or late acute-phase specimens among the archived specimens or the underrepresentation of certain serotypes during prospective collection at the individual sites. For the purposes of this publication, study result claims were limited to broader insights on sensitivity and specificity that have adequate statistical rigor to be universally generalizable and directly attributable to use of the device for dengue diagnosis.
In conclusion, we conducted a quality systems controlled multicountry prospective evaluation of four commercially available dengue diagnostic devices. The results of this assessment are reported herein in accordance with Standards for Reporting of Diagnostic Accuracy (STARD) guidelines for diagnostic evaluation (58). We conclude that the clinical sensitivity and specificity of the SD Bioline Dengue Duo make it a useful tool at the point-of-need for diagnosing patients suspected of having dengue in low-resource settings. We also establish that the low specificity of the Panbio Dengue Duo Cassette may necessitate confirmatory testing of all RDT-positive specimens. The Panbio dengue IgM ELISA may serve as a reliable assay for such confirmatory testing. The use of an inexpensive, reliable RDT at the point of need enhances the ability of front-line health care providers to utilize their limited resources better, thereby reducing the economic burden of this globally pervasive disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank MDC Associates, LLC, for providing regulatory and logistical support. We are also grateful to the many health care professionals and laboratory staff who made this study possible, including Sophea Sot and Hip Phireak in Cambodia, Carolina Guevara and Stalin Vilcarromero in Peru, and Andrea Contreras and Eduardo Guerra in Venezuela. We also thank the following people in Iquitos, Peru, for facilitating the study there: Geraldine Ocmin Galan, Zoila Reátegui Chota, Rubiela Rubio Briceño, Zenith Tamani Guerrero, Lucy Navarro Sánchez, Johnni Mozombite Flores, Iris Reátegui Carrión, Juan Flores Michi, Leny Curico Manihuari, Nora Marin Romero, Magaly Ochoa Isuiza, Nadia Montes Criollo, Isabel Ruiz Berger, Rebeca Carrión Torres, Regina Fernández Montano, Wieslawa Álava Flores, Leslye Angulo Meléndez, Guadalupe Flores Ancajima, and Patricia Barrera Bardales. Thanks are due to the IRB staff at NAMRU-6 and those who helped coordinate the submissions, specifically Roxana Lescano and Gloria Talledo. We are grateful to Myriam Battistutta of Alere for her support and for providing the devices for this evaluation. Peifang Sun and Theron Gilliland, Jr., helped review the manuscript.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of the Army, the Department of Defense, or the U.S. government. The work described here was covered under a Cooperative Research and Development Agreement NCRADA-NMRC-10-3488 among the Navy (NMRC, NAMRU-2, NAMRU-6), the Army (AFRIMS, WRAIR), and Alere Inc.
Andrea Valks is employed by Alere, Inc. Allison Dauner and Subhamoy Pal are employed by the Henry M. Jackson Foundation for the Advancement of Military Medicine and are funded to do this work by the U.S. government. Shuenn-Jue Wu, LCDR Tadeusz Kochel, Lt. Col. Eric Halsey, Maj. Richard Jarman, Maj. Stefan Fernandez, Lou Jasper, and LCDR Chadwick Yasuda are military service members or employees of the U.S. government. This work was prepared as part of their official duties.
This work was funded by the Military Infectious Diseases Research Program, proposal numbers L0091_07_NM, L010011_09_NM, L0185_10_NM, L0186_10_NM, L0257_12_NM, and L0262_12_NM. The work was supported by Work Unit Number 6000.RAD1.L.A1220.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03042-14.
REFERENCES
- 1.Henchal EA, Putnak JR. 1990. The dengue viruses. Clin Microbiol Rev 3:376–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. 1998. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11:480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. 1997. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem, p 1−22. In Gubler DJ, Kuno G (ed), Dengue and dengue hemorrhagic fever. CAB International, New York, NY. [Google Scholar]
- 4.Lee MS, Hwang KP, Chen TC, Lu PL, Chen TP. 2006. Clinical characteristics of dengue and dengue hemorrhagic fever in a medical center of southern Taiwan during the 2002 epidemic. J Microbiol Immunol Infect 39:121–129. [PubMed] [Google Scholar]
- 5.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead SB, Deen J. 2002. The future of dengue vaccines. Lancet 360:1243–1245. doi: 10.1016/S0140-6736(02)11276-1. [DOI] [PubMed] [Google Scholar]
- 7.Halstead SB. 2002. Dengue. Curr Opin Infect Dis 15:471–476. doi: 10.1097/00001432-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed World Health Organization, Geneva Switzerland. [Google Scholar]
- 9.World Health Organization. 2009. Dengue guidelines for diagnosis, treatment, prevention and control, new ed World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 10.Guzmán MG, Kouri G. 1996. Advances in dengue diagnosis. Clin Diagn Lab Immunol 3:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu PY, Huang JH. 2004. Current advances in dengue diagnosis. Clin Diagn Lab Immunol 11:642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teles FR, Prazeres DM, Lima-Filho JL. 2005. Trends in dengue diagnosis. Rev Med Virol 15:287–302. doi: 10.1002/rmv.461. [DOI] [PubMed] [Google Scholar]
- 13.Tun-Lin W, Lenhart A, Nam VS, Rebollar-Tellez E, Morrison AC, Barbazan P, Cote M, Midega J, Sanchez F, Manrique-Saide P, Kroeger A, Nathan MB, Meheus F, Petzold M. 2009. Reducing costs and operational constraints of dengue vector control by targeting productive breeding places: a multi-country non-inferiority cluster randomized trial. Trop Med Int Health 14:1143–1153. doi: 10.1111/j.1365-3156.2009.02341.x. [DOI] [PubMed] [Google Scholar]
- 14.Guzmán MG, Kouri G. 2004. Dengue diagnosis, advances and challenges. Int J Infect Dis 8:69–80. doi: 10.1016/j.ijid.2003.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Vorndam V, Kuno G. 1997. Laboratory diagnosis of dengue virus infections. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 16.Kuno G, Gubler DJ, Santiago de Weil NS. 1985. Antigen capture ELISA for the identification of dengue viruses. J Virol Methods 12:93–103. doi: 10.1016/0166-0934(85)90011-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuno G, Gomez I, Gubler DJ. 1991. An ELISA procedure for the diagnosis of dengue infections. J Virol Methods 33:101–113. doi: 10.1016/0166-0934(91)90011-N. [DOI] [PubMed] [Google Scholar]
- 18.Tricou V, Vu HT, Quynh NV, Nguyen CV, Tran HT, Farrar J, Wills B, Simmons CP. 2010. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis 10:142. doi: 10.1186/1471-2334-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh CJ, Chen MJ. 2009. The commercial dengue NS1 antigen-capture ELISA may be superior to IgM detection, virus isolation and RT-PCR for rapid laboratory diagnosis of acute dengue infection based on a single serum sample. J Clin Virol 44:102. doi: 10.1016/j.jcv.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Bessoff K, Delorey M, Sun W, Hunsperger E. 2008. Comparison of two commercially available dengue virus (DENV) NS1 capture enzyme-linked immunosorbent assays using a single clinical sample for diagnosis of acute DENV infection. Clin Vaccine Immunol 15:1513–1518. doi: 10.1128/CVI.00140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal S, Dauner AL, Mitra I, Forshey BM, Garcia P, Morrison AC, Halsey ES, Kochel TJ, Wu SJ. 2014. Evaluation of dengue NS1 antigen rapid tests and ELISA kits using clinical samples. PLoS One 9:e113411. doi: 10.1371/journal.pone.0113411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigau-Perez JG. 1997. Clinical manifestations of dengue hemorrhagic fever in Puerto Rico, 1990-1991. Puerto Rico Association of Epidemiologists. Rev Panam Salud Publica 1:381–388. doi: 10.1590/S1020-49891997000500007. [DOI] [PubMed] [Google Scholar]
- 23.Chen LH, Wilson ME. 2010. Dengue and chikungunya infections in travelers. Curr Opin Infect Dis 23:438–444. doi: 10.1097/QCO.0b013e32833c1d16. [DOI] [PubMed] [Google Scholar]
- 24.Ranjit S, Kissoon N. 2011. Dengue hemorrhagic fever and shock syndromes. Pediatr Crit Care Med 12:90–100. doi: 10.1097/PCC.0b013e3181e911a7. [DOI] [PubMed] [Google Scholar]
- 25.Antunes AC, Oliveira GL, Nunes LI, Guedes Filho LA, Prado RS, Henriques HR, Vieira AJ. 2013. Evaluation of the diagnostic value of the tourniquet test in predicting severe dengue cases in a population from Belo Horizonte, State of Minas Gerais, Brazil. Rev Soc Bras Med Trop 46:542–546. doi: 10.1590/0037-8682-0161-2013. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez G, Gresh L, Perez MA, Elizondo D, Aviles W, Kuan G, Balmaseda A, Harris E. 2013. Evaluation of the diagnostic utility of the traditional and revised WHO dengue case definitions. PLoS Negl Trop Dis 7:e2385. doi: 10.1371/journal.pntd.0002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peeling RW, Artsob H, Pelegrino JL, Buchy P, Cardosa MJ, Devi S, Enria DA, Farrar J, Gubler DJ, Guzman MG, Halstead SB, Hunsperger E, Kliks S, Margolis HS, Nathanson CM, Nguyen VC, Rizzo N, Vazquez S, Yoksan S. 2010. Evaluation of diagnostic tests: dengue. Nat Rev Microbiol 8:S30–38. doi: 10.1038/nrmicro2459. [DOI] [PubMed] [Google Scholar]
- 28.Singh KR, Paul SD. 1969. Isolation of dengue viruses in Aedes albopictus cell cultures. Bull World Health Organ 40:982–983. [PMC free article] [PubMed] [Google Scholar]
- 29.Tesh RB. 1979. A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am J Trop Med Hyg 28:1053–1059. [DOI] [PubMed] [Google Scholar]
- 30.Innis BL, Nisalak A, Nimmannitya S, Kusalerdchariya S, Chongswasdi V, Suntayakorn S, Puttisri P, Hoke CH. 1989. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am J Trop Med Hyg 40:418–427. [DOI] [PubMed] [Google Scholar]
- 31.Jarman RG, Nisalak A, Anderson KB, Klungthong C, Thaisomboonsuk B, Kaneechit W, Kalayanarooj S, Gibbons RV. 2011. Factors influencing dengue virus isolation by C6/36 cell culture and mosquito inoculation of nested PCR-positive clinical samples. Am J Trop Med Hyg 84:218–223. doi: 10.4269/ajtmh.2011.09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell PK, Nisalak A. 1967. Dengue virus identification by the plaque reduction neutralization test. J Immunol 99:291–296. [PubMed] [Google Scholar]
- 33.Russell PK, Nisalak A, Sukhavachana P, Vivona S. 1967. A plaque reduction test for dengue virus neutralizing antibodies. J Immunol 99:285–290. [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention. 2012. Laboratory guidance and diagnostic testing. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/dengue/clinicalLab/laboratory.html. [Google Scholar]
- 35.Sadon N, Delers A, Jarman RG, Klungthong C, Nisalak A, Gibbons RV, Vassilev V. 2008. A new quantitative RT-PCR method for sensitive detection of dengue virus in serum samples. J Virol Methods 153:1–6. doi: 10.1016/j.jviromet.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Lin L. 2008. Overview of agreement statistics for medical devices. J Biopharm Stat 18:126–144. doi: 10.1080/10543400701668290. [DOI] [PubMed] [Google Scholar]
- 37.Clopper CJ, Pearson ES. 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413. doi: 10.1093/biomet/26.4.404. [DOI] [Google Scholar]
- 38.Lowry R. 2015. The significance of the difference between two independent proportions. http://vassarstats.net/propdiff_ind.html.
- 39.Cohen J. 1968. Weighted kappa: nominal scale agreement with provision for scaled disagreement or partial credit. Psychol Bull 70:213–220. doi: 10.1037/h0026256. [DOI] [PubMed] [Google Scholar]
- 40.Simmons CP, Chau TN, Thuy TT, Tuan NM, Hoang DM, Thien NT, Lien le B, Quy NT, Hieu NT, Hien TT, McElnea C, Young P, Whitehead S, Hung NT, Farrar J. 2007. Maternal antibody and viral factors in the pathogenesis of dengue virus in infants. J Infect Dis 196:416–424. doi: 10.1086/519170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fry SR, Meyer M, Semple MG, Simmons CP, Sekaran SD, Huang JX, McElnea C, Huang CY, Valks A, Young PR, Cooper MA. 2011. The diagnostic sensitivity of dengue rapid test assays is significantly enhanced by using a combined antigen and antibody testing approach. PLoS Negl Trop Dis 5:e1199. doi: 10.1371/journal.pntd.0001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tricou V, Minh NN, Farrar J, Tran HT, Simmons CP. 2011. Kinetics of viremia and NS1 antigenemia are shaped by immune status and virus serotype in adults with dengue. PLoS Negl Trop Dis 5:e1309. doi: 10.1371/journal.pntd.0001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan VC, Tan LK, Lye DC, Pok KY, Mok SQ, Chua RC, Leo YS, Ng LC. 2014. Diagnosing dengue at the point-of-care: utility of a rapid combined diagnostic kit in Singapore. PLoS One 9:e90037. doi: 10.1371/journal.pone.0090037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang SM, Sekaran SD. 2010. Evaluation of a commercial SD dengue virus NS1 antigen capture enzyme-linked immunosorbent assay kit for early diagnosis of dengue virus infection. J Clin Microbiol 48:2793–2797. doi: 10.1128/JCM.02142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andries AC, Duong V, Ngan C, Ong S, Huy R, Sroin KK, Te V, Try Y BPL, Buchy P. 2012. Field evaluation and impact on clinical management of a rapid diagnostic kit that detects dengue NS1, IgM and IgG. PLoS Negl Trop Dis 6:e1993. doi: 10.1371/journal.pntd.0001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blacksell SD, Jarman RG, Gibbons RV, Tanganuchitcharnchai A, Mammen MP Jr, Nisalak A, Kalayanarooj S, Bailey MS, Premaratna R, de Silva HJ, Day NP, Lalloo DG. 2012. Comparison of seven commercial antigen and antibody enzyme-linked immunosorbent assays for detection of acute dengue infection. Clin Vaccine Immunol 19:804–810. doi: 10.1128/CVI.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blacksell SD, Jarman RG, Bailey MS, Tanganuchitcharnchai A, Jenjaroen K, Gibbons RV, Paris DH, Premaratna R, de Silva HJ, Lalloo DG, Day NP. 2011. Evaluation of six commercial point-of-care tests for the diagnosis of acute dengue infections: The need for combining NS1 antigen and IgM/IgG antibody detection to achieve acceptable levels of accuracy. Clin Vaccine Immunol 18:2095−2101. doi: 10.1128/CVI.05285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osorio L, Ramirez M, Bonelo A, Villar LA, Parra B. 2010. Comparison of the diagnostic accuracy of commercial NS1-based diagnostic tests for early dengue infection. Virol J 7:361. doi: 10.1186/1743-422X-7-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaughn DW, Nisalak A, Kalayanarooj S, Solomon T, Dung NM, Cuzzubbo A, Devine PL. 1998. Evaluation of a rapid immunochromatographic test for diagnosis of dengue virus infection. J Clin Microbiol 36:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuzzubbo AJ, Vaughn DW, Nisalak A, Solomon T, Kalayanarooj S, Aaskov J, Dung NM, Devine PL. 1999. Comparison of PanBio dengue duo enzyme-linked immunosorbent assay (ELISA) and MRL dengue fever virus immunoglobulin M capture ELISA for diagnosis of dengue virus infections in Southeast Asia. Clin Diagn Lab Immunol 6:705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam SK, Devine PL. 1998. Evaluation of capture ELISA and rapid immunochromatographic test for the determination of IgM and IgG antibodies produced during dengue infection. Clin Diagn Virol 10:75–81. doi: 10.1016/S0928-0197(98)00002-6. [DOI] [PubMed] [Google Scholar]
- 52.Blacksell SD, Mammen MP Jr, Thongpaseuth S, Gibbons RV, Jarman RG, Jenjaroen K, Nisalak A, Phetsouvanh R, Newton PN, Day NP. 2008. Evaluation of the Panbio dengue virus nonstructural 1 antigen detection and immunoglobulin M antibody enzyme-linked immunosorbent assays for the diagnosis of acute dengue infections in Laos. Diagn Microbiol Infect Dis 60:43–49. doi: 10.1016/j.diagmicrobio.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Vazquez S, Hafner G, Ruiz D, Calzada N, Guzman MG. 2007. Evaluation of immunoglobulin M and G capture enzyme-linked immunosorbent assay Panbio kits for diagnostic dengue infections. J Clin Virol 39:194–198. doi: 10.1016/j.jcv.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Groen J, Koraka P, Velzing J, Copra C, Osterhaus AD. 2000. Evaluation of six immunoassays for detection of dengue virus-specific immunoglobulin M and G antibodies. Clin Diagn Lab Immunol 7:867–871. doi: 10.1128/CDLI.7.6.867-871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blacksell SD, Newton PN, Bell D, Kelley J, Mammen MP Jr, Vaughn DW, Wuthiekanun V, Sungkakum A, Nisalak A, Day NP. 2006. The comparative accuracy of 8 commercial rapid immunochromatographic assays for the diagnosis of acute dengue virus infection. Clin Infect Dis 42:1127–1134. doi: 10.1086/501358. [DOI] [PubMed] [Google Scholar]
- 56.Hang VT, Nguyet NM, Trung DT, Tricou V, Yoksan S, Dung NM, Van Ngoc T, Hien TT, Farrar J, Wills B, Simmons CP. 2009. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl Trop Dis 3:e360. doi: 10.1371/journal.pntd.0000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Centers for Disease Control and Prevention. Symptoms and what to do if you think you have dengue. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/dengue/symptoms/. [Google Scholar]
- 58.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig LM, Lijmer JG, Moher D, Rennie D, de Vet HC, Standards for Reporting of Diagnostic Accuracy Group . 2003. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clin Biochem 36:2–7. doi: 10.1016/S0009-9120(02)00443-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.