Abstract
Colonization with Candida species is an independent risk factor for invasive candidiasis (IC), but the minimum and most practicable parameters for prediction of IC have not been optimized. We evaluated Candida colonization in a prospective cohort of 6,015 nonneutropenic, critically ill patients. Throat, perineum, and urine were sampled 72 h post-intensive care unit (ICU) admission and twice weekly until discharge or death. Specimens were cultured onto chromogenic agar, and a subset underwent molecular characterization. Sixty-three (86%) patients who developed IC were colonized prior to infection; 61 (97%) tested positive within the first two time points. The median time from colonization to IC was 7 days (range, 0 to 35). Colonization at any site was predictive of IC, with the risk of infection highest for urine colonization (relative risk [RR] = 2.25) but with the sensitivity highest (98%) for throat and/or perineum colonization. Colonization of ≥2 sites and heavy colonization of ≥1 site were significant independent risk factors for IC (RR = 2.25 and RR = 3.7, respectively), increasing specificity to 71% to 74% but decreasing sensitivity to 48% to 58%. Molecular testing would have prompted a resistance-driven decision to switch from fluconazole treatment in only 11% of patients infected with C. glabrata, based upon species-level identification alone. Positive predictive values (PPVs) were low (2% to 4%) and negative predictive values (NPVs) high (99% to 100%) regardless of which parameters were applied. In the Australian ICU setting, culture of throat and perineum within the first two time points after ICU admission captures 84% (61/73 patients) of subsequent IC cases. These optimized parameters, in combination with clinical risk factors, should strengthen development of a setting-specific risk-predictive model for IC.
INTRODUCTION
Invasive candidiasis (IC), particularly, candidemia, is a significant cause of mortality in critically ill patients, accounting for almost a third of nosocomial infections in intensive care units (ICUs) (1, 2). Early antifungal therapy reduces IC-related mortality and approximately halves the incidence of IC (3–8). Untargeted prophylactic antifungal use, however, is expensive, has the potential to cause adverse drug reactions, and may select for resistant fungal species (9).
Clinical risk prediction rules that identify the high-risk patients most likely to benefit from prophylaxis have been developed (10–16), but those studies have used different sets of predictors, including (a) clinical risk factors only and (b) clinical risk factors in combination with colonization indices (CIs). Those rules have been applied in various settings and have included assessment at different times postadmission to the ICU and different types of ICU (surgical ICUs only or mixed medical/surgical ICUs—defined as units that house both medical and surgical populations). These differences may explain why predictive models and algorithms have performed poorly outside their derivative populations (17).
Since IC is preceded by colonization of mucosal surfaces with the infecting strain (14, 18–20) and since colonization is an independent risk factor for IC (14, 19, 21, 22), it is logical that it should be incorporated into predictive models (15, 16). Using data from mixed medical/surgical ICUs in Australia, we demonstrated that the post hoc addition of colonization parameters to two published clinical risk factor-only predictive models improved their performance characteristics (17).
Although several studies examining colonization as a risk factor for IC have been published, the methodology has not been standardized and most were confined to surgical ICUs (14, 23–25). Interstudy differences existed with respect to sites of sampling, use of convenience and/or specified sample sites, timing of first samples and frequency of sampling, quantification of colonization density, and culture media and methods used for species identification. Furthermore, nucleic acid-based methods of fungal identification were not included.
A systematic evaluation of Candida colonization in a prospective multicenter cohort study of Australian ICU patients was conducted between June 2007 and January 2012. The overall project aimed to develop and validate a clinical culture and colonization-based risk-predictive model suitable for use in mixed Australian medical and surgical ICUs. In this study, we determined the utility of culture on CHROMagar for semiquantitative estimates of Candida colonization (with parallel molecular testing performed on a subset of patients), the optimal timing of sample collection, and the minimum number of sites to be sampled. Our overall aim was to develop a protocol to optimize detection of colonization status for inclusion into risk-predictive models of IC in critically ill patients.
MATERIALS AND METHODS
Study design.
A total of 6,015 nonneutropenic critically ill patients admitted consecutively to mixed medical and surgical ICUs in seven Australian hospitals (Westmead, Sydney [n = 1,163]; Princess Alexandra, Brisbane [n = 1,231]; Royal Brisbane Women's and Children's, Brisbane [n = 896]; St Vincent's Hospital, Sydney [n = 611]; Royal Melbourne Hospital, Melbourne [n = 1,227]; Concord Hospital, Sydney [n = 371]; and Nepean Hospital, Sydney [n = 516]) for at least 72 h were studied prospectively from 15 June 2007 to 1 January 2012. Patients with neutropenia (absolute neutrophil count < 0.5 × 109/liter) within the first 72 h of ICU admission were excluded. Approval for the study was obtained from each respective Human Research Ethics Committee.
Surveillance specimen collection.
Published literature (23) and a preliminary study at the Westmead Hospital site suggested that the throat, the perineum (rather than the groin or rectum), and urine were the most accessible, feasible, and highest-yield sites for the assessment of Candida colonization. Sampling techniques were standardized across sites. In brief, standard sterile red-top culturette swabs containing liquid Amies media (Copan, Murrieta, CA, USA) were used to sample the oropharynx and perineum (just anterior to the anus), and catheter (or midstream) urine specimens were obtained. Specimens were obtained independently of any clinically indicated samples. Preliminary experiments (performed prior to the commencement of this study) showed that sampling the throat and perineum with dry swabs followed by immersion in 2 ml nutrient broth containing 10% glycerol at the bedside was a suitable but less practical alternative (data not shown). Eswabs (Copan Diagnostics and Becton, Dickinson and Co., Franklin Lakes, NJ), which allow automated specimen processing and improved microbial transport and recovery (26, 27), were not evaluated in this study due to their lack of availability in Australia during the study period. All three surveillance sites were sampled at 72 h following ICU admission and twice weekly thereafter until ICU discharge or death.
Candida surveillance cultures.
Swabs of the throat and perineum and 10 μl of urine were plated onto Candida chromogenic agar (CHROMagar, Paris, France) using a half-plate quantitative urine streaking method. Plates were incubated at 35°C for 48 h. Candida albicans, Candida tropicalis, and Candida krusei were differentiated by color as described by the manufacturer. All other yeast isolates (nearly all were Candida glabrata or Candida parapsilosis as confirmed by multiplex tandem PCR [MT-PCR]) were collectively designated “other Candida spp.” Growth of each morphologically distinct yeast isolate was enumerated semiquantitatively, and the extent of growth was classified as none, light (<10 colonies), moderate (10 to 100 colonies), or heavy (>100 colonies).
MT-PCR.
Swabs of the throat and perineum obtained from the first 205 patients sampled at Westmead Hospital underwent multiplex tandem PCR (MT-PCR) analysis in addition to culture. Following culture, the swabs were placed into 3 ml saline solution containing 10% nutrient broth (Oxoid, Adelaide, South Australia) and stored at 4°C until nucleic acid was extracted. DNA was isolated from 1 ml of the suspension using a nucliSENS easyMAG instrument (bioMérieux, Baulkham Hills, New South Wales, Australia) according to the manufacturer's instructions. The elution volume was 110 μl. As previously described (28–30), the MT-PCR method enabled identification of seven Candida species, including C. albicans, Candida dubliniensis, C. glabrata, Candida guilliermondii, C. krusei, C. parapsilosis complex, and C. tropicalis, as well as Saccharomyces cerevisiae. An internal positive control (artificial DNA) was included with each specimen to test for PCR inhibition, and a negative water control was included in each run to monitor contamination. Urine specimens were not tested by MT-PCR, as the application of this technology to urine samples had not yet been validated during the study period.
Definitions.
Colonization was defined as the isolation of a Candida species from at least one surveillance site. Positive samples collected as part of routine clinical management were not included in the analysis. The colonization index (CI) was defined as the ratio of the number of sites colonized by Candida spp. to the number of body sites surveyed (14). The corrected colonization index (CCI) was defined as the ratio of the number of sites heavily colonized by Candida spp. to the number of body sites surveyed (14). In this present study, the CCI threshold was reduced from ≥0.5 (14) to ≥0.3 to accommodate the reduced number of sites surveyed (three as opposed to five).
Surveillance specimens were collected at 72 h following ICU admission and twice weekly thereafter until ICU discharge or death. Time points 1, 2, and 3 were defined as 3 to 4 days, 6 to 8 days, and 9 to 11 days post-ICU admission, respectively.
IC episodes were defined using modified European Organization for Research and Treatment of Cancer/Mycoses Study Group (EORTC/MSG) criteria (12, 17). Patients with candiduria alone were excluded because this was not considered an invasive infection. IC episodes detected at least 72 h following ICU admission and within 72 h after ICU discharge were considered to have been ICU acquired.
Statistical analysis.
Surveillance culture results from ICU patients who did develop IC and from those who did not were compared with respect to categorical variables that included the anatomical site of colonization, the colonization indices (CI and CCI), the density of colonization, the number of colonizing species, and the time of surveillance. The relative risk of developing IC based on colonization status was calculated using Fisher's exact test. A P value of <0.05 was considered significant. The predictive performance characteristics (including sensitivity, specificity, positive predictive value [PPV], and negative predictive value [NPV]) were calculated for each variable studied. MT-PCR and culture results were compared for the detection of colonization status and the time to detection of colonization status using Fisher's exact test. All statistical calculations were performed using MedCalc (www.medcalc.org).
RESULTS
Patient characteristics.
Of the 6,015 patients studied, 73 (1%) developed IC in the ICU. Fifty-eight (79%) of these had proven infection, i.e., candidemia (n = 43) or noncandidemic IC (n = 15); the remaining 15 (21%) patients were classified as probable cases (12, 17). Infections were caused by C. albicans (n = 45, 62%), C. glabrata (n = 8, 11%), C. tropicalis (n = 8, 11%), C. parapsilosis (n = 6, 8%), Candida spp. (n = 5, 7%), and C. krusei (n = 1, 1%). The median time from ICU admission to the development of ICU-acquired IC was 11 days (range, 4 to 41; average, 14 days).
Systemic (oral or parenteral) antifungal drugs had been received by five (7%) patients prior to ICU admission (days −7 to 0) or at time point 1 (days 3 to 4); by an additional seven (10%) between days 5 and 7; and by an additional six (8%) between days 8 and 10. Fluconazole therapy was administered in 16 cases of IC and caspofungin in 2 cases. Twelve (16%) of 73 patients who developed IC had received empirical antifungal therapy (none had received antifungal prophylaxis) prior to diagnosis; 9 (7 with infection due to C. albicans, 1 with infection due to C. lusitaniae, and 1 with infection due to C. tropicalis) (75%) of the 12 had been treated with fluconazole.
Colonization (culture) versus IC: anatomical site of colonization.
Table 1 summarizes the risk of developing IC by site of colonization over the first three time points. A total of 3,511 (60%), 2,025 (63%), and 1,162 (64%) patients were colonized at time points 1 (days 3 to 4 post-ICU admission), 2 (days 6 to 8 post-ICU admission), and 3 (days 9 to 11 post-ICU admission), respectively.
TABLE 1.
Risk of invasive candidiasis based on screening time point and anatomical site of colonization (derived from culture results only; entire patient cohort included)
| Variablea | n | RRb | P value | 95% confidence interval (low) | 95% confidence interval (high) | Sensitivity (%) | Specificity (%) | PPVc (%) | NPVd (%) |
|---|---|---|---|---|---|---|---|---|---|
| Time point 1 (n studied = 6,015) | |||||||||
| Any site colonized (CIe ≥ 0.3) | 3,511 | 2.35 | 0.002 | 1.35 | 4.08 | 78 | 40 | 2 | 99 |
| At least throat colonized | 2,927 | 2.04 | 0.004 | 1.25 | 3.34 | 68 | 49 | 2 | 99 |
| At least perineum colonized | 1,967 | 1.76 | 0.0155 | 1.1 | 2.77 | 48 | 66 | 2 | 99 |
| At least urine colonized | 630 | 2.25 | 0.003 | 1.32 | 3.84 | 23 | 88 | 3 | 99 |
| Time point 2 (n studied = 3,244) | |||||||||
| Any site colonized (CI ≥ 0.3) | 2,025 | 3.69 | 0.001 | 1.66 | 8.17 | 86 | 38 | 2 | 99 |
| At least throat colonized | 1,555 | 2.97 | 0.0007 | 1.58 | 5.56 | 74 | 51 | 2 | 99 |
| At least perineum colonized | 1,313 | 1.54 | NSf | 0.89 | 2.67 | 52 | 59 | 2 | 99 |
| At least urine colonized | 450 | 1.81 | NS | 0.95 | 3.44 | 24 | 85 | 3 | 99 |
| Time point 3 (n studied = 1,820) | |||||||||
| Any site colonized (CI ≥ 0.3) | 1,162 | 1.41 | <0.0001 | 1.23 | 1.61 | 89 | 37 | 2 | 100 |
| At least throat colonized | 837 | 5.04 | 0.001 | 1.93 | 13.21 | 82 | 53 | 3 | 99 |
| At least perineum colonized | 762 | 2.04 | NS | 0.96 | 4.34 | 61 | 57 | 2 | 99 |
| At least urine colonized | 284 | NS |
Time point 1, days 3 to 4 post-ICU admission; time point 2, days 6 to 8 post-ICU admission; time point 3, days 9 to 11 post-ICU admission.
RR, relative risk.
PPV, positive predictive value.
NPV, negative predictive value.
CI, colonization index.
NS, not significant.
Of the 73 patients who developed IC, Candida colonization was detected in 63 (86%) prior to infection: 56 (89%) of these at time point 1, an additional 5 at time point 2, and a further 2 at time point 3. Patients with detected colonization in the throat, perineum, or urine at time point 1 were all at increased risk of developing IC (relative risk [RR] = 2.35, P value = 0.002, sensitivity = 78%), with colonization of the urine being associated with the highest relative risk of IC (RR = 2.25) compared with throat and perineum (RR = 2.04 and 1.76, respectively; Table 1). At time points 2 and 3, patients colonized in the throat or in at least one of the three sites sampled (CI ≥ 0.3) were at increased risk of developing IC. NPVs were consistently high (99% to 100%; Table 1).
Of the 63 patients in whom Candida colonization was detected prior to infection, 38 were colonized in at least two sites, most commonly the throat and perineum (n = 32, 84%). Nineteen (30%) patients with IC were colonized in the throat alone, five (8%) in the perineum alone, and one (2%) in the urine alone.
The median time from the first positive surveillance specimen to the first positive clinical culture was 7 days (range, 0 to 35; average, 9 days).
Ten patients (14%) had negative surveillance cultures prior to the development of IC. Seven (70%) of the 10 patients had only one set of surveillance cultures obtained before discharge from the ICU. The remaining three had two, five, or seven sets of surveillance cultures obtained; the times from the last surveillance culture to the development of IC were 2, 15, and 19 days, respectively. All but 1 of the 10 patients who had negative surveillance cultures prior to developing IC had undergone surgery just prior to or during ICU admission. In six instances where the type of surgery was specified, five had involved penetration of the gastrointestinal tract.
Colonization index (CI), corrected colonization index (CCI), and invasive candidasis (IC).
Of the 58% patients (n = 3,511) in whom Candida colonization was detected at the first time point, 1,671 (48%) had Candida spp. isolated from at least two surveillance sites (CI ≥ 0.5; Table 2). Almost half (n = 35, 48%) of the patients who developed IC had a CI of ≥0.5 (RR = 2.25, P value = 0.0005; Table 2).
TABLE 2.
Performance characteristics determined using a colonization index value of >0.5 and a corrected colonization index value of ≥0.3 (derived from culture results only; entire patient cohort included)
| Variablea | n | RRb | P value | 95% confidence interval (low) | 95% confidence interval (high) | Sensitivity (%) | Specificity (%) | PPVc (%) | NPVd (%) |
|---|---|---|---|---|---|---|---|---|---|
| Time point 1 (n studied = 6,015) | |||||||||
| At least 2 sites colonized (CIe ≥ 0.5) | 1,671 | 2.25 | 0.0005 | 1.4 | 3.5 | 48 | 71 | 2 | 99 |
| All 3 sites colonized | 342 | 2.25 | 0.016 | 1.16 | 4.34 | 14 | 94 | 3 | 99 |
| At least one site heavy density (CCIf ≥ 0.3) | 1,549 | 3.7 | <0.0001 | 2.36 | 5.93 | 58 | 74 | 3 | 99 |
| At least 2 sites heavy density | 448 | 3.1 | 0.001 | 1.77 | 5.4 | 21 | 92 | 3 | 99 |
| At least throat heavy density | 1,025 | 3.77 | <0.0001 | 2.39 | 5.94 | 45 | 82 | 3 | 99 |
| At least perineum heavy density | 703 | 2 | 0.01 | 1.15 | 3.46 | 22 | 88 | 2 | 99 |
| At least urine heavy density | 327 | NSg | |||||||
| Time point 2 (n studied = 3,244) | |||||||||
| At least 2 sites colonized (CI ≥ 0.5) | 1,056 | 2.4 | 0.002 | 1.38 | 4.16 | 54 | 67 | 3 | 99 |
| All 3 sites colonized | 238 | NS | |||||||
| At least one site heavy density (CCI ≥ 0.3) | 915 | 3.24 | <0.0001 | 1.86 | 5.63 | 56 | 72 | 3 | 99 |
| At least 2 sites heavy density | 287 | 2.9 | 0.001 | 1.5 | 5.6 | 22 | 91 | 4 | 99 |
| At least throat heavy density | 530 | 3.93 | <0.0001 | 2.26 | 6.81 | 44 | 84 | 4 | 99 |
| At least perineum heavy density | 481 | NS | |||||||
| At least urine heavy density | 233 | NS | |||||||
| Time point 3 (n studied = 1,820) | |||||||||
| At least 2 sites colonized (CI ≥ 0.5) | 596 | 3.12 | 0.003 | 1.47 | 6.62 | 61 | 67 | 3 | 99 |
| All 3 sites colonized | 122 | NS | |||||||
| At least one site heavy density (CCI ≥ 0.3) | 480 | 2.1 | 0.05 | 0.99 | 4.39 | 43 | 74 | 3 | 99 |
| At least 2 sites heavy density | 143 | NS | |||||||
| At least throat heavy density | 253 | 2.83 | 0.009 | 1.3 | 6.2 | 32 | 86 | 4 | 99 |
| At least perineum heavy density | 263 | NS | |||||||
| At least urine heavy density | 126 | NS |
Time point 1, days 3 to 4 post-ICU admission; time point 2, days 6 to 8 post-ICU admission; time point 3, days 9 to 11 post-ICU admission.
RR, relative risk.
PPV, positive predictive value.
NPV, negative predictive value.
CI, uncorrected colonization index.
CCI, corrected colonization index.
NS, not significant.
The sensitivity, specificity, and risk of developing IC increased slightly when heavy colonization was taken into account (CCI ≥ 0.3; Table 2). At time point 1, 42 (58%) patients who developed IC were heavily colonized in at least one site (RR = 3.7, P value < 0.0001). Specificity increased (74% to 92%) when two sites were heavily colonized, but this in turn reduced sensitivity from 58% to 21% (Table 2).
Prevalence of Candida colonization and incidence of IC over time in the ICU.
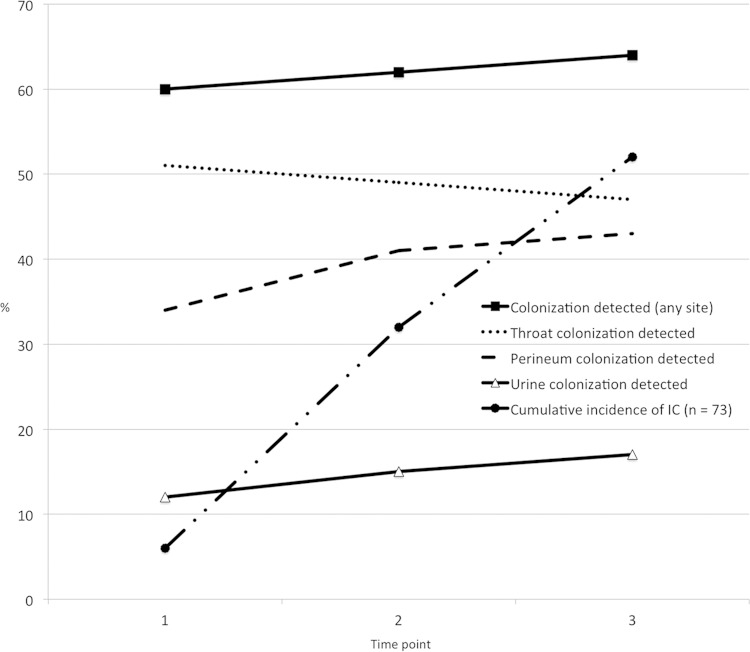
Detection of Candida colonization at any site was maintained at 60% over the first three time points (Fig. 1). Detection of perineum colonization increased the most over the first two screens, while the likelihood of Candida colonization in the urine increased steadily with prolonged ICU stay. Detection of Candida colonization in the throat was highest at time point 1 and decreased with time in the ICU. Of the 73 infected patients, 6, 17, and 15 patients developed IC within time points 1, 2, and 3, respectively, leading to cumulative incidences of IC of 8%, 32%, and 52% within the first three time points (Fig. 1).
FIG 1.
Cumulative incidence of IC and percentage of patients colonized at any site or specifically in the throat, the perineum, or urine over the first three time points.
Distribution of colonizing species.
Most patients were colonized with C. albicans (n = 2,854, 81%), followed by “other” Candida spp. (n = 1,059, 30%), C. tropicalis (n = 343, 10%), and C. krusei (n = 123, 4%). The majority (80%) of patients were colonized with a single Candida species. Mixed cultures generally consisted of two species (n = 662), typically C. albicans with C. glabrata or C. parapsilosis (n = 467, 67%).
All 63 colonized patients who developed IC were colonized with the species that subsequently caused infection. Twenty (32%) patients were colonized with two or more Candida species; the dominant colonizing species caused subsequent infection. The remaining 43 patients were colonized with one species (33 with C. albicans, 8 with “other” Candida spp., and 1 each with C. krusei and C. tropicalis). The number of colonizing species was not associated with the development of IC (data not shown).
Comparison of MT-PCR and culture for the detection of Candida colonization in patients with or without IC.
In addition to culture, swabs of the throat and perineum from 205 patients underwent MT-PCR analysis. The median time of IC acquisition in this patient cohort was 5 days (range, 4 to 19 days) post-ICU admission. MT-PCR detected Candida DNA in an additional seven patients (n = 173, 84%) compared with culture (n = 166, 81%). No significant difference was found between MT-PCR and culture for detecting colonization status or between the sampling periods in which colonization was detected (91% and 93% of colonized patients were positive by MT-PCR and culture, respectively, at time point 1).
While only C. albicans, C. tropicalis, and C. krusei were distinguished with certainty on CHROMagar (based on the manufacturer's protocol), a further five species, including C. dubliniensis, C. glabrata, C. guilliermondii, C. parapsilosis, and S. cerevisiae, were identified by MT-PCR. In this cohort, C. albicans was cultured from 134 (65%) patients whereas C. albicans DNA was detected in only 124 (60%) patients; C. dubliniensis DNA was detected in these 10 discrepant cases. Non-Candida albicans spp. detected by MT-PCR included C. dubliniensis (n = 27, 13%), C. glabrata (n = 29, 14%), C. guilliermondii (n = 3, 1%), C. krusei (n = 12, 6%), C. parapsilosis (n = 17, 8%), C. tropicalis (n = 15, 7%), and S. cerevisiae (n = 35, 17%). MT-PCR provided species identification in 73 cases where the species of the yeast could not be determined by CHROMagar.
The turnaround time for MT-PCR (3 h [but, in practice, 8 to 24 h, allowing for specimen transport and incorporation into routine laboratory workload]) was shorter than the 48 h required to obtain results from culture-based surveillance.
DISCUSSION
In this prospective multicenter study, we systematically determined the most effective and practical protocol to measure colonization status and predict IC in nonneutropenic patients in mixed medical/surgical Australian ICUs. As expected, colonization was detected in most patients (86%) prior to development of IC. Standardized cultures of the throat and perineum at time points 1 and 2 captured 97% (61/63) of colonized patients who developed IC, a median of 7 days prior to the diagnosis of IC (designated the diagnostic specimen collection date). Notably, 14% of the patients (n = 10) had negative surveillance cultures prior to the diagnosis of IC, similarly to a Spanish study of mixed medical/surgical ICUs (19). Based on our prior observation (17), we expect that this subgroup will be captured when clinical parameters such as recent gastrointestinal surgery are built into derivation of our model.
Prior Candida colonization is a major risk factor for developing IC; however, the sites surveyed have differed considerably (14, 19, 21, 31–33). Furthermore, proponents of “clinical only” prediction rules have argued for exclusion of colonization data because the process is resource intensive and adds to laboratory costs. Our study determined that collection of samples from three sites alone (throat, perineum, and urine) was practical and feasible but was not necessary for routine surveillance, since 98% (62/63) of colonized patients who subsequently developed IC were captured by throat and perineum surveillance cultures. In contrast to our findings, others have indicated that the mouth is not a good site for routine Candida surveillance due to lower positivity rates (23); the reason for this difference is not clear but may be linked to changes in the oropharyngeal niche and differences in oral hygiene practices among ICUs.
To optimize the timing of surveillance cultures, we compared results of cultures at time points 1, 2, and 3 (Tables 1 and 2 and Fig. 1). In a trend similar to those previously reported by others (21, 34), the majority of colonized patients who subsequently developed IC tested positive on the first screen (n = 56, 89%), indicating that new colonizing strains are acquired in a minority of patients after 72 h in the ICU (21). Detection of throat colonization unexpectedly decreased with time in the ICU (Fig. 1). Selective decontamination of the digestive tract (SDD) was not used in the present cohort, although changes to the oropharyngeal flora upon ICU admission are expected, considering variables such as patient intubation status, nutritional components, and different oral hygiene methods. Since 97% (61/63) of colonized patients who developed IC had positive surveillance in the throat and/or perineum within the first two time points and since the median time from ICU admission to IC was 11 days, we suggest that surveillance cultures taken at 72 h and 7 days post-ICU admission are sufficient for determining Candida colonization status for inclusion into risk-predictive models.
Multifocal colonization (at least two sites, CI > 0.5) and heavy colonization (at least one site, CCI ≥ 0.3) were both significant independent risk factors in our cohort (Table 2), but the decrease (78% to 58%) in sensitivity when CCI was applied indicates its poor utility for predicting IC in our mixed medical/surgical ICU population. In the original study by Pittet et al. (14), the CCI resulted in a 100% PPV compared with the 2% to 4% in our study (Table 2). This large discrepancy is likely because the CCI (in the Pittet study) was derived from a small cohort of critically ill surgical patients who were selected as “high risk for IC” based on multiple clinical risk factors and because colonization status was determined through a variable number of “convenience” samples received in the laboratory at any time prior to the development of IC (14). This contrasts with populations, such as ours, that were selected on the basis of ICU length of stay, resulting in a low PPV and a high NPV. Defined sampling protocols and the mixed medical/surgical populations in our ICUs may also have contributed to the considerable difference with respect to CCI performance.
As in other studies (20, 23), low PPVs were obtained when Candida colonization parameters were applied as the sole criterion for predicting IC—mainly due to its low incidence (1% to 2%, as in our study). With a median time from ICU admission to infection of 11 days (range, 4 to 41), initial screening at 72 h post-ICU admission was necessary to predict the 25% of cases that would have been missed had we used the 7-day entry point proposed by others (16, 22). Excellent NPVs were generated for all Candida colonization parameters investigated, indicating that patients with negative surveillance cultures are highly unlikely to develop disease and would not benefit from early antifungal intervention. An approach to patient care based on these observations would in turn reduce hospital costs, drug-related toxicities, and the likelihood of emerging antifungal resistance.
In a trend similar to those observed in European and North American studies (2, 19, 21, 22, 34), C. albicans was the dominant colonizing (81%) and infecting (77%) species. A rising prevalence of IC due to non-albicans Candida species has been attributed to azole prophylaxis and empirical treatment (6, 35–38). Others, however, have reported negligible effects of systemic antifungal use on colonization status (including that by C. glabrata, specifically) (24, 25). In the present study, only 12 of the 63 colonized IC patients had received antifungal therapy (all empirical; 10 fluconazole); hence, associations could not be tested.
MT-PCR was evaluated on a subset of patients (n = 205). Overall, results of detection of colonization status were similar between MT-PCR and culture (81% and 84%, respectively), although instances of culture misidentification were found (namely, C. dubliniensis was misidentified as C. albicans by CHROMagar). Other than providing species identification in cases where the yeast could not be identified on CHROMagar (14% C. glabrata and 8% C. parapsilosis), MT-PCR did not provide sufficient additional information to justify faster results and a 5-fold cost increase. Furthermore, yeast species determinations can now be performed rapidly and inexpensively from culture using technologies such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (this was not available in clinical laboratories in Australia during the study period).
A potential limitation of this study was its multicenter characteristic, which encompassed possible variations in diagnostic and therapeutic decision making. Australian ICUs, however, are comparable with respect to case mix and management approaches, and a center effect was not demonstrated when data from individual hospitals were analyzed and compared (data not shown). In addition, a large sample size was employed to minimize disparities and improve the generalizability of results. The possibility of systematic bias was eliminated by the inclusion of patients who were admitted consecutively into the ICUs.
In conclusion, after performing a large-scale, multicenter, prospective, systemic evaluation of a variety of Candida colonization parameters, we propose a simplified protocol to optimize detection of colonization status for inclusion into risk-predictive models of IC in critically ill patients. This protocol, consisting of throat and perineum surveillance sampling at 72 h post-ICU admission and 3 to 4 days later, captures most patients likely to develop IC. It should be noted that colonization might not be detected prior to infection in a subset of patients who proceed to develop IC (14% in our cohort), but we expect that these patients will be captured by incorporation of clinical risk factors along with these colonization data into our final risk-predictive model.
ACKNOWLEDGMENTS
We particularly acknowledge Maureen Puhlmann, Research Nurse at the Concord Hospital site, for her enthusiasm, hard work, and support of this project. Maureen passed away during the course of the study, a great loss to her profession.
We also acknowledge the following contributors to this study: Claire Kesby, June Kelly, and Catriona Halliday, Westmead Hospital, Sydney; Neil Underwood and Joan Faoagali, Princess Alexandra Hospital, Brisbane; Quoc Nguyen and Sam Rudham, St Vincent's Hospital, Sydney; Graeme Nimmo, Alex Dank, Melissa Lassig-Smith, Janine Stuart, and Renae Deans, Royal Brisbane Women's and Children's Hospital, Brisbane; Leanne Redl and Caroline Marshall, Royal Melbourne Hospital, Melbourne; Jocelynne McRae, David Milliss, Evanthia Tambosis, and Charlotte Webster, Concord Hospital, Sydney; James Branley and Sarah Whereat, Nepean Hospital, Sydney; staff at the Victorian Infectious Diseases Reference Laboratories, Melbourne; and statistician Brian O'Toole, the Centre for Infectious Diseases and Microbiology Public Health, Westmead Hospital, Sydney.
T.C.S. is a Sydney Medical School Foundation fellow.
This project was funded by grants from the National Health and Medical Research Council of Australia (Project Grant no. 512307 and Centre of Clinical Research Excellence Grant no. 264625) and an Australian Universities Post-graduate Award to A.F.L.
REFERENCES
- 1.Vazquez JA. 2010. Invasive fungal infections in the intensive care unit. Semin Respir Crit Care Med 31:79–86. doi: 10.1055/s-0029-1246289. [DOI] [PubMed] [Google Scholar]
- 2.Leroy O, Gangneux JP, Montravers P, Mira JP, Gouin F, Sollet JP, Carlet J, Reynes J, Rosenheim M, Regnier B, Lortholary O. 2009. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006). Crit Care Med 37:1612–1618. doi: 10.1097/CCM.0b013e31819efac0. [DOI] [PubMed] [Google Scholar]
- 3.Garey KW, Rege M, Pai MP, Mingo DE, Suda KJ, Turpin RS, Bearden DT. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis 43:25–31. doi: 10.1086/504810. [DOI] [PubMed] [Google Scholar]
- 4.Morrell M, Fraser VJ, Kollef MH. 2005. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 49:3640–3645. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruciani M, de Lalla F, Mengoli C. 2005. Prophylaxis of Candida infections in adult trauma and surgical intensive care patients: a systematic review and meta-analysis. Intensive Care Med 31:1479–1487. doi: 10.1007/s00134-005-2794-y. [DOI] [PubMed] [Google Scholar]
- 6.Playford EG, Webster AC, Sorrell TC, Craig JC. 2006. Antifungal agents for preventing fungal infections in nonneutropenic critically ill and surgical patients: systematic review and meta-analysis of randomized clinical trials. J Antimicrob Chemother 57:628–638. doi: 10.1093/jac/dki491. [DOI] [PubMed] [Google Scholar]
- 7.Shorr AF, Chung K, Jackson WL, Waterman PE, Kollef MH. 2005. Fluconazole prophylaxis in critically ill surgical patients: a meta-analysis. Crit Care Med 33:1928–1935; quiz, 1936. doi: 10.1097/01.CCM.0000178352.14703.49. [DOI] [PubMed] [Google Scholar]
- 8.Vardakas KZ, Samonis G, Michalopoulos A, Soteriades ES, Falagas ME. 2006. Antifungal prophylaxis with azoles in high-risk, surgical intensive care unit patients: a meta-analysis of randomized, placebo-controlled trials. Crit Care Med 34:1216–1224. doi: 10.1097/01.CCM.0000208357.05675.C3. [DOI] [PubMed] [Google Scholar]
- 9.Hsueh PR, Graybill JR, Playford EG, Watcharananan SP, Oh MD, Ja'alam K, Huang S, Nangia V, Kurup A, Padiglione AA. 2009. Consensus statement on the management of invasive candidiasis in intensive care units in the Asia-Pacific region. Int J Antimicrob Agents 34:205–209. doi: 10.1016/j.ijantimicag.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Ostrosky-Zeichner L, Sable C, Sobel J, Alexander BD, Donowitz G, Kan V, Kauffman CA, Kett D, Larsen RA, Morrison V, Nucci M, Pappas PG, Bradley ME, Major S, Zimmer L, Wallace D, Dismukes WE, Rex JH. 2007. Multicenter retrospective development and validation of a clinical prediction rule for nosocomial invasive candidiasis in the intensive care setting. Eur J Clin Microbiol Infect Dis 26:271–276. doi: 10.1007/s10096-007-0270-z. [DOI] [PubMed] [Google Scholar]
- 11.Dupont H, Bourichon A, Paugam-Burtz C, Mantz J, Desmonts JM. 2003. Can yeast isolation in peritoneal fluid be predicted in intensive care unit patients with peritonitis? Crit Care Med 31:752–757. doi: 10.1097/01.CCM.0000053525.49267.77. [DOI] [PubMed] [Google Scholar]
- 12.Paphitou NI, Ostrosky-Zeichner L, Rex JH. 2005. Rules for identifying patients at increased risk for candidal infections in the surgical intensive care unit: approach to developing practical criteria for systematic use in antifungal prophylaxis trials. Med Mycol 43:235–243. doi: 10.1080/13693780410001731619. [DOI] [PubMed] [Google Scholar]
- 13.Ibàñez-Nolla J, Nolla-Salas M, León MA, García F, Marrugat J, Soria G, Díaz RM, Torres-Rodríguez JM. 2004. Early diagnosis of candidiasis in non-neutropenic critically ill patients. J Infect 48:181–192. doi: 10.1016/S0163-4453(03)00120-8. [DOI] [PubMed] [Google Scholar]
- 14.Pittet D, Monod M, Suter PM, Frenk E, Auckenthaler R. 1994. Candida colonization and subsequent infections in critically ill surgical patients. Ann Surg 220:751–758. doi: 10.1097/00000658-199412000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.León C, Ruiz-Santana S, Saavedra P, Almirante B, Nolla-Salas J, Alvarez-Lerma F, Garnacho-Montero J, León MA; EPCAN Study Group. 2006. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with Candida colonization. Crit Care Med 34:730–737. doi: 10.1097/01.CCM.0000202208.37364.7D. [DOI] [PubMed] [Google Scholar]
- 16.León C, Ruiz-Santana S, Saavedra P, Galván B, Blanco A, Castro C, Balasini C, Utande-Vázquez A, González de Molina FJ, Blasco-Navalproto MA, López MJ, Charles PE, Martín E, Hernández-Viera MA, Cava Study Group . 2009. Usefulness of the “Candida score” for discriminating between Candida colonization and invasive candidiasis in non-neutropenic critically ill patients: a prospective multicenter study. Crit Care Med 37:1624–1633. doi: 10.1097/CCM.0b013e31819daa14. [DOI] [PubMed] [Google Scholar]
- 17.Playford EG, Lipman J, Kabir M, McBryde ES, Nimmo GR, Lau A, Sorrell TC. 2009. Assessment of clinical risk predictive rules for invasive candidiasis in a prospective multicentre cohort of ICU patients. Intensive Care Med 35:2141–2145. doi: 10.1007/s00134-009-1619-9. [DOI] [PubMed] [Google Scholar]
- 18.Mann PA, McNicholas PM, Chau AS, Patel R, Mendrick C, Ullmann AJ, Cornely OA, Patino H, Black TA. 28 September 2009. Impact of antifungal prophylaxis on colonization and azole susceptibility of Candida species. Antimicrob Agents Chemother doi: 10.1128/AAC.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordà-Marcos R, Alvarez-Lerma F, Jurado M, Palomar M, Nolla-Salas J, León MA, León C; EPCAN Study Group. 2007. Risk factors for candidaemia in critically ill patients: a prospective surveillance study. Mycoses 50:302–310. doi: 10.1111/j.1439-0507.2007.01366.x. [DOI] [PubMed] [Google Scholar]
- 20.Pelz RK, Lipsett PA, Swoboda SM, Diener-West M, Hammond JM, Hendrix CW. 2000. The diagnostic value of fungal surveillance cultures in critically ill patients. Surg Infect (Larchmt) 1:273–281. doi: 10.1089/109629600750067200. [DOI] [PubMed] [Google Scholar]
- 21.Charles PE, Dalle F, Aube H, Doise JM, Quenot JP, Aho LS, Chavanet P, Blettery B. 2005. Candida spp. colonization significance in critically ill medical patients: a prospective study. Intensive Care Med 31:393–400. doi: 10.1007/s00134-005-2571-y. [DOI] [PubMed] [Google Scholar]
- 22.León C, Alvarez-Lerma F, Ruiz-Santana S, León MA, Nolla J, Jordá R, Saavedra P, Palomar M, EPCAN Study Group . 2009. Fungal colonization and/or infection in non-neutropenic critically ill patients: results of the EPCAN observational study. Eur J Clin Microbiol Infect Dis 28:233–242. doi: 10.1007/s10096-008-0618-z. [DOI] [PubMed] [Google Scholar]
- 23.Magill SS, Swoboda SM, Johnson EA, Merz WG, Pelz RK, Lipsett PA, Hendrix CW. 2006. The association between anatomic site of Candida colonization, invasive candidiasis, and mortality in critically ill surgical patients. Diagn Microbiol Infect Dis 55:293–301. doi: 10.1016/j.diagmicrobio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Magill SS, Swoboda SM, Shields CE, Colantuoni EA, Fothergill AW, Merz WG, Lipsett PA, Hendrix CW. 2009. The epidemiology of Candida colonization and invasive candidiasis in a surgical intensive care unit where fluconazole prophylaxis is utilized: follow-up to a randomized clinical trial. Ann Surg 249:657–665. doi: 10.1097/SLA.0b013e31819ed914. [DOI] [PubMed] [Google Scholar]
- 25.Piarroux R, Grenouillet F, Balvay P, Tran V, Blasco G, Millon L, Boillot A. 2004. Assessment of preemptive treatment to prevent severe candidiasis in critically ill surgical patients. Crit Care Med 32:2443–2449. doi: 10.1097/01.CCM.0000147726.62304.7F. [DOI] [PubMed] [Google Scholar]
- 26.Van Horn KG, Audette CD, Tucker KA, Sebeck D. 2008. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn Microbiol Infect Dis 62:471–473. doi: 10.1016/j.diagmicrobio.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Van Horn KG, Audette CD, Sebeck D, Tucker KA. 2008. Comparison of the Copan ESwab system with two Amies agar swab transport systems for maintenance of microorganism viability. J Clin Microbiol 46:1655–1658. doi: 10.1128/JCM.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau A, Sorrell TC, Chen S, Stanley K, Iredell J, Halliday C. 2008. Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogens from blood culture specimens. J Clin Microbiol 46:3021–3027. doi: 10.1128/JCM.00689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau A, Sorrell TC, Lee O, Stanley K, Halliday C. 2008. Colony multiplex-tandem PCR for rapid, accurate identification of fungal cultures. J Clin Microbiol 46:4058–4060. doi: 10.1128/JCM.01411-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau A, Halliday C, Chen SCA, Playford EG, Stanley K, Sorrell TC. 2010. Comparison of whole blood, serum and plasma for early detection of candidemia by multiplex-tandem PCR. J Clin Microbiol 48:811–816. doi: 10.1128/JCM.01650-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karabinis A, Hill C, Leclercq B, Tancrede C, Baume D, Andremont A. 1988. Risk factors for candidemia in cancer patients: a case-control study. J Clin Microbiol 26:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wey SB, Mori M, Pfaller MA, Woolson RF, Wenzel RP. 1989. Risk factors for hospital-acquired candidemia. A matched case-control study. Arch Intern Med 149:2349–2353. [PubMed] [Google Scholar]
- 33.Garbino J, Lew DP, Romand JA, Hugonnet S, Auckenthaler R, Pittet D. 2002. Prevention of severe Candida infections in nonneutropenic, high-risk, critically ill patients: a randomized, double-blind, placebo-controlled trial in patients treated by selective digestive decontamination. Intensive Care Med 28:1708–1717. doi: 10.1007/s00134-002-1540-y. [DOI] [PubMed] [Google Scholar]
- 34.Agvald-Ohman C, Klingspor L, Hjelmqvist H, Edlund C. 2008. Invasive candidiasis in long-term patients at a multidisciplinary intensive care unit: Candida colonization index, risk factors, treatment and outcome. Scand J Infect Dis 40:145–153. doi: 10.1080/00365540701534509. [DOI] [PubMed] [Google Scholar]
- 35.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassetti M, Righi E, Costa A, Fasce R, Molinari MP, Rosso R, Pallavicini FB, Viscoli C. 2006. Epidemiological trends in nosocomial candidemia in intensive care. BMC Infect Dis 6:21. doi: 10.1186/1471-2334-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comert F, Kulah C, Aktas E, Eroglu O, Ozlu N. 2007. Identification of Candida species isolated from patients in intensive care unit and in vitro susceptibility to fluconazole for a 3-year period. Mycoses 50:52–57. doi: 10.1111/j.1439-0507.2006.01309.x. [DOI] [PubMed] [Google Scholar]
- 38.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]