Abstract
Isolates of Paracoccidioides brasiliensis and Paracoccidioides lutzii, previously characterized by molecular techniques, were identified for the first time by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). All isolates were correctly identified, with log score values of >2.0. Thus, MALDI-TOF MS is a new tool for differentiating species of the genus Paracoccidioides.
TEXT
Two species of the genus Paracoccidioides are now considered the causal agents of paracoccidioidomycosis (PCM) (1), the most important systemic mycosis in nonimmunocompromised hosts in Latin America (2, 3). Epidemiological surveys in areas where the parasite is endemic suggest that >10 million people are infected by these fungi (3). Paracoccidioides spp. have been recovered from human clinical samples, soil, and tissues from certain armadillo species, such as Dasypus novemcinctus (4).
Early observations from our group (5, 6) and others (C. J. Fontes and A. P. Vicentini, unpublished data) have reported a lack of reactivity to Paracoccidioides brasiliensis antigens in routine serological assays by sera from PCM patients living in areas of Brazil outside the southern and southeastern regions endemic for the parasite. These observations suggest that fungi from different regions endemic for the parasite might present significant antigenic differences, and these differences have only recently been elucidated. Multilocus phylogenetic analysis showed that P. brasiliensis, which was previously considered the single causal agent of PCM, in fact comprises a complex of cryptic species (7, 8), one of which was separated into a very divergent phylogenetic branch and is currently being proposed as a new species, Paracoccidioides lutzii sp. nov. (9). This new species appears to occur in the central, southwestern, and northwestern regions of Brazil, but the ecology, boundaries, and specific clinical aspects of infections related to P. lutzii remain unclear (9). For example, a recent report identified a P. lutzii-like isolate in a PCM patient who lived only in the southern and southeastern areas endemic for the parasite, and the identification of this isolate required the utilization of molecular techniques (10). Although molecular techniques are able to accurately identify the two Paracoccidioides species, these methods are time-consuming and labor-intensive.
Thus, only technically accessible routine species differentiation can provide the proper tools to epidemiologists and clinicians to address the unresolved issues of PCM. Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has been successfully applied in clinical laboratories worldwide and has also provided a rapid and accurate alternative methodology for fungal identification (11–13). The aim of this study was to differentiate between P. lutzii and P. brasiliensis using MALDI-TOF MS analysis.
A total of 22 strains representing the two species previously identified by molecular techniques, including multilocus sequence typing, PCR of the hsp70 gene, and internal transcribed space (ITS) rRNA gene sequencing (7, 9, 14), were analyzed (Table 1). Due to biohazard issues, the protocol was performed with Paracoccidioides yeast cells, the noninfective parasitic form observed within patient lesions. The colonies were grown on Fava-Netto solid medium at 37°C (15) and subcultured once a week. A standard protein extraction protocol, with some modifications, was carried out (16). The yeast cell suspensions were heated for 30 min at 95°C in a dry bath, and absolute ethanol (Merck, Darmstadt, Germany) and glass beads were then added to the suspensions before vortexing. After centrifugation, extractions were performed using 85% formic acid and acetonitrile (Sigma, St. Louis, MO, USA), and the clear supernatants were spotted in quadruplicate onto the MALDI target. After air-drying, each sample was overlaid with 1.2 μl of cyano-4-hydroxycinnamic acid matrix solution (Sigma) and dried completely before MALDI-TOF MS measurement in an autoflex MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Calibration was performed before each experiment using a Bruker bacterial test standard (Bruker Daltonics GmbH).
TABLE 1.
Paracoccidioides strains and their identification by MALDI-TOF mass spectrometry according to the newly created MSP library
| Strain name | Origin | Best match log score for main spectra of: |
|
|---|---|---|---|
| P. brasiliensis Pb18 | P. lutzii Pb01 | ||
| P. brasiliensis | |||
| D03 | Southeast Brazil | 2.283 | 0.805 |
| T15LN1 | Southeast Brazil | 2.502 | 0.949 |
| Pb113 | North Brazil | 2.163 | 0.946 |
| Pb339 | Brazil | 2.247 | 1.297 |
| Pbdog | South Brazil | 2.483 | 1.512 |
| Pb262 | Southeast Brazil | 2.368 | 1.953 |
| Pb927 | Uruguay | 2.537 | 1.643 |
| Pb03 | Southeast Brazil | 2.287 | 1.685 |
| BACR | Colombia | 2.455 | 1.187 |
| BAT | Southeast Brazil | 2.470 | 1.722 |
| CNH | Colombia | 2.381 | 1.736 |
| SMA | Southeast Brazil | 2.382 | 1.487 |
| CA | Colombia | 2.114 | 0.718 |
| DM | Southeast Brazil | 2.353 | 1.637 |
| 192 | Southeast Brazil | 2.362 | 1.327 |
| P. lutzii | |||
| Pb8334 | Central-West Brazil | 1.452 | 2.107 |
| 1578 | Central-West Brazil | 1.936 | 2.641 |
| Pb66 | Central-West Brazil | 1.556 | 2.590 |
| EE1 | Central-West Brazil | 1.663 | 2.022 |
| ED01 | Central-West Brazil | 1.722 | 2.419 |
More than 60 spectra were acquired for each of the two strains, P. brasiliensis Pb18 and P. lutzii Pb01, from three independent culture extracts (20 spectra per culture extract). The quality of each spectrum was assessed with the Flex analysis 3.5 software (Bruker Daltonics). Flat-liners and spectra with peak variations (outliers) were removed from the collection, and additional measurements were performed to complete the 60 spectra from each strain. The raw spectra were then loaded into Biotyper 3.1 (Bruker Daltonics), and the creation of main spectra (MSPs) was carried out with the default settings of the Biotyper software.
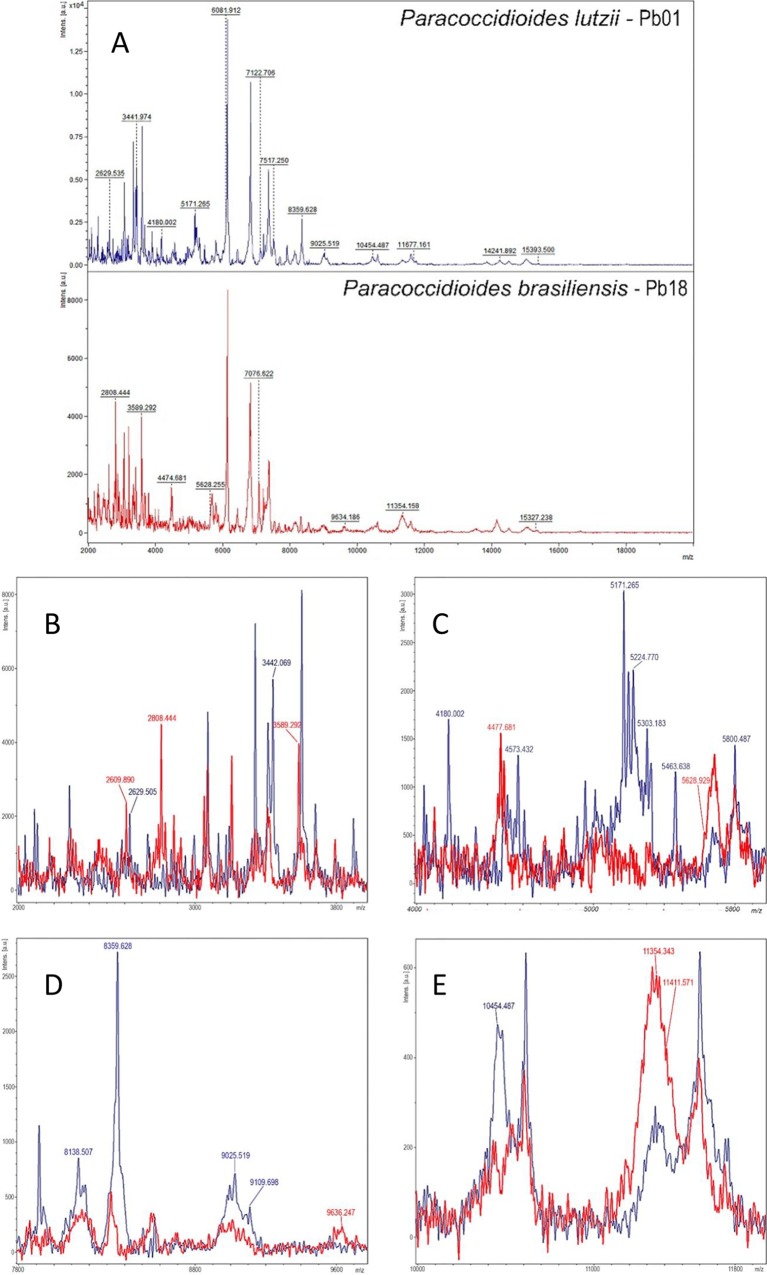
A total of six MSPs were created: three MSPs for Pb18, the standard type strain of P. brasiliensis, and three for Pb01, the standard type strain of P. lutzii (9). To check the specificity of the newly created MSPs, identifications were carried out against all MSPs available in the Bruker database. Finally, to evaluate the performances of the created MSPs, the remaining 15 strains of P. brasiliensis and five of P. lutzii were extracted and subjected to MALDI-TOF MS identification with the automated option in the Biotyper software. To ensure reproducibility, all tests were performed in triplicate. A selection of the specific mass spectra from the two species of Paracoccidioides delineates two different protein profiles (Fig. 1).
FIG 1.
(A) Representative mass spectra (smoothed and baseline subtracted) from P. lutzii Pb01 (blue) and P. brasiliensis Pb18 (red) with potential species-specific ion peaks retrieved from the main spectra peak list. The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratio. Amplified images highlighting overlaying mass spectra of P. lutzii (blue) and P. brasiliensis (red) are shown at 2 to 4 kDa (B), 4 to 6 kDa (C), 8 to 10 kDa (D), and 10 to 12 kDa (E).
The created Paracoccidioides MSPs were found to be unique and suitable for MALDI-TOF MS-based identification, as no misidentifications (log score, ≥2.0) with the Biotyper database were observed. All strains had a correct species assignment, with best match log score values in the range of 2.022 to 2.641 for the P. lutzii strains and 2.114 to 2.537 for the P. brasiliensis strains. The final identifications of the strains by the new MSP library are summarized in Table 1.
Paracoccidioides is the second causal agent of a severe endemic deep mycosis to have its genus split in two different species through recent advances in phylogenetic analyses. More than 10 years ago, the causative agent of coccidioidomycosis was also split in two species: Coccidioides immitis and Coccidioides posadasii (17). It is thought that these species cause the same spectrum of clinical manifestations in humans, although there are few data regarding the features of the diseases caused by each species. Nonetheless, this division is of morphological and epidemiological relevance, because differences in phenotype and geographical distribution were found between the two species (18). In contrast, differences in virulence, response to chemotherapy, and the clinical and laboratorial characteristics of the illnesses caused by the two Paracoccidioides species have been suggested (9). However, these issues are still subject to debate, in part due of the lack of rapid and straightforward methods for species identification of patient isolates. Indeed, micromorphological differentiation between P. brasiliensis and P. lutzii is not reliable. Early studies of some isolates suggested differences in conidial morphology, such as size and shape (8); however, as more P. lutzii isolates became available, the variability in these characteristics increased and overlapped (1). Thus, differentiation based on these characteristics can lead to misinterpretations. MALDI-TOF MS for microorganism identification is a technique that does not require extensive training and is well adapted to routine laboratories. Our results demonstrate that it can be the method of choice for species differentiation of the genus Paracoccidioides, benefiting clinical and laboratorial studies aiming to determine possible differences between the diseases caused by these two species.
ACKNOWLEDGMENTS
We thank Carlos P. Taborda and Marcus M. Teixeira for their kind donation of the Paracoccidioides strains.
Anna S. S. Levin and Silvia F. Costa carefully edited the English language in the manuscript.
The Paracoccidioides MSPs produced in this work are freely available by contacting the corresponding author.
REFERENCES
- 1.Theodoro RC, de Melo Teixeira M, Felipe MMSS, Paduan KdS, Ribolla PM, San-Blas G, Bagagli E. 2012. Genus Paracoccidioides: species recognition and biogeographic aspects. PLoS One 7:e37694. doi: 10.1371/journal.pone.0037694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Restrepo A, McEwen JG, Castañeda E. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med Mycol 39:233–241. doi: 10.1080/714031028. [DOI] [PubMed] [Google Scholar]
- 3.Restrepo A, Benard G, de Castro CC, Agudelo CA, Tobón AM. 2008. Pulmonary paracoccidioidomycosis. Semin Respir Crit Care Med 29:182–197. doi: 10.1055/s-2008-1063857. [DOI] [PubMed] [Google Scholar]
- 4.Bagagli E, Franco M, Bosco Sde MG, Hebeler-Barbosa F, Trinca LA, Montenegro MR. 2003. High frequency of Paracoccidioides brasiliensis infection in armadillos (Dasypus novemcinctus): an ecological study. Med Mycol 41:217–223. doi: 10.1080/13693780310001597368. [DOI] [PubMed] [Google Scholar]
- 5.del Negro GM, Benard G, de Assis CM, Vidal MS, Garcia NM, Otani C, Shikanai-Yasuda MA, da S Lacaz C. 1995. Lack of reactivity of paracoccidioidomycosis sera in the double immunodiffusion test with the gp43 antigen: report of two cases. J Med Vet Mycol 33:113–116. [DOI] [PubMed] [Google Scholar]
- 6.Vidal MSM, Benard G, de Brito T, Dantas KC, Pereira CN, França FOS, da Silva AMG, Martins JEC. 2005. Atypical serological response marked by a lack of detectable anti-gp43 antibodies in a patient with disseminated paracoccidioidomycosis. J Clin Microbiol 43:3014–3016. doi: 10.1128/JCM.43.6.3014-3016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matute DR, Sepulveda VE, Quesada LM, Goldman GH, Taylor JW, Restrepo A, McEwen JG. 2006. Microsatellite analysis of three phylogenetic species of Paracoccidioides brasiliensis. J Clin Microbiol 44:2153–2157. doi: 10.1128/JCM.02540-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teixeira MM, Theodoro RC, de Carvalho MJA, Fernandes L, Paes HC, Hahn RC, Mendoza L, Bagagli E, San-Blas G, Felipe MSS. 2009. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol 52:273–283. doi: 10.1016/j.ympev.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira M de M, Theodoro RC, Oliveira FFM de, Machado GC, Hahn RC, Bagagli E, San-Blas G, Soares Felipe MS. 2014. Paracoccidioides lutzii sp. nov.: biological and clinical implications. Med Mycol 52:19–28. doi: 10.3109/13693786.2013.794311. [DOI] [PubMed] [Google Scholar]
- 10.Takayama A, Itano EN, Sano A, Ono MA, Kamei K. 2010. An atypical Paracoccidioides brasiliensis clinical isolate based on multiple gene analysis. Med Mycol 48:64–72. doi: 10.3109/13693780902718065. [DOI] [PubMed] [Google Scholar]
- 11.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev Proteomics 10:151–164. doi: 10.1586/epr.13.8. [DOI] [PubMed] [Google Scholar]
- 12.Lau AF, Drake SK, Calhoun LB, Henderson CM, Zelazny AM. 2013. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:828–834. doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Almeida Júnior JN, Figueiredo DSY, Toubas D, Del Negro GMB, Motta AL, Rossi F, Guitard J, Morio F, Bailly E, Angoulvant A, Mazier D, Benard G, Hennequin C. 2014. Usefulness of matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry for identifying clinical Trichosporon isolates. Clin Microbiol Infect 20:784–790. doi: 10.1111/1469-0691.12502. [DOI] [PubMed] [Google Scholar]
- 14.Hahn RC, Rodrigues AM, Fontes CJF, Nery AF, Tadano T, Queiroz L de P Jr, de Camargo ZP. 2014. Fatal fungemia due to Paracoccidioides lutzii. Am J Trop Med Hyg 91:394–398. doi: 10.4269/ajtmh.13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fava-Netto C. 1961. Contribuição para o estudo imunológico da blastomicose de Lutz. Rev Inst Adolfo Lutz 21:99–194. [Google Scholar]
- 16.Verroken A, Janssens M, Berhin C, Bogaerts P, Huang T-D, Wauters G, Glupczynski Y. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry for identification of Nocardia species. J Clin Microbiol 48:4015–4021. doi: 10.1128/JCM.01234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher MC, Koenig GL, White TJ, Taylor JW. 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia 94:73–84. doi: 10.2307/3761847. [DOI] [PubMed] [Google Scholar]
- 18.Cox RA, Magee DM. 2004. Coccidioidomycosis: host response and vaccine development. Clin Microbiol Rev 17:804–839. doi: 10.1128/CMR.17.4.804-839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]