Abstract
Clostridium difficile is the most commonly identified pathogen among health care-associated infections in the United States. There is a need for accurate and low-cost typing tools that produce comparable data across studies (i.e., portable data) to help characterize isolates during epidemiologic investigations of C. difficile outbreaks and sporadic cases of disease. The most popular C. difficile-typing technique is PCR ribotyping, and we previously developed methods using fluorescent PCR primers and amplicon sizing on a Sanger-style sequencer to generate fluorescent PCR ribotyping data. This technique has been used to characterize tens of thousands of C. difficile isolates from cases of disease. Here, we present validation of a protocol for the cost-effective generation of fluorescent PCR ribotyping data. A key component of this protocol is the ability to accurately identify PCR ribotypes against an online database (http://walklab.rcg.montana.edu) at no cost. We present results from a blinded multicenter study to address data portability across four different laboratories and three different sequencing centers. Our standardized protocol and centralized database for typing of C. difficile pathogens will increase comparability between studies so that important epidemiologic linkages between cases of disease and patterns of emergence can be rapidly identified.
INTRODUCTION
Clostridium difficile infection (CDI) has emerged as the most frequently encountered nosocomial infection in the United States (1). The clinical manifestations of CDI range from acute, self-limiting diarrhea to fulminant and sometimes fatal colitis (2). Over the past decade, there has been a doubling of CDI-related discharge diagnoses and a 10-fold increase in CDI-attributable mortality in the United States (3). Because of its clinical importance, a number of C. difficile-typing techniques, including pulsed-field gel electrophoresis (PFGE), restriction endonuclease analysis (REA) typing, and PCR ribotyping, have been implemented to differentiate between C. difficile strains and address important epidemiologic questions.
PCR ribotyping is the most commonly cited method for typing C. difficile isolates. This technique quantifies the differences in length between 16S rRNA and 23S rRNA encoding genes at approximately 11 rRNA-encoding operons around the C. difficile genome (4). Like other popular typing techniques (REA and PFGE), PCR ribotyping is gel based, and the data are not easily portable between laboratories. Sequence-based methods, like multilocus sequence type (MLST) and genome sequencing, provide highly portable data but are expensive in comparison to gel-based methods. A significant improvement on traditional PCR ribotyping came with the use of a fluorescently labeled PCR primer and sizing of the resulting amplicons using a Sanger-style sequencer (i.e., fluorescent PCR ribotyping) (5). We developed a similar approach and have applied the method to C. difficile isolates from cases of disease and those that circulate in the community (6–11).
Fluorescent PCR ribotyping data are digital and, in theory, should be portable between laboratories if a common protocol and a common data source (i.e., database) are used for identification. The goals of this study were to minimize the costs associated with fluorescent PCR ribotyping and to test whether our protocol and database produced portable data across sequencing centers and laboratories. To help minimize costs, we compared data generated using reagents that vary in cost. For sequencing center comparisons, we analyzed the same fluorescent PCR amplicons at three different sequencing centers. We then conducted a blinded, multicenter study in which the collaborating labs were sent the same template DNA from representative C. difficile isolates and were asked to generate data using a standardized protocol. All data were analyzed using a free, online analysis tool. Collectively, our results suggest that the protocol produces highly portable data and identifies C. difficile ribotypes with high accuracy and precision across different labs and sequencing centers. Trends in the data suggest that user (human) error played a significant role, and ways to minimize this error are discussed.
MATERIALS AND METHODS
Isolate cultivation and DNA extraction.
A list of isolates, including the source of each isolate used in this study, is provided in Table S1 in the supplemental material. The cultivation of C. difficile isolates was performed in a vinyl anaerobic chamber (Coy Laboratories Products, Inc., Grass Lake, MI, USA). The isolates were grown overnight under anaerobic conditions (85% nitrogen, 10% hydrogen, 5% carbon dioxide) at 37°C in prereduced brain heart infusion broth with 5 g/liter yeast extract and 0.1% l-cysteine, as recommended (12). The genomic DNA was extracted using the Easy-DNA kit (K1800-01; Thermo Fisher Scientific, Inc.), quantified using a NanoDrop 1000 (NanoDrop Technologies, Houston, TX, USA), and diluted to a concentration of 100 ng/μl using sterile DNase/RNase free water.
Fluorescent PCR ribotyping.
A list of the recommended equipment for successful PCR ribotyping is provided in Table S2 in the supplemental material. Each PCR was carried out in 25-μl total volumes, and a master mix was used for all reactions. Individual reagents were as follows: 12.5 μl master mix (AmpliTaq Gold 360 master mix, 4398881, Thermo Fisher Scientific, Inc.; GoTaq Hot Start Colorless master mix, M5132, Promega, Inc.; PCR master mix, M7502, Promega, Inc.), 0.5 μl forward primer (GTGCGGCTGGATCACCTCCT) (4), 0.5 μl 6-carboxyfluorescein (FAM)-labeled reverse primer (56-FAM/CCCTGCACCCTTAATAACTTGACC) (10), 10.5 μl DNase/RNase free water, and 100 ng (1 μl) template DNA. Conditions for all PCRs were as previously reported (10). Because fluorescent PCR amplicons are sensitive to light and multiple freeze/thaw events, the PCRs were covered (with foil) and frozen (−20°C) a single time. For the fragment analysis, samples were thawed and diluted 1:1,000 in sterile DNase/RNase free water. A 12-μl mixture of ROX 1000 size standard (BioVentures, Inc.) and Hi-Di Formamide (4311320; Thermo Fisher Scientific, Inc.) in a ratio of 1:240 was added to 5 μl of diluted, fluorescent PCR amplicons in barcoded, semiskirted 96-well plates (GeneMate, T-3107-1; BioExpress, Inc.). For example, 5 μl of size standard was mixed with 1.2 ml of formamide, and 12 μl was aliquoted into wells containing 5 μl fluorescent amplicons. Plates were sealed with caps (83009-684; VWR International), centrifuged at a low speed to ensure that the liquid was at the bottom of each well, and shipped with an ice pack to sequencing centers.
Analysis pipeline.
An online analysis tool was generated using the Python computer program and is available for use at http://walklab.rcg.montana.edu. Peaks in the chromatogram files (.fsa file format) were identified using the freely available Peak Scanner software version 1.0 (Applied Biosystems, Inc.). A text file containing the peak size (in base pairs) and area was exported from Peak Scanner and uploaded for analysis at the website named above. Peaks were normalized as a percentage of the total peak area, as previously published (10). The presence/absence of peaks contained in 5-bp bins (between 200 bp and 1,000 bp in size) and their relative abundance (normalized peak areas) were compared to profiles in a curated database using a Bray-Curtis dissimilarity (BCD) metric. BCDs range between 0 and 1, where 0 is assigned to identical matches and 1 is assigned to completely different peak profiles. The online analysis tool returns a list of lowest BCD matches, and a threshold of 0.2 (see the Results section) was generally found to differentiate matches (<0.2) from mismatches (>0.2).
Statistical analyses.
A three-way analysis of variance (ANOVA) was used to compare Bray-Curtis dissimilarities based on a complete block design of Taq (AmpliTaq Gold, Promega GoTaq, and standard Promega), primer purification (HPLC and standard desalting), and ribotype (F001, F012, and F003). The analysis of variance [aov()] function in the R statistical software package (version 3.0.0) (13) was used for main effects with Tukey's honestly significant different (HSD) for post hoc comparisons. Two separate two-way ANOVAs were used to compare Bray-Curtis dissimilarities between different sequencing centers and ribotypes and between different laboratories and ribotypes using GraphPad Prism version 6.00 (GraphPad Software, Inc.). Tukey's correction was used to adjust P values for multiple comparisons.
RESULTS
F ribotype database.
Previously published fluorescent PCR ribotyping data from clinical cases of disease were assembled as .fsa (chromatogram) files. Data were generated using a single ABI3730xl sequencer at the University of Michigan DNA sequencing core facility, and all chromatograms were visually inspected to ensure identical peak profiles (i.e., presence/absence of peaks). A total of 113 distinct ribotypes were identified and assigned an alpha-numeric code beginning with the letter F. This code is meant to differentiate fluorescent PCR ribotypes from previously reported PCR ribotypes generated by traditional gel-electrophoresis methods. Twelve F ribotypes in the database correspond to previously reported ribotypes based on the analysis of reference strains obtained from collaborators, so these ribotype numbers were maintained for comparability to historic data (e.g., F027 indicates ribotype 027). The remaining ribotypes were assigned the prefix FP to distinguish proposed F ribotyping numbers. As more data are generated on historic and/or reference isolates, the FP ribotype designations can be easily changed to F ribotypes according to their previously assigned numbers. In total, our database included 1,038 .fsa files representing 113 distinct F ribotypes (12 F and 101 FP ribotypes).
Influence of PCR reagents on F ribotype identification and database matching.
The costs involved in fluorescent PCR ribotyping can be divided into three parts: PCR reagents, standard reagents/supplies, and fragment analysis (Table 1). Of these, PCR reagents have the potential for significant cost savings, whereas there are few, if any, alternatives for standard reagents/supplies or fragment analysis. We therefore investigated whether data generated using low-cost PCR reagents were comparable to data generated using more expensive PCR reagents. We generated data on three C. difficile isolates representing three different F ribotypes (F001, F012, and F003) using three Taq polymerases associated with high (AmpliTaq Gold), medium (Promega GoTaq), and low (standard Promega) cost and using PCR primers that were purified using either HPLC (high cost) or standard desalting (low cost). We then determined whether the expected F ribotypes were identified and how well the data matched to the F ribotyping database using the Bray-Curtis dissimilarity (BCD) metric.
TABLE 1.
Minimized cost of fluorescent PCR ribotyping reagents
| Purchased item | Vendor | List price ($) | Units per 96-well plate | Shipping cost ($) | Cost per 96-well plate ($) |
|---|---|---|---|---|---|
| PCR reagent | |||||
| Forward primer (standard desalting) | IDTDNAa | 7.00 | 46.04 | 3.75 | 0.23 |
| FAM-labeled reverse primer (standard desalting) | IDTDNA | 73.20 | 132.29 | 3.75 | 0.58 |
| 96-well PCR plate | VWR International | 315.10 | 100 | 3.15 | |
| Promega PCR master mix | Fisher Scientific | 88.40 | 2.08 | 42.43 | |
| Standard reagent and supplies | |||||
| Bar-coded 96-well plate | BioExpress | 1,412.35 | 500 | 67.50 | 2.96 |
| Hi-Di Formamide | Life Technologies | 36.45 | 21.48 | 61.50 | 4.56 |
| MapMarker X-Rhodamine size standard | BioVentures | 335.00 | 80 | 29.38 | 4.55 |
| Fragment analysis | |||||
| Shipping to sequencing core facility | 16.17 | ||||
| CE fragment analysis on ABI3730XL | 52.50 | ||||
| Total cost per 96-well plate | 127.15 | ||||
| Total cost per well | 1.32 |
IDTDNA, Integrated DNA Technologies.
In all cases, regardless of the Taq used or how the PCR primers were purified, the expected F ribotypes were identified. Also, no significant differences were detected in BCDs between data generated using HPLC and standard desalting purification of primers (P = 0.799), regardless of the Taq polymerase used or ribotype considered. However, the cheapest Taq tested (Promega Taq master mix) produced significantly lower BCDs than either of the other Taqs for all three F ribotypes (P < 0.0001 for all comparisons, Fig. 1). Because standard desalting of primers did not affect BCDs and because the Promega Taq master mix produced significantly better matches to the database (i.e., lower BCDs), these reagents were used to compare data generated at multiple sequencing facilities and across multiple laboratories.
FIG 1.
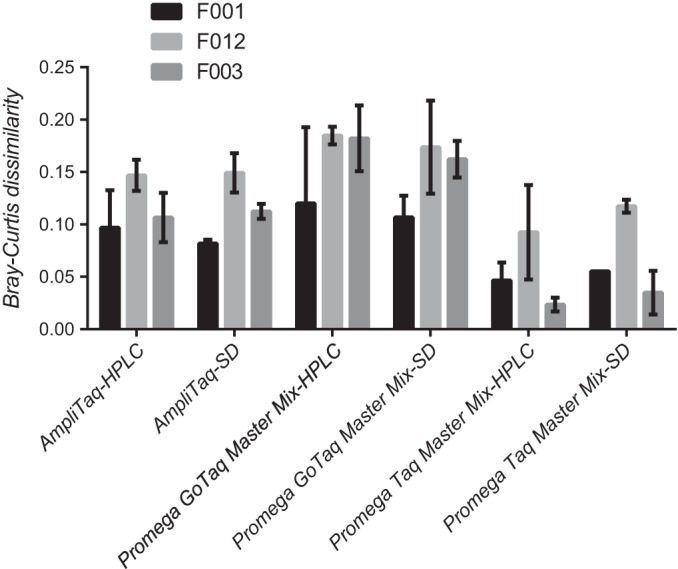
Influence of three different Taq polymerases and two different PCR primer purification techniques on database matching for three different F ribotypes. Bars represent mean Bray-Curtis dissimilarity for three different F ribotypes. Error bars represent 95% confidence limits.
Data portability across different sequencing centers.
To evaluate the comparability of fluorescent PCR ribotyping data generated at different sequencing centers, we prepared DNA from 19 C. difficile isolates representing 12 different F ribotypes and aliquoted this DNA into quadruplicate wells of a 96-well plate, along with sterile water as negative controls so that all wells of the plate were used (i.e., [19 isolates × 4] + 20 water). Fluorescent PCR ribotyping amplicons were generated, and the same amplicons were submitted to three different sequencing centers for capillary electrophoresis (CE) fragment analysis on an ABI3730xl Sanger-style sequencer.
As with the PCR reagent analysis above, only the expected F ribotypes were identified, regardless of the sequencing center. Data were missing in two wells analyzed from center 1 (2 of 76, 2.6%) and in three wells from center 3 (3 of 76, 3.9%). No signal was detected in wells containing water (negative control). Significant differences in BCDs were detected between the different sequencing centers (P < 0.0001, Fig. 2). BCDs from center 2 were significantly greater than (P < 0.05) BCDs from both of the other two centers for 4 of the 12 F ribotypes. In contrast, BCDs at centers 1 and 2 were indistinguishable (P ≥ 0.05) for 11 of 12 F ribotypes. With few exceptions, BCDs at centers 1 and 2 were below 0.2 (Fig. 2, dotted line).
FIG 2.
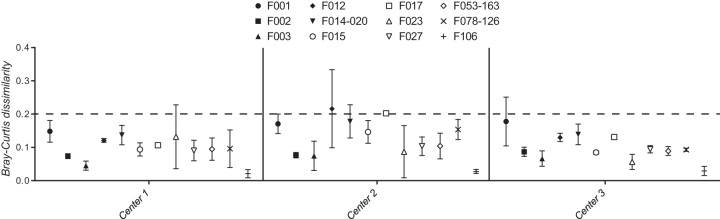
Comparison of CE fragment analysis of the same fluorescent PCR amplicons from 12 F ribotypes of C. difficile at three different sequencing centers. Mean Bray-Curtis dissimilarities at each sequencing center are shown for different F ribotypes (symbols) along with 95% confidence limits (error bars).
Data portability across different laboratories and sequencing centers.
To evaluate the data comparability across the different laboratories and sequencing centers, we prepared DNA from a total of 45 C. difficile isolates representing 18 different F ribotypes and aliquoted the DNA into duplicate wells of a 96-well plate. Sterile water was placed into six wells as negative controls so that all wells were used (i.e., [45 isolates × 2] + 6 water). Four replicate 96-well plates were then made from the primary sample plate using a multichannel pipette. Three of these replicate plates were sent to different laboratories for generation of fluorescent PCR amplicons, and one plate was analyzed in-house for the comparison (Fig. 3). Sites 1, 2, and 3 submitted amplicons to a single sequencing center (center 1; Fig. 1), and site 4 submitted amplicons to a different sequencing center (center 3) for fragment analysis.
FIG 3.
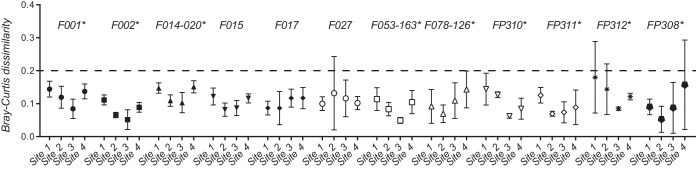
Comparison of fluorescent PCR ribotyping data for 12 F ribotypes across four different laboratories (sites) and two different sequencing centers. Mean Bray-Curtis dissimilarities (BCDs) are shown for different F ribotypes (symbols) along with 95% confidence limits (error bars). Asterisks indicate F ribotypes for which BCDs from at least one site differed from those from at least one other site.
A total of 384 (i.e., 96 × 4) data points were available from this design. A fluorescent signal was detected above the background noise in 3 of the 24 (12.5%) negative-control samples, i.e., in two samples at site 2 and in one sample at site 3, suggesting that cross-contamination may have occurred. No fluorescent signal was detected in negative-control wells at sites 1 and 4. Only 2 (0.5%) negative PCRs were observed among the 360 wells that were expected to have a fluorescent signal, and both incidents occurred at site 2. Also, only 2 (0.5%) instances of possible contamination were observed (i.e., a different PCR ribotype than expected was identified), and both of these incidents occurred at site 2. Overall, a deviation from the expected result was observed in only 7 of the 384 (1.8%) data points, and 6 of these 7 events occurred in data generated by a single laboratory.
Of the 18 F ribotypes analyzed, 12 were represented by at least two different isolates and could be used to statistically test for differences in BCDs across sites. Significant differences were detected between sites (P < 0.0001), and BCDs from at least one site significantly differed (P < 0.05) from the other three for 9 of the 12 F ribotypes analyzed (Fig. 3). Site 3 produced the lowest mean BCDs for 6 of the 12 F ribotypes, but no other site-specific patterns were easily discernible. No significant differences were observed in BCDs produced by site 4 (sequencing center 3) and those produced by sites 1 to 3 (sequencing center 1) for any of the 12 F ribotypes. As for the results of the sequencing center comparison, nearly all BCDs were below 0.2 (Fig. 3, dotted line).
DISCUSSION
PCR ribotyping has been used to genotype C. difficile isolates for nearly 2 decades (14), and it remains the most commonly used genotyping technique for C. difficile pathogens. In 2008, Indra et al. published a method for fluorescent PCR ribotyping using capillary gel electrophoresis (i.e., fragment analysis on a Sanger-style sequencer) and pointed out that the technique decreased the required hands-on time and had the potential to overcome problems associated with interlaboratory comparisons using traditional (gel-based) PCR ribotyping data (5). Since this initial report, little progress has been made in comparing fluorescent PCR ribotyping data across sequencing centers or developing a standardized protocol across laboratories.
Using methods similar to but distinct from those of Indra et al., we previously generated fluorescent PCR ribotyping data on C. difficile isolates from a variety of clinical and community settings (6–11). These data have been curated and compiled into an online database for the scientific community (http://walklab.rcg.montana.edu). Users can download step-by-step instructions for generating comparable data and can identify F ribotypes that are in the database. Users are also encouraged to submit isolates and/or data files so that the database is more representative of the C. difficile isolate diversity in circulation. A number of other techniques and protocols have been developed for genotyping C. difficile isolates (15), and these data can be easily incorporated into the online database as they become available (e.g., North American pulsed field types, restriction endonuclease types, and multilocus sequence types).
To maintain a high-quality database, a single person (S.T.W.) is responsible for adding data to and curating the database described in this study. If users believe they have discovered a ribotype not already in the database, it is recommended that a stock of the isolate be sent to the curator for verification (at no cost to the user). In lieu of an actual isolate, only high-quality chromatograms represented by at least two documented, independent PCRs and fragment analyses will be considered for incorporation. Such F ribotypes (i.e., those not verified by the curator) will be labeled with an asterisk and reported as such so that all users can easily recognize them. Metadata (data other than F ribotype chromatograms) can be linked to isolates by contacting the curator using the contact form on the website (http://walklab.rcg.montana.edu/contacts.html). A data form is available containing various fields (e.g., data of isolation, location, previous typing methods). All data, including metadata associated with isolates, will be made publicly available.
The comparison of different PCR reagents described above identified a decreased overall cost of fluorescent PCR ribotyping to $1.32 per isolate. This price includes the listed cost of reagents, which may be further reduced by ordering reagents in bulk and utilizing discounts commonly offered by large vendors. A direct consequence of lowering the cost of typing is that more isolates can be evaluated with the fluorescent PCR technique than with other techniques, with similar discriminatory power. For example, the cost of multilocus sequence typing (MLST) is ∼40 U.S. dollars per isolate, so approximately 30 C. difficile isolates could be typed by fluorescent PCR ribotyping for every isolate typed by MLST.
The real advantage of low-cost fluorescent PCR ribotyping, however, is simply to limit the use of more discriminant but higher-cost techniques. For example, multiple-locus variable number tandem repeat analysis (MLVA) and genome sequencing are more discriminant (i.e., resolve more recent evolutionary changes) than fluorescent PCR ribotyping and can be used to investigate nosocomial CDI outbreaks. MLVA (16) and the recent modified version (17) are similar to PCR ribotyping but involve more PCRs that increase the reagent cost. A recent comparison of MLVA and genome sequencing reported the reagent costs were $42 and $65, respectively, per isolate (18). It should be noted that these prices do not include the cost of analysis, which represents a significant investment in bioinformatics and computer software for some laboratories. We suggest that fluorescent PCR ribotyping can be used first to narrow down the number of isolates requiring more discriminant characterization. This cost savings will allow for more detailed epidemiologic investigations, because more isolates can be considered.
The emergence of CDI as the most common nosocomial infection in the United States underlies the need for active C. difficile surveillance by clinical laboratories and infection control authorities. There is an urgent need to understand the patterns of C. difficile emergence across different clinical centers, including the patterns of antimicrobial resistance (19, 20). For these types of analyses, genotyping techniques that produce easily comparable data are essential. The fluorescent PCR ribotyping protocol tested here provides the technical standardization for the comparison of data across four laboratories and three sequencing centers. However, some of the observed results were likely due to human error, as nearly all of the unexpected results in the multicenter study (6 of 7) were produced at a single laboratory. Similarly, we were able to detect center-specific sequencing differences in how well the data matched to the database (i.e., differences in BCDs), although these differences were not great enough to confound correct identification. Collectively, these observations suggest that there is a learning curve for using the protocol and that the inclusion of data from multiple sequencing centers into the database will increase the quality of F ribotype matching.
Manual inspection of fragment analysis chromatograms (.fsa files) may still be required in some situations. For example, nearly all BCDs for data across two of the sequencing centers and across all four laboratories were below 0.2. Since a match is defined in our analysis as the smallest BCD, it was somewhat safe to assume that any mismatches will have greater BCDs. Therefore, this value (0.2) seems appropriate for differentiating between high- and low-quality matches and/or potential mismatches. We suggest that chromatograms from matches producing BCDs that are ≥0.2 should be visually compared to reference chromatograms (also available at the database website named above).
In summary, the fluorescent PCR ribotyping protocol used in this study can be done at a low per-sample cost. The data can be analyzed online, and the F ribotypes can be identified free of charge. Data from previously named and newly identified C. difficile genotypes can be easily added to the database for broader comparisons between laboratories and different studies. Finally, we recommend the use of this technique during epidemiologic investigations to maximize the number of isolates considered while minimizing costs associated with more discriminant techniques, such as MLVA or genome sequencing.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant K01AI09728101 from the National Institutes of Health (to S.T.W.), the Claude D. Pepper Older American Independence Center grant AG-024824 (to K.R.), and the Texas Department of State Health Services grant 2014-045577 (to K.W.G.).
We thank Judy Opp (University of Michigan, Ann Arbor, MI) and Jane M. Marsh (University of Pittsburgh, Pittsburgh, PA) for contributing time and effort to this project and also Dale Gerding (Edward Hines, Jr., VA Hospital, Chicago, IL) and Trevor Lawley (Wellcome Trust Sanger Institute, Hinxton, Cambridge, United Kingdom) for providing reference strains.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03591-14.
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated I, Antimicrobial Use Prevalence Survey T . 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuijper EJ, Coignard B, Tüll P, ESCMID Study Group for Clostridium difficile, EU Member States, European Centre for Disease Prevention and Control . 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12(suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 3.Lessa FC, Gould CV, McDonald LC. 2012. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 55:S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 175:261–266. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 5.Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ. 2008. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57:1377–1382. doi: 10.1099/jmm.0.47714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alam MJ, Anu A, Walk ST, Garey KW. 2014. Investigation of potentially pathogenic Clostridium difficile contamination in household environs. Anaerobe 27:31–33. doi: 10.1016/j.anaerobe.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Behroozian AA, Chludzinski JP, Lo ES, Ewing SA, Waslawski S, Newton DW, Young VB, Aronoff DM, Walk ST. 2013. Detection of mixed populations of Clostridium difficile from symptomatic patients using capillary-based polymerase chain reaction ribotyping. Infect Control Hosp Epidemiol 34:961–966. doi: 10.1086/671728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson PE Jr, Walk ST, Bourgis AE, Liu MW, Kopliku F, Lo E, Young VB, Aronoff DM, Hanna PC. 2013. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe 24:109–116. doi: 10.1016/j.anaerobe.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao K, Walk ST, Micic D, Chenoweth E, Deng L, Galecki AT, Jain R, Trivedi I, Yu M, Santhosh K, Ring C, Young VB, Huffnagle GB, Aronoff DM. 2013. Procalcitonin levels associate with severity of Clostridium difficile infection. PLoS One 8:e58265. doi: 10.1371/journal.pone.0058265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walk ST, Micic D, Jain R, Lo ES, Trivedi I, Liu EW, Almassalha LM, Ewing SA, Ring C, Galecki AT, Rogers MA, Washer L, Newton DW, Malani PN, Young VB, Aronoff DM. 2012. Clostridium difficile ribotype does not predict severe infection. Clin Infect Dis 55:1661–1668. doi: 10.1093/cid/cis786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waslawski S, Lo ES, Ewing SA, Young VB, Aronoff DM, Sharp SE, Novak-Weekley SM, Crist AE Jr, Dunne WM, Hoppe-Bauer J, Johnson M, Brecher SM, Newton DW, Walk ST. 2013. Clostridium difficile ribotype diversity at six health care institutions in the United States. J Clin Microbiol 51:1938–1941. doi: 10.1128/JCM.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 13.R Project for Statistical Computing. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 14.Cartwright CP, Stock F, Beekmann SE, Williams EC, Gill VJ. 1995. PCR amplification of rRNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J Clin Microbiol 33:184–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, Frost EH, Savelkoul P, Nicholson B, van den Berg RJ, Kato H, Sambol SP, Zukowski W, Woods C, Limbago B, Gerding DN, McDonald LC. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol 46:431–437. doi: 10.1128/JCM.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh JW, O'Leary MM, Shutt KA, Pasculle AW, Johnson S, Gerding DN, Muto CA, Harrison LH. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J Clin Microbiol 44:2558–2566. doi: 10.1128/JCM.02364-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broukhanski G, Low DE, Pillai DR. 2011. Modified multiple-locus variable-number tandem-repeat analysis for rapid identification and typing of Clostridium difficile during institutional outbreaks. J Clin Microbiol 49:1983–1986. doi: 10.1128/JCM.02359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eyre DW, Fawley WN, Best EL, Griffiths D, Stoesser NE, Crook DW, Peto TE, Walker AS, Wilcox MH. 2013. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51:4141–4149. doi: 10.1128/JCM.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover FC, Tickler IA, Persing DH. 2012. Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob Agents Chemother 56:2929–2932. doi: 10.1128/AAC.00220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terhes G, Maruyama A, Latkoczy K, Szikra L, Konkoly-Thege M, Princz G, Nagy E, Urban E. 2014. In vitro antibiotic susceptibility profile of Clostridium difficile excluding PCR ribotype 027 outbreak strain in Hungary. Anaerobe 30:41–44. doi: 10.1016/j.anaerobe.2014.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


